PCR PCR Primer design The Polymerase Chain Reaction







- Slides: 21

PCR & PCR Primer design

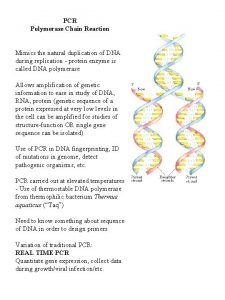
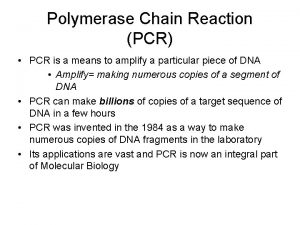
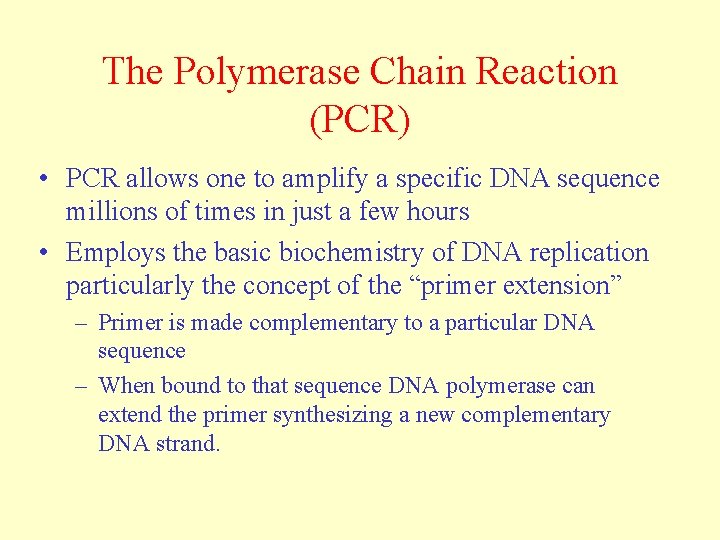
The Polymerase Chain Reaction (PCR) • PCR allows one to amplify a specific DNA sequence millions of times in just a few hours • Employs the basic biochemistry of DNA replication particularly the concept of the “primer extension” – Primer is made complementary to a particular DNA sequence – When bound to that sequence DNA polymerase can extend the primer synthesizing a new complementary DNA strand.

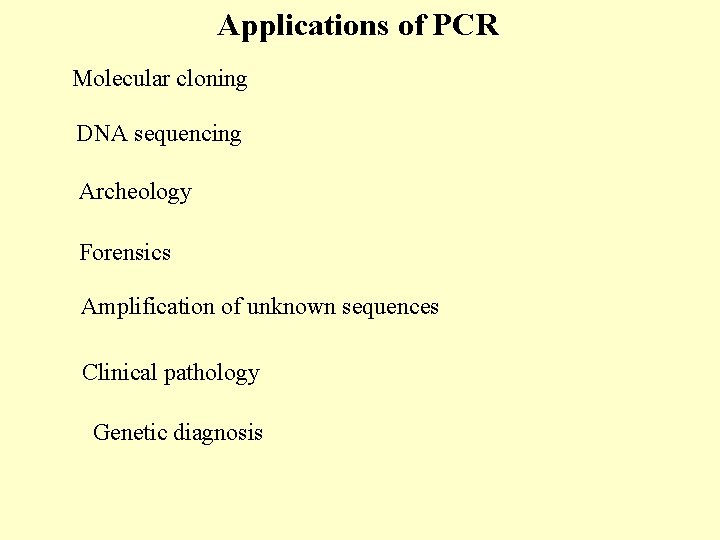
Applications of PCR Molecular cloning DNA sequencing Archeology Forensics Amplification of unknown sequences Clinical pathology Genetic diagnosis

