Data augmentation using learned transforms for oneshot medical










- Slides: 59

Data augmentation using learned transforms for one-shot medical image segmentation Amy Zhao, Guha Balakrishnan, Fre do Durand, John V. Guttag, Adrian V. Dalca MIT CVPR 2019 Ruohua Shi 2019. 04. 08 1

Motivation o Motivation n Segmentation methods based on CNN rely on supervised training with large labeled datasets. n Labeling datasets of medical images requires significant expertise and time, and is infeasible at large scales. data augmentation hand-engineered hand-tuned preprocessing architectures dataset-specific steps

Motivation o Motivation n Segmentation methods based on CNN rely on supervised training with large labeled datasets. n Labeling datasets of medical images requires significant expertise and time, and is infeasible at large scales. Automated data augmentation method for medical images

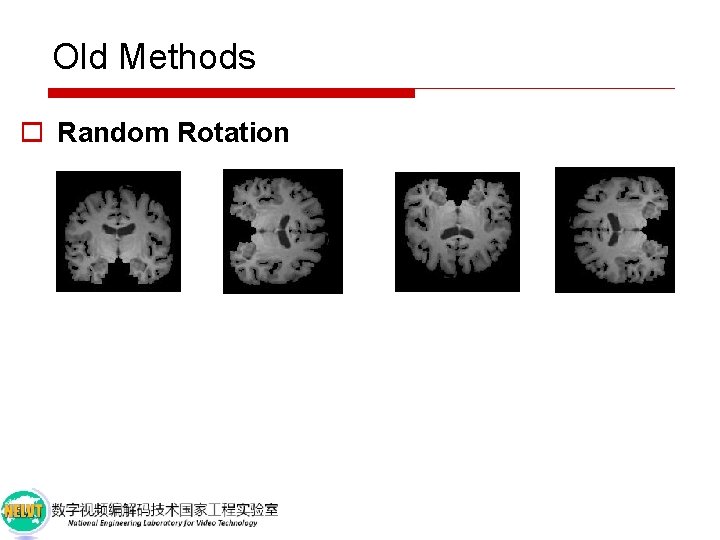
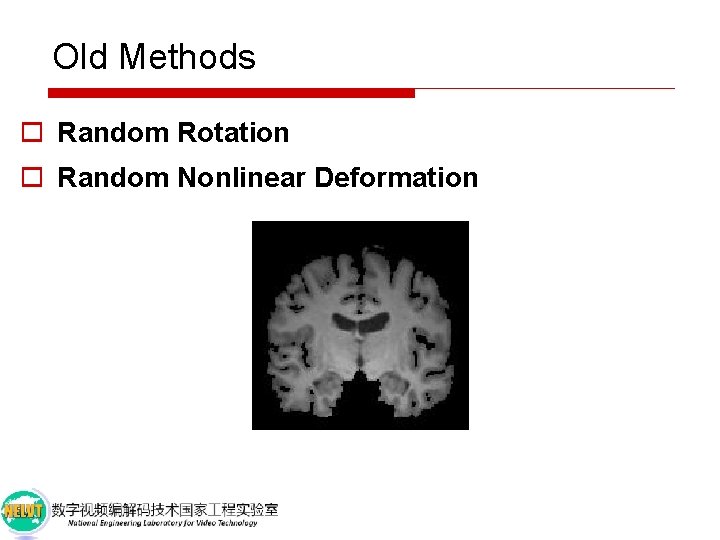
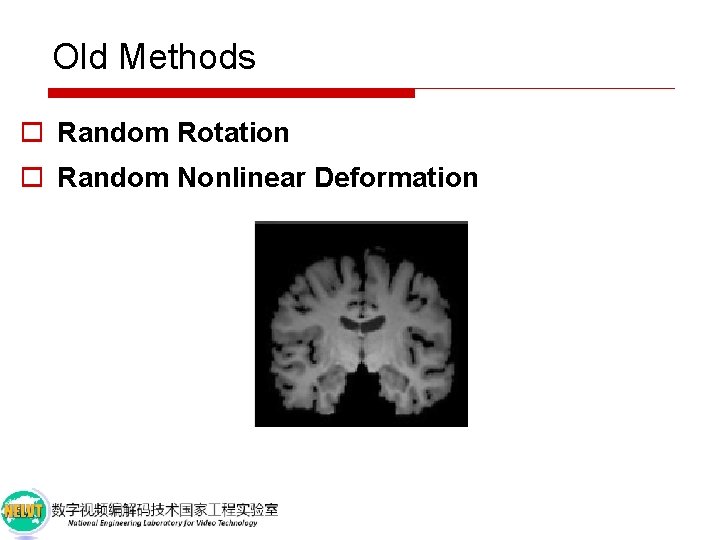
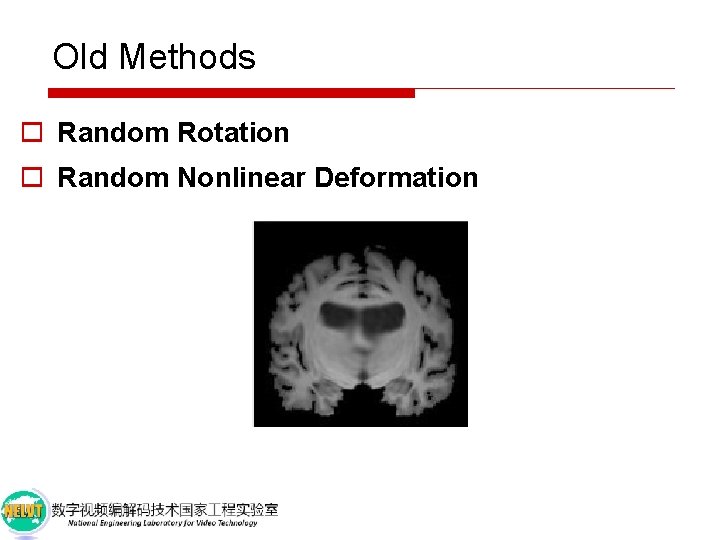
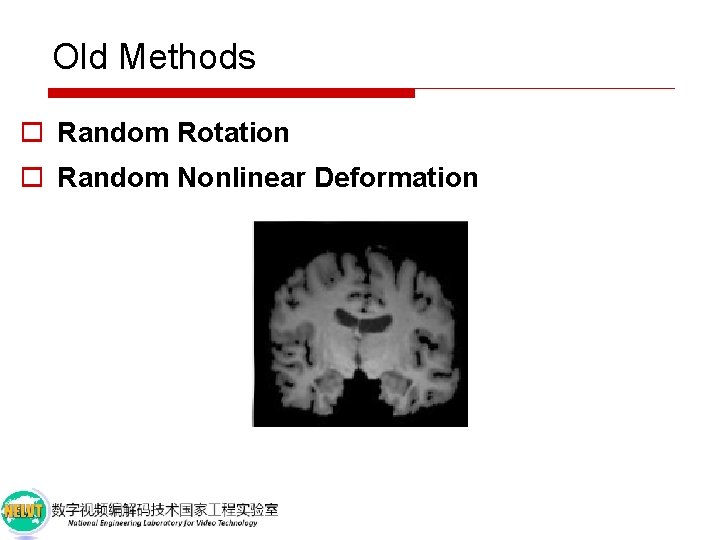
Old Methods o Random Rotation

