What is in a genome Genome The genome














- Slides: 56

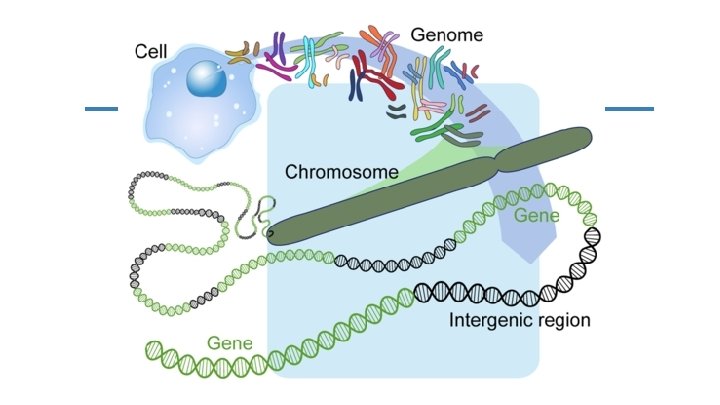
What is in a genome?


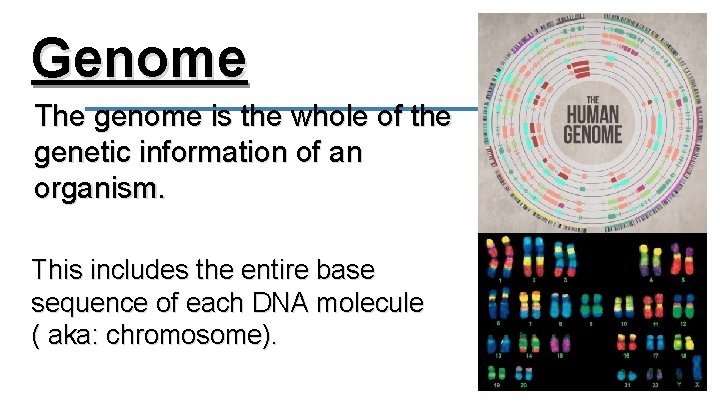
Genome The genome is the whole of the genetic information of an organism. This includes the entire base sequence of each DNA molecule ( aka: chromosome).

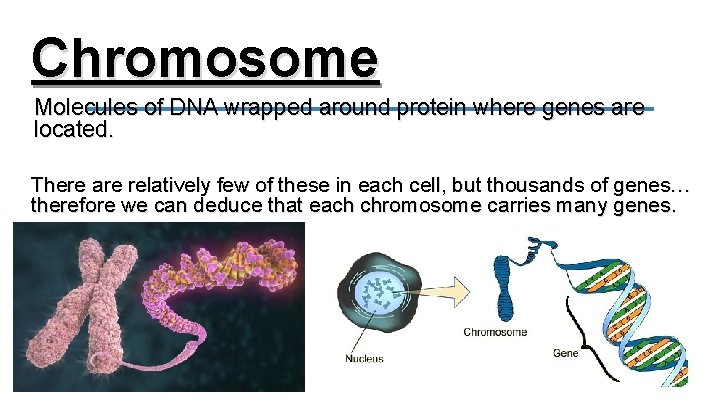
Chromosome Molecules of DNA wrapped around protein where genes are located. There are relatively few of these in each cell, but thousands of genes… therefore we can deduce that each chromosome carries many genes.

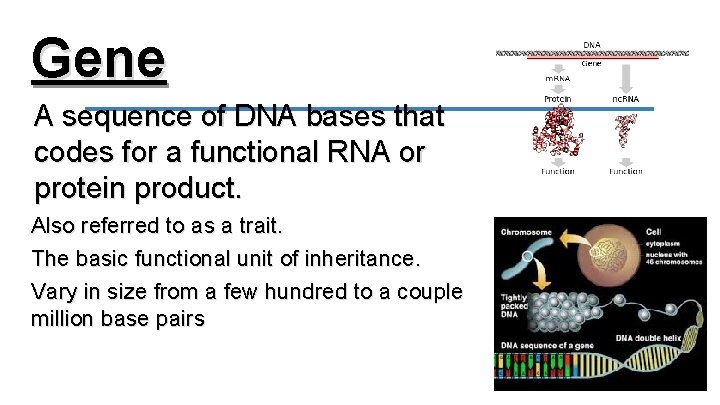
Gene A sequence of DNA bases that codes for a functional RNA or protein product. Also referred to as a trait. The basic functional unit of inheritance. Vary in size from a few hundred to a couple million base pairs

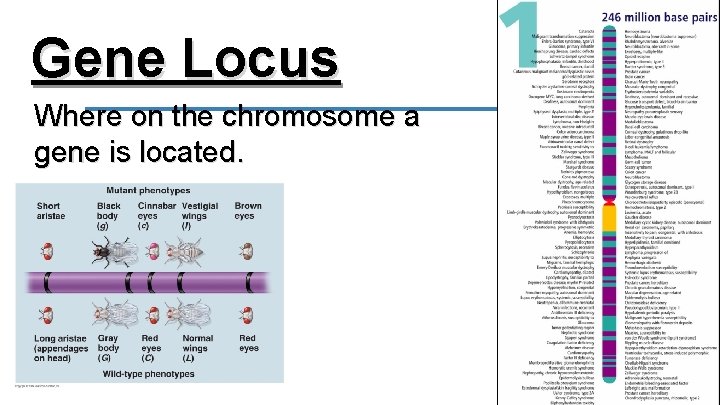
Gene Locus Where on the chromosome a gene is located.

