Disease Genomics What is genomics Looking at the











- Slides: 50

Disease Genomics

What is genomics? • Looking at the properties of the genome as a whole – “seeing the wood for the trees”; identifying patterns by considering many data points at once. – Examining large-scale properties requires a model of what is expected just by chance, the null hypothesis.

What is disease genomics? • OED: A condition of the body, or of some part or organ of the body, in which its functions are disturbed or deranged; • So disease genomics is about taking a whole-genome view to genetic disorders so we can discover: – – The identification of the underlying genetic determinants insights into the pathoetiology of the disease How to select the appropriate treatment How to prevent disease


Preventive Medicine • Empower people to make the appropriate life-style choices – 23 and. Me, Coriell Study • Treat the cause of the disease rather than the symptoms – E. g. peptic ulcers • “All medicine may become pediatrics” Paul Wise, Professor of Pediatrics, Stanford Medical School, 2008 • Effects of environment, accidents, aging, penetrance … – Somatic change, understanding how the genome changes over a lifetime – cancer • Health care costs can be greatly reduced if – Invest in preventive medicine – Target the cause of disease rather than symptoms

23 and. Me © 23 and. Me 2009

23 and. Me Spittoon

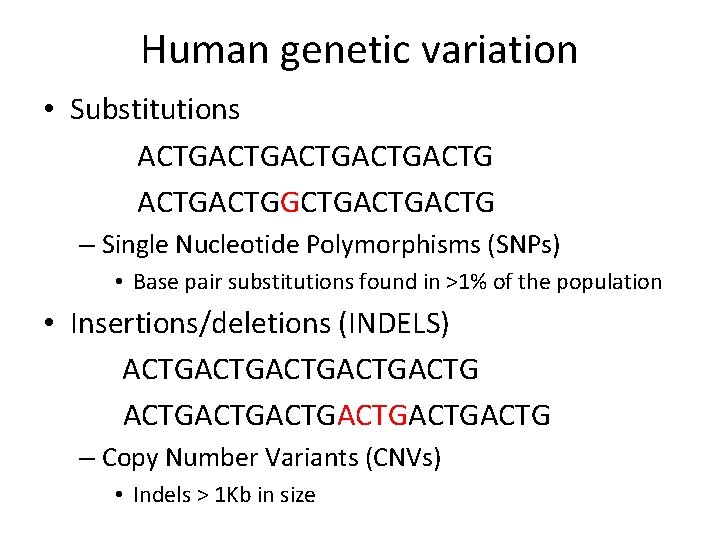
23 and. Me Research Reports

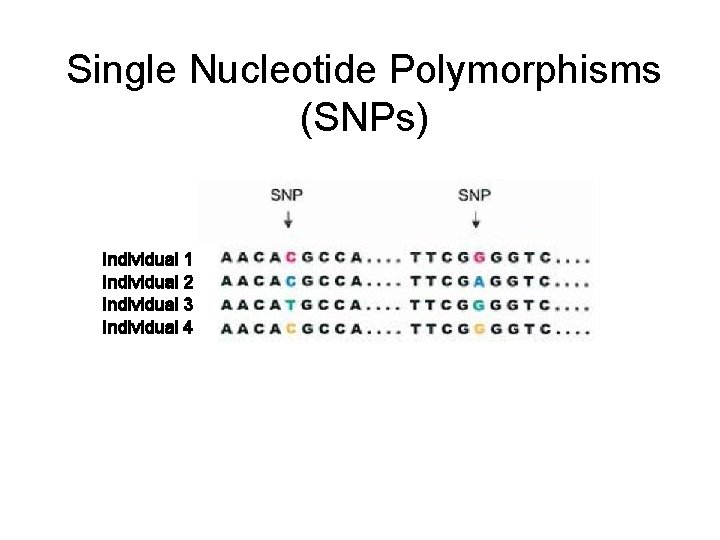
Human genetic variation • Substitutions ACTGACTGACTGGCTGACTG – Single Nucleotide Polymorphisms (SNPs) • Base pair substitutions found in >1% of the population • Insertions/deletions (INDELS) ACTGACTGACTGACTGACTGACTG – Copy Number Variants (CNVs) • Indels > 1 Kb in size

Human genetic variation • Variation can have an effect on function – Non-synonymous substitutions can change the amino acid encoded by a codon or give rise to premature stop codons – Indels can cause frame-shifts – Mutations may affect splice sites or regulatory sequence outside of genes or within introns

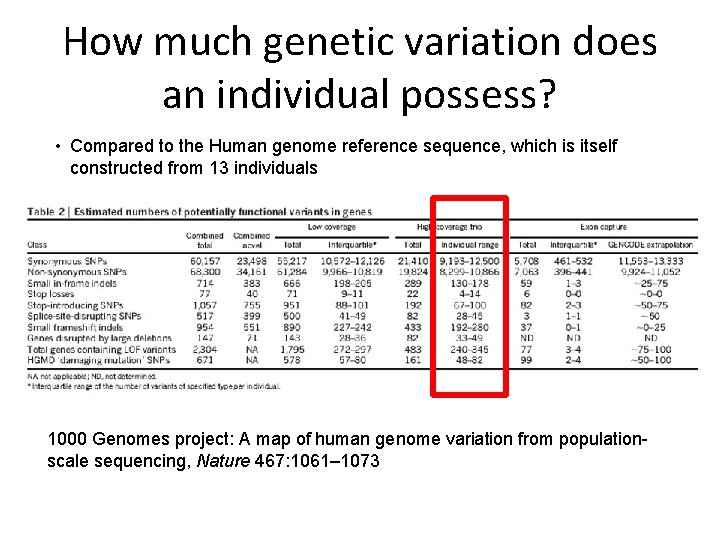
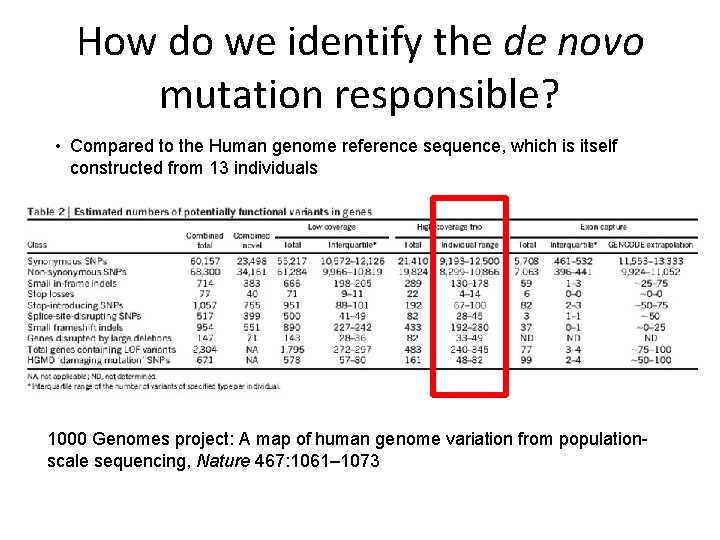
How much genetic variation does an individual possess? • Compared to the Human genome reference sequence, which is itself constructed from 13 individuals 1000 Genomes project: A map of human genome variation from populationscale sequencing, Nature 467: 1061– 1073