Old Methods o Random Rotation o Random Nonlinear Deformation

Old Methods o Random Rotation o Random Nonlinear Deformation

Old Methods o Random Rotation o Random Nonlinear Deformation

Old Methods o Random Rotation o Random Nonlinear Deformation

Old Methods o Disadvantages n Have limited ability to emulate diverse and realistic examples. n Be highly sensitive to the choice of parameters. o Solution Synthesize new labeled training examples.

Old Methods o Disadvantages n Have limited ability to emulate diverse and realistic examples. n Be highly sensitive to the choice of parameters. o Solution Synthesize new labeled training examples.

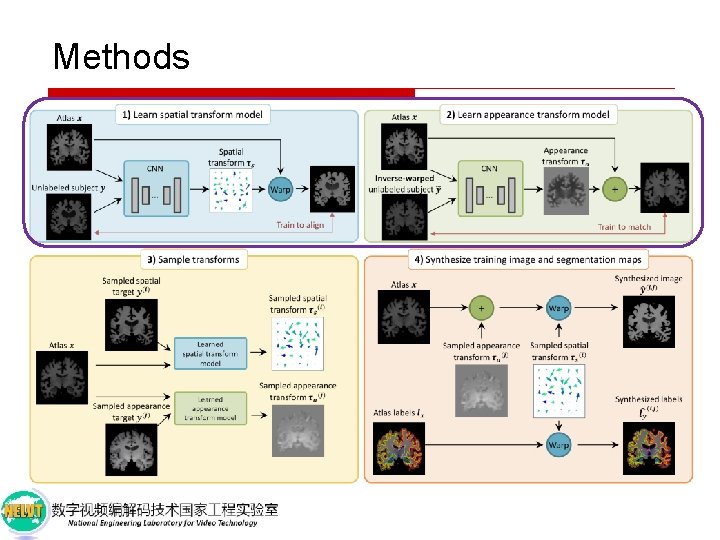
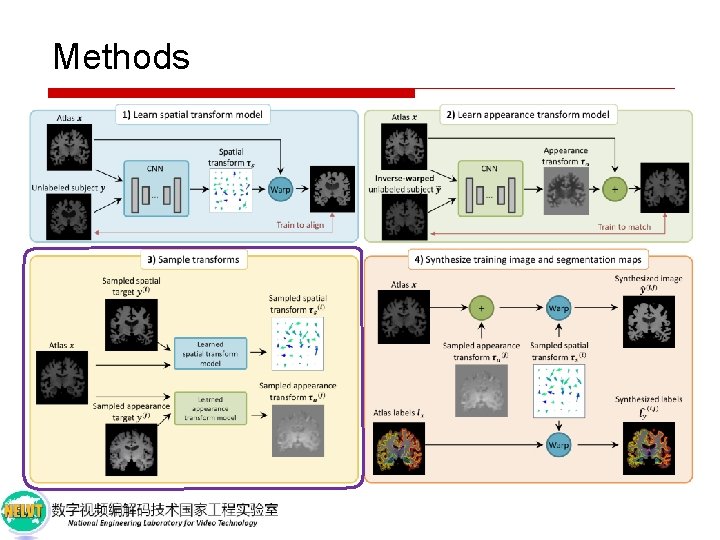
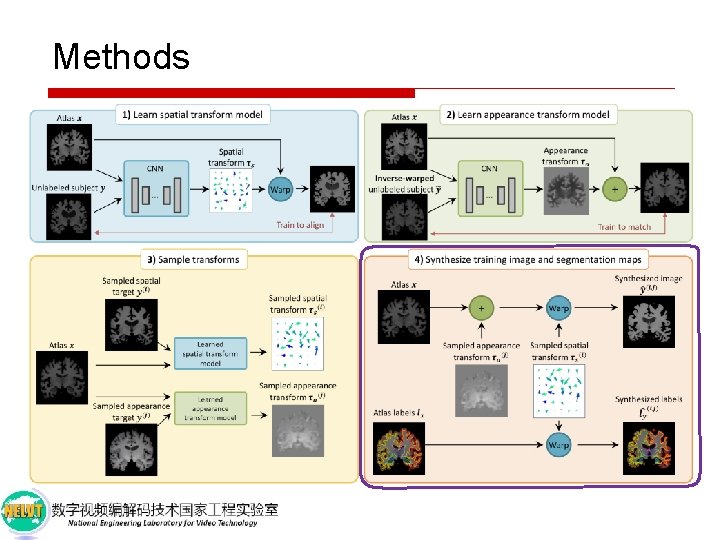
Methods

Methods

Methods

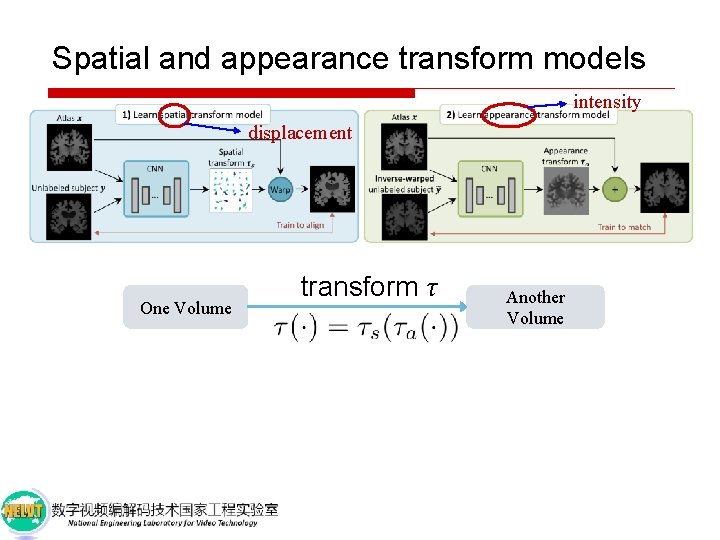
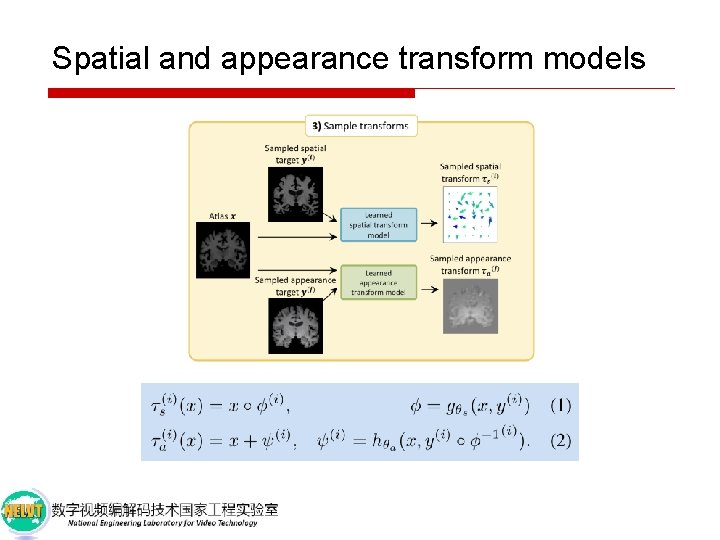
Spatial and appearance transform models intensity displacement One Volume transform τ Another Volume