Alleles: Variations of A Gene

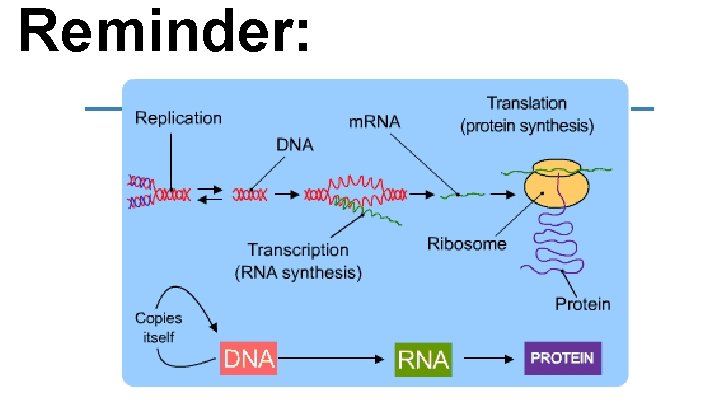
Reminder:


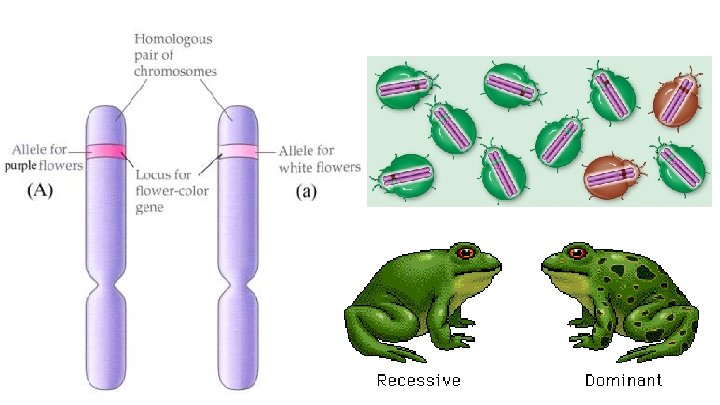
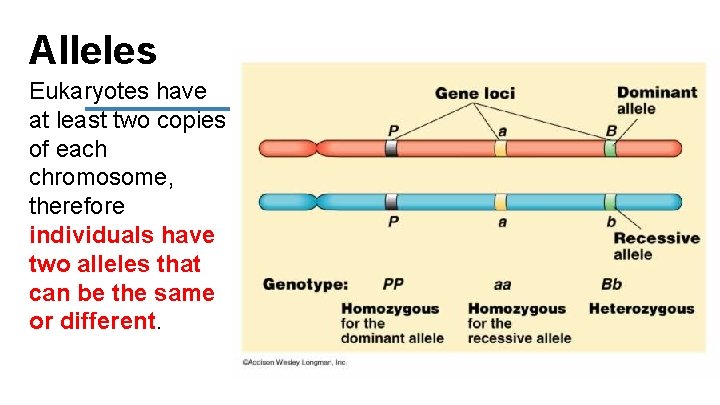
Alleles are alternative versions of the same gene and therefore occupy the same locus on the chromosome. Only one allele can occupy the locus of the gene on a chromosome.

Alleles Eukaryotes have at least two copies of each chromosome, therefore individuals have two alleles that can be the same or different.

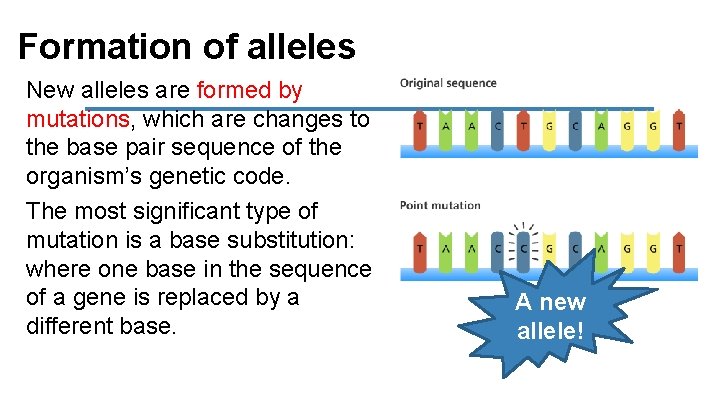
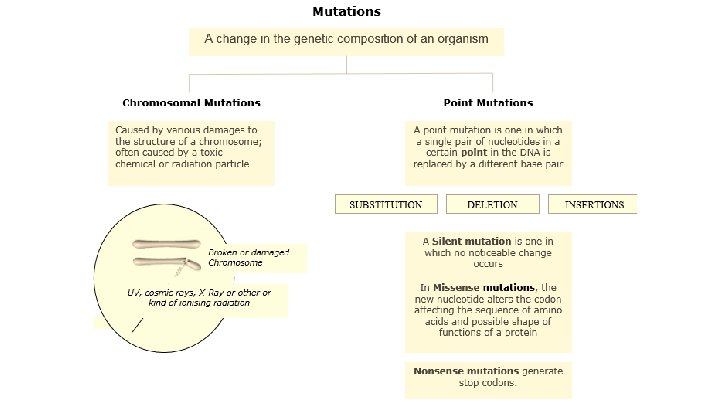
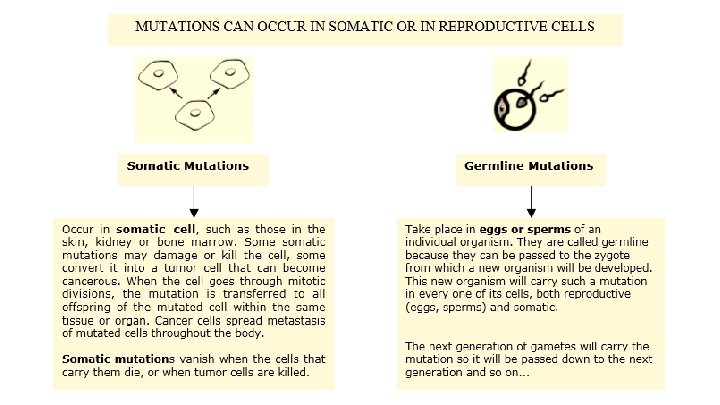
Formation of alleles New alleles are formed by mutations, which are changes to the base pair sequence of the organism’s genetic code. The most significant type of mutation is a base substitution: where one base in the sequence of a gene is replaced by a different base. A new allele!

