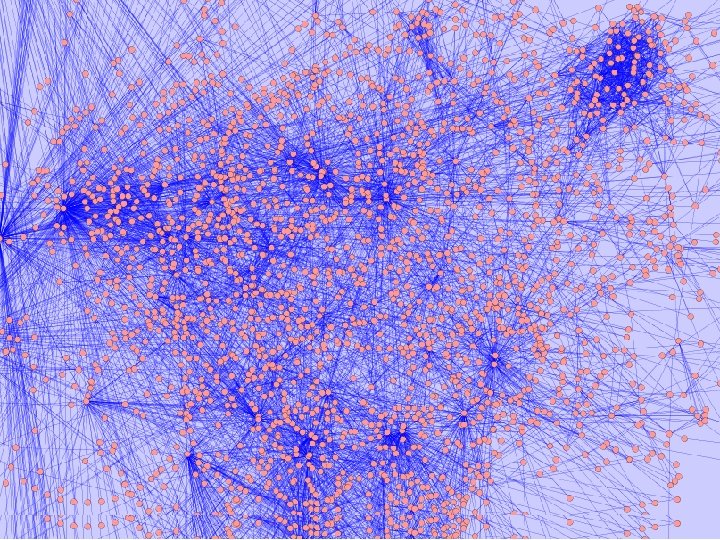
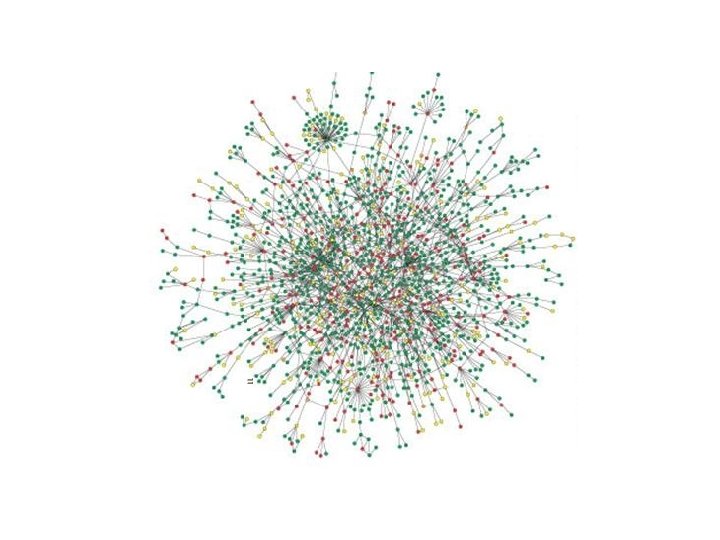
Network Clustering 7000 Yeast interactions among 3000 proteins







- Slides: 69

Network Clustering 7000 Yeast interactions among 3000 proteins

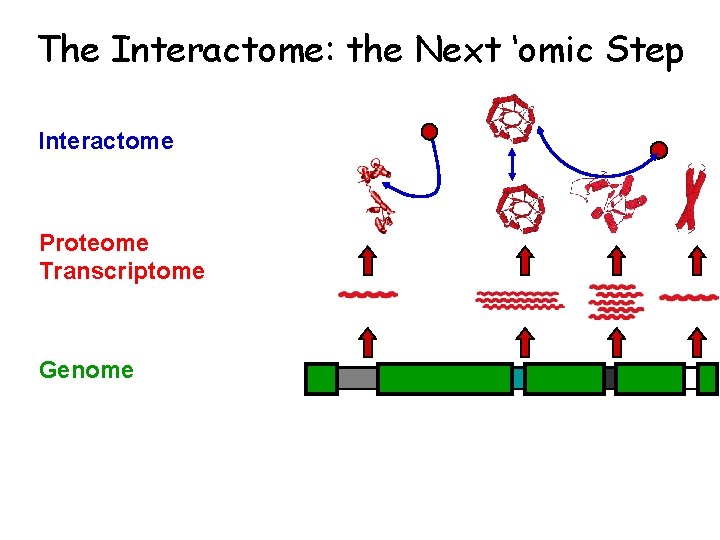
The Interactome: the Next ‘omic Step Interactome Proteome Transcriptome Genome

