Oligo 7 Primer Analysis Software Rules for PCR


- Slides: 42

Oligo 7 Primer Analysis Software Rules for PCR Primer Selection and Presentation of the Software Ⓒ 2010 Molecular Biology Insights, Inc.

Contents • • • 1. PCR Primer Selection Criteria - explanation of primer stability calculations - what makes a good PCR primer - other factors besides individual primer characteristics influencing PCR reaction 2. Oligo 7 Analyze Features - the Sequence window - general information (the Key Info) - duplex and hairpin formation - data about primers (Composition and Melting Temperatures) - false priming sites and homology analysis - oligonucleotide stability graph (Tm, DG) - internal stability and its importance - sequence frequency - other DNA analysis options: ORF, restriction sites, DNA calculator 3. Search Options - search for primers and probes - other searches - batch processing 4. Oligonucleotide Database - multiplexing 5. Concluding Remarks

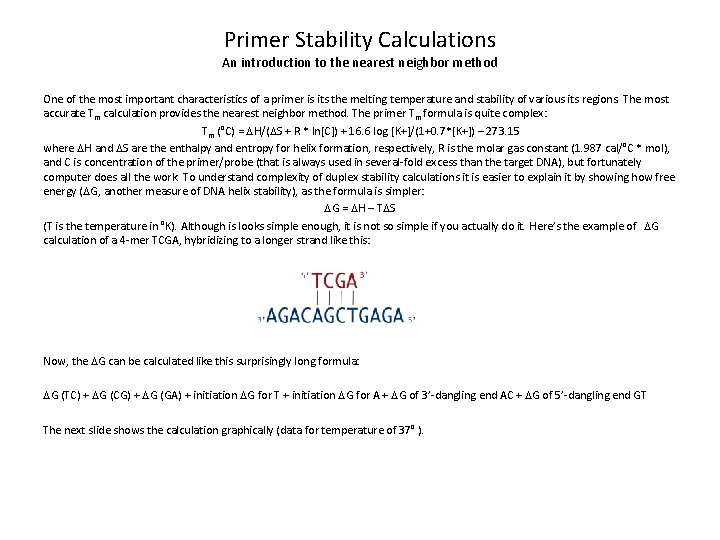
Primer Stability Calculations An introduction to the nearest neighbor method One of the most important characteristics of a primer is its the melting temperature and stability of various its regions. The most accurate Tm calculation provides the nearest neighbor method. The primer Tm formula is quite complex: Tm (°C) = DH/(DS + R * ln[C]) + 16. 6 log [K+]/(1+0. 7*[K+]) – 273. 15 where DH and DS are the enthalpy and entropy for helix formation, respectively, R is the molar gas constant (1. 987 cal/°C * mol), and C is concentration of the primer/probe (that is always used in several-fold excess than the target DNA), but fortunately computer does all the work. To understand complexity of duplex stability calculations it is easier to explain it by showing how free energy (DG, another measure of DNA helix stability), as the formula is simpler: DG = DH – TDS (T is the temperature in °K). Although is looks simple enough, it is not so simple if you actually do it. Here’s the example of DG calculation of a 4 -mer TCGA, hybridizing to a longer strand like this: Now, the DG can be calculated like this surprisingly long formula: DG (TC) + DG (CG) + DG (GA) + initiation DG for T + initiation DG for A + DG of 3’-dangling end AC + DG of 5’-dangling end GT The next slide shows the calculation graphically (data for temperature of 37° ).

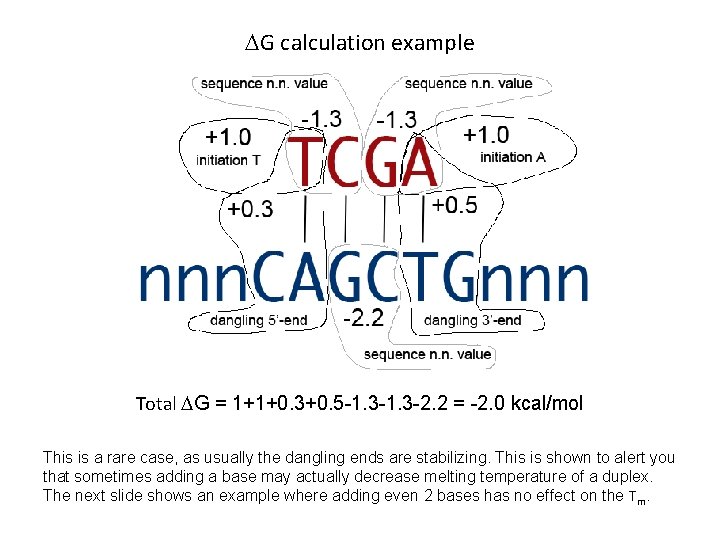
DG calculation example Total DG = 1+1+0. 3+0. 5 -1. 3 -2. 2 = -2. 0 kcal/mol This is a rare case, as usually the dangling ends are stabilizing. This is shown to alert you that sometimes adding a base may actually decrease melting temperature of a duplex. The next slide shows an example where adding even 2 bases has no effect on the Tm.

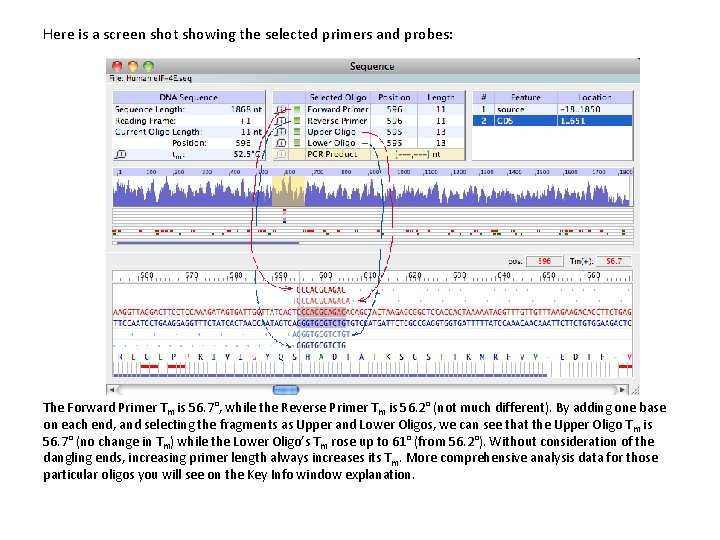
Here is a screen shot showing the selected primers and probes: The Forward Primer Tm is 56. 7°, while the Reverse Primer Tm is 56. 2° (not much different). By adding one base on each end, and selecting the fragments as Upper and Lower Oligos, we can see that the Upper Oligo T m is 56. 7° (no change in Tm) while the Lower Oligo’s Tm rose up to 61° (from 56. 2°). Without consideration of the dangling ends, increasing primer length always increases its Tm. More comprehensive analysis data for those particular oligos you will see on the Key Info window explanation.

What makes a good primer 1. High priming efficiency to achieve this, a good primer should be - of a reasonably high Tm, - without dimers, especially on their 3’-ends (to prevent self-extension), - without hairpin stems, especially on their 3’-ends (to prevent self-priming), - lacking repetitive sequences to ensure quick and correct annealing. - all primers/probes in one incubation mixture should not form significant 3’dimers between each other. 2. High specificity to achieve this, a good primer should be - long enough to increase specificity, - unique, especially at its 3’-end, to avoid false priming, - moderately stable at its 3’-end (as opposed to highly GC-rich) to ensure that a very short fragment won’t initialize the extension (too low 3’-end stability hurts the priming efficiency).

Other factors besides individual primer characteristics influencing PCR reaction Besides the primer properties listed on the previous page, PCR efficiency also depends on the template. Regions with high secondary structures and GC islands amplify poorly. Oligo 7 penalizes primers that would amplify strong hairpin structures, decreasing overall scores for PCR primer sets. If you have to make hard to amplify products, select primers with high scores (usually with higher Tm and GC contents than standard), and adjust composition of your PCR mix by adding solvents, additives or reducing/changing salts. Increasing primer concentration above 1 mmol is not recommended due to unspecific primer/primer extensions (even if the software does not show any 3’-dimers).