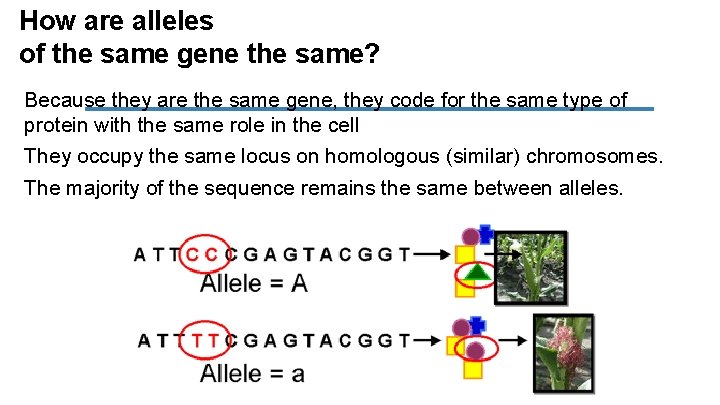
How are alleles of the same gene the same? Because they are the same gene, they code for the same type of protein with the same role in the cell They occupy the same locus on homologous (similar) chromosomes. The majority of the sequence remains the same between alleles.

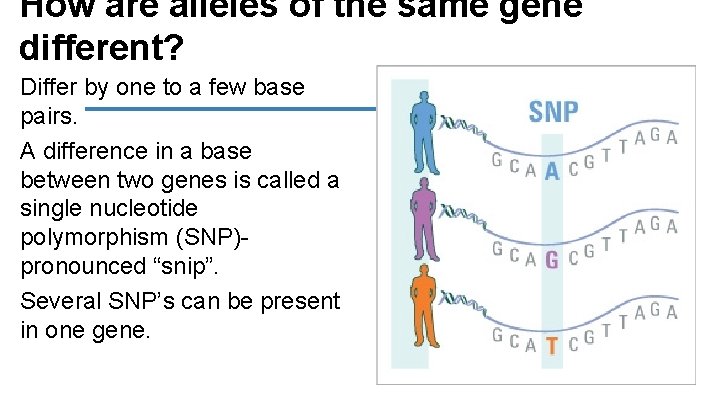
How are alleles of the same gene different? Differ by one to a few base pairs. A difference in a base between two genes is called a single nucleotide polymorphism (SNP)- pronounced “snip”. Several SNP’s can be present in one gene.

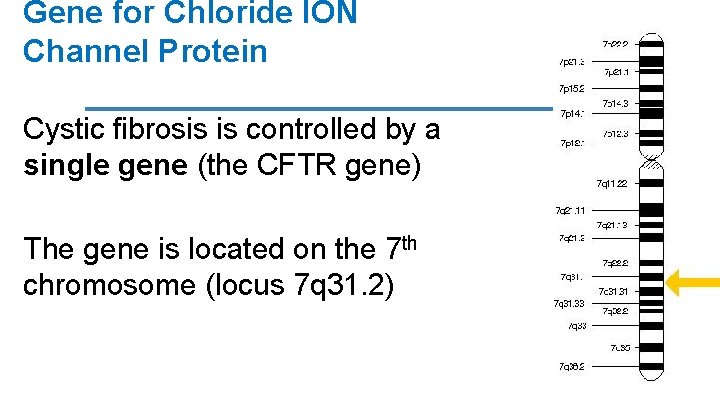
Gene for Chloride ION Channel Protein Cystic fibrosis is controlled by a single gene (the CFTR gene) The gene is located on the 7 th chromosome (locus 7 q 31. 2)

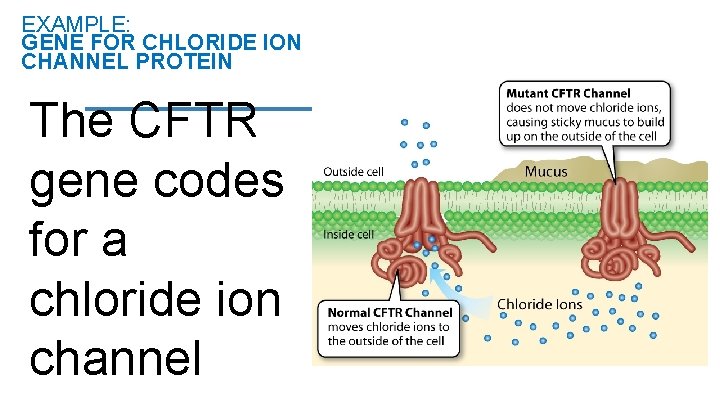
EXAMPLE: GENE FOR CHLORIDE ION CHANNEL PROTEIN The CFTR gene codes for a chloride ion channel

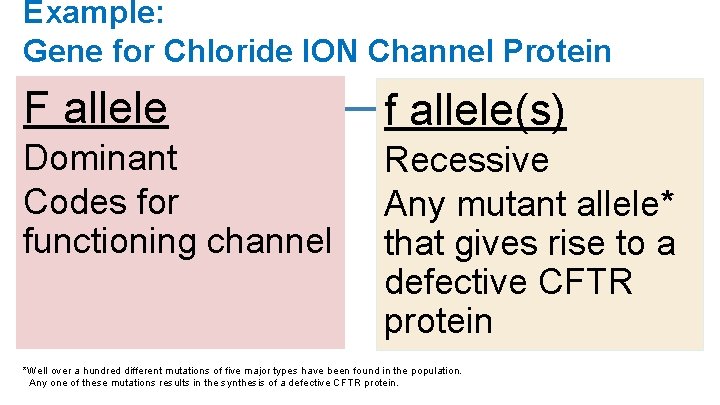
Example: Gene for Chloride ION Channel Protein F allele f allele(s) Dominant Codes for functioning channel Recessive Any mutant allele* that gives rise to a defective CFTR protein *Well over a hundred different mutations of five major types have been found in the population. Any one of these mutations results in the synthesis of a defective CFTR protein.