Penetrance of genetic variants • Highly penetrant Mendelian single gene diseases – Huntington’s Disease caused by excess CAG repeats in huntingtin’s protein gene – Autosomal dominant, 100% penetrant, invariably lethal • Reduced penetrance, some genes lead to a predisposition to a disease – BRCA 1 & BRCA 2 genes can lead to a familial breast or ovarian cancer – Disease alleles lead to 80% overall lifetime chance of a cancer, but 20% of patients with the rare defective genes show no cancers • Complex diseases requiring alleles in multiple genes – Many cancers (solid tumors) require somatic mutations that induce cell proliferation, mutations that inhibit apoptosis, mutations that induce angiogenesis, and mutations that cause metastasis – Cancers are also influenced by environment (smoking, carcinogens, exposure to UV) – Atherosclerosis (obesity, genetic and nutritional cholesterol) • Some complex diseases have multiple causes – Genetic vs. spontaneous vs. environment vs. behavior • Some complex diseases can be caused by multiple pathways – Type 2 Diabetes can be caused by reduced beta-cells in pancreas, reduced production of insulin, reduced sensitivity to insulin (insulin resistance) as well as environmental conditions (obesity, sedentary lifestyle, smoking etc. ).

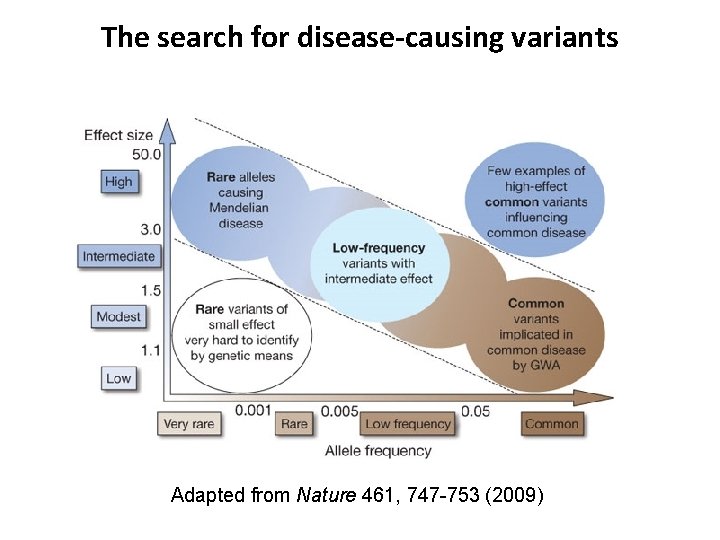
The search for disease-causing variants Adapted from Nature 461, 747 -753 (2009)

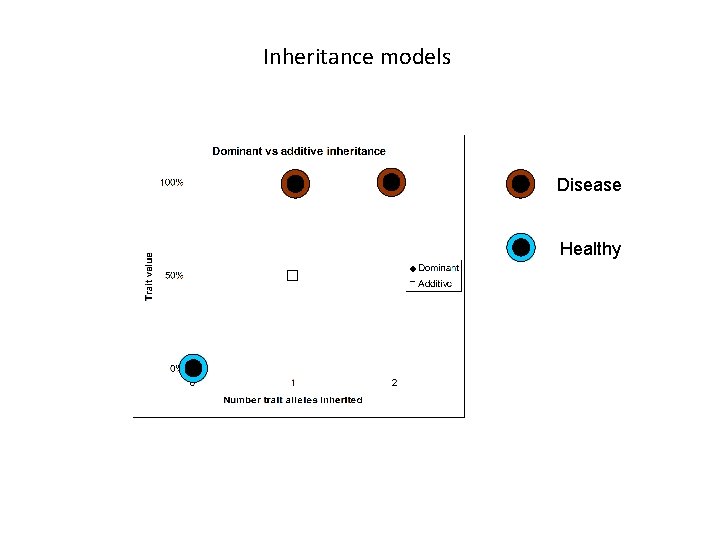
Inheritance models

Inheritance models Disease Healthy

Identifying the genetic causes of highly penetrant disorders • de novo mutations • Mendelian disorders

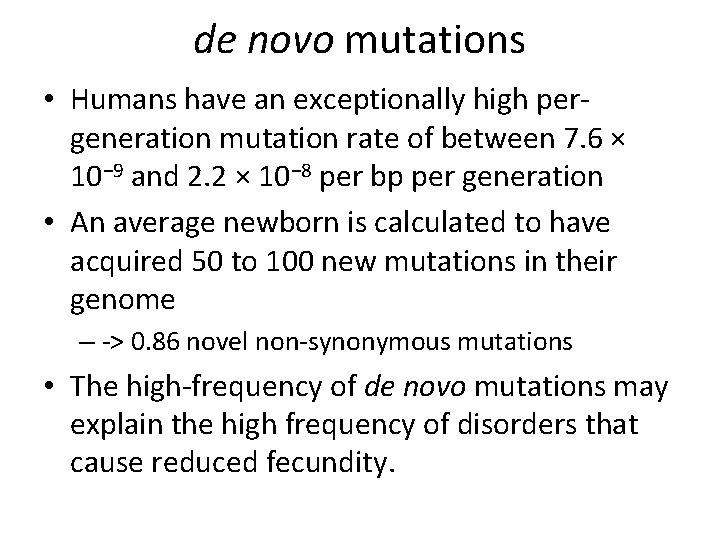
de novo mutations • Humans have an exceptionally high pergeneration mutation rate of between 7. 6 × 10− 9 and 2. 2 × 10− 8 per bp per generation • An average newborn is calculated to have acquired 50 to 100 new mutations in their genome – -> 0. 86 novel non-synonymous mutations • The high-frequency of de novo mutations may explain the high frequency of disorders that cause reduced fecundity.