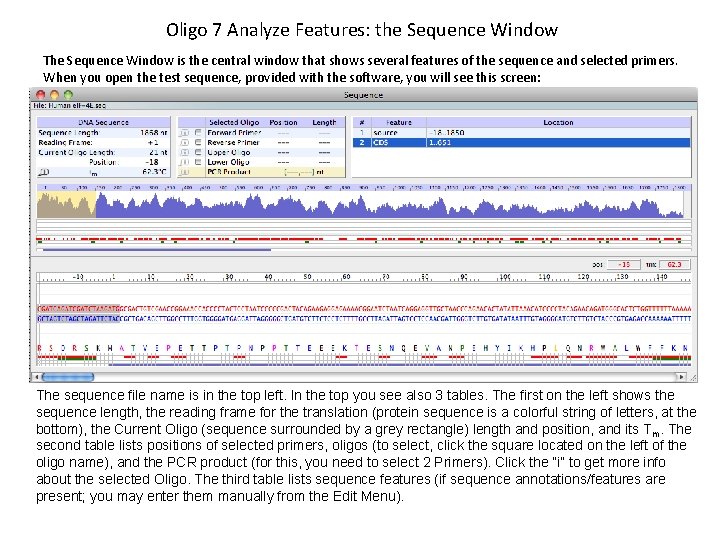
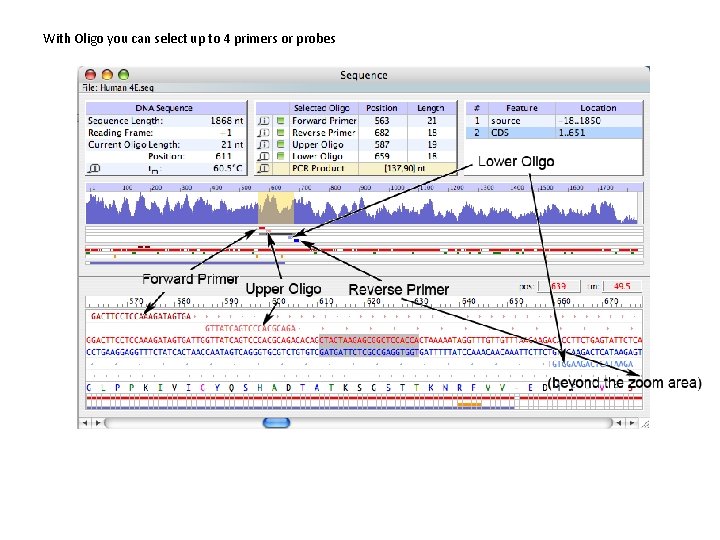
Oligo 7 Analyze Features: the Sequence Window The Sequence Window is the central window that shows several features of the sequence and selected primers. When you open the test sequence, provided with the software, you will see this screen: The sequence file name is in the top left. In the top you see also 3 tables. The first on the left shows the sequence length, the reading frame for the translation (protein sequence is a colorful string of letters, at the bottom), the Current Oligo (sequence surrounded by a grey rectangle) length and position, and its T m. The second table lists positions of selected primers, oligos (to select, click the square located on the left of the oligo name), and the PCR product (for this, you need to select 2 Primers). Click the “i” to get more info about the selected Oligo. The third table lists sequence features (if sequence annotations/features are present; you may enter them manually from the Edit Menu).

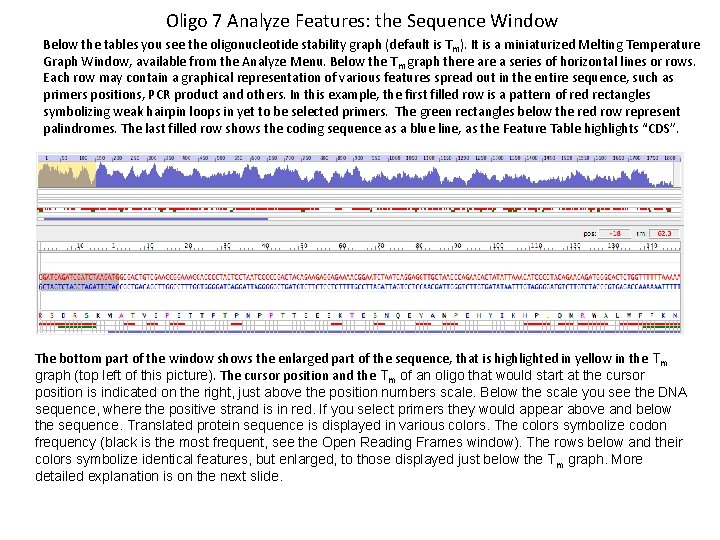
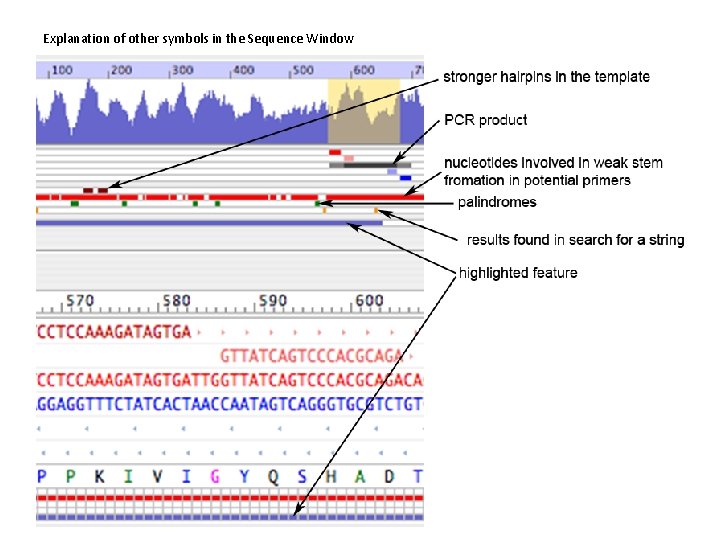
Oligo 7 Analyze Features: the Sequence Window Below the tables you see the oligonucleotide stability graph (default is T m). It is a miniaturized Melting Temperature Graph Window, available from the Analyze Menu. Below the Tm graph there a series of horizontal lines or rows. Each row may contain a graphical representation of various features spread out in the entire sequence, such as primers positions, PCR product and others. In this example, the first filled row is a pattern of red rectangles symbolizing weak hairpin loops in yet to be selected primers. The green rectangles below the red row represent palindromes. The last filled row shows the coding sequence as a blue line, as the Feature Table highlights “CDS”. The bottom part of the window shows the enlarged part of the sequence, that is highlighted in yellow in the Tm graph (top left of this picture). The cursor position and the Tm of an oligo that would start at the cursor position is indicated on the right, just above the position numbers scale. Below the scale you see the DNA sequence, where the positive strand is in red. If you select primers they would appear above and below the sequence. Translated protein sequence is displayed in various colors. The colors symbolize codon frequency (black is the most frequent, see the Open Reading Frames window). The rows below and their colors symbolize identical features, but enlarged, to those displayed just below the Tm graph. More detailed explanation is on the next slide.

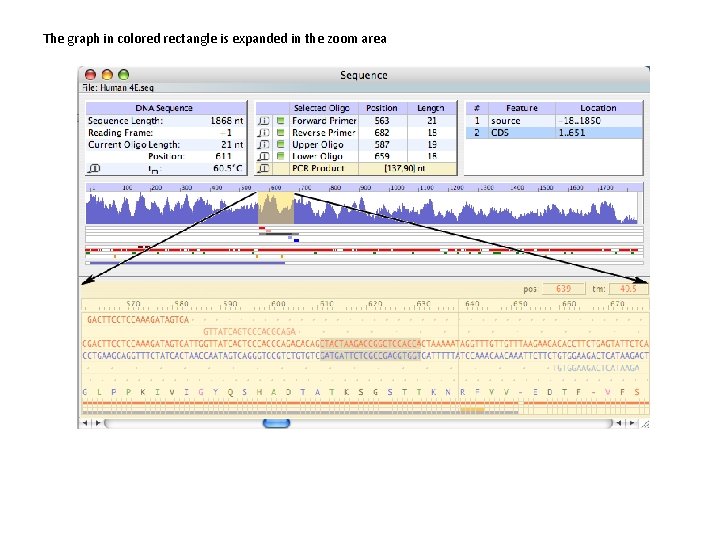
The graph in colored rectangle is expanded in the zoom area

With Oligo you can select up to 4 primers or probes

Explanation of other symbols in the Sequence Window

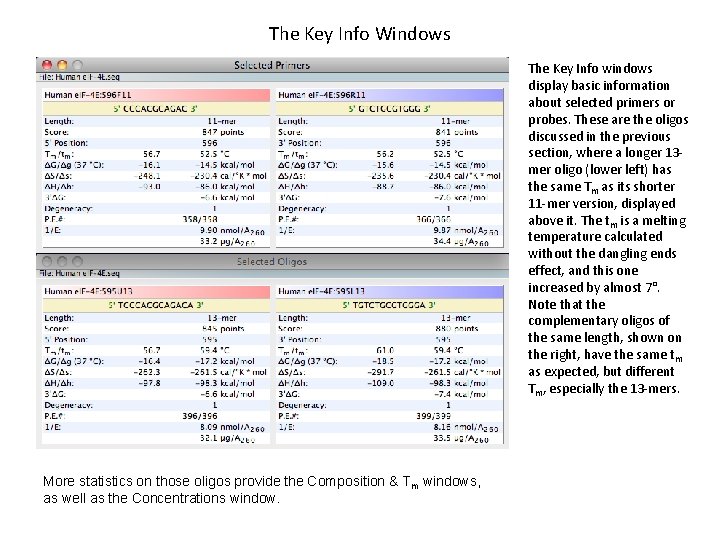
The Key Info Windows The Key Info windows display basic information about selected primers or probes. These are the oligos discussed in the previous section, where a longer 13 mer oligo (lower left) has the same Tm as its shorter 11 -mer version, displayed above it. The tm is a melting temperature calculated without the dangling ends effect, and this one increased by almost 7°. Note that the complementary oligos of the same length, shown on the right, have the same tm as expected, but different Tm, especially the 13 -mers. More statistics on those oligos provide the Composition & Tm windows, as well as the Concentrations window.