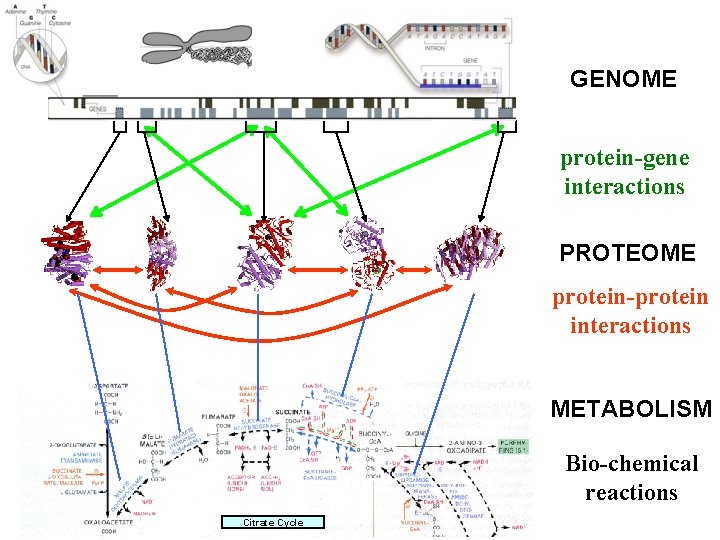
GENOME protein-gene interactions PROTEOME protein-protein interactions METABOLISM Bio-chemical reactions Citrate Cycle

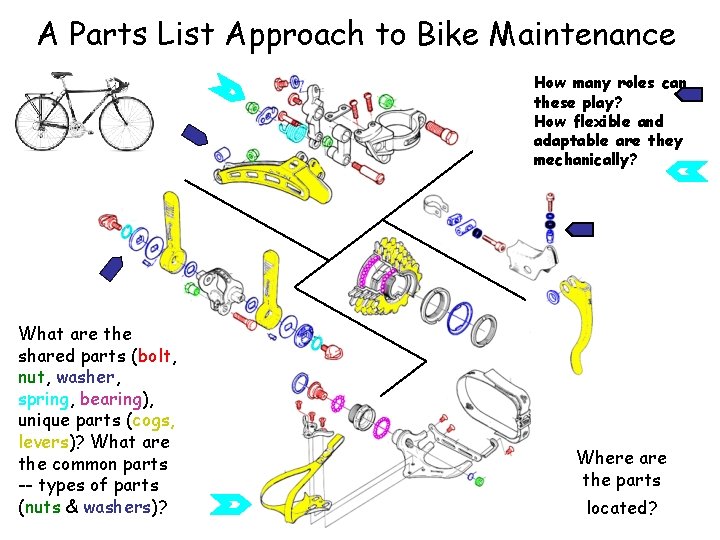
A Parts List Approach to Bike Maintenance How many roles can these play? How flexible and adaptable are they mechanically? What are the shared parts (bolt, nut, washer, spring, bearing), unique parts (cogs, levers)? What are the common parts -- types of parts (nuts & washers)? Where are the parts located?


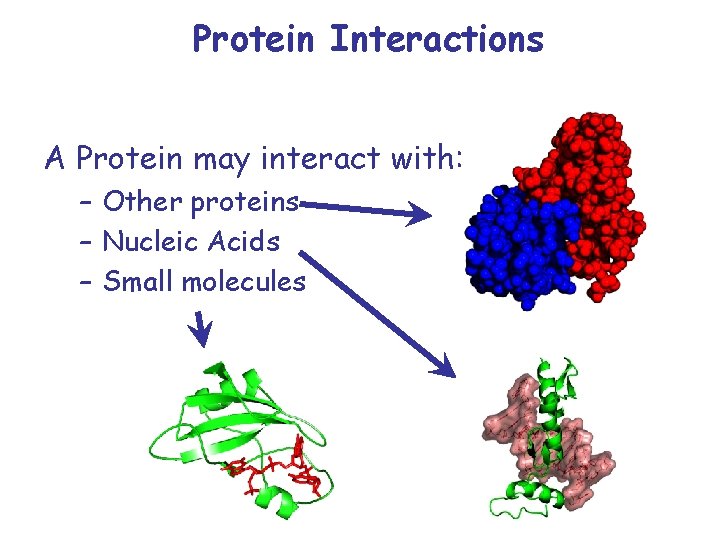
Protein Interactions A Protein may interact with: – Other proteins – Nucleic Acids – Small molecules