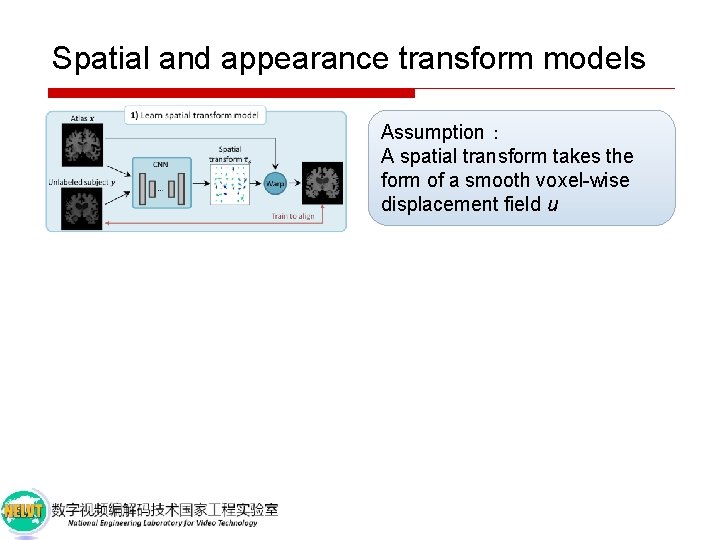
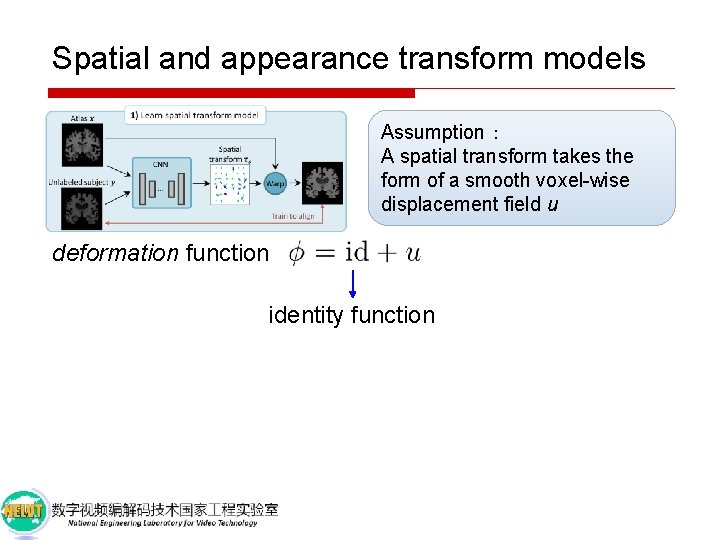
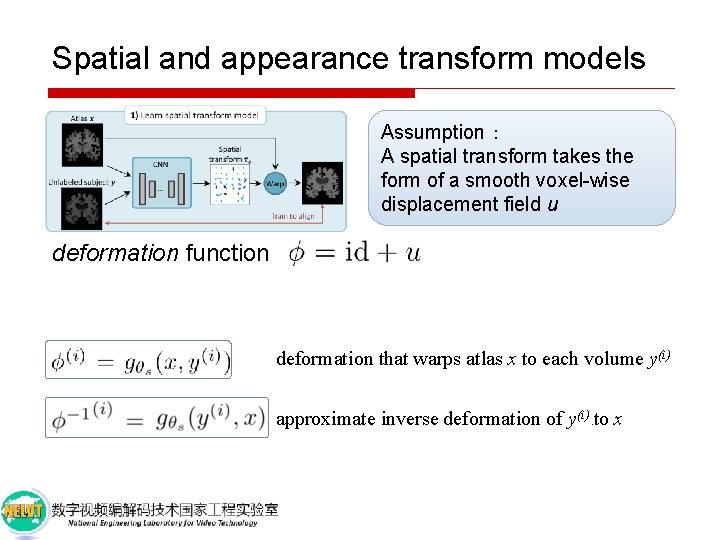
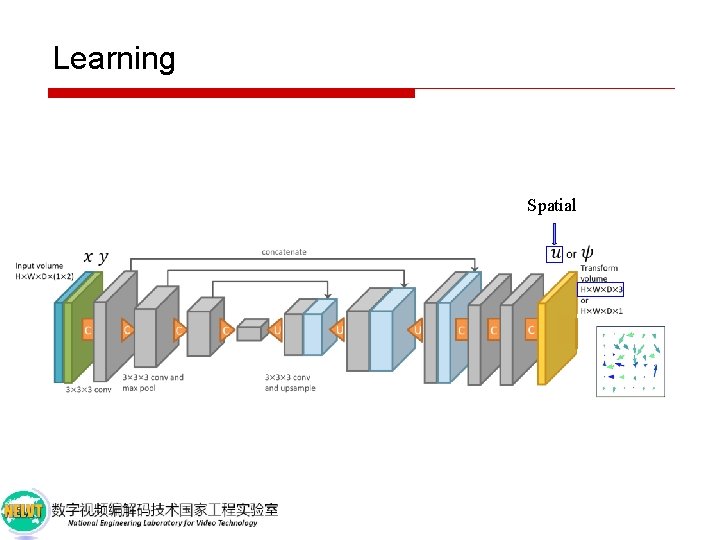
Spatial and appearance transform models Assumption: A spatial transform takes the form of a smooth voxel-wise displacement field u

Spatial and appearance transform models Assumption: A spatial transform takes the form of a smooth voxel-wise displacement field u deformation function identity function

Spatial and appearance transform models Assumption: A spatial transform takes the form of a smooth voxel-wise displacement field u deformation function deformation that warps atlas x to each volume y(i) approximate inverse deformation of y(i) to x )