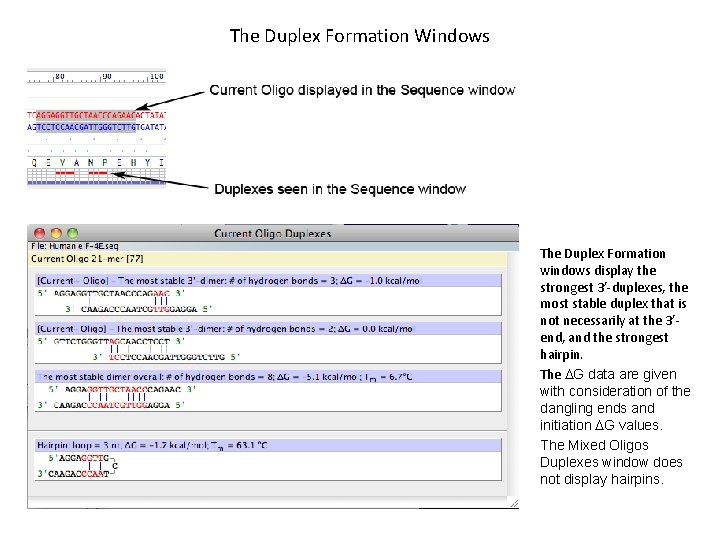
The Duplex Formation Windows The Duplex Formation windows display the strongest 3’-duplexes, the most stable duplex that is not necessarily at the 3’end, and the strongest hairpin. The DG data are given with consideration of the dangling ends and initiation DG values. The Mixed Oligos Duplexes window does not display hairpins.

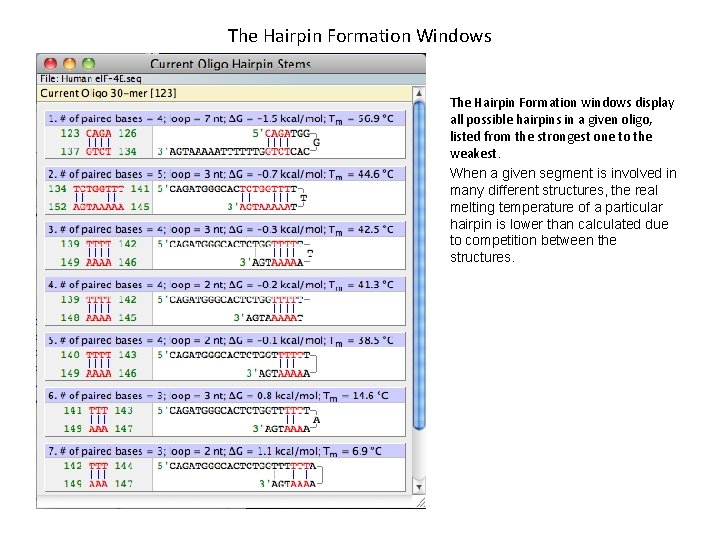
The Hairpin Formation Windows The Hairpin Formation windows display all possible hairpins in a given oligo, listed from the strongest one to the weakest. When a given segment is involved in many different structures, the real melting temperature of a particular hairpin is lower than calculated due to competition between the structures.

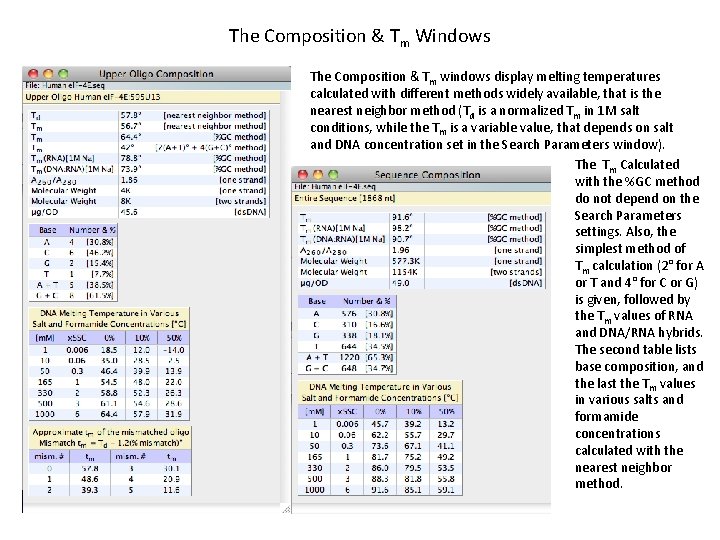
The Composition & Tm Windows The Composition & Tm windows display melting temperatures calculated with different methods widely available, that is the nearest neighbor method (Td is a normalized Tm in 1 M salt conditions, while the Tm is a variable value, that depends on salt and DNA concentration set in the Search Parameters window). The Tm Calculated with the %GC method do not depend on the Search Parameters settings. Also, the simplest method of Tm calculation (2° for A or T and 4° for C or G) is given, followed by the Tm values of RNA and DNA/RNA hybrids. The second table lists base composition, and the last the Tm values in various salts and formamide concentrations calculated with the nearest neighbor method.

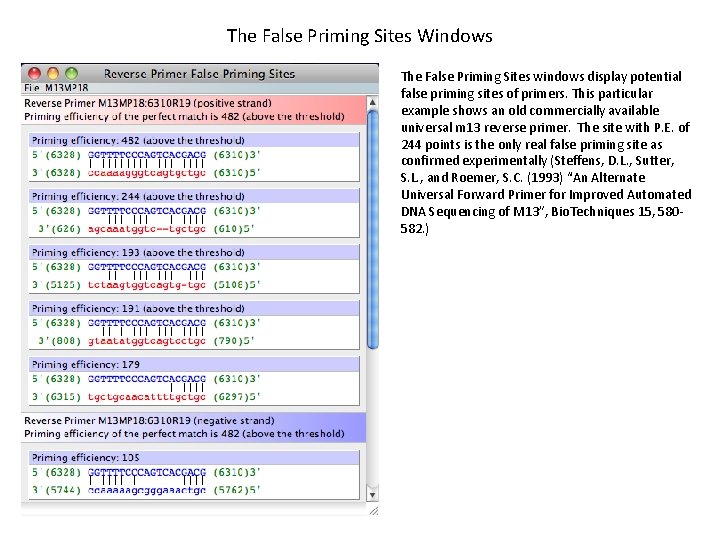
The False Priming Sites Windows The False Priming Sites windows display potential false priming sites of primers. This particular example shows an old commercially available universal m 13 reverse primer. The site with P. E. of 244 points is the only real false priming site as confirmed experimentally (Steffens, D. L. , Sutter, S. L. , and Roemer, S. C. (1993) “An Alternate Universal Forward Primer for Improved Automated DNA Sequencing of M 13”, Bio. Techniques 15, 580582. )

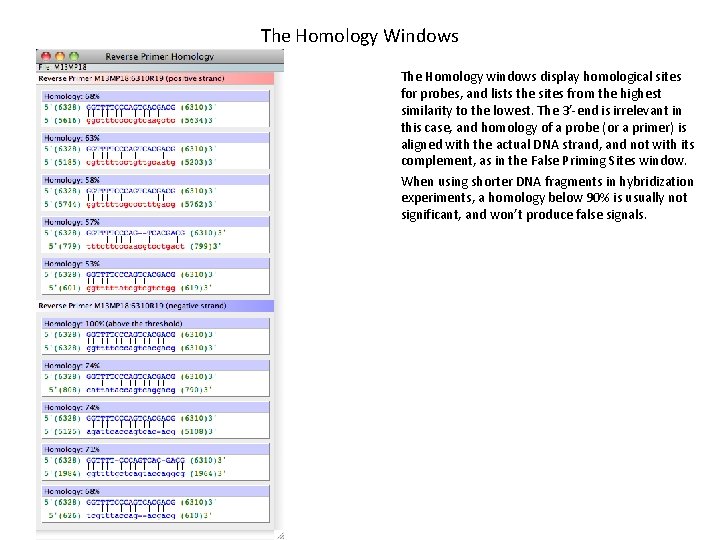
The Homology Windows The Homology windows display homological sites for probes, and lists the sites from the highest similarity to the lowest. The 3’-end is irrelevant in this case, and homology of a probe (or a primer) is aligned with the actual DNA strand, and not with its complement, as in the False Priming Sites window. When using shorter DNA fragments in hybridization experiments, a homology below 90% is usually not significant, and won’t produce false signals.