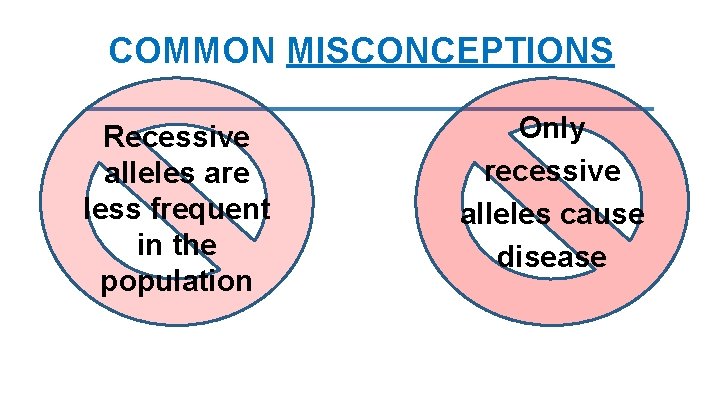
COMMON MISCONCEPTIONS Recessive alleles are less frequent in the population Only recessive alleles cause disease

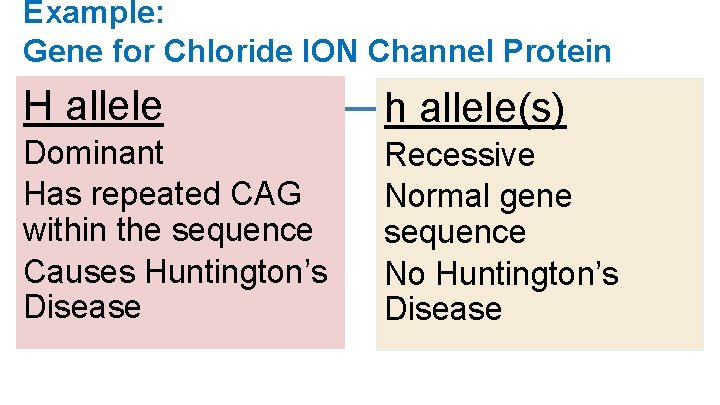
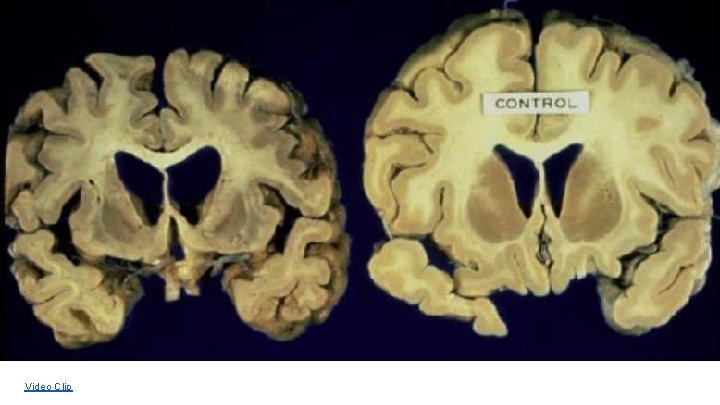
Example: Gene for Huntingtins Huntington's Disease is controlled by a single gene (the HTT gene) The gene is located on the 4 th chromosome (locus 4 p 16. 3)

Example: Gene for Huntingtins The HTT gene provides instructions for making a protein called huntingtin. The function of this protein is unknown; it appears to play an important role in nerve cells (neurons) in the brain.

Example: Gene for Chloride ION Channel Protein H allele h allele(s) Dominant Has repeated CAG within the sequence Causes Huntington’s Disease Recessive Normal gene sequence No Huntington’s Disease

Video Clip

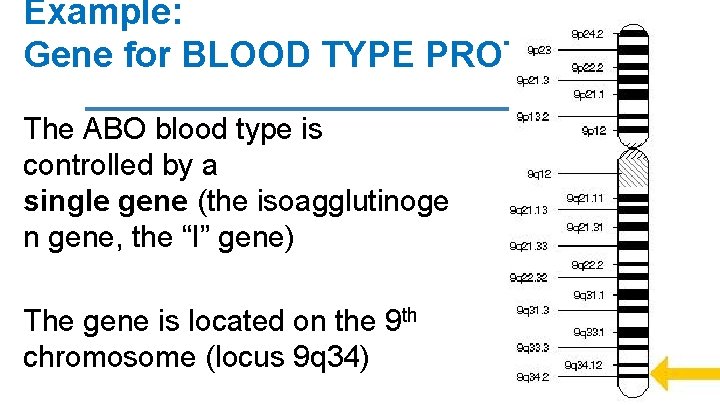
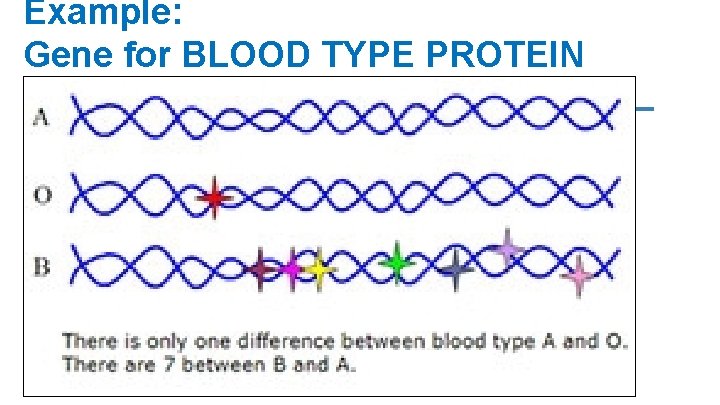
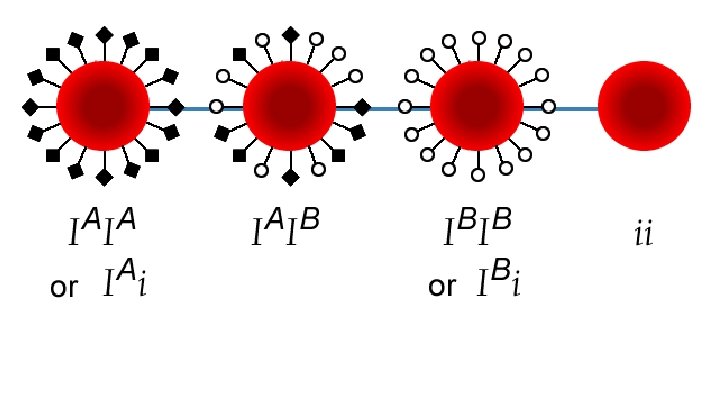
Example: Gene for BLOOD TYPE PROTEIN The ABO blood type is controlled by a single gene (the isoagglutinoge n gene, the “I” gene) The gene is located on the 9 th chromosome (locus 9 q 34)