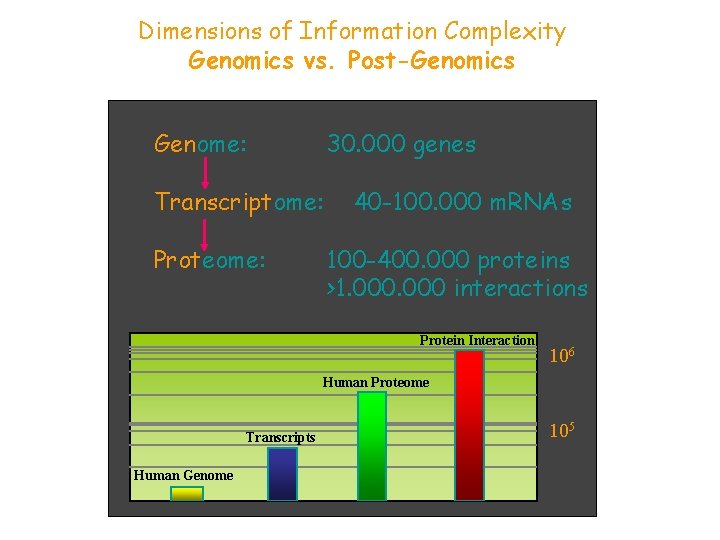
Dimensions of Information Complexity Genomics vs. Post-Genomics Genome: 30. 000 genes Transcriptome: Proteome: 40 -100. 000 m. RNAs 100 -400. 000 proteins >1. 000 interactions Protein Interaction 106 Human Proteome Transcripts Human Genome 105


Comprehensive Analysis of Complex Protein Structures in the Cell Multiprotein Complex/Organelle Total Protein Characterization • Protein Identification: What’s there • Post Translational Modifications: Regulation • Quantification: Dynamics

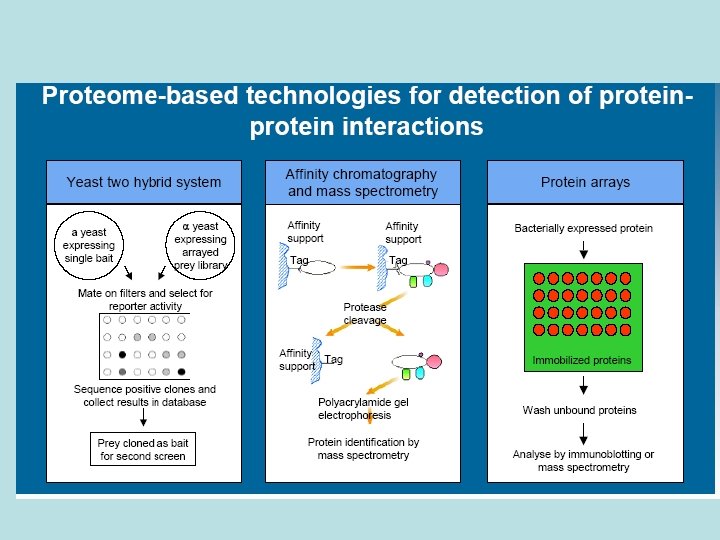
Protein-Protein Interactions: The “Interactome” Experimental methods: Mass Spec, yeast 2 -hybrid system, microarrays, … Computational techniques: phylogenic profiles, sequence analysis, … 2 challenges: - find which proteins interact (the partners) - find which residues participate in the interactions