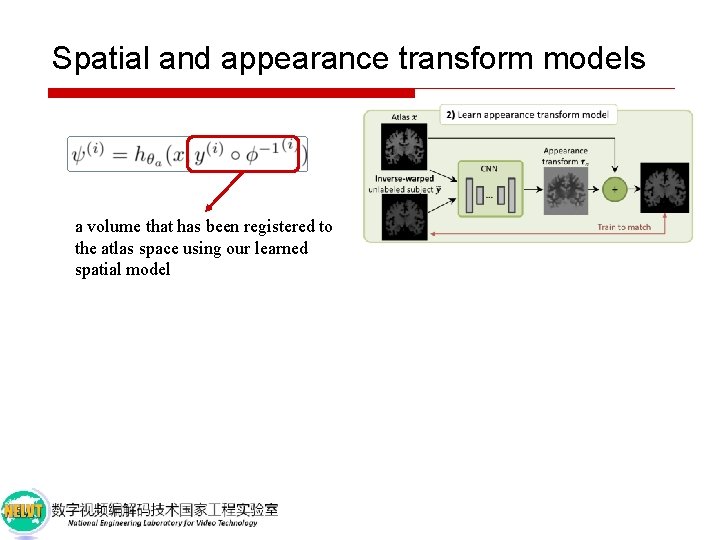
Spatial and appearance transform models a volume that has been registered to the atlas space using our learned spatial model

Spatial and appearance transform models

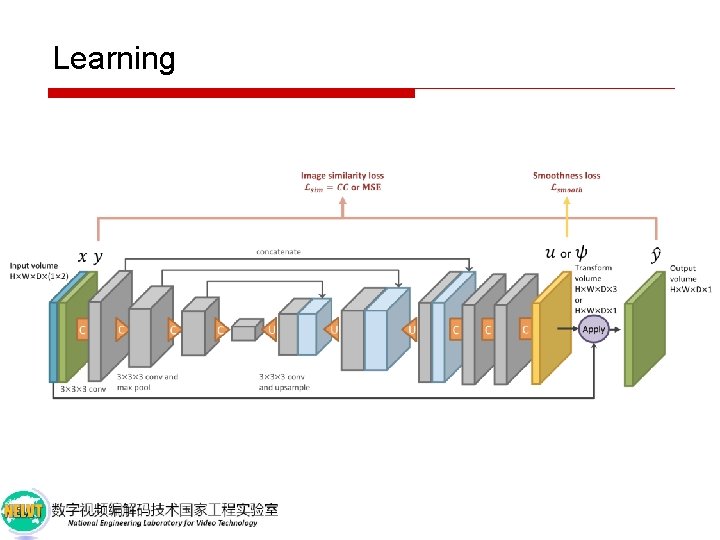
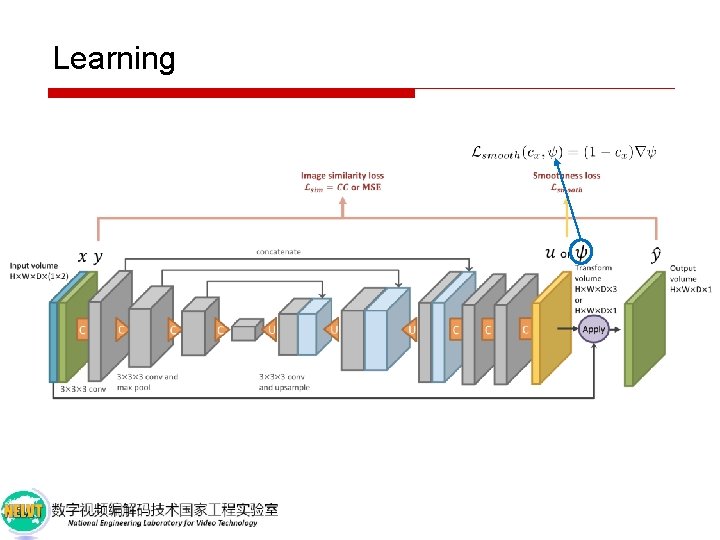
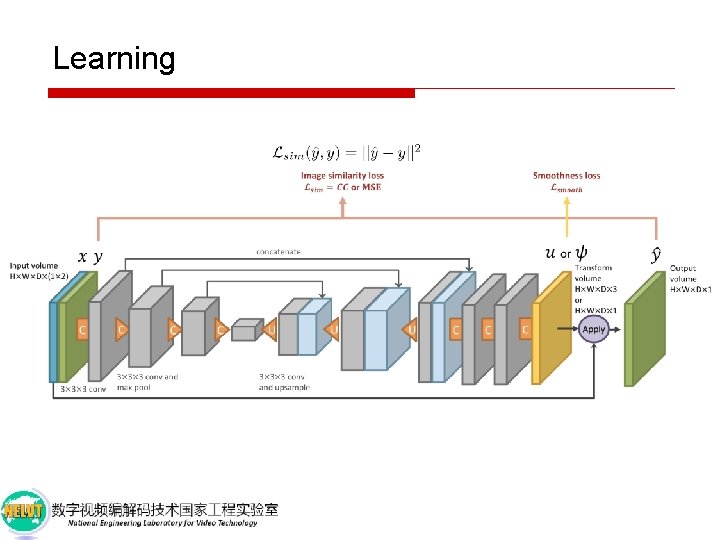
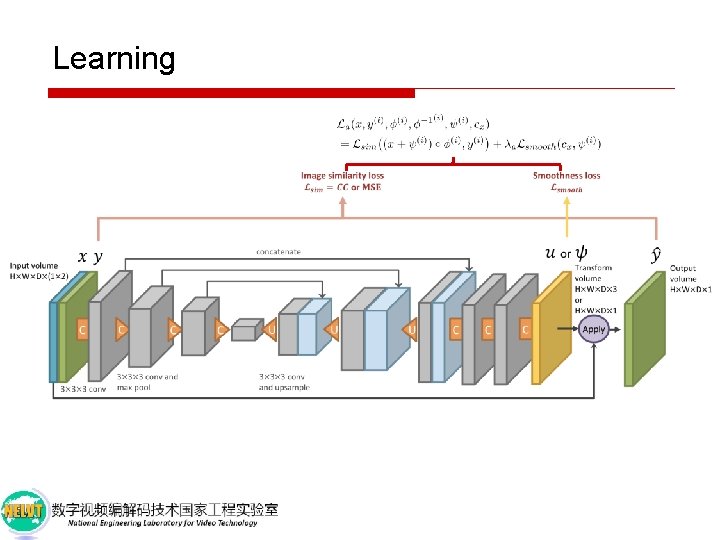
Learning