Example: Gene for BLOOD TYPE PROTEIN The I gene has three alleles A I (dominant – codes for type A proteins) B I (dominant – codes for type B proteins) i (recessive – codes for no protein)

Example: Gene for BLOOD TYPE PROTEIN




Effects of Radiation

Effects of Radiation Vertus Hardiman

Hiroshima

Chernobyl

DNA Sequencing Di-deoxynucleotide Chain Termination

. . . ponder. . . 1. What does it mean to “sequence” DNA? 2. What are some applications of DNA sequencing?

Step 1 The DNA to be sequenced is prepared as a single strand by applying heat.

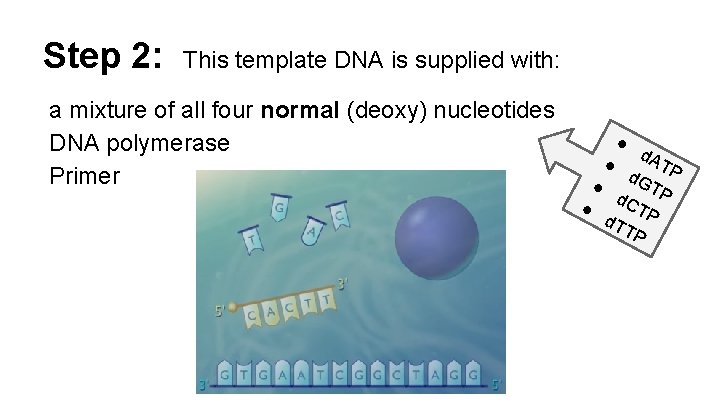
Step 2: This template DNA is supplied with: a mixture of all four normal (deoxy) nucleotides DNA polymerase Primer ● d. A T d. G P ● T d. C P ● T d. T P TP ●

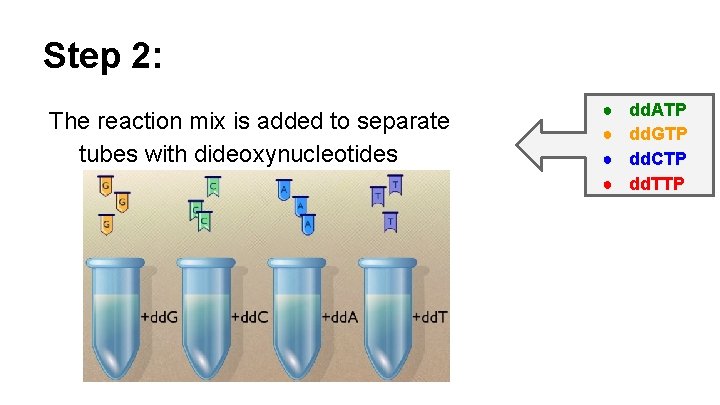
Step 2: The reaction mix is added to separate tubes with dideoxynucleotides ● ● dd. ATP dd. GTP dd. CTP dd. TTP

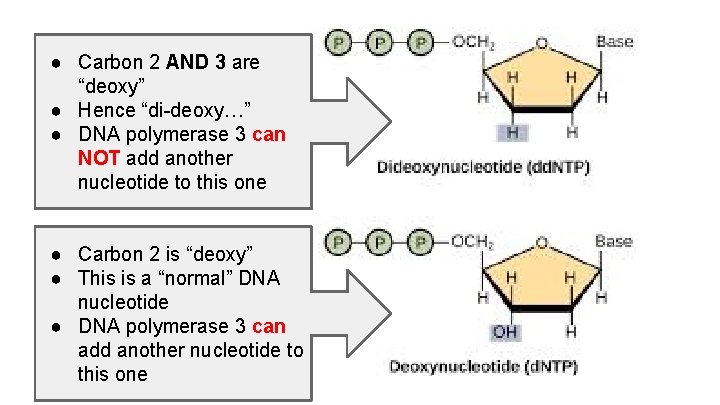
● Carbon 2 AND 3 are “deoxy” ● Hence “di-deoxy…” ● DNA polymerase 3 can NOT add another nucleotide to this one ● Carbon 2 is “deoxy” ● This is a “normal” DNA nucleotide ● DNA polymerase 3 can add another nucleotide to this one

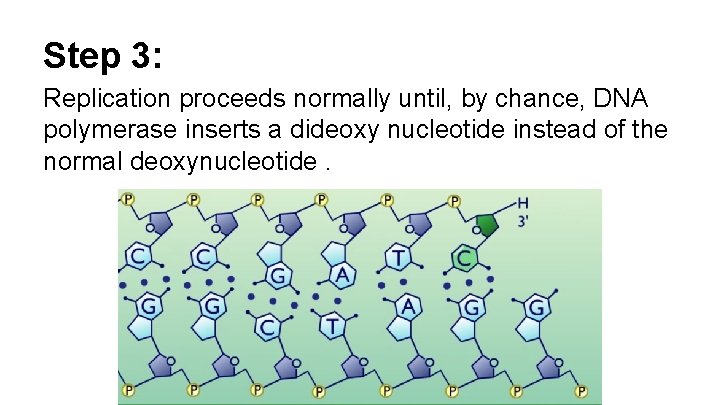
Step 3: Replication proceeds normally until, by chance, DNA polymerase inserts a dideoxy nucleotide instead of the normal deoxynucleotide.