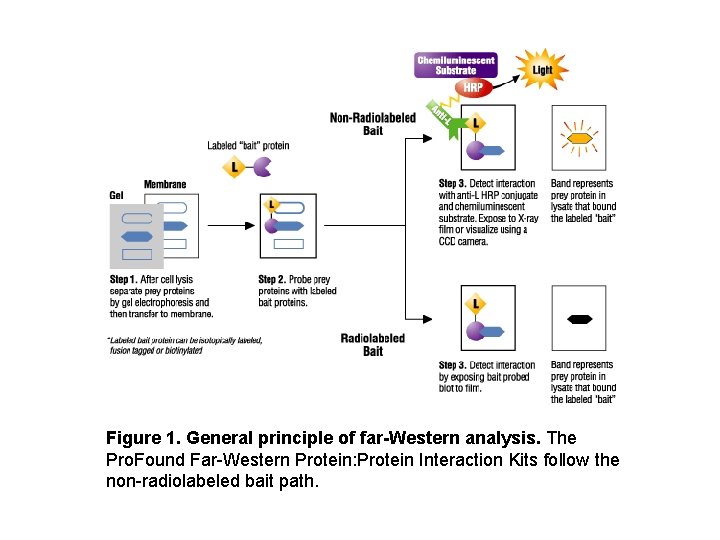
Figure 1. General principle of far-Western analysis. The Pro. Found Far-Western Protein: Protein Interaction Kits follow the non-radiolabeled bait path.

Finding Protein Partners

Approaches

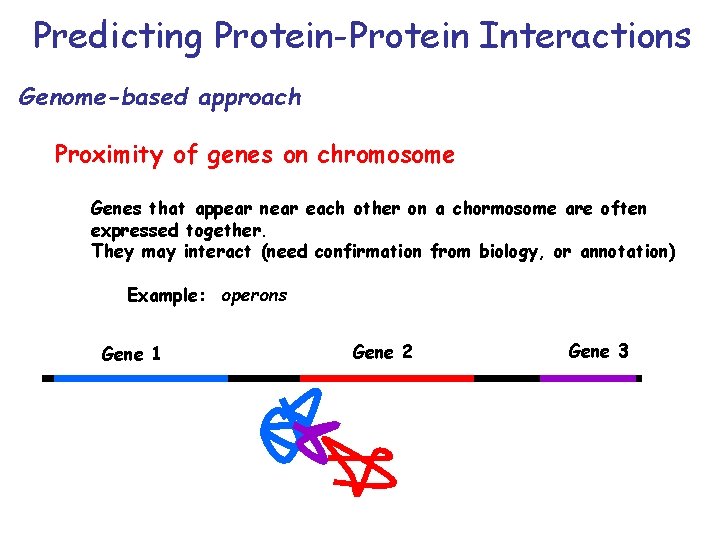
Predicting Protein-Protein Interactions Genome-based approach Proximity of genes on chromosome Genes that appear near each other on a chormosome are often expressed together. They may interact (need confirmation from biology, or annotation) Example: operons Gene 1 Gene 2 Gene 3

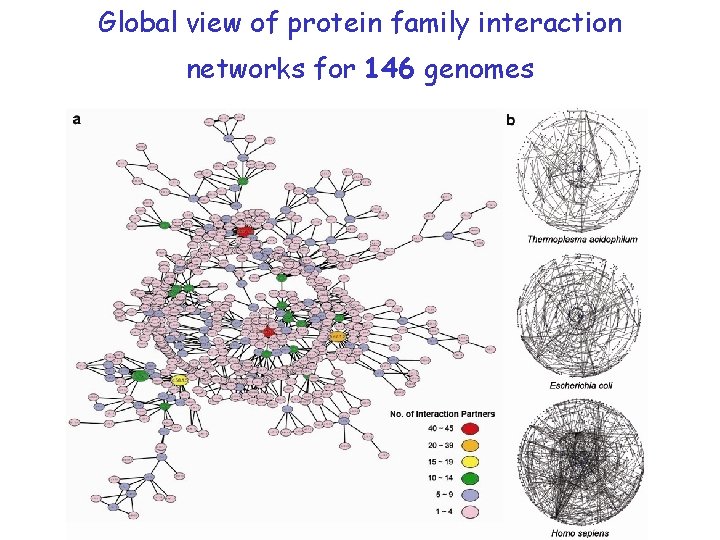
Global view of protein family interaction networks for 146 genomes