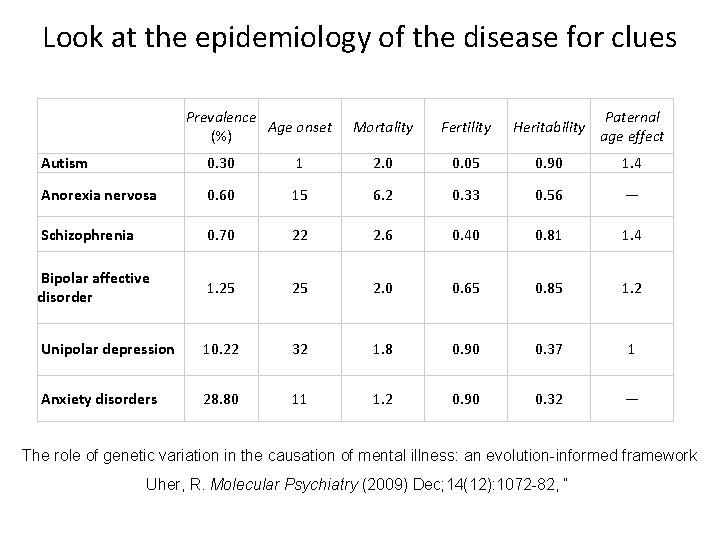
Look at the epidemiology of the disease for clues Prevalence Age onset (%) Mortality Fertility Heritability Paternal age effect Autism 0. 30 1 2. 0 0. 05 0. 90 1. 4 Anorexia nervosa 0. 60 15 6. 2 0. 33 0. 56 — Schizophrenia 0. 70 22 2. 6 0. 40 0. 81 1. 4 Bipolar affective disorder 1. 25 25 2. 0 0. 65 0. 85 1. 2 Unipolar depression 10. 22 32 1. 8 0. 90 0. 37 1 Anxiety disorders 28. 80 11 1. 2 0. 90 0. 32 — The role of genetic variation in the causation of mental illness: an evolution-informed framework Uher, R. Molecular Psychiatry (2009) Dec; 14(12): 1072 -82, “

How do we identify the de novo mutation responsible? • Compared to the Human genome reference sequence, which is itself constructed from 13 individuals 1000 Genomes project: A map of human genome variation from populationscale sequencing, Nature 467: 1061– 1073

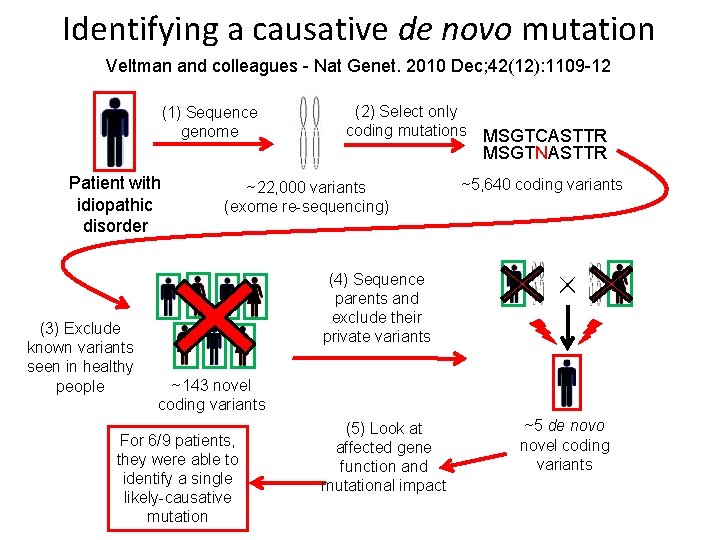
Identifying a causative de novo mutation Veltman and colleagues - Nat Genet. 2010 Dec; 42(12): 1109 -12 (1) Sequence genome (2) Select only coding mutations MSGTCASTTR MSGTNASTTR Patient with idiopathic disorder (3) Exclude known variants seen in healthy people ~22, 000 variants (exome re-sequencing) ~5, 640 coding variants (4) Sequence parents and exclude their private variants ~143 novel coding variants For 6/9 patients, they were able to identify a single likely-causative mutation (5) Look at affected gene function and mutational impact ~5 de novo novel coding variants

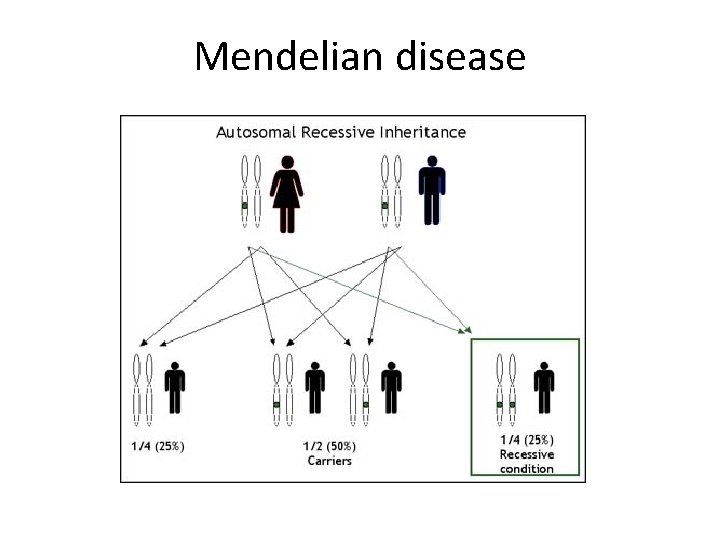
Mendelian disease • Definition: Diseases in which the phenotypes are largely determined by the action, lack of action, of mutations at individual loci. • Rare 1% of all live born individuals • 4 types of inheritance : Autosomal dominant : Autosomal recessive : X linked dominant : X linked recessive

Mendelian disease

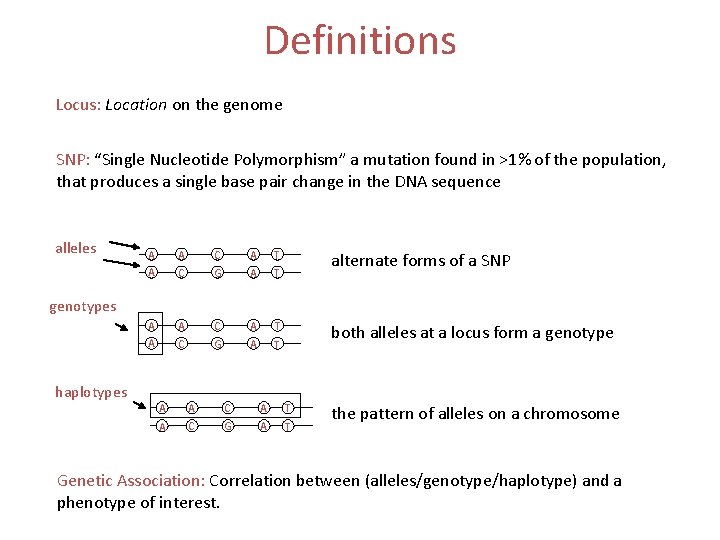
Definitions Locus: Location on the genome SNP: “Single Nucleotide Polymorphism” a mutation found in >1% of the population, that produces a single base pair change in the DNA sequence alleles A A A C C A G A A A C A T T alternate forms of a SNP C A T G A T both alleles at a locus form a genotypes haplotypes A A C A T A C G A T the pattern of alleles on a chromosome Genetic Association: Correlation between (alleles/genotype/haplotype) and a phenotype of interest.