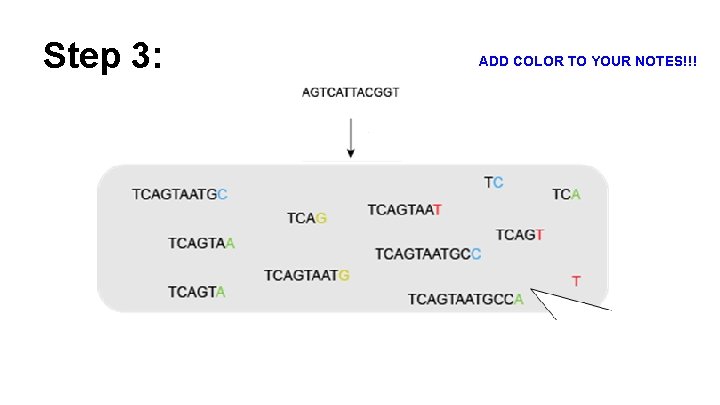
Step 3: ADD COLOR TO YOUR NOTES!!!

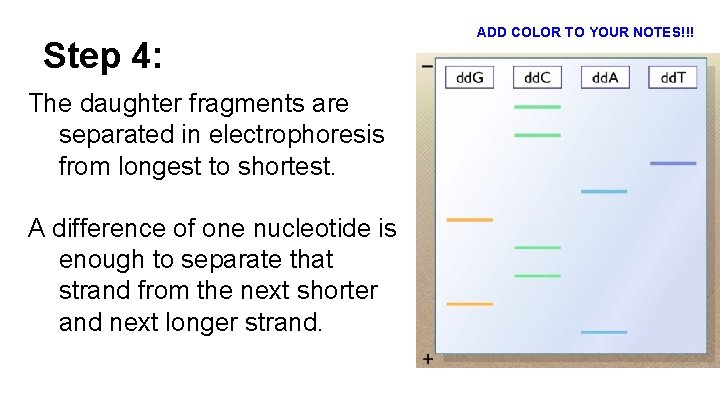
Step 4: The daughter fragments are separated in electrophoresis from longest to shortest. A difference of one nucleotide is enough to separate that strand from the next shorter and next longer strand. ADD COLOR TO YOUR NOTES!!!

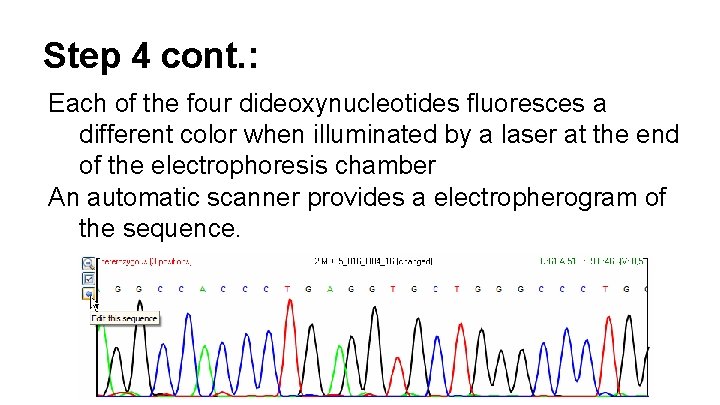
Step 4 cont. : Each of the four dideoxynucleotides fluoresces a different color when illuminated by a laser at the end of the electrophoresis chamber An automatic scanner provides a electropherogram of the sequence.

Helpful links 1. Sanger Method reading 2. DNA Sequencing reading with animation 3. DNA Sequencing animation #1 4. DNA Sequencing animation #2 5. DNA Sequencing animation #3 with quiz

Bioinformatics &DNA Sequence Alignment

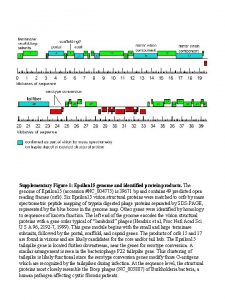
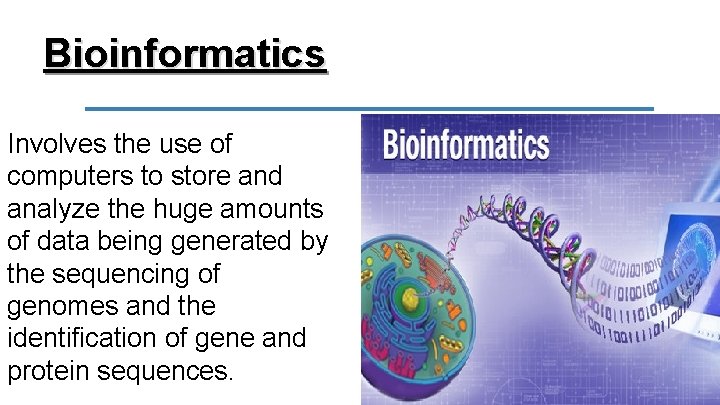
Bioinformatics Involves the use of computers to store and analyze the huge amounts of data being generated by the sequencing of genomes and the identification of gene and protein sequences.