Learning

Learning Spatial

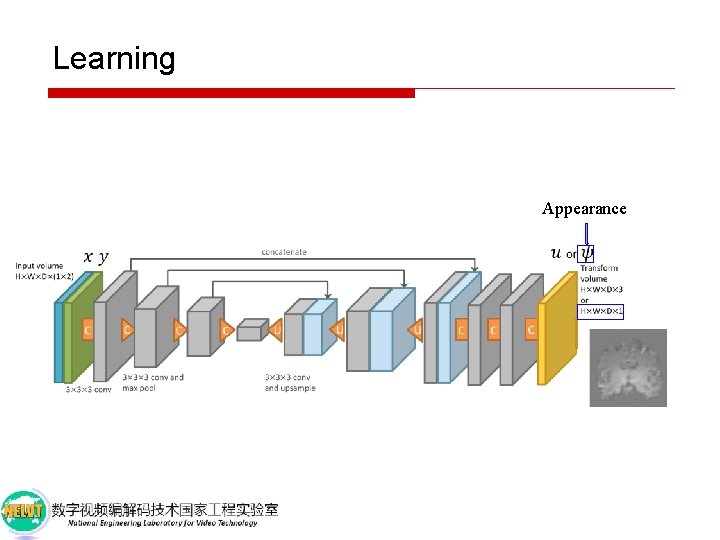
Learning Appearance

Learning

Learning

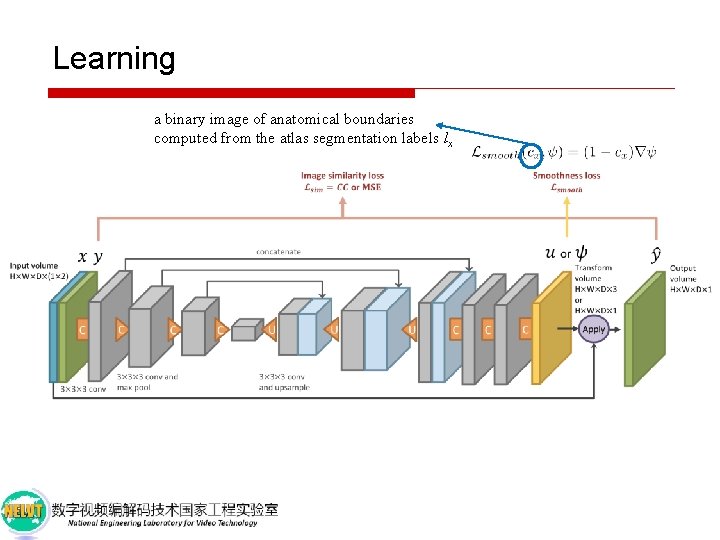
Learning a binary image of anatomical boundaries computed from the atlas segmentation labels lx x

Learning

Learning

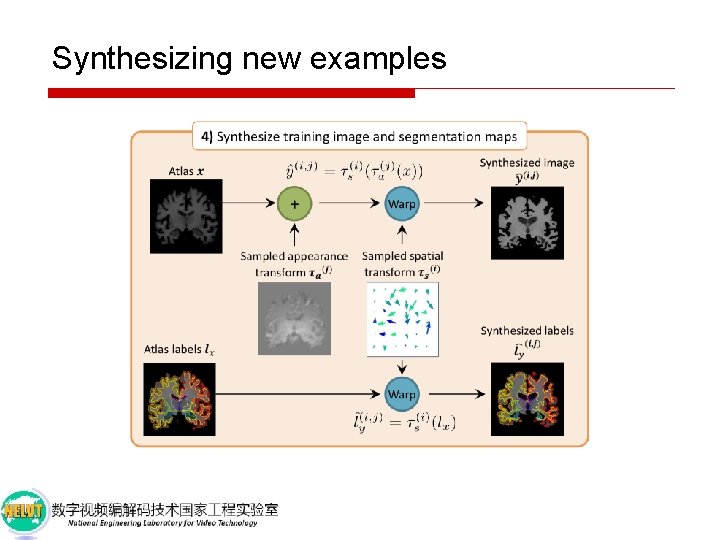
Synthesizing new examples

Synthesizing new examples

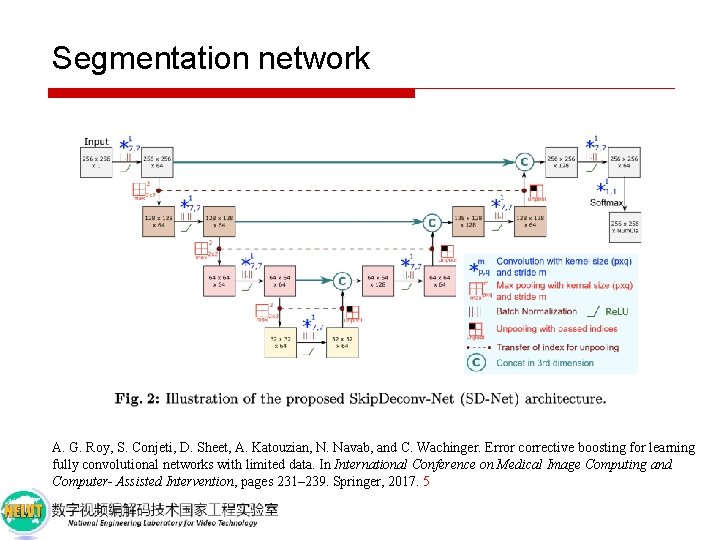
Segmentation network A. G. Roy, S. Conjeti, D. Sheet, A. Katouzian, N. Navab, and C. Wachinger. Error corrective boosting for learning fully convolutional networks with limited data. In International Conference on Medical Image Computing and Computer- Assisted Intervention, pages 231– 239. Springer, 2017. 5

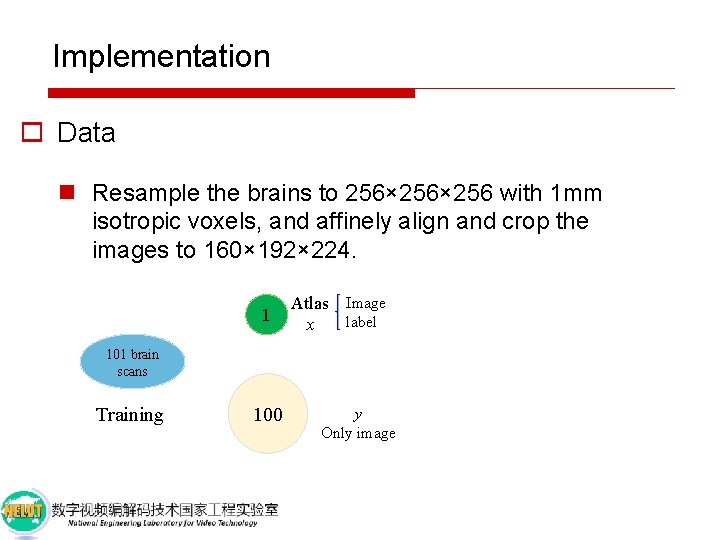
Implementation o Data n Publicly available dataset of T 1 -weighted MRI brain scans contains 8 databases: n ADNI, OASIS, ABIDE, ADHD 200, MCIC, PPMI, HABS, and Harvard GSP n Resample the brains to 256× 256 with 1 mm isotropic voxels, and affinely align and crop the images to 160× 192× 224.