Single Nucleotide Polymorphisms (SNPs) Individual 1 Individual 2 Individual 3 Individual 4

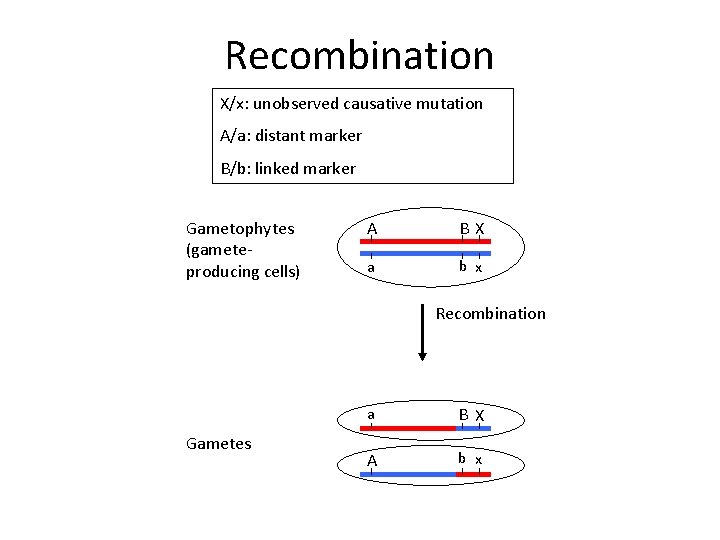
Recombination X/x: unobserved causative mutation A/a: distant marker B/b: linked marker Gametophytes (gameteproducing cells) A BX a b x Recombination Gametes a B X A b x

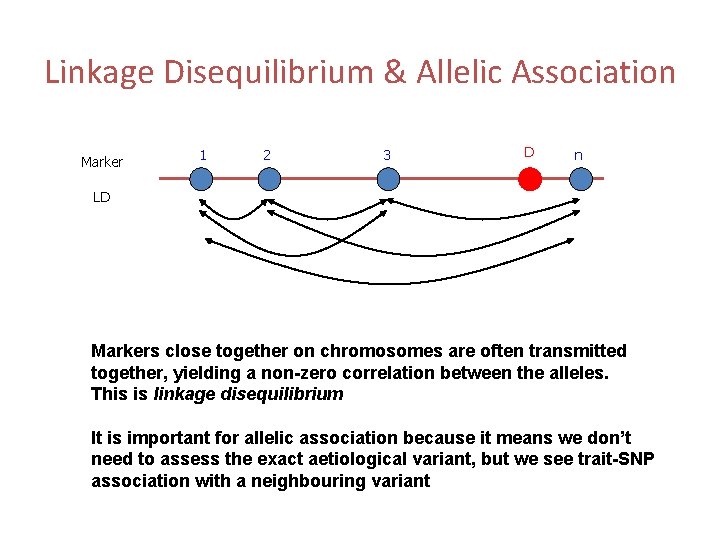
Linkage Disequilibrium & Allelic Association Marker 1 2 3 D n LD Markers close together on chromosomes are often transmitted together, yielding a non-zero correlation between the alleles. This is linkage disequilibrium It is important for allelic association because it means we don’t need to assess the exact aetiological variant, but we see trait-SNP association with a neighbouring variant

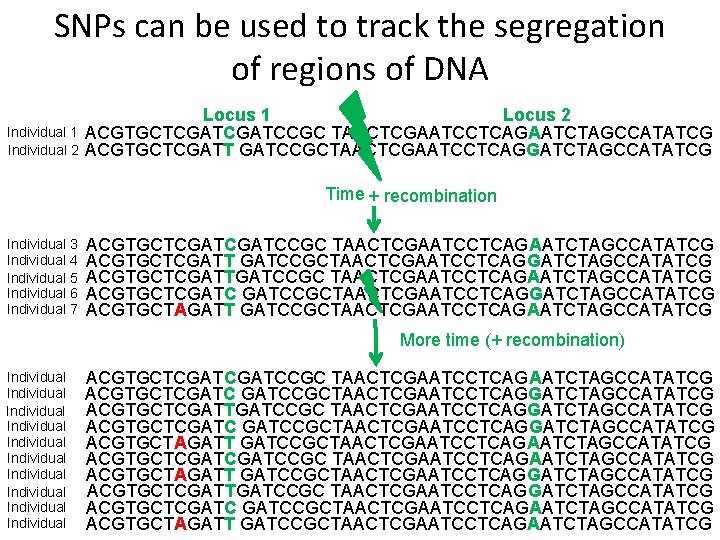
SNPs can be used to track the segregation of regions of DNA Individual 1 Individual 2 Locus 1 Locus 2 ACGTGCTCGATCCGC TAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTCGATT GATCCGCTAACTCGAATCCTCAGGATCTAGCCATATCG Time + recombination Individual 3 Individual 4 Individual 5 Individual 6 Individual 7 ACGTGCTCGATCCGC TAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTCGATT GATCCGCTAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTCGATTGATCCGC TAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTCGATCCGCTAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTAGATT GATCCGCTAACTCGAATCCTCAGAATCTAGCCATATCG More time (+ recombination) Individual Individual Individual ACGTGCTCGATCCGC TAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTCGATCCGCTAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTCGATTGATCCGC TAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTCGATCCGCTAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTAGATT GATCCGCTAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTCGATCCGC TAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTAGATT GATCCGCTAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTCGATTGATCCGC TAACTCGAATCCTCAGGATCTAGCCATATCG ACGTGCTCGATCCGCTAACTCGAATCCTCAGAATCTAGCCATATCG ACGTGCTAGATT GATCCGCTAACTCGAATCCTCAGAATCTAGCCATATCG

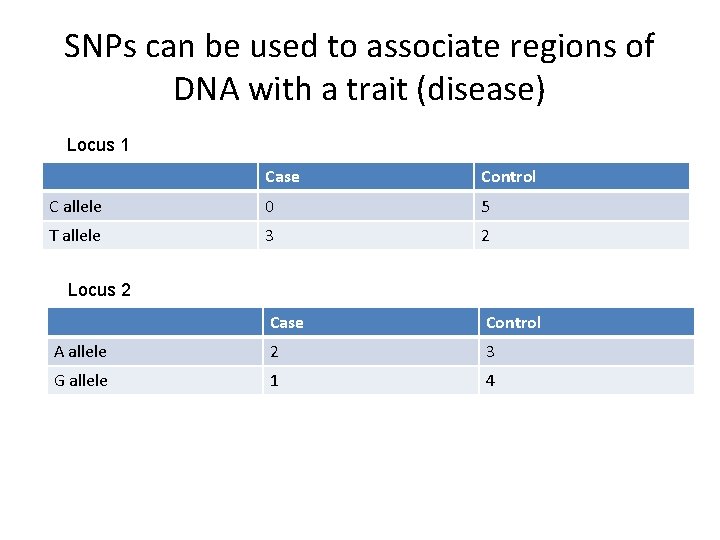
SNPs can be used to associate regions of DNA with a trait (disease) Locus 1 Case Control C allele 0 5 T allele 3 2 Case Control A allele 2 3 G allele 1 4 Locus 2