PCR • During PCR, high temperature is used to separate the DNA molecules into single strands. • Synthetic single stranded DNA sequences of about 20 nucleotides complementary to specific DNA sequences serve as primers. – Two primers are used to bracket the target sequence to be amplified • One comp to one strand at the beginning (sense) • One comp to the other strand at the end (anti sense)

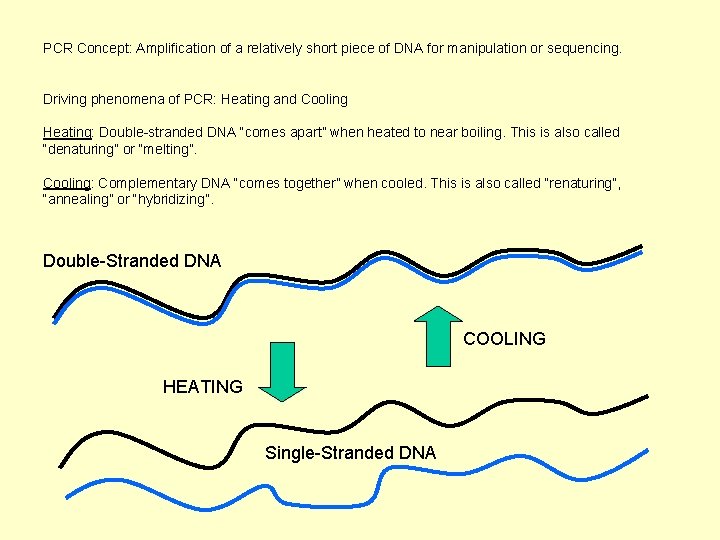
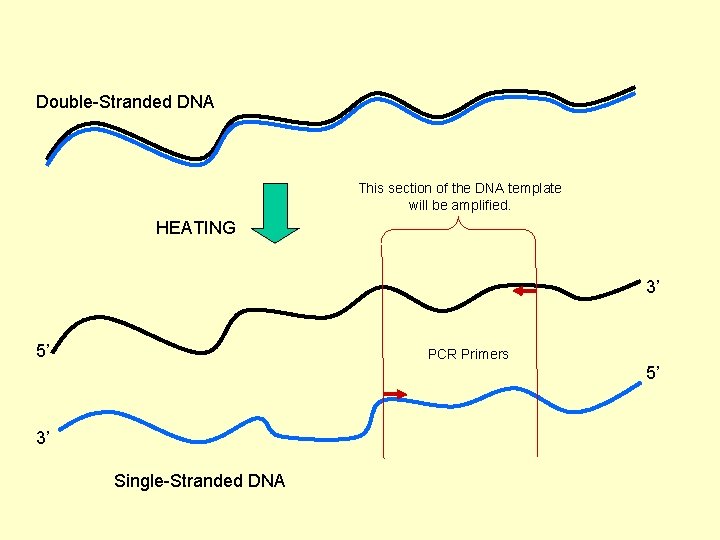
PCR Concept: Amplification of a relatively short piece of DNA for manipulation or sequencing. Driving phenomena of PCR: Heating and Cooling Heating: Double-stranded DNA “comes apart” when heated to near boiling. This is also called “denaturing” or “melting”. Cooling: Complementary DNA “comes together” when cooled. This is also called “renaturing”, “annealing” or “hybridizing”. Double-Stranded DNA COOLING HEATING Single-Stranded DNA