Implementation o Data n Publicly available dataset of T 1 -weighted MRI brain scans contains 8 databases: n ADNI, OASIS, ABIDE, ADHD 200, MCIC, PPMI, HABS, and Harvard GSP n Resample the brains to 256× 256 with 1 mm isotropic voxels, and affinely align and crop the images to 160× 192× 224.

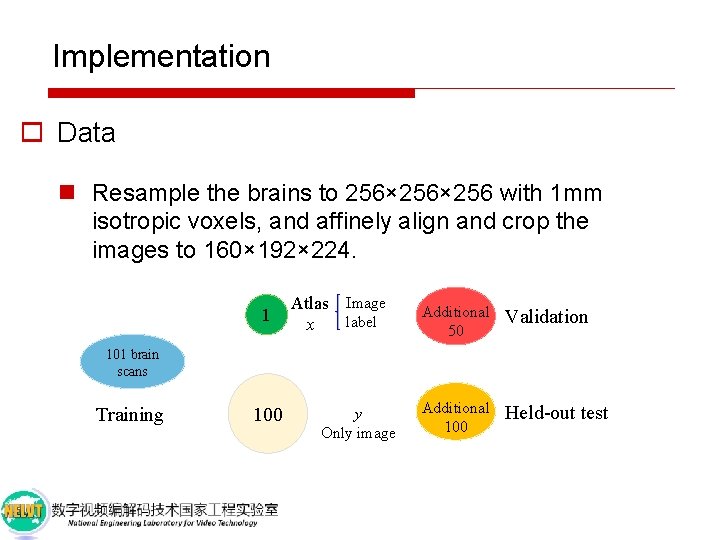
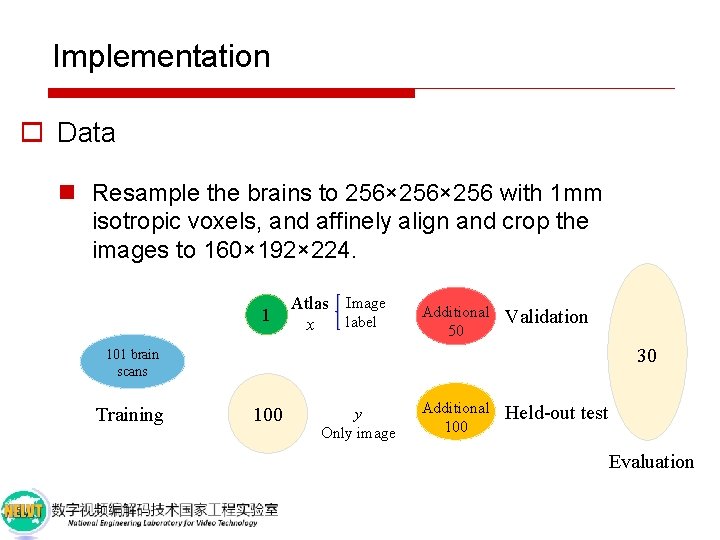
Implementation o Data n Resample the brains to 256× 256 with 1 mm isotropic voxels, and affinely align and crop the images to 160× 192× 224. 101 brain scans Training

Implementation o Data n Resample the brains to 256× 256 with 1 mm isotropic voxels, and affinely align and crop the images to 160× 192× 224. 1 Atlas Image label x 101 brain scans Training 100 y Only image

Implementation o Data n Resample the brains to 256× 256 with 1 mm isotropic voxels, and affinely align and crop the images to 160× 192× 224. 1 Atlas Image label x Additional 50 Validation Additional 100 Held-out test 101 brain scans Training 100 y Only image

Implementation o Data n Resample the brains to 256× 256 with 1 mm isotropic voxels, and affinely align and crop the images to 160× 192× 224. 1 Atlas Image label x Additional 50 Validation 30 101 brain scans Training 100 y Only image Additional 100 Held-out test Evaluation

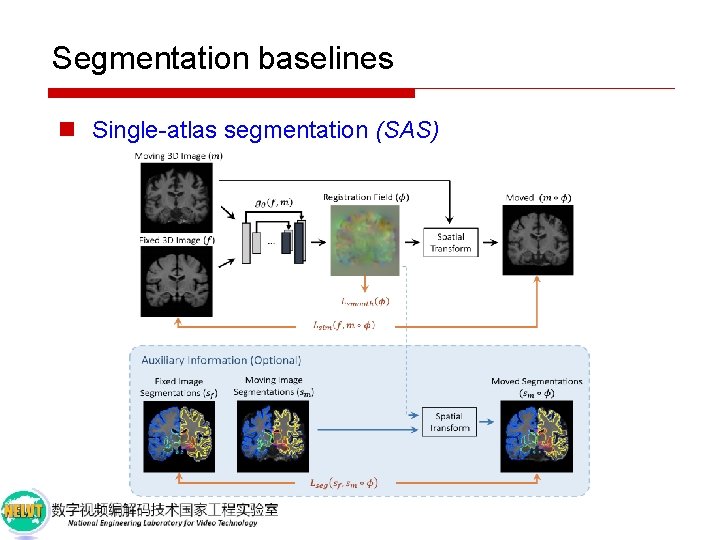
Segmentation baselines n Single-atlas segmentation (SAS)

Segmentation baselines n Single-atlas segmentation (SAS) n Data augmentation usingle-atlas segmentation (SAS-aug) p Use SAS results as labels for the unannotated training brains, which we then include as training examples for supervised segmentation. p This adds 100 new training examples to the segmenter training set. n Hand-tuned random data augmentation (rand-aug) n Supervised

Segmentation baselines n Single-atlas segmentation (SAS) n Data augmentation usingle-atlas segmentation (SAS-aug) n Hand-tuned random data augmentation (rand-aug) p 1. Create a random smooth deformation field by sampling random displacement vectors on a sparse grid. p 2. Applying bilinear interpolation and spatial blurring. n Supervised

Segmentation baselines n Single-atlas segmentation (SAS) n Data augmentation usingle-atlas segmentation (SAS-aug) n Hand-tuned random data augmentation (rand-aug) n Supervised p Train a fully-supervised segmentation network that uses ground truth labels for all 101 examples in training dataset. p Apart from the atlas labels, these labels are not available for any of the other methods. This method serves as an upper bound.