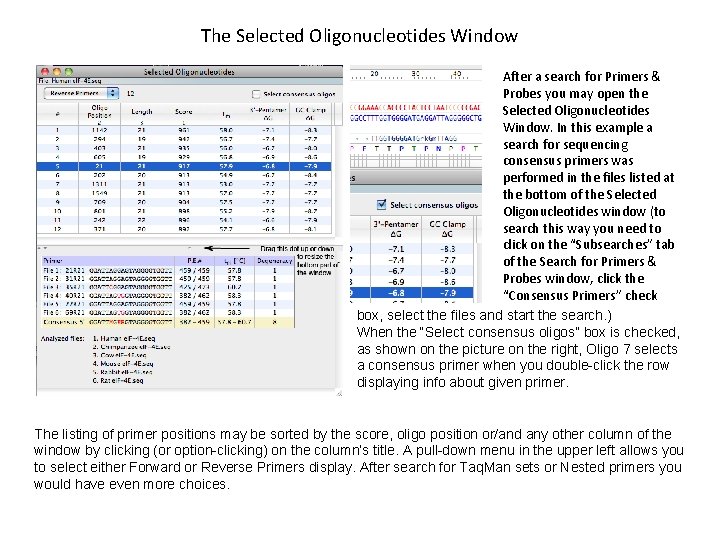
The Selected Oligonucleotides Window After a search for Primers & Probes you may open the Selected Oligonucleotides Window. In this example a search for sequencing consensus primers was performed in the files listed at the bottom of the Selected Oligonucleotides window (to search this way you need to click on the “Subsearches” tab of the Search for Primers & Probes window, click the “Consensus Primers” check box, select the files and start the search. ) When the “Select consensus oligos” box is checked, as shown on the picture on the right, Oligo 7 selects a consensus primer when you double-click the row displaying info about given primer. The listing of primer positions may be sorted by the score, oligo position or/and any other column of the window by clicking (or option-clicking) on the column’s title. A pull-down menu in the upper left allows you to select either Forward or Reverse Primers display. After search for Taq. Man sets or Nested primers you would have even more choices.

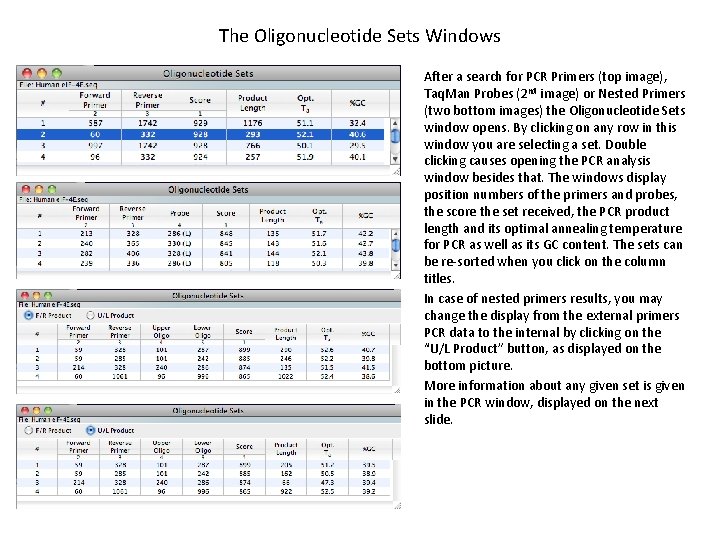
The Oligonucleotide Sets Windows After a search for PCR Primers (top image), Taq. Man Probes (2 nd image) or Nested Primers (two bottom images) the Oligonucleotide Sets window opens. By clicking on any row in this window you are selecting a set. Double clicking causes opening the PCR analysis window besides that. The windows display position numbers of the primers and probes, the score the set received, the PCR product length and its optimal annealing temperature for PCR as well as its GC content. The sets can be re-sorted when you click on the column titles. In case of nested primers results, you may change the display from the external primers PCR data to the internal by clicking on the “U/L Product” button, as displayed on the bottom picture. More information about any given set is given in the PCR window, displayed on the next slide.

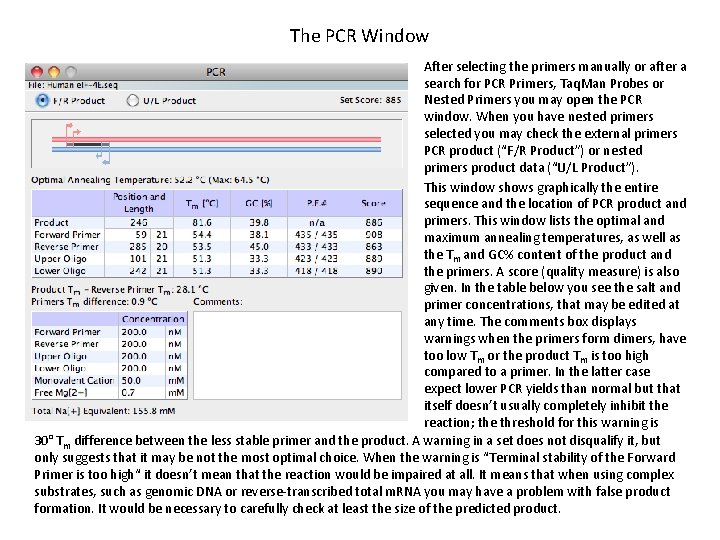
The PCR Window After selecting the primers manually or after a search for PCR Primers, Taq. Man Probes or Nested Primers you may open the PCR window. When you have nested primers selected you may check the external primers PCR product (“F/R Product”) or nested primers product data (“U/L Product”). This window shows graphically the entire sequence and the location of PCR product and primers. This window lists the optimal and maximum annealing temperatures, as well as the Tm and GC% content of the product and the primers. A score (quality measure) is also given. In the table below you see the salt and primer concentrations, that may be edited at any time. The comments box displays warnings when the primers form dimers, have too low Tm or the product Tm is too high compared to a primer. In the latter case expect lower PCR yields than normal but that itself doesn’t usually completely inhibit the reaction; the threshold for this warning is 30° Tm difference between the less stable primer and the product. A warning in a set does not disqualify it, but only suggests that it may be not the most optimal choice. When the warning is “Terminal stability of the Forward Primer is too high“ it doesn’t mean that the reaction would be impaired at all. It means that when using complex substrates, such as genomic DNA or reverse-transcribed total m. RNA you may have a problem with false product formation. It would be necessary to carefully check at least the size of the predicted product.

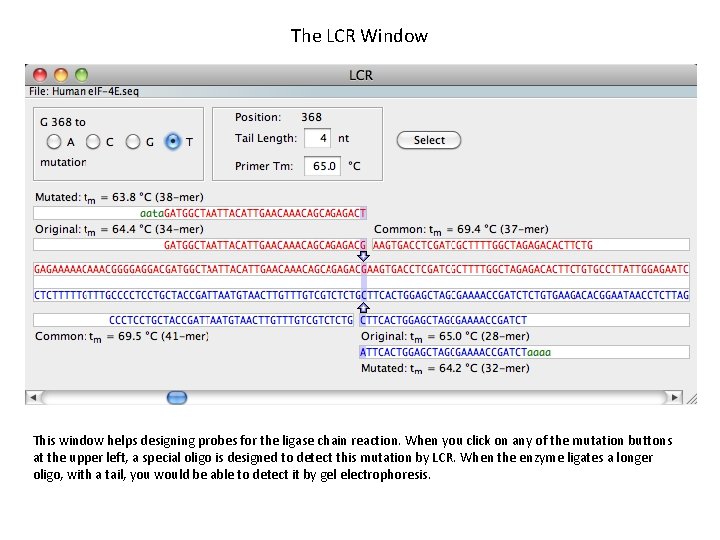
The LCR Window This window helps designing probes for the ligase chain reaction. When you click on any of the mutation buttons at the upper left, a special oligo is designed to detect this mutation by LCR. When the enzyme ligates a longer oligo, with a tail, you would be able to detect it by gel electrophoresis.

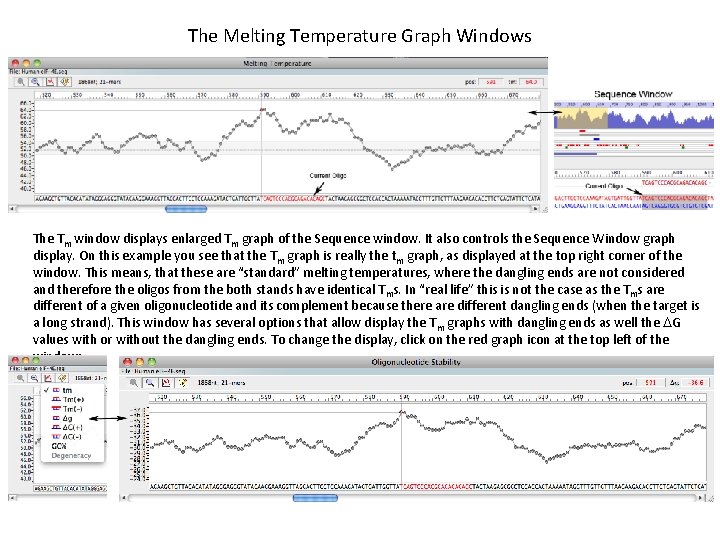
The Melting Temperature Graph Windows The Tm window displays enlarged Tm graph of the Sequence window. It also controls the Sequence Window graph display. On this example you see that the Tm graph is really the tm graph, as displayed at the top right corner of the window. This means, that these are “standard” melting temperatures, where the dangling ends are not considered and therefore the oligos from the both stands have identical Tms. In “real life” this is not the case as the Tms are different of a given oligonucleotide and its complement because there are different dangling ends (when the target is a long strand). This window has several options that allow display the Tm graphs with dangling ends as well the DG values with or without the dangling ends. To change the display, click on the red graph icon at the top left of the window:

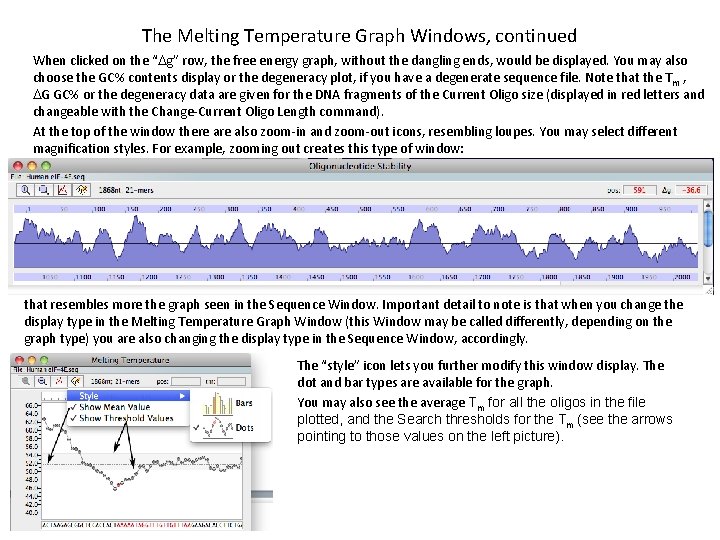
The Melting Temperature Graph Windows, continued When clicked on the “Dg” row, the free energy graph, without the dangling ends, would be displayed. You may also choose the GC% contents display or the degeneracy plot, if you have a degenerate sequence file. Note that the T m , DG GC% or the degeneracy data are given for the DNA fragments of the Current Oligo size (displayed in red letters and changeable with the Change-Current Oligo Length command). At the top of the window there also zoom-in and zoom-out icons, resembling loupes. You may select different magnification styles. For example, zooming out creates this type of window: that resembles more the graph seen in the Sequence Window. Important detail to note is that when you change the display type in the Melting Temperature Graph Window (this Window may be called differently, depending on the graph type) you are also changing the display type in the Sequence Window, accordingly. The “style” icon lets you further modify this window display. The dot and bar types are available for the graph. You may also see the average Tm for all the oligos in the file plotted, and the Search thresholds for the Tm (see the arrows pointing to those values on the left picture).

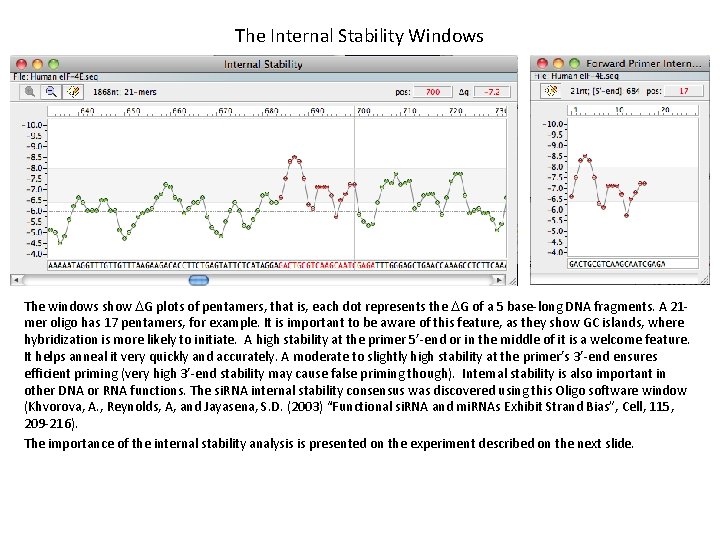
The Internal Stability Windows The windows show DG plots of pentamers, that is, each dot represents the DG of a 5 base-long DNA fragments. A 21 mer oligo has 17 pentamers, for example. It is important to be aware of this feature, as they show GC islands, where hybridization is more likely to initiate. A high stability at the primer 5’-end or in the middle of it is a welcome feature. It helps anneal it very quickly and accurately. A moderate to slightly high stability at the primer’s 3’-end ensures efficient priming (very high 3’-end stability may cause false priming though). Internal stability is also important in other DNA or RNA functions. The si. RNA internal stability consensus was discovered using this Oligo software window (Khvorova, A. , Reynolds, A, and Jayasena, S. D. (2003) “Functional si. RNA and mi. RNAs Exhibit Strand Bias”, Cell, 115, 209 -216). The importance of the internal stability analysis is presented on the experiment described on the next slide.

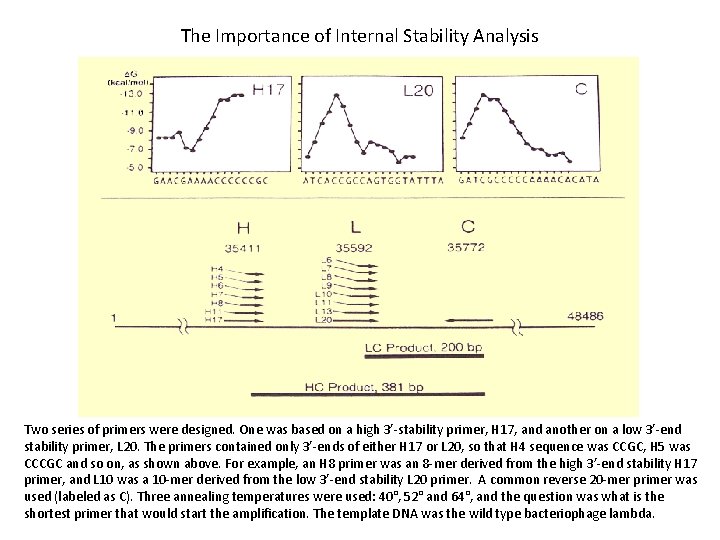
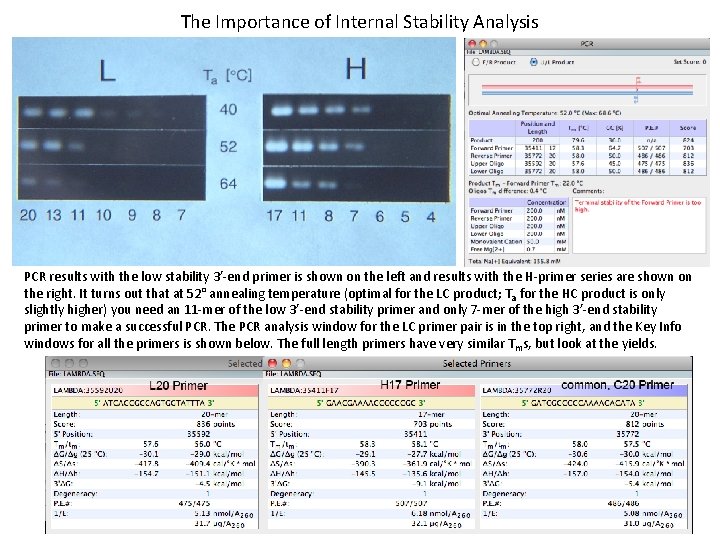
The Importance of Internal Stability Analysis Two series of primers were designed. One was based on a high 3’-stability primer, H 17, and another on a low 3’-end stability primer, L 20. The primers contained only 3’-ends of either H 17 or L 20, so that H 4 sequence was CCGC, H 5 was CCCGC and so on, as shown above. For example, an H 8 primer was an 8 -mer derived from the high 3’-end stability H 17 primer, and L 10 was a 10 -mer derived from the low 3’-end stability L 20 primer. A common reverse 20 -mer primer was used (labeled as C). Three annealing temperatures were used: 40°, 52° and 64°, and the question was what is the shortest primer that would start the amplification. The template DNA was the wild type bacteriophage lambda.

The Importance of Internal Stability Analysis PCR results with the low stability 3’-end primer is shown on the left and results with the H-primer series are shown on the right. It turns out that at 52° annealing temperature (optimal for the LC product; T a for the HC product is only slightly higher) you need an 11 -mer of the low 3’-end stability primer and only 7 -mer of the high 3’-end stability primer to make a successful PCR. The PCR analysis window for the LC primer pair is in the top right, and the Key Info windows for all the primers is shown below. The full length primers have very similar T ms, but look at the yields.