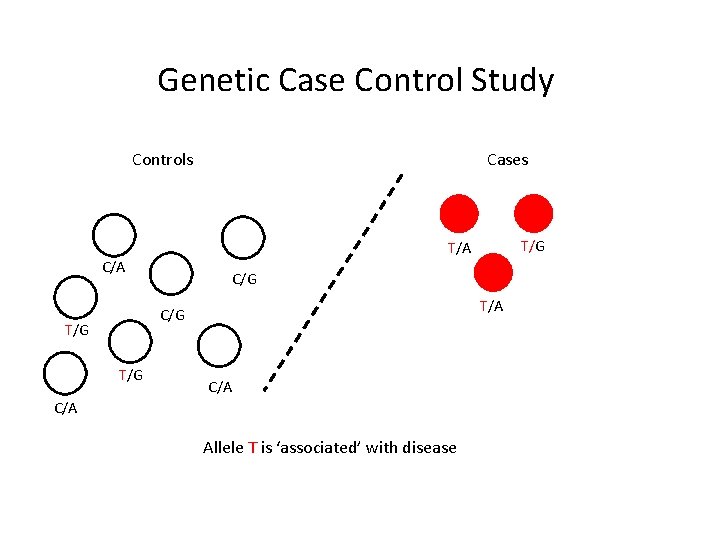
Genetic Case Control Study Controls Cases C/A C/G T/G T/G T/A C/A Allele T is ‘associated’ with disease

Measures of Association: The Odds Ratio • Odds are related to probability: odds = p/(1 -p) – If probability of horse winning race is 50%, odds are 1/1 – If probability of horse winning race is 25%, odds are 1/3 for win or 3 to 1 against win • If probability of exposed person getting disease is 25%, odds = p/(1 -p) = 25/75 = 1/3 • We can calculate an odds ratio = cross-product ratio (“ad/bc”)

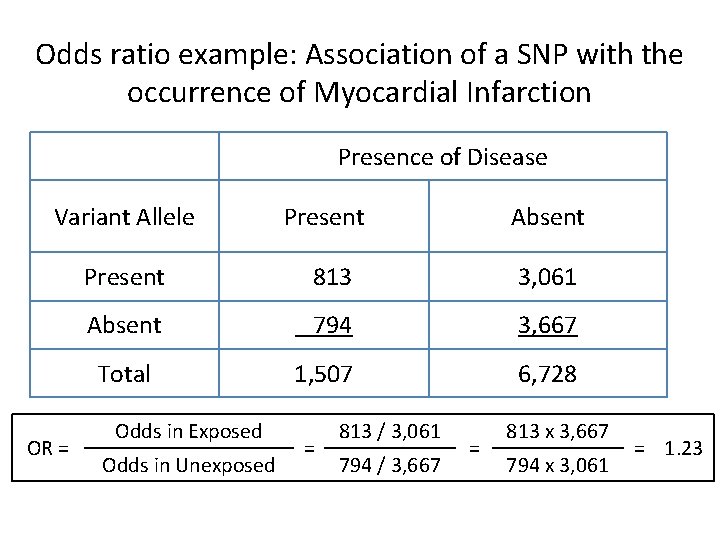
Odds ratio example: Association of a SNP with the occurrence of Myocardial Infarction Presence of Disease Variant Allele Absent Present 813 3, 061 Absent 794 3, 667 1, 507 6, 728 Total OR = Present Odds in Exposed Odds in Unexposed = 813 / 3, 061 794 / 3, 667 = 813 x 3, 667 794 x 3, 061 = 1. 23

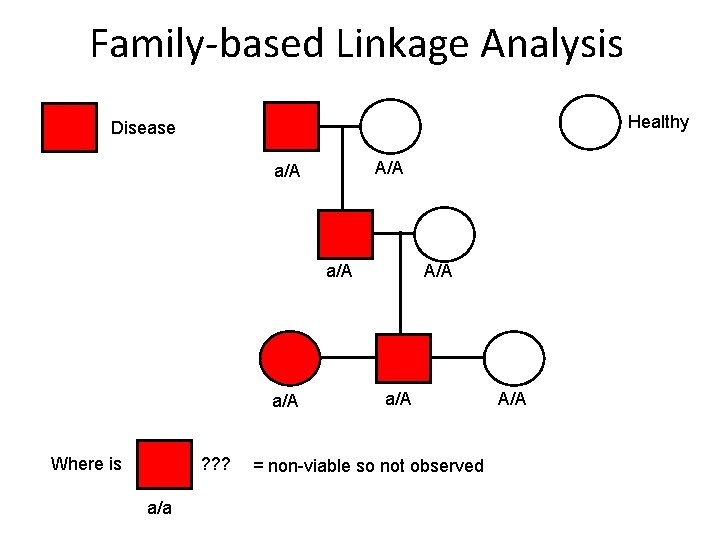
Family-based Linkage Analysis Healthy Disease A/A a/A a/A Where is ? ? ? a/a A/A a/A = non-viable so not observed A/A

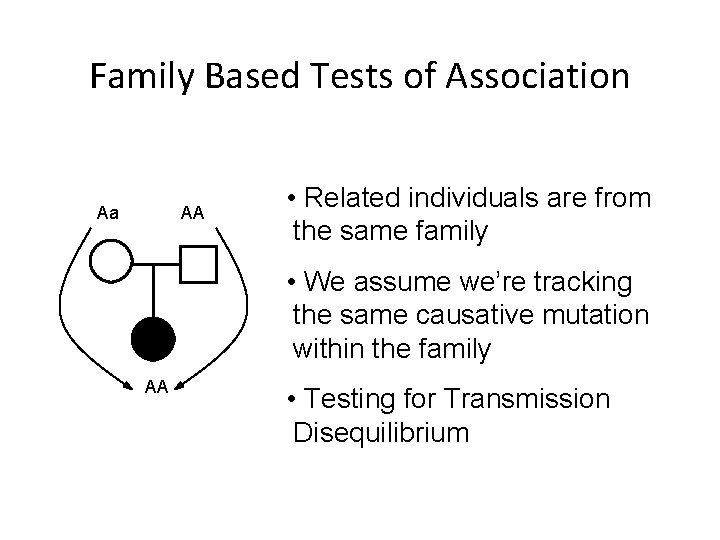
Family Based Tests of Association Aa AA • Related individuals are from the same family • We assume we’re tracking the same causative mutation within the family AA • Testing for Transmission Disequilibrium