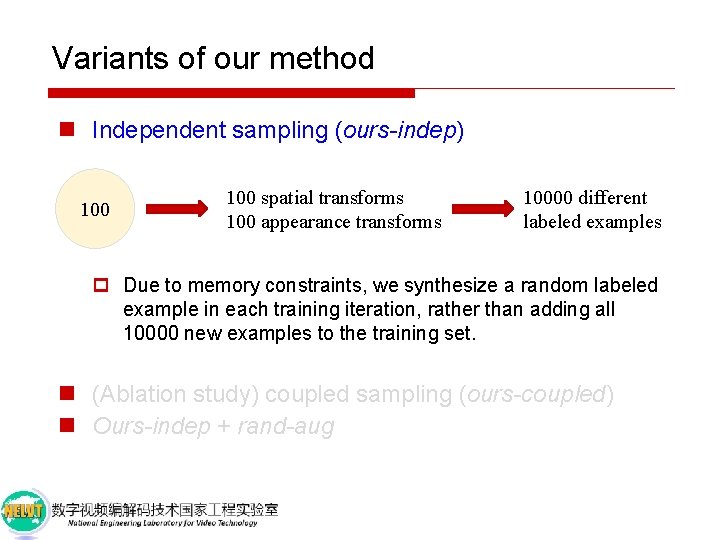
Variants of our method n Independent sampling (ours-indep) 100 spatial transforms 100 appearance transforms 10000 different labeled examples p Due to memory constraints, we synthesize a random labeled example in each training iteration, rather than adding all 10000 new examples to the training set. n (Ablation study) coupled sampling (ours-coupled) n Ours-indep + rand-aug

Variants of our method n Independent sampling (ours-indep) n (Ablation study) coupled sampling (ours-coupled) p Sample each of the spatial and appearance transforms from the same target image. This results in 100 possible synthetic examples. p As in ours-indep, we synthesize a random example in each training iteration. y(i, i) n Ours-indep + rand-aug

Variants of our method n Independent sampling (ours-indep) n (Ablation study) coupled sampling (ours-coupled) n Ours-indep + rand-aug training

Variants of our method n Independent sampling (ours-indep) n (Ablation study) coupled sampling (ours-coupled) n Ours-indep + rand-aug training

Variants of our method n Independent sampling (ours-indep) n (Ablation study) coupled sampling (ours-coupled) n Ours-indep + rand-aug training

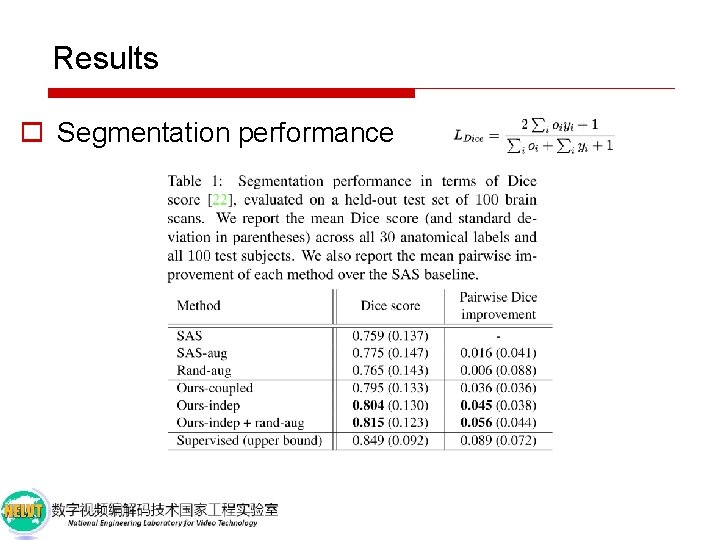
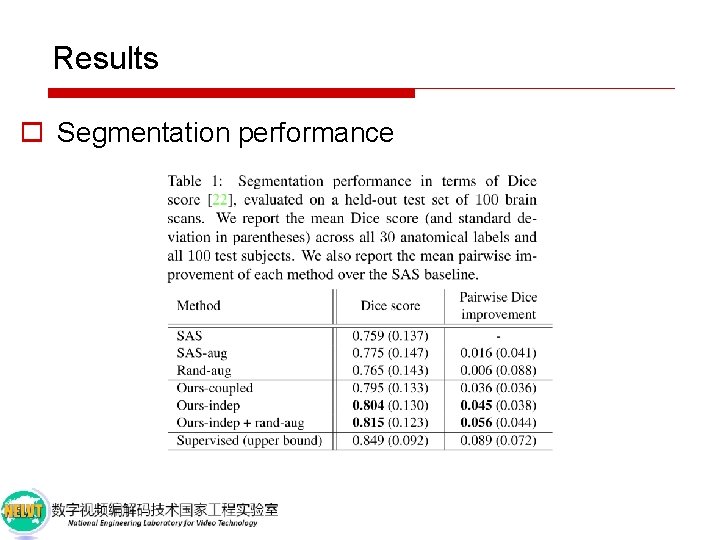
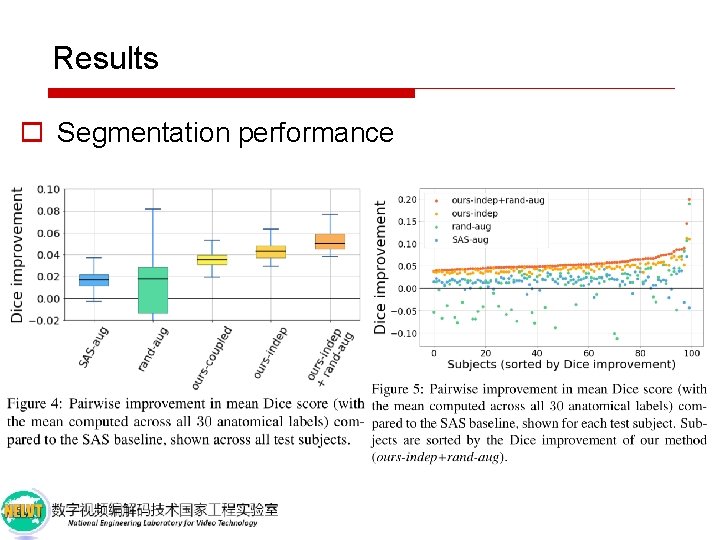
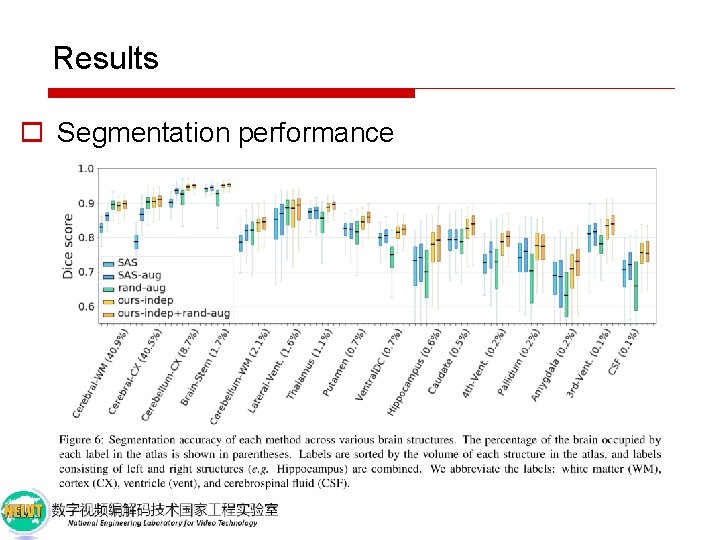
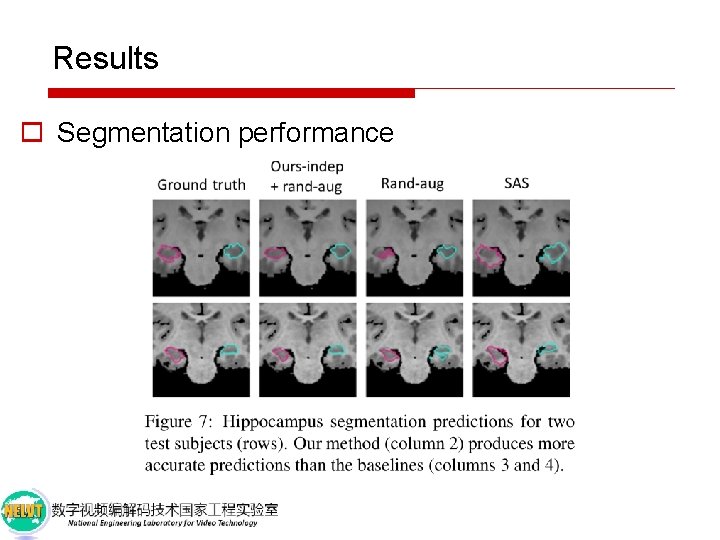
Results o Segmentation performance