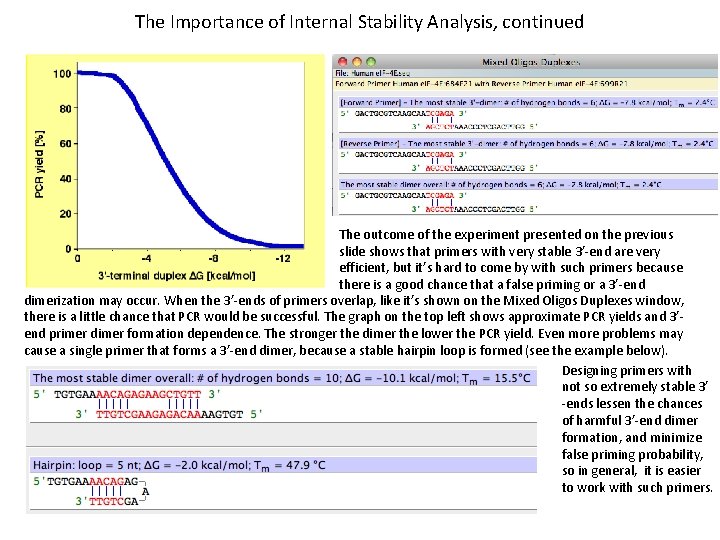
The Importance of Internal Stability Analysis, continued The outcome of the experiment presented on the previous slide shows that primers with very stable 3’-end are very efficient, but it’s hard to come by with such primers because there is a good chance that a false priming or a 3’-end dimerization may occur. When the 3’-ends of primers overlap, like it’s shown on the Mixed Oligos Duplexes window, there is a little chance that PCR would be successful. The graph on the top left shows approximate PCR yields and 3’end primer dimer formation dependence. The stronger the dimer the lower the PCR yield. Even more problems may cause a single primer that forms a 3’-end dimer, because a stable hairpin loop is formed (see the example below). Designing primers with not so extremely stable 3’ -ends lessen the chances of harmful 3’-end dimer formation, and minimize false priming probability, so in general, it is easier to work with such primers.

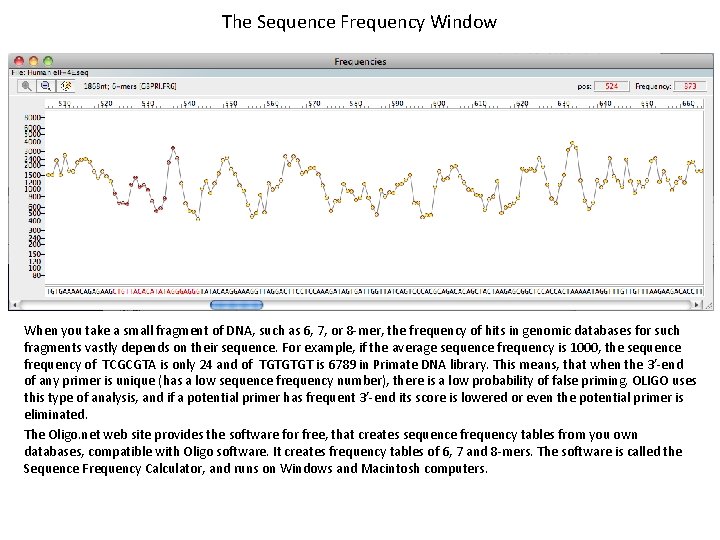
The Sequence Frequency Window When you take a small fragment of DNA, such as 6, 7, or 8 -mer, the frequency of hits in genomic databases for such fragments vastly depends on their sequence. For example, if the average sequence frequency is 1000, the sequence frequency of TCGCGTA is only 24 and of TGTGTGT is 6789 in Primate DNA library. This means, that when the 3’-end of any primer is unique (has a low sequence frequency number), there is a low probability of false priming. OLIGO uses this type of analysis, and if a potential primer has frequent 3’-end its score is lowered or even the potential primer is eliminated. The Oligo. net web site provides the software for free, that creates sequence frequency tables from you own databases, compatible with Oligo software. It creates frequency tables of 6, 7 and 8 -mers. The software is called the Sequence Frequency Calculator, and runs on Windows and Macintosh computers.

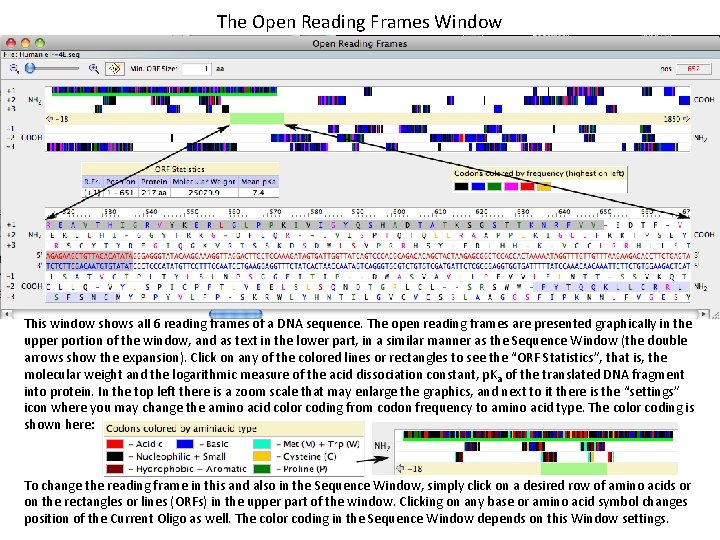
The Open Reading Frames Window This window shows all 6 reading frames of a DNA sequence. The open reading frames are presented graphically in the upper portion of the window, and as text in the lower part, in a similar manner as the Sequence Window (the double arrows show the expansion). Click on any of the colored lines or rectangles to see the “ORF Statistics”, that is, the molecular weight and the logarithmic measure of the acid dissociation constant, p. K a of the translated DNA fragment into protein. In the top left there is a zoom scale that may enlarge the graphics, and next to it there is the “settings” icon where you may change the amino acid color coding from codon frequency to amino acid type. The color coding is shown here: To change the reading frame in this and also in the Sequence Window, simply click on a desired row of amino acids or on the rectangles or lines (ORFs) in the upper part of the window. Clicking on any base or amino acid symbol changes position of the Current Oligo as well. The color coding in the Sequence Window depends on this Window settings.

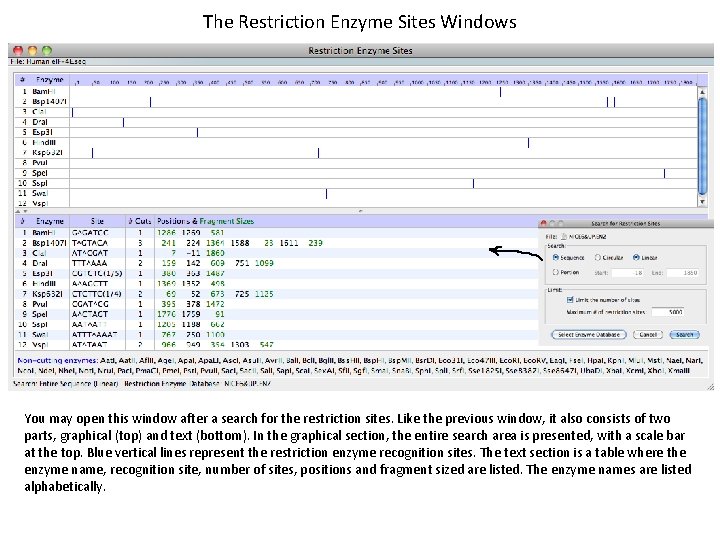
The Restriction Enzyme Sites Windows You may open this window after a search for the restriction sites. Like the previous window, it also consists of two parts, graphical (top) and text (bottom). In the graphical section, the entire search area is presented, with a scale bar at the top. Blue vertical lines represent the restriction enzyme recognition sites. The text section is a table where the enzyme name, recognition site, number of sites, positions and fragment sized are listed. The enzyme names are listed alphabetically.

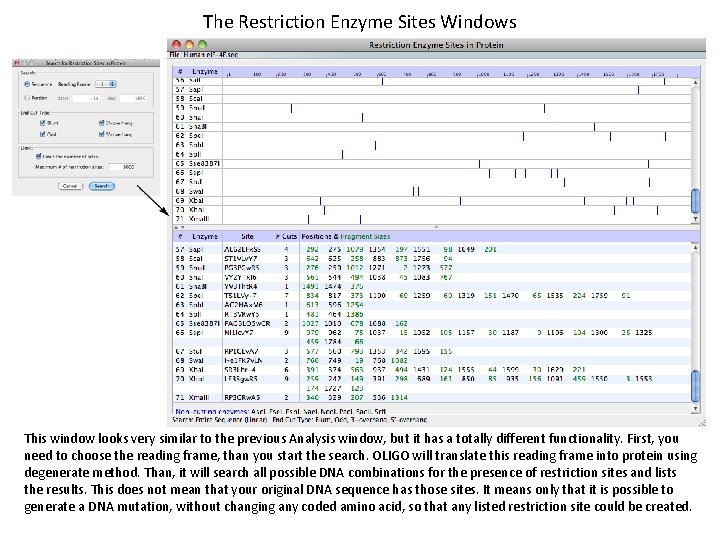
The Restriction Enzyme Sites Windows This window looks very similar to the previous Analysis window, but it has a totally different functionality. First, you need to choose the reading frame, than you start the search. OLIGO will translate this reading frame into protein using degenerate method. Than, it will search all possible DNA combinations for the presence of restriction sites and lists the results. This does not mean that your original DNA sequence has those sites. It means only that it is possible to generate a DNA mutation, without changing any coded amino acid, so that any listed restriction site could be created.