Bioinformatics These sequences are stored in databases such as Gen. Bank (a US database) which are accessible worldwide to the general public and other scientists. You visited this database to identify your mystery genes!

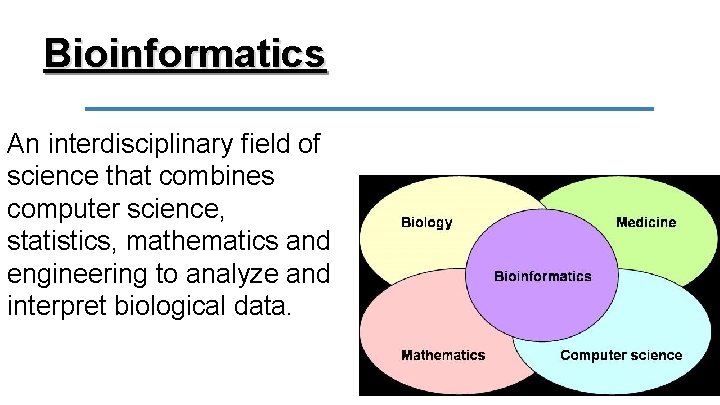
Bioinformatics An interdisciplinary field of science that combines computer science, statistics, mathematics and engineering to analyze and interpret biological data.

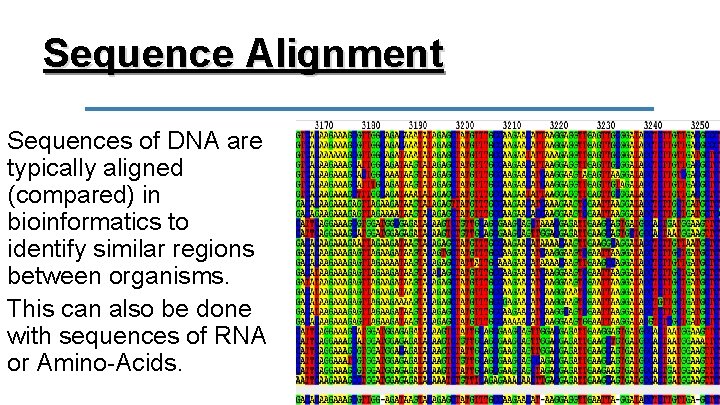
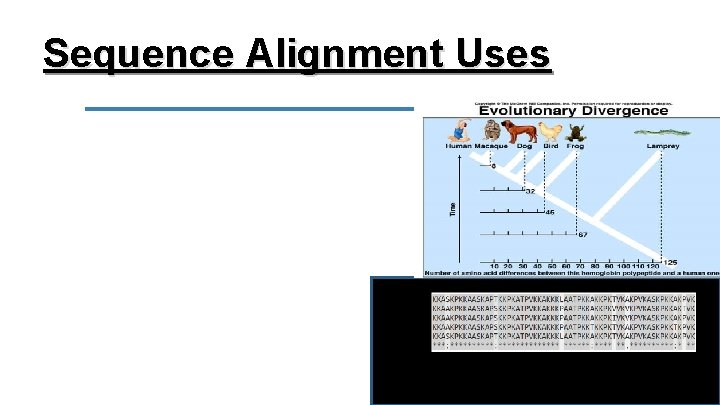
Sequence Alignment Sequences of DNA are typically aligned (compared) in bioinformatics to identify similar regions between organisms. This can also be done with sequences of RNA or Amino-Acids.

Sequence Alignment This can be used to: Compare SNP’s between individuals in a population. Identify sequence similarities in people with a genetic disorder. Create/refine phylogenetic (evolutionary) trees by comparing sequences between different species.

Sequence Alignment Computers are needed to do these types of searches, because of the enormous amount of information being accessed. By using different algorithims, we can fine tune computers to find similarities and differences between genomic sequences much quicker and with better accuracy than humans.

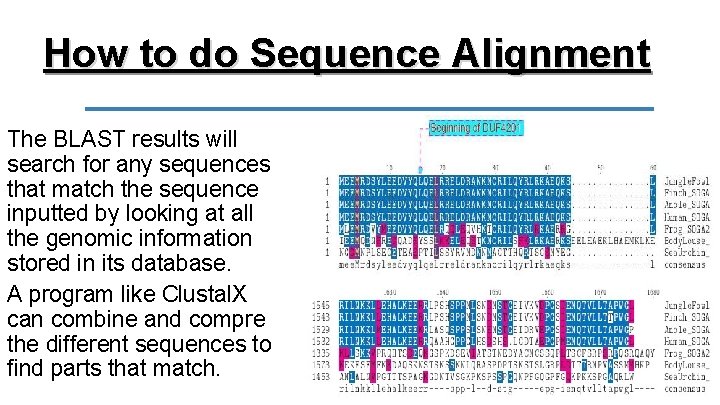
How to do Sequence Alignment To carry out a search for a homologous nucleotide or amino acid sequence, a scientist would conduct a BLAST search. This stands for Basic Local Alignment Search Tool

How to do Sequence Alignment The BLAST results will search for any sequences that match the sequence inputted by looking at all the genomic information stored in its database. A program like Clustal. X can combine and compre the different sequences to find parts that match.

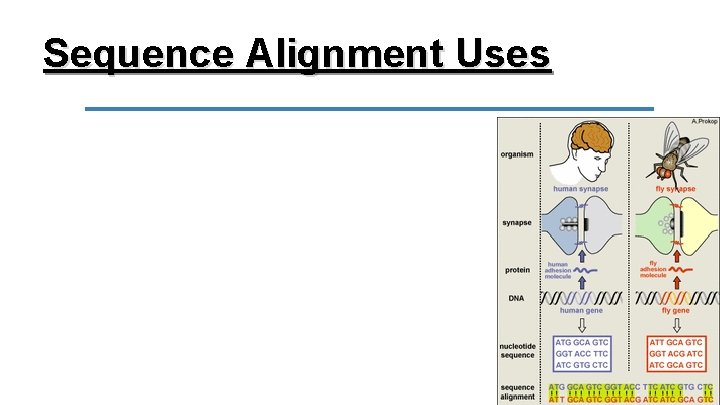
Sequence Alignment Uses DNA/amino acid sequences that match are called homologous/conserved. If they are homologous across species, they most likely play a similar functional role.