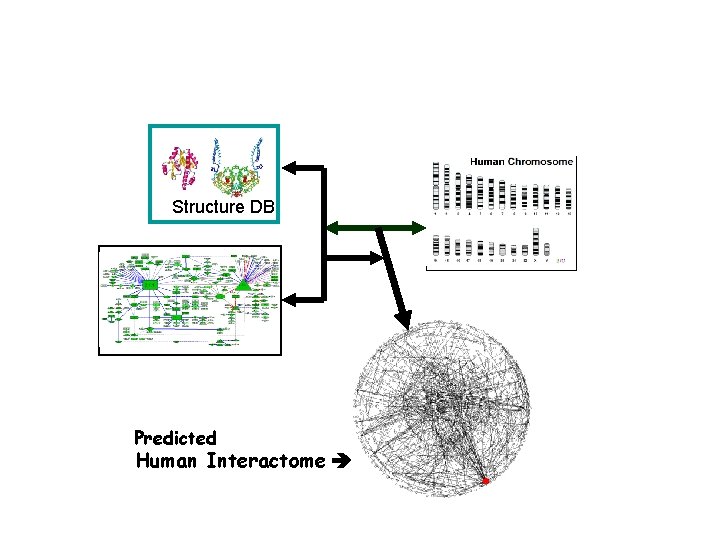
Structure DB Predicted Human Interactome


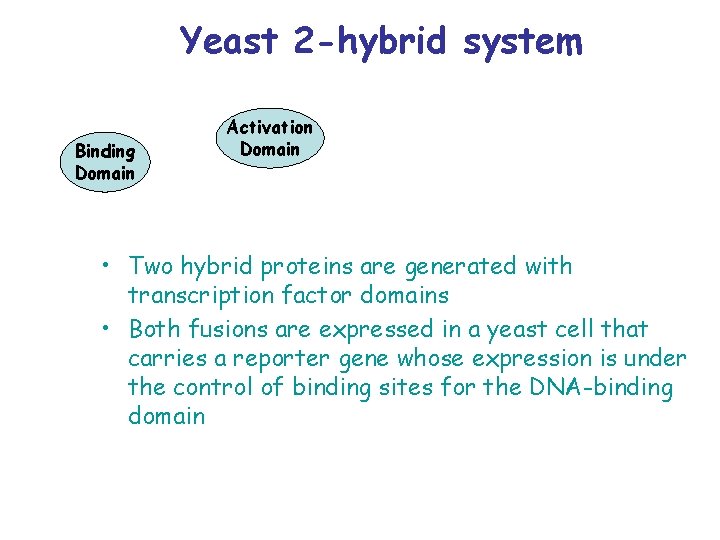
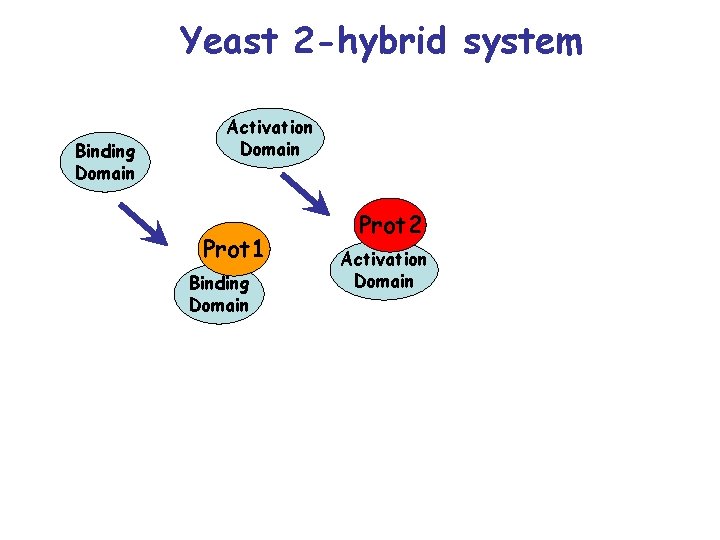
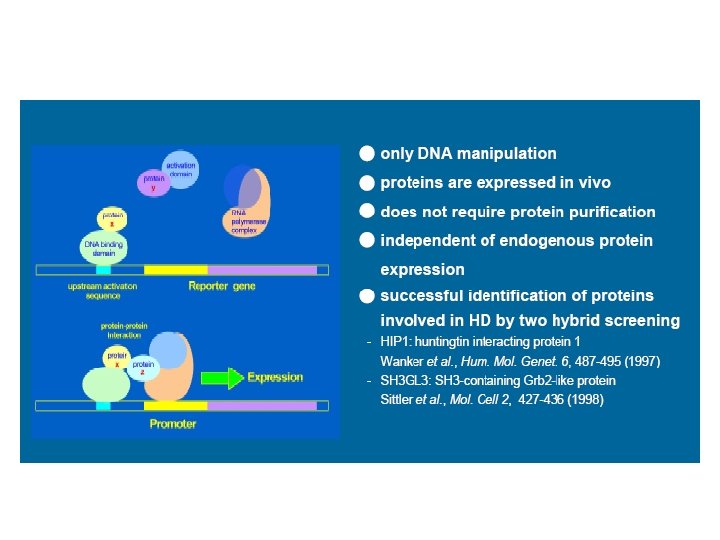
Yeast 2 -hybrid system Binding Domain Activation Domain • Two hybrid proteins are generated with transcription factor domains • Both fusions are expressed in a yeast cell that carries a reporter gene whose expression is under the control of binding sites for the DNA-binding domain