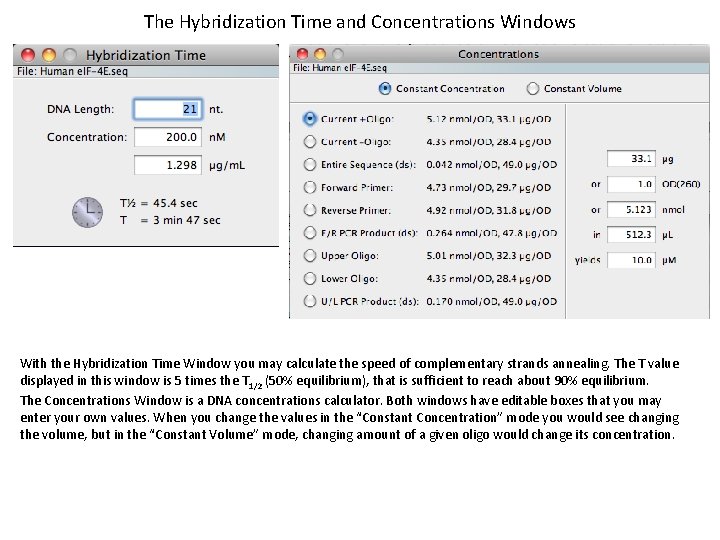
The Hybridization Time and Concentrations Windows With the Hybridization Time Window you may calculate the speed of complementary strands annealing. The T value displayed in this window is 5 times the T 1/2 (50% equilibrium), that is sufficient to reach about 90% equilibrium. The Concentrations Window is a DNA concentrations calculator. Both windows have editable boxes that you may enter your own values. When you change the values in the “Constant Concentration” mode you would see changing the volume, but in the “Constant Volume” mode, changing amount of a given oligo would change its concentration.

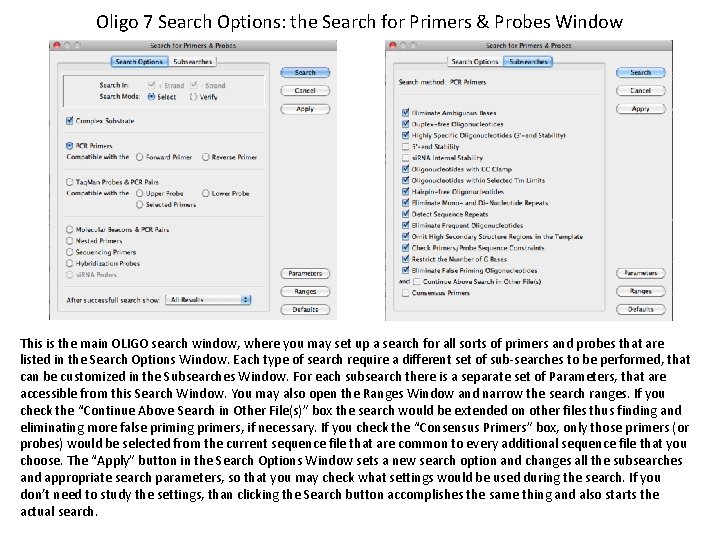
Oligo 7 Search Options: the Search for Primers & Probes Window This is the main OLIGO search window, where you may set up a search for all sorts of primers and probes that are listed in the Search Options Window. Each type of search require a different set of sub-searches to be performed, that can be customized in the Subsearches Window. For each subsearch there is a separate set of Parameters, that are accessible from this Search Window. You may also open the Ranges Window and narrow the search ranges. If you check the “Continue Above Search in Other File(s)” box the search would be extended on other files thus finding and eliminating more false priming primers, if necessary. If you check the “Consensus Primers” box, only those primers (or probes) would be selected from the current sequence file that are common to every additional sequence file that you choose. The “Apply” button in the Search Options Window sets a new search option and changes all the subsearches and appropriate search parameters, so that you may check what settings would be used during the search. If you don’t need to study the settings, than clicking the Search button accomplishes the same thing and also starts the actual search.

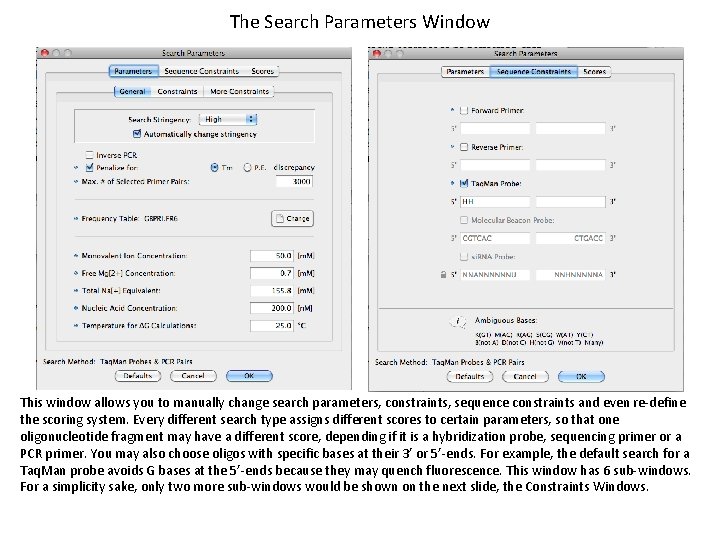
The Search Parameters Window This window allows you to manually change search parameters, constraints, sequence constraints and even re-define the scoring system. Every different search type assigns different scores to certain parameters, so that one oligonucleotide fragment may have a different score, depending if it is a hybridization probe, sequencing primer or a PCR primer. You may also choose oligos with specific bases at their 3’ or 5’-ends. For example, the default search for a Taq. Man probe avoids G bases at the 5’-ends because they may quench fluorescence. This window has 6 sub-windows. For a simplicity sake, only two more sub-windows would be shown on the next slide, the Constraints Windows.

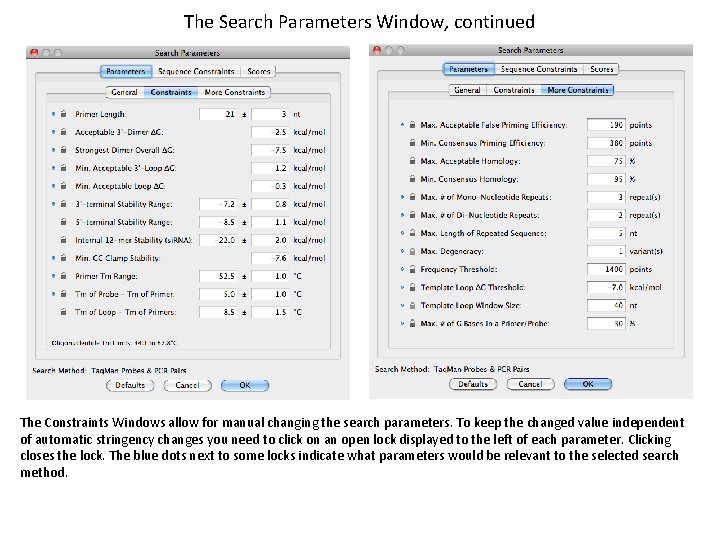
The Search Parameters Window, continued The Constraints Windows allow for manual changing the search parameters. To keep the changed value independent of automatic stringency changes you need to click on an open lock displayed to the left of each parameter. Clicking closes the lock. The blue dots next to some locks indicate what parameters would be relevant to the selected search method.

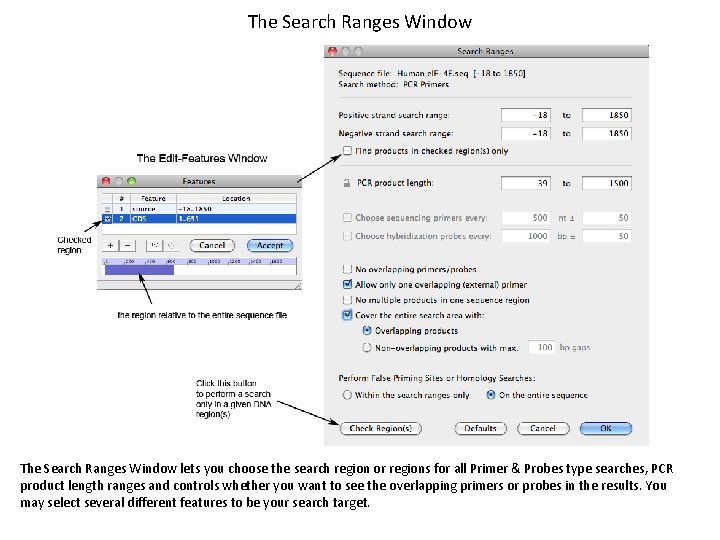
The Search Ranges Window lets you choose the search region or regions for all Primer & Probes type searches, PCR product length ranges and controls whether you want to see the overlapping primers or probes in the results. You may select several different features to be your search target.