Sequence Alignment Uses We can also interpret the homology of sequences to infer relatedness. The more similar the sequences the more closely related the two species are and the less time has passed since there was a common ancestor between the two.

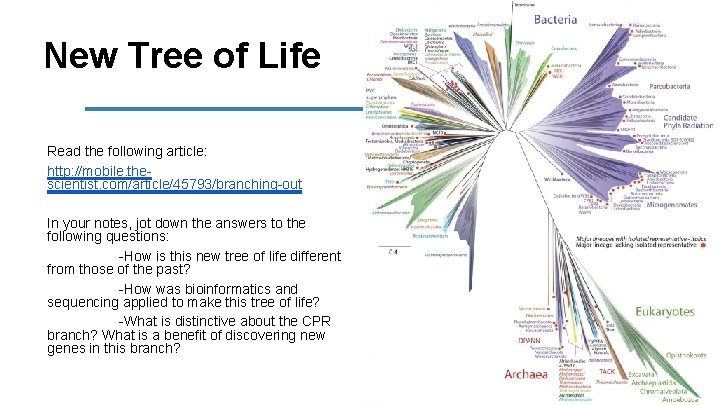
New Tree of Life Read the following article: http: //mobile. thescientist. com/article/45793/branching-out In your notes, jot down the answers to the following questions: -How is this new tree of life different from those of the past? -How was bioinformatics and sequencing applied to make this tree of life? -What is distinctive about the CPR branch? What is a benefit of discovering new genes in this branch?
 Semi-global alignment
Semi-global alignment Khi nào hổ mẹ dạy hổ con săn mồi
Khi nào hổ mẹ dạy hổ con săn mồi Giọng cùng tên là
Giọng cùng tên là Làm thế nào để 102-1=99
Làm thế nào để 102-1=99 Các loại đột biến cấu trúc nhiễm sắc thể
Các loại đột biến cấu trúc nhiễm sắc thể Thể thơ truyền thống
Thể thơ truyền thống Bài hát chúa yêu trần thế alleluia
Bài hát chúa yêu trần thế alleluia đại từ thay thế
đại từ thay thế Thiếu nhi thế giới liên hoan
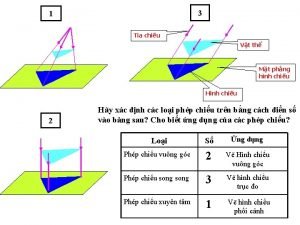
Thiếu nhi thế giới liên hoan Vẽ hình chiếu vuông góc của vật thể sau
Vẽ hình chiếu vuông góc của vật thể sau Công thức tiính động năng
Công thức tiính động năng Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Vẽ hình chiếu đứng bằng cạnh của vật thể
Vẽ hình chiếu đứng bằng cạnh của vật thể Hệ hô hấp
Hệ hô hấp Quá trình desamine hóa có thể tạo ra
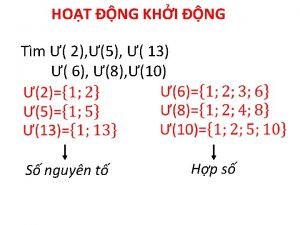
Quá trình desamine hóa có thể tạo ra Số nguyên tố là số gì
Số nguyên tố là số gì Các môn thể thao bắt đầu bằng tiếng chạy
Các môn thể thao bắt đầu bằng tiếng chạy Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay điện thế nghỉ
điện thế nghỉ ưu thế lai là gì
ưu thế lai là gì Nguyên nhân của sự mỏi cơ sinh 8
Nguyên nhân của sự mỏi cơ sinh 8 Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Tư thế ngồi viết
Tư thế ngồi viết Phản ứng thế ankan
Phản ứng thế ankan Chó sói
Chó sói Vẽ hình chiếu vuông góc của vật thể sau
Vẽ hình chiếu vuông góc của vật thể sau Từ ngữ thể hiện lòng nhân hậu
Từ ngữ thể hiện lòng nhân hậu Một số thể thơ truyền thống
Một số thể thơ truyền thống Tư thế ngồi viết
Tư thế ngồi viết Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Slidetodoc
Slidetodoc Sự nuôi và dạy con của hổ
Sự nuôi và dạy con của hổ đặc điểm cơ thể của người tối cổ
đặc điểm cơ thể của người tối cổ Mật thư tọa độ 5x5
Mật thư tọa độ 5x5 Diễn thế sinh thái là
Diễn thế sinh thái là Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Tư thế worms-breton
Tư thế worms-breton Thế nào là mạng điện lắp đặt kiểu nổi
Thế nào là mạng điện lắp đặt kiểu nổi Thẻ vin
Thẻ vin Lời thề hippocrates
Lời thề hippocrates Bàn tay mà dây bẩn
Bàn tay mà dây bẩn Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Bổ thể
Bổ thể Act artemis
Act artemis Biologists search the volumes of the human genome using
Biologists search the volumes of the human genome using Human genome size
Human genome size Gene annotation
Gene annotation Genome assembly
Genome assembly Human genome project
Human genome project Genome adalah
Genome adalah Human genome structure
Human genome structure Genomics
Genomics Stanford
Stanford Genome mapping
Genome mapping Genome identification
Genome identification Yale university poster
Yale university poster Chapter 14 the human genome
Chapter 14 the human genome