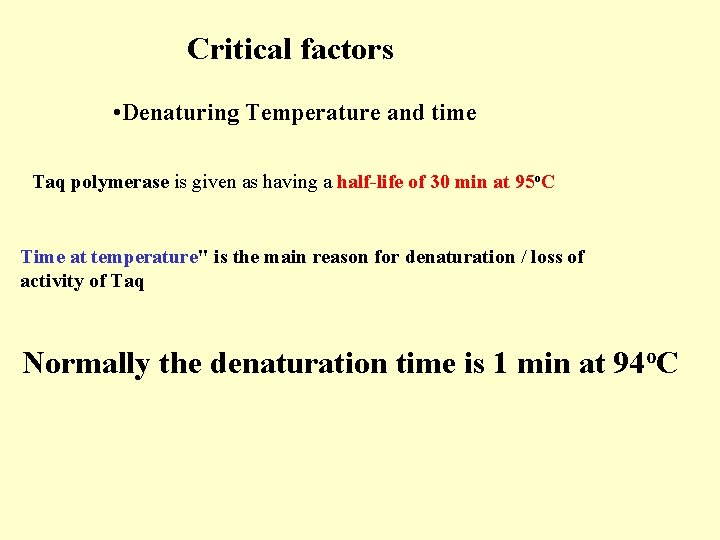
Critical factors • Denaturing Temperature and time Taq polymerase is given as having a half-life of 30 min at 95 o. C Time at temperature" is the main reason for denaturation / loss of activity of Taq Normally the denaturation time is 1 min at 94 o. C

What is Primer? : A short stretch of single stranded DNA : Which is artificially synthesized by user : It is required by Taq polymerase to initiate elongation : Taq uses it as a scaffold to start the reaction

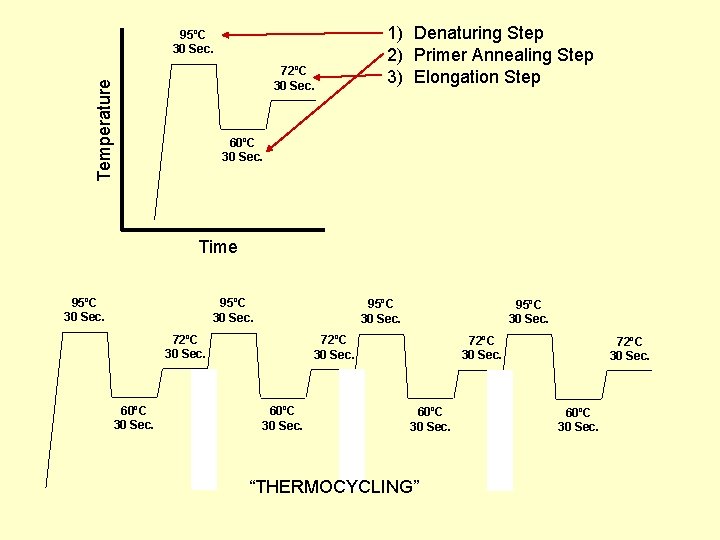
Performing PCR • Very small amount of DNA sample is mixed with a buffer solution containing DNA polymerase and primers – Polymerase (TAQ) is isolated from a thermophylic bacteria so it will not be denatured at high temperature • The mixture is taken through replication cycles consisting of – Time at 94 C to denature DNA – Time at 50 -60 C to allow primers to “Anneal” – Time at 72 C during which DNA polymerase can extend the primers producing complementary strands • The DNA sequence between the primers doubles after each cycle.

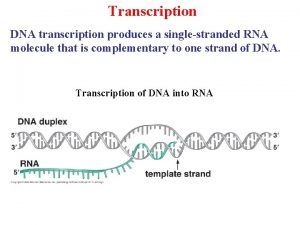
Molecular Basis of PCR: Polymerase Activity A Polymerase is an enzyme that synthesizes DNA. 1) DNA can ONLY be synthesized using the complementary strand! 2) Polymerases synthesize DNA in the 5’ 3’ direction! 5’-GTCGATGTCTGATCAATTGGGCTGATCATGTCGATGATGCTAGAAT-3’ 3’CTACGATCTTA-5’ 5’-GTCGATGTCTGATCAATTGGGCTGATCATGTCGATGATGCTAGAAT-3’ ACTAGTACAGCTACTACGATCTTA-5’

Double-Stranded DNA This section of the DNA template will be amplified. HEATING 3’ 5’ PCR Primers 5’ 3’ Single-Stranded DNA