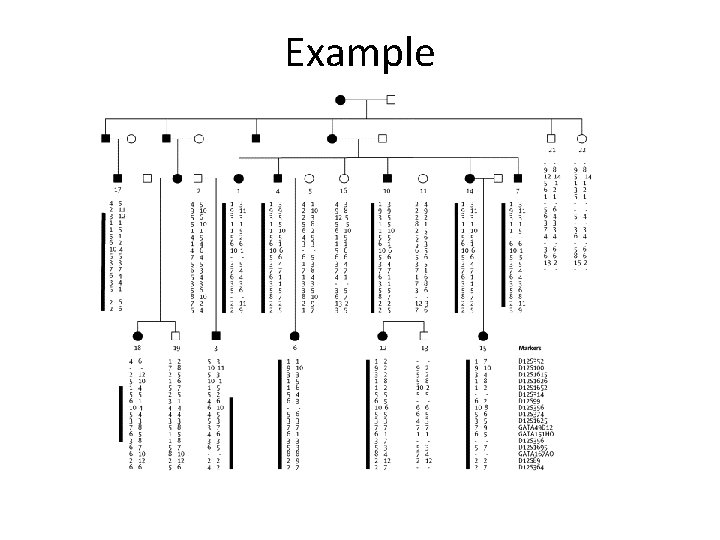
Example

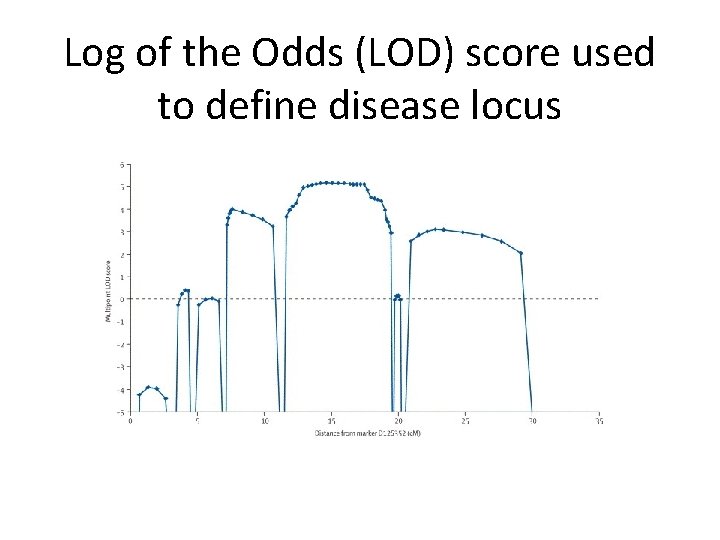
Log of the Odds (LOD) score used to define disease locus

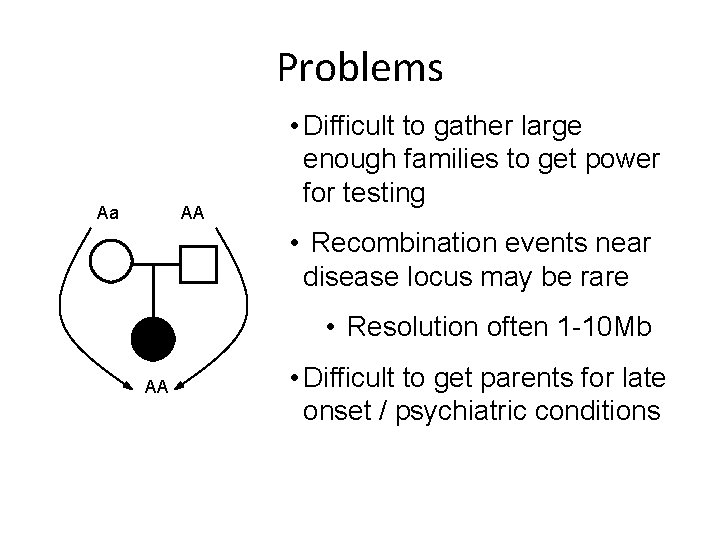
Problems Aa AA • Difficult to gather large enough families to get power for testing • Recombination events near disease locus may be rare • Resolution often 1 -10 Mb AA • Difficult to get parents for late onset / psychiatric conditions

Genome-wide Association Studies (GWAS) • Looking for the segregation of disease (case/control) with particular genotypes across a whole population • A lot of recombination within the population so you can very finely map loci • Based on the common-disease, common-variant hypothesis – Only makes sense for moderate effect sizes (odds ratio < 1. 5)

GWAS • Technology makes it feasible -- Affymetrix: 500 K; 1 M chip arrived 2007. (Randomly distributed SNPs) -- Illumina: 550 K chip costs (gene-based) w w Good for moderate effect sizes ( odds ratio < 1. 5). Particularly useful in finding genetic variations that contribute to common, complex diseases.

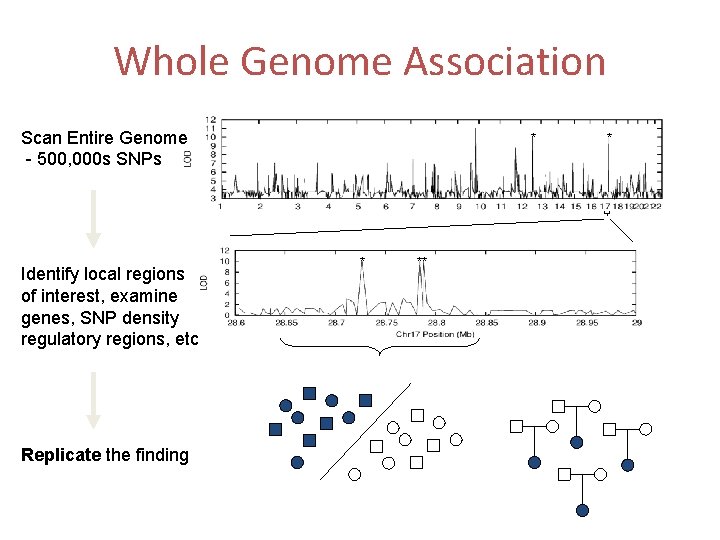
Whole Genome Association Scan Entire Genome - 500, 000 s SNPs Identify local regions of interest, examine genes, SNP density regulatory regions, etc Replicate the finding * * ** *

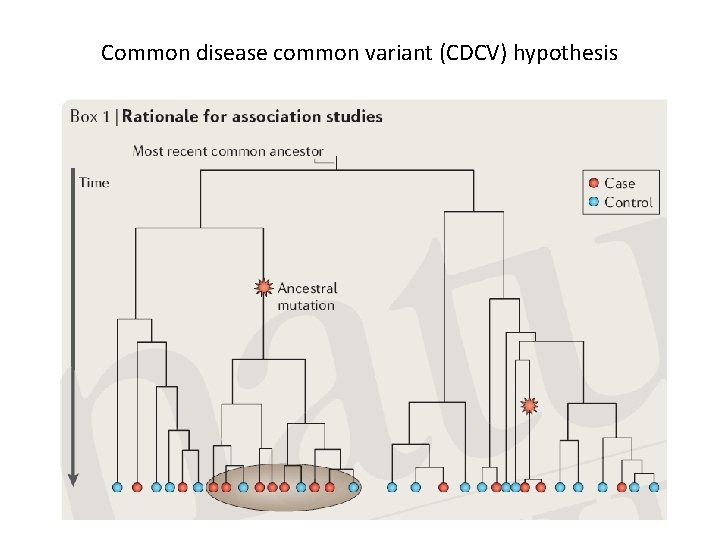
Common disease common variant (CDCV) hypothesis