Yeast 2 -hybrid system Binding Domain Activation Domain Prot 1 Binding Domain Prot 2 Activation Domain

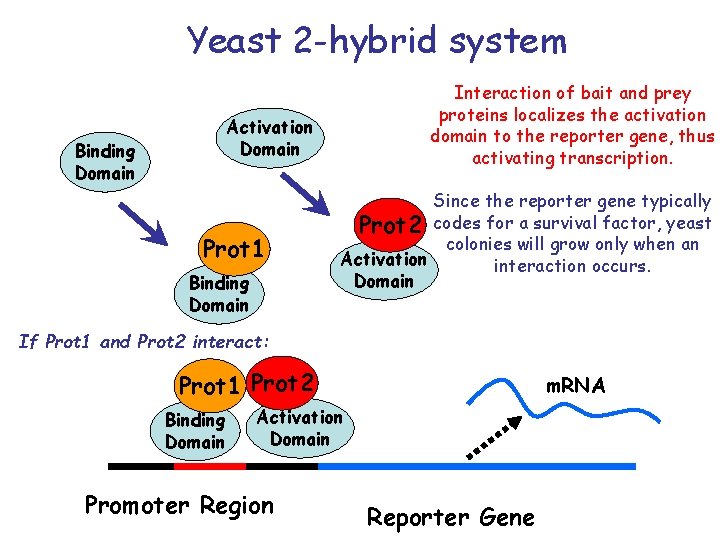
Yeast 2 -hybrid system Interaction of bait and prey proteins localizes the activation domain to the reporter gene, thus activating transcription. Activation Domain Binding Domain Prot 1 Binding Domain Since the reporter gene typically Prot 2 codes for a survival factor, yeast colonies will grow only when an Activation interaction occurs. Domain If Prot 1 and Prot 2 interact: Prot 1 Prot 2 Binding Domain m. RNA Activation Domain Promoter Region Reporter Gene