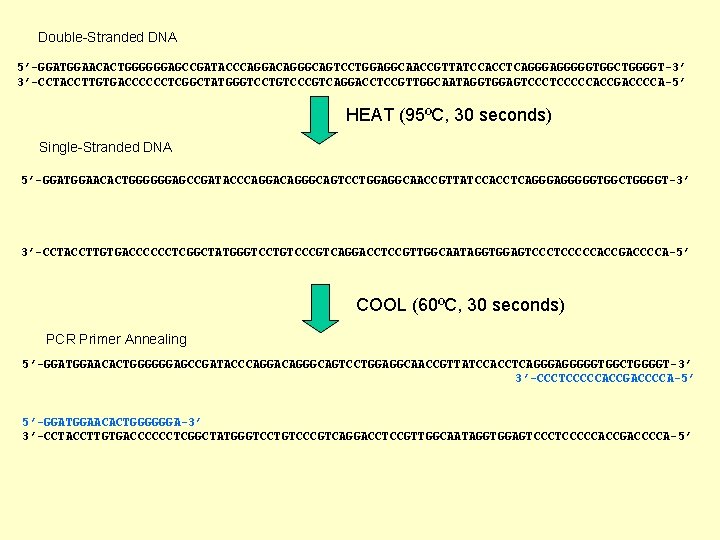
Double-Stranded DNA 5’-GGATGGAACACTGGGGGGAGCCGATACCCAGGACAGGGCAGTCCTGGAGGCAACCGTTATCCACCTCAGGGGGTGGCTGGGGT-3’ 3’-CCTACCTTGTGACCCCCCTCGGCTATGGGTCCTGTCCCGTCAGGACCTCCGTTGGCAATAGGTGGAGTCCCCCACCGACCCCA-5’ HEAT (95ºC, 30 seconds) Single-Stranded DNA 5’-GGATGGAACACTGGGGGGAGCCGATACCCAGGACAGGGCAGTCCTGGAGGCAACCGTTATCCACCTCAGGGGGTGGCTGGGGT-3’ 3’-CCTACCTTGTGACCCCCCTCGGCTATGGGTCCTGTCCCGTCAGGACCTCCGTTGGCAATAGGTGGAGTCCCCCACCGACCCCA-5’ COOL (60ºC, 30 seconds) PCR Primer Annealing 5’-GGATGGAACACTGGGGGGAGCCGATACCCAGGACAGGGCAGTCCTGGAGGCAACCGTTATCCACCTCAGGGGGTGGCTGGGGT-3’ 3’-CCCTCCCCCACCGACCCCA-5’ 5’-GGATGGAACACTGGGGGGA-3’ 3’-CCTACCTTGTGACCCCCCTCGGCTATGGGTCCTGTCCCGTCAGGACCTCCGTTGGCAATAGGTGGAGTCCCCCACCGACCCCA-5’

HEAT (72ºC, 30 seconds) Polymerase Elongation 5’-GGATGGAACACTGGGGGGAGCCGATACCCAGGACAGGGCAGTCCTGGAGGCAACCGTTATCCACCTCAGGGGGTGGCTGGGGT-3’ CTCGGCTATGGGTCCTGTCCCGTCAGGACCTCCGTTGGCAATAGGTGGAGTCCCCCACCGACCCCA-5’ 5’-GGATGGAACACTGGGGGGAGCCGATACCCAGGACAGGGCAGTCCTGGAGGCAACCGTTATCCACCTCAGGGA 3’-CCTACCTTGTGACCCCCCTCGGCTATGGGTCCTGTCCCGTCAGGACCTCCGTTGGCAATAGGTGGAGTCCCCCACCGACCCCA-5’ DNA Synthesis after 1 “cycle” of PCR = 1 double stranded DNA is now 2 “copies” 5’-GGATGGAACACTGGGGGGAGCCGATACCCAGGACAGGGCAGTCCTGGAGGCAACCGTTATCCACCTCAGGGAGGGGGTGGCTGGGGT-3’ 3’-CCTACCTTGTGACCCCCCTCGGCTATGGGTCCTGTCCCGTCAGGACCTCCGTTGGCAATAGGTGGAGTCCCTCCCCCACCGACCCCA-5’