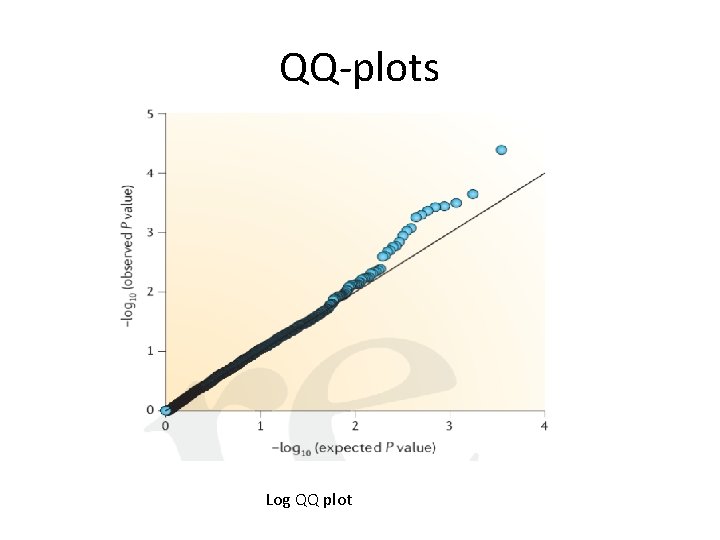
QQ-plots Log QQ plot

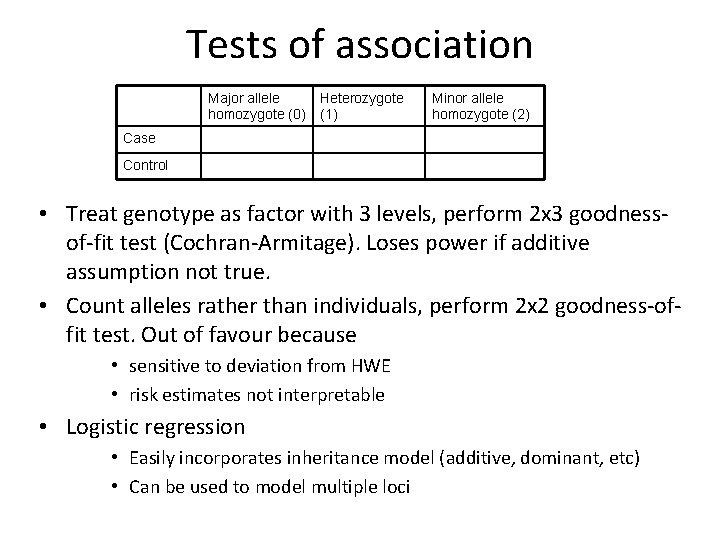
Tests of association Major allele homozygote (0) Heterozygote (1) Minor allele homozygote (2) Case Control • Treat genotype as factor with 3 levels, perform 2 x 3 goodnessof-fit test (Cochran-Armitage). Loses power if additive assumption not true. • Count alleles rather than individuals, perform 2 x 2 goodness-offit test. Out of favour because • sensitive to deviation from HWE • risk estimates not interpretable • Logistic regression • Easily incorporates inheritance model (additive, dominant, etc) • Can be used to model multiple loci

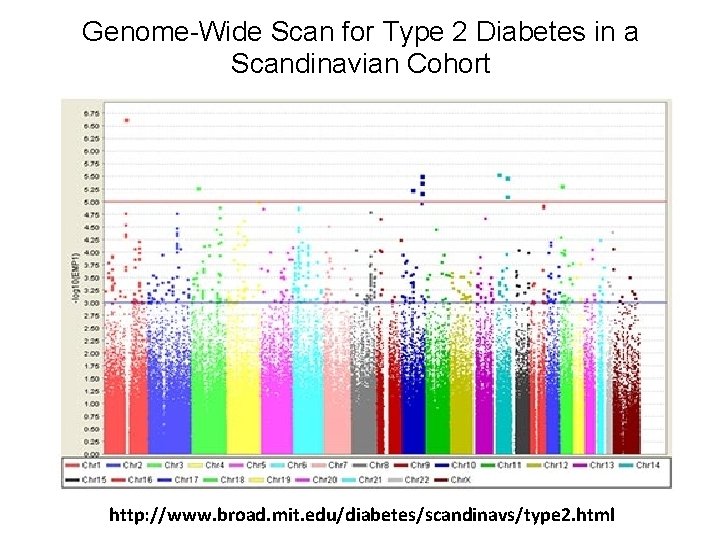
Genome-Wide Scan for Type 2 Diabetes in a Scandinavian Cohort http: //www. broad. mit. edu/diabetes/scandinavs/type 2. html

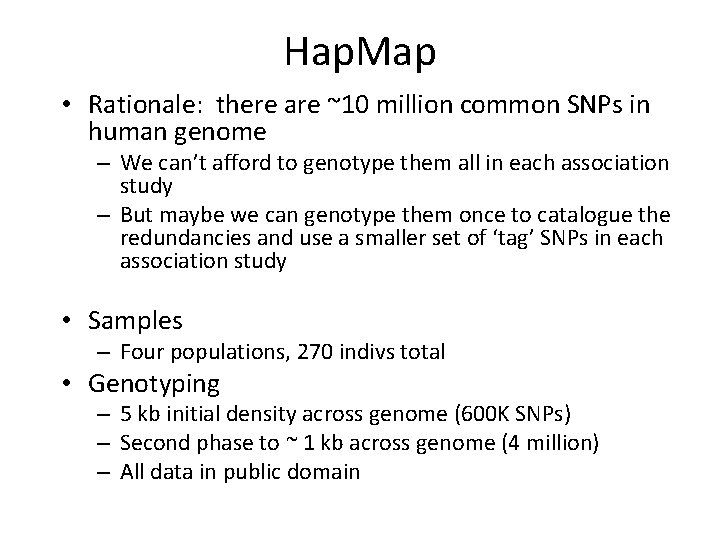
Hap. Map • Rationale: there are ~10 million common SNPs in human genome – We can’t afford to genotype them all in each association study – But maybe we can genotype them once to catalogue the redundancies and use a smaller set of ‘tag’ SNPs in each association study • Samples – Four populations, 270 indivs total • Genotyping – 5 kb initial density across genome (600 K SNPs) – Second phase to ~ 1 kb across genome (4 million) – All data in public domain