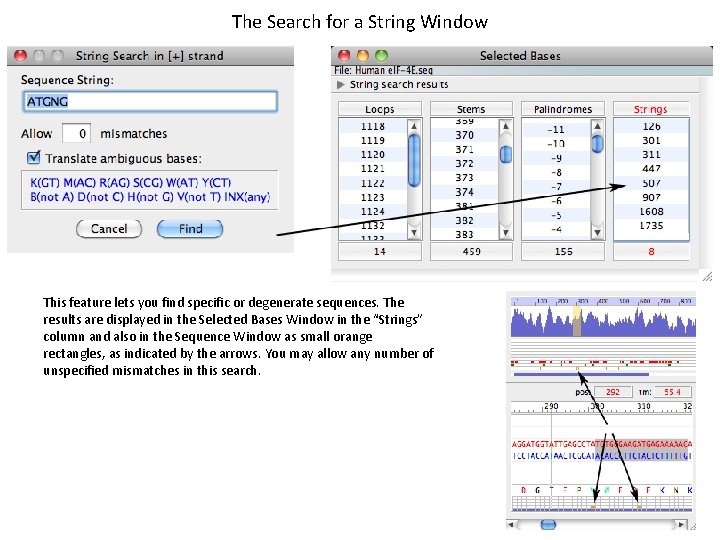
The Search for a String Window This feature lets you find specific or degenerate sequences. The results are displayed in the Selected Bases Window in the “Strings” column and also in the Sequence Window as small orange rectangles, as indicated by the arrows. You may allow any number of unspecified mismatches in this search.

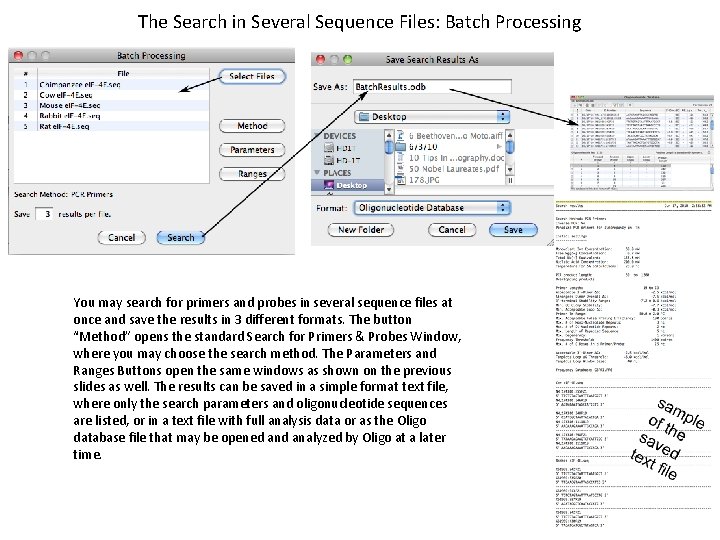
The Search in Several Sequence Files: Batch Processing You may search for primers and probes in several sequence files at once and save the results in 3 different formats. The button “Method” opens the standard Search for Primers & Probes Window, where you may choose the search method. The Parameters and Ranges Buttons open the same windows as shown on the previous slides as well. The results can be saved in a simple format text file, where only the search parameters and oligonucleotide sequences are listed, or in a text file with full analysis data or as the Oligo database file that may be opened analyzed by Oligo at a later time.

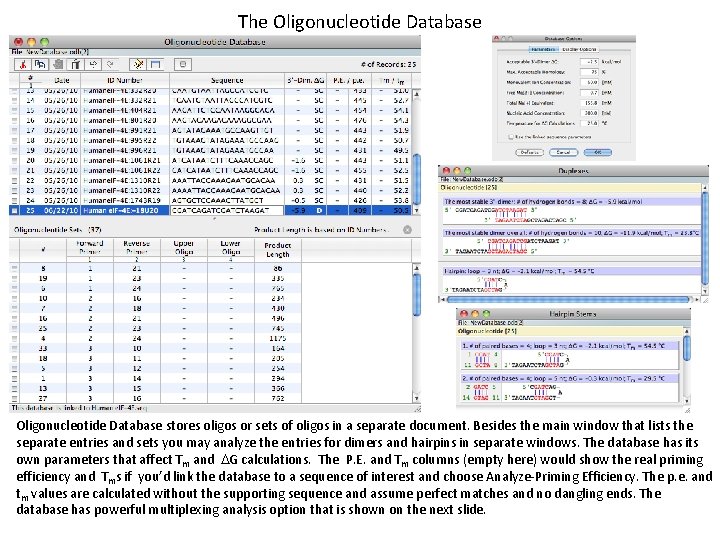
The Oligonucleotide Database stores oligos or sets of oligos in a separate document. Besides the main window that lists the separate entries and sets you may analyze the entries for dimers and hairpins in separate windows. The database has its own parameters that affect Tm and DG calculations. The P. E. and Tm columns (empty here) would show the real priming efficiency and Tms if you’d link the database to a sequence of interest and choose Analyze-Priming Efficiency. The p. e. and tm values are calculated without the supporting sequence and assume perfect matches and no dangling ends. The database has powerful multiplexing analysis option that is shown on the next slide.

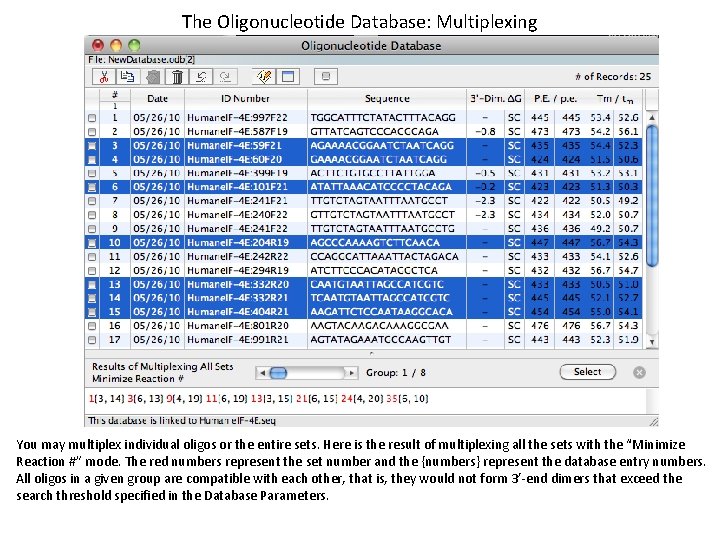
The Oligonucleotide Database: Multiplexing You may multiplex individual oligos or the entire sets. Here is the result of multiplexing all the sets with the “Minimize Reaction #” mode. The red numbers represent the set number and the {numbers} represent the database entry numbers. All oligos in a given group are compatible with each other, that is, they would not form 3’-end dimers that exceed the search threshold specified in the Database Parameters.

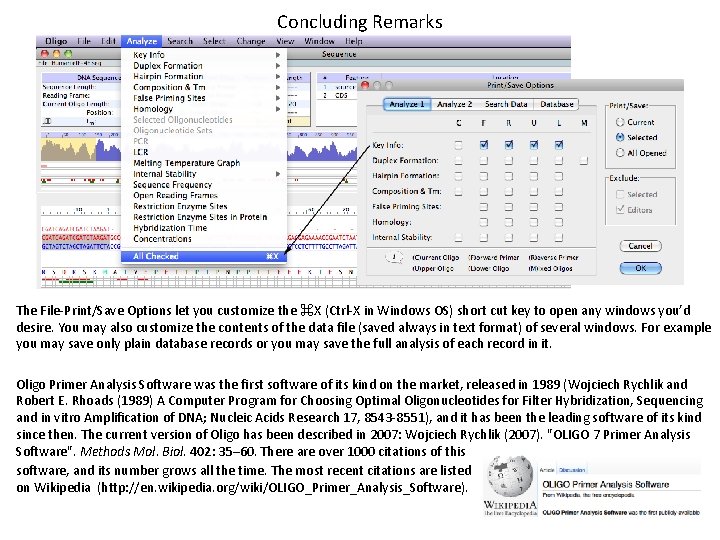
Concluding Remarks The File-Print/Save Options let you customize the ⌘X (Ctrl-X in Windows OS) short cut key to open any windows you’d desire. You may also customize the contents of the data file (saved always in text format) of several windows. For example you may save only plain database records or you may save the full analysis of each record in it. Oligo Primer Analysis Software was the first software of its kind on the market, released in 1989 (Wojciech Rychlik and Robert E. Rhoads (1989) A Computer Program for Choosing Optimal Oligonucleotides for Filter Hybridization, Sequencing and in vitro Amplification of DNA; Nucleic Acids Research 17, 8543 -8551), and it has been the leading software of its kind since then. The current version of Oligo has been described in 2007: Wojciech Rychlik (2007). "OLIGO 7 Primer Analysis Software". Methods Mol. Biol. 402: 35– 60. There are over 1000 citations of this software, and its number grows all the time. The most recent citations are listed on Wikipedia (http: //en. wikipedia. org/wiki/OLIGO_Primer_Analysis_Software).