95ºC 30 Sec. Temperature 72ºC 30 Sec. 1) Denaturing Step 2) Primer Annealing Step 3) Elongation Step 60ºC 30 Sec. Time 95ºC 30 Sec. 72ºC 30 Sec. 60ºC 30 Sec. “THERMOCYCLING” 72ºC 30 Sec. 60ºC 30 Sec.

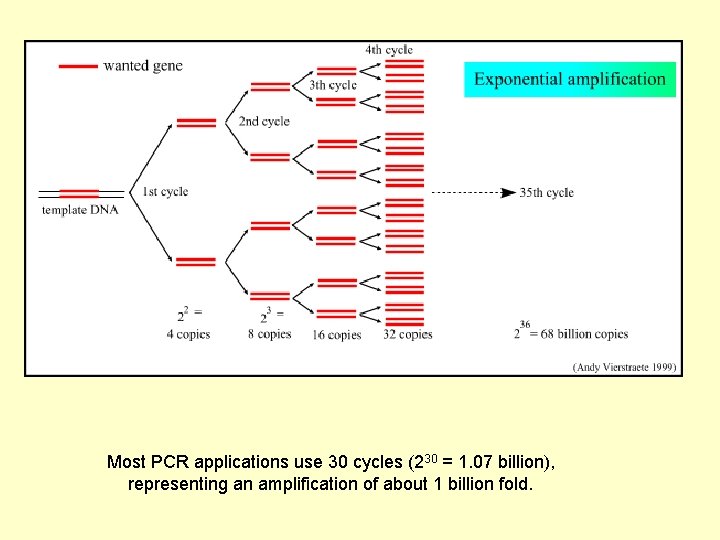
Most PCR applications use 30 cycles (230 = 1. 07 billion), representing an amplification of about 1 billion fold.

PCR Primers Design 1) Requires knowledge of the target sequence (i. e. we can’t use PCR to amplify an unknown sequence) 2) Primers must have similar “melting temperatures” (e. g. 60ºC)

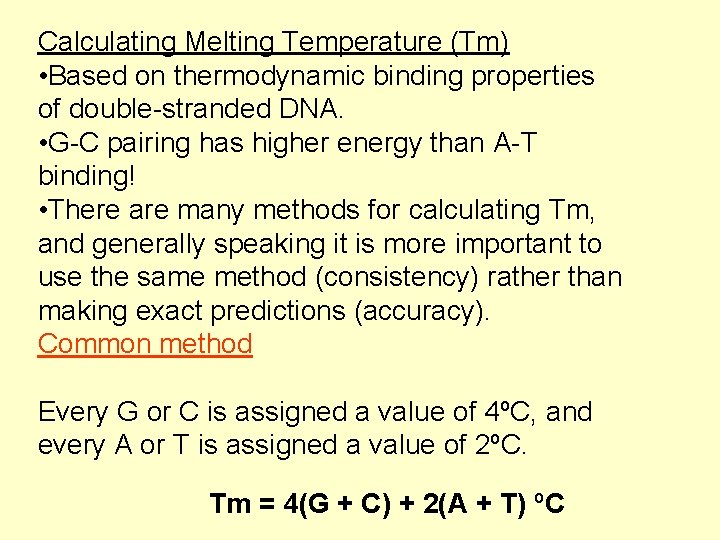
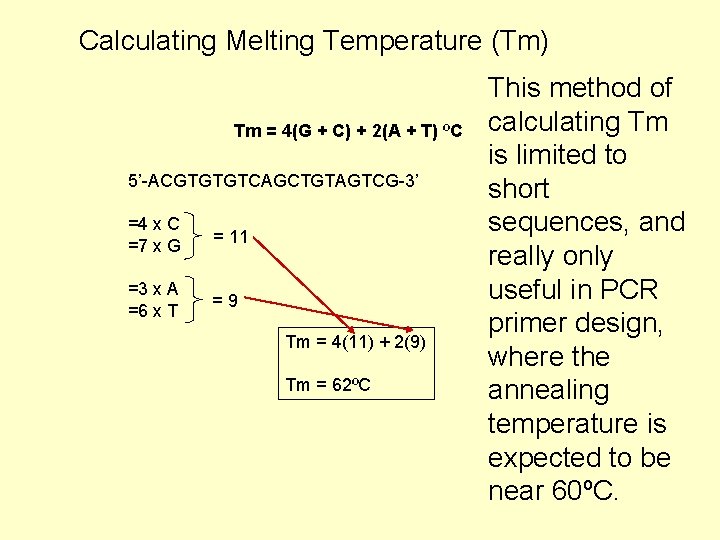
Calculating Melting Temperature (Tm) • Based on thermodynamic binding properties of double-stranded DNA. • G-C pairing has higher energy than A-T binding! • There are many methods for calculating Tm, and generally speaking it is more important to use the same method (consistency) rather than making exact predictions (accuracy). Common method Every G or C is assigned a value of 4ºC, and every A or T is assigned a value of 2ºC. Tm = 4(G + C) + 2(A + T) ºC