Results o Segmentation performance

Results o Segmentation performance

Results o Segmentation performance

Results o Segmentation performance

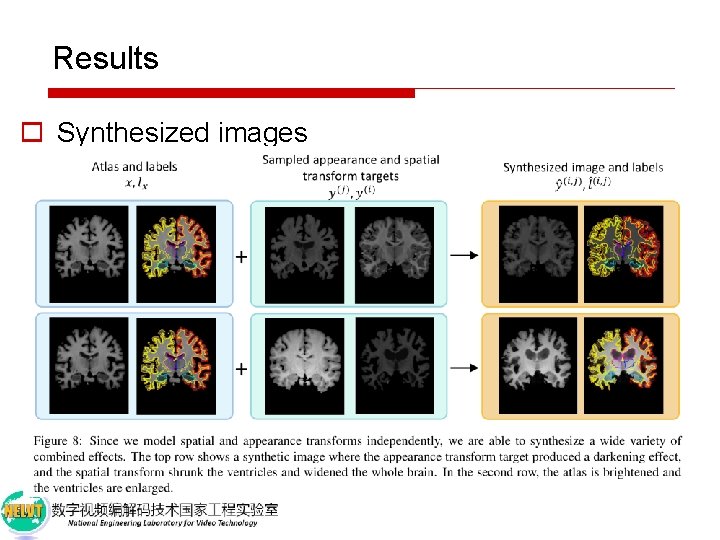
Results o Synthesized images

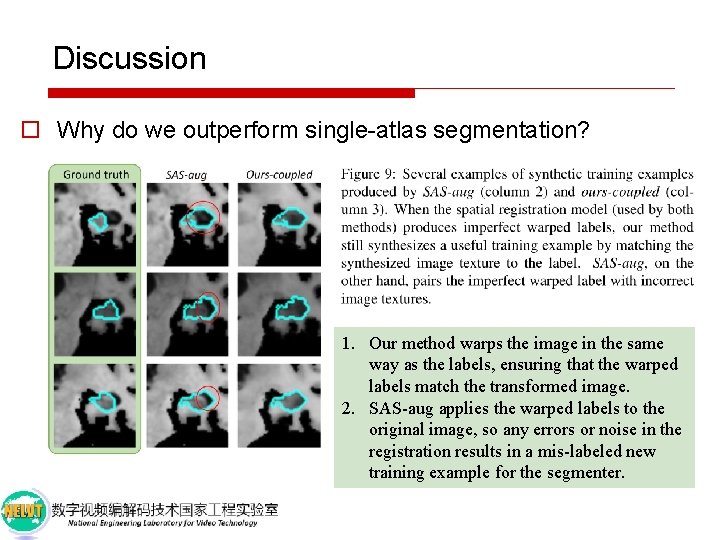
Discussion o Why do we outperform single-atlas segmentation? 1. Our method warps the image in the same way as the labels, ensuring that the warped labels match the transformed image. 2. SAS-aug applies the warped labels to the original image, so any errors or noise in the registration results in a mis-labeled new training example for the segmenter.

Discussion o Extensions n Learning a separate inverse spatial transform model ☞ diffeomorphic registration n Sample transforms from a discrete set of spatial and appearance transforms. ☞ Span the space of transforms more richly, e. g. , through interpolation between transforms, or using compositions of transforms. n On brain MRIs ☞ Other anatomy or imaging modalities, such as CT.

Discussion o Extensions n Learning a separate inverse spatial transform model ☞ diffeomorphic registration n Sample transforms from a discrete set of spatial and appearance transforms. ☞ Span the space of transforms more richly, e. g. , through interpolation between transforms, or using compositions of transforms. n On brain MRIs ☞ Other anatomy or imaging modalities, such as CT.

Discussion o Extensions n Learning a separate inverse spatial transform model ☞ diffeomorphic registration n Sample transforms from a discrete set of spatial and appearance transforms. ☞ Span the space of transforms more richly, e. g. , through interpolation between transforms, or using compositions of transforms. n On brain MRIs ☞ Other anatomy or imaging modalities, such as CT.

Conclusion o In summary, this work shows that: n Learning independent models of spatial and appearance transforms from unlabeled images enables the synthesis of diverse and realistic labeled examples n These synthesized examples can be used to train a segmentation model that out-performs existing methods in a one-shot scenario.

Conclusion o In summary, this work shows that: n Learning independent models of spatial and appearance transforms from unlabeled images enables the synthesis of diverse and realistic labeled examples n These synthesized examples can be used to train a segmentation model that out-performs existing methods in a one-shot scenario.

Thanks!