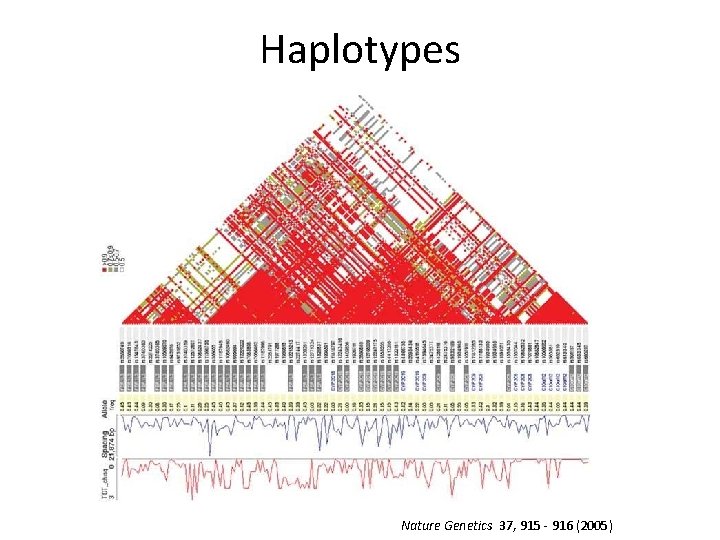
Haplotypes Nature Genetics 37, 915 - 916 (2005)

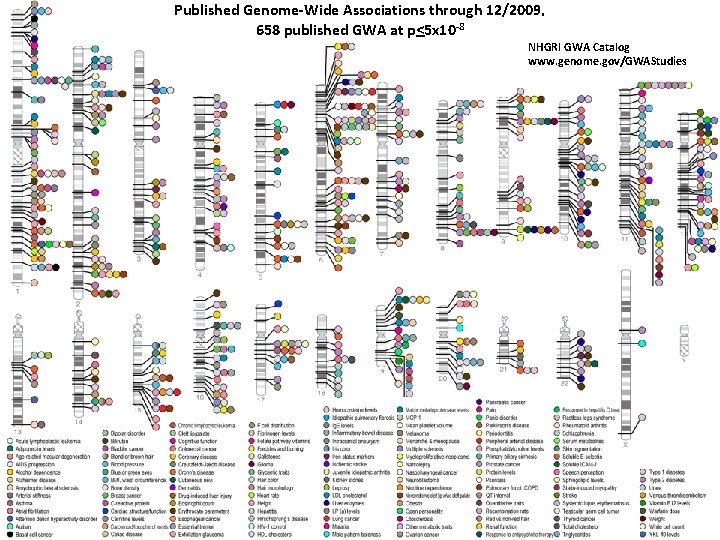
Published Genome-Wide Associations through 12/2009, 658 published GWA at p<5 x 10 -8 NHGRI GWA Catalog www. genome. gov/GWAStudies

Population Stratification can be a problem • Imagine a sample of individuals drawn from a population consisting of two distinct subgroups which differ in allele frequency. • If the prevalence of disease is greater in one sub-population, then this group will be over-represented amongst the cases. • Any marker which is also of higher frequency in that subgroup will appear to be associated with the disease

Traditional Issues Persist Allelic heterogeneity – When multiple disease variants exist at the same gene, a single marker may not capture them well enough. – Haplotype-based association analysis is good theoretically, but it hasn’t shown its advantage in practice. Locus heterogeneity – Multiple genes may influence the disease risk independently. As a result, for any single gene, a fraction of the cases may be no different from the controls. Effect modification (a. k. a. interaction) between two genes may exist with weak/no marginal effects. – It is unknown how often this happens in reality. But when this happens, analyses that only look at marginal effects won’t be useful. – It often requires larger sample size to have reasonable power to detect interaction effects than the sample size needed to detect marginal effects.

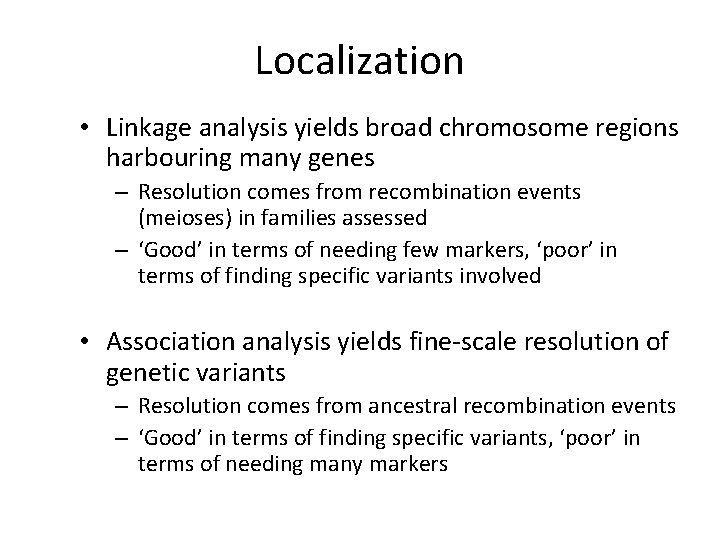
Localization • Linkage analysis yields broad chromosome regions harbouring many genes – Resolution comes from recombination events (meioses) in families assessed – ‘Good’ in terms of needing few markers, ‘poor’ in terms of finding specific variants involved • Association analysis yields fine-scale resolution of genetic variants – Resolution comes from ancestral recombination events – ‘Good’ in terms of finding specific variants, ‘poor’ in terms of needing many markers

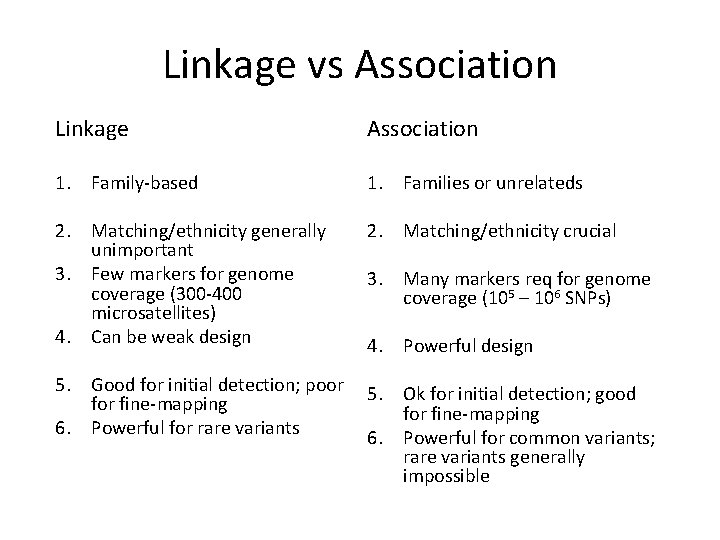
Linkage vs Association Linkage Association 1. Family-based 1. Families or unrelateds 2. Matching/ethnicity generally unimportant 3. Few markers for genome coverage (300 -400 microsatellites) 4. Can be weak design 2. Matching/ethnicity crucial 5. Good for initial detection; poor fine-mapping 6. Powerful for rare variants 5. Ok for initial detection; good for fine-mapping 6. Powerful for common variants; rare variants generally impossible 3. Many markers req for genome coverage (105 – 106 SNPs) 4. Powerful design