Calculating Melting Temperature (Tm) Tm = 4(G + C) + 2(A + T) ºC 5’-ACGTGTGTCAGCTGTAGTCG-3’ =4 x C =7 x G = 11 =3 x A =6 x T =9 Tm = 4(11) + 2(9) Tm = 62ºC This method of calculating Tm is limited to short sequences, and really only useful in PCR primer design, where the annealing temperature is expected to be near 60ºC.

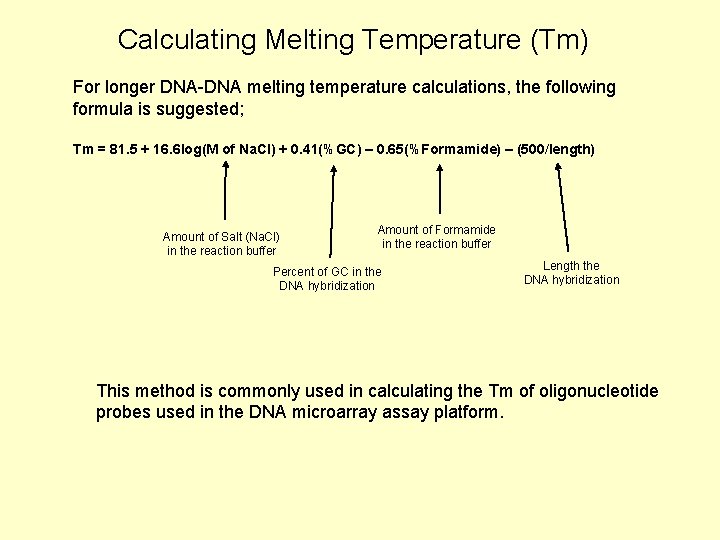
Calculating Melting Temperature (Tm) For longer DNA-DNA melting temperature calculations, the following formula is suggested; Tm = 81. 5 + 16. 6 log(M of Na. Cl) + 0. 41(%GC) – 0. 65(%Formamide) – (500/length) Amount of Salt (Na. Cl) in the reaction buffer Amount of Formamide in the reaction buffer Percent of GC in the DNA hybridization Length the DNA hybridization This method is commonly used in calculating the Tm of oligonucleotide probes used in the DNA microarray assay platform.