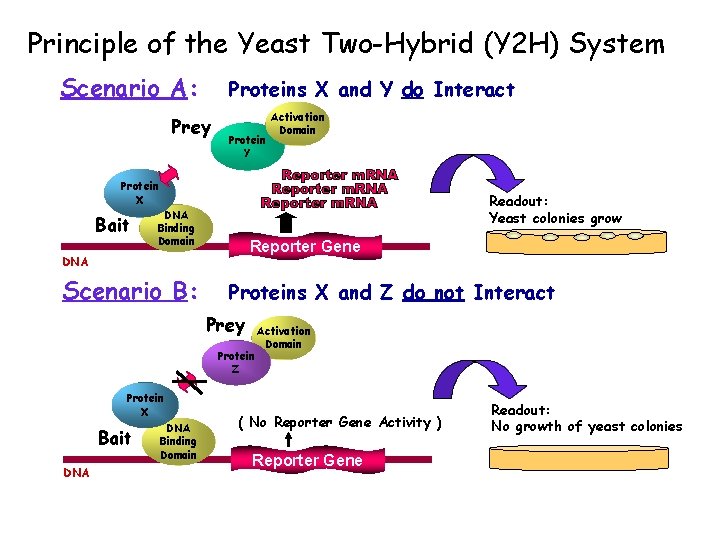
Principle of the Yeast Two-Hybrid (Y 2 H) System Scenario A: Proteins X and Y do Interact Prey Protein Y Activation Domain Protein X Bait Readout: Yeast colonies grow DNA Binding Domain Reporter Gene DNA Scenario B: Proteins X and Z do not Interact Prey Protein Z Protein X Bait DNA Binding Domain Activation Domain ( No Reporter Gene Activity ) Reporter Gene Readout: No growth of yeast colonies

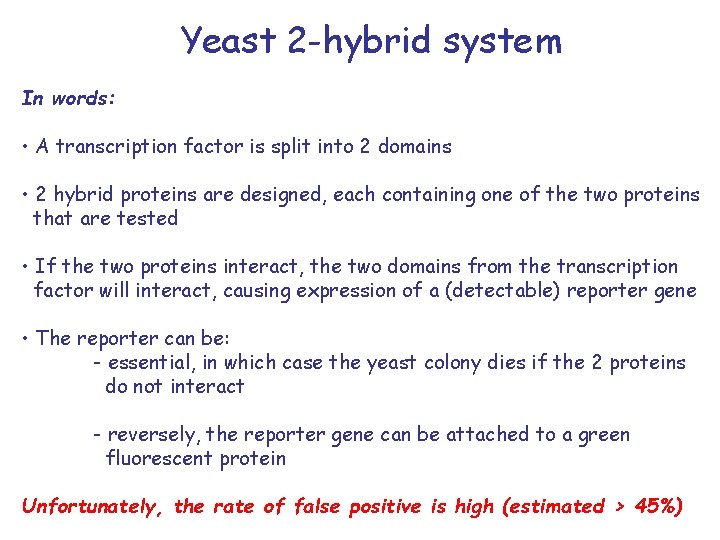
Yeast 2 -hybrid system In words: • A transcription factor is split into 2 domains • 2 hybrid proteins are designed, each containing one of the two proteins that are tested • If the two proteins interact, the two domains from the transcription factor will interact, causing expression of a (detectable) reporter gene • The reporter can be: - essential, in which case the yeast colony dies if the 2 proteins do not interact - reversely, the reporter gene can be attached to a green fluorescent protein Unfortunately, the rate of false positive is high (estimated > 45%)


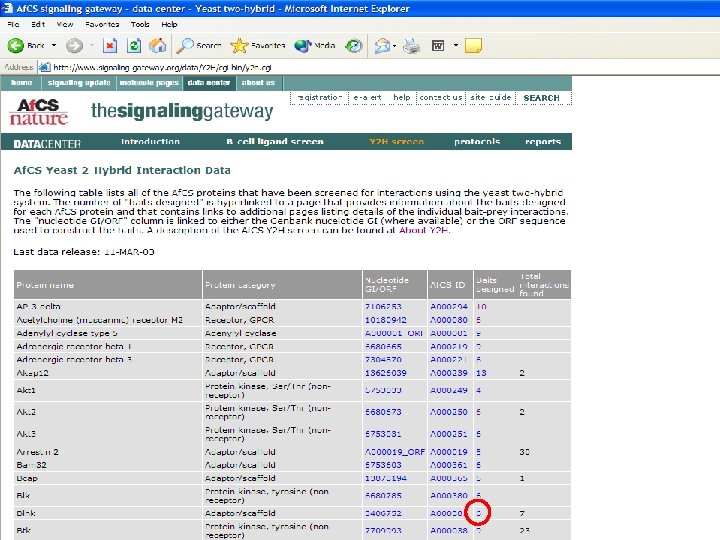