Computationally, PCR Primer design involves the coordination of; 1) Selecting 2 different primers that are: a) Any distance apart (there are length limits in PCR!) b) -OR- A specified distance apart (e. g. “PCR product will be 300 base-pairs long”) c) -OR- Span a specific subsequence or “motif” (e. g. “PCR product includes exons #1 and #2”) 2) Selecting 2 different primers that are nearly/exactly the same melting temperature (Tm). This is usually 60ºC. 3) Many times PCR Primers are designed with a “GC Clamp”. This simply means that the 3’ end of the primers is a G or a C, which binds more tightly (than AT) and the Polymerase initiates synthesis more efficiently. 4) Finally, better results from PCR reactions occur if the two primers are not complementary to each other, or contain extensive palindrome sequence.

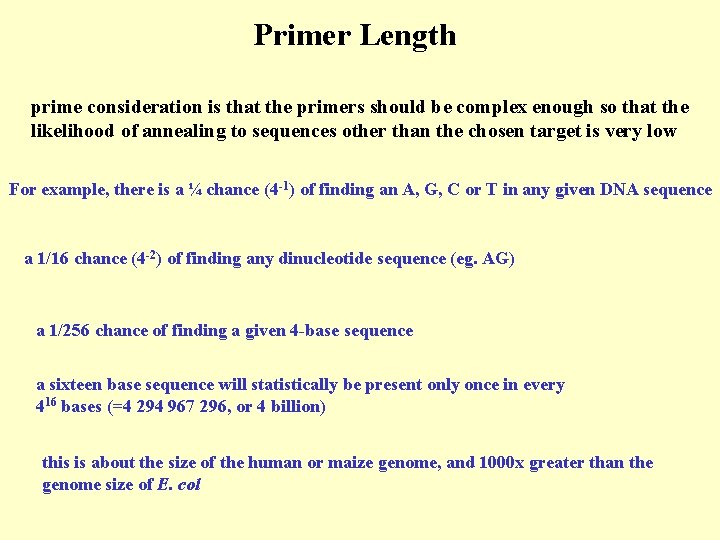
Primer Length prime consideration is that the primers should be complex enough so that the likelihood of annealing to sequences other than the chosen target is very low For example, there is a ¼ chance (4 -1) of finding an A, G, C or T in any given DNA sequence a 1/16 chance (4 -2) of finding any dinucleotide sequence (eg. AG) a 1/256 chance of finding a given 4 -base sequence a sixteen base sequence will statistically be present only once in every 416 bases (=4 294 967 296, or 4 billion) this is about the size of the human or maize genome, and 1000 x greater than the genome size of E. col

Elongation Temperature and Time This is normally 70 - 72 o. C, for 0. 5 - 3 min. elongation occurs from the moment of annealing, even if this is transient, which results in considerably greater stability primer extension occurs at up to 100 bases/sec. About 1 min is sufficient for reliable amplification of 2 kb sequences 3 min is a good bet for 3 kb and longer products
 What are applications of pcr
What are applications of pcr Polymerase chain reaction
Polymerase chain reaction Polymerase chain reaction
Polymerase chain reaction Polymerase chain reaction application
Polymerase chain reaction application Pcr
Pcr Polymerase chain reaction
Polymerase chain reaction The three steps of polymerase chain reaction
The three steps of polymerase chain reaction Clonaggio
Clonaggio Rt pcr primer design
Rt pcr primer design Sequence of food chain
Sequence of food chain Rna polymerase 1 2 3
Rna polymerase 1 2 3 Proofreading and repair of a dna strand occurs during:
Proofreading and repair of a dna strand occurs during: Dna primerase
Dna primerase Replication fork
Replication fork Dna replication eukaryotes
Dna replication eukaryotes Types of dna polymerase in eukaryotes
Types of dna polymerase in eukaryotes Types of dna polymerase in eukaryotes
Types of dna polymerase in eukaryotes Dna polymerase
Dna polymerase Transcription initiation in eukaryotes
Transcription initiation in eukaryotes Dna polymerase
Dna polymerase Rna polymerase
Rna polymerase Adn polymérase
Adn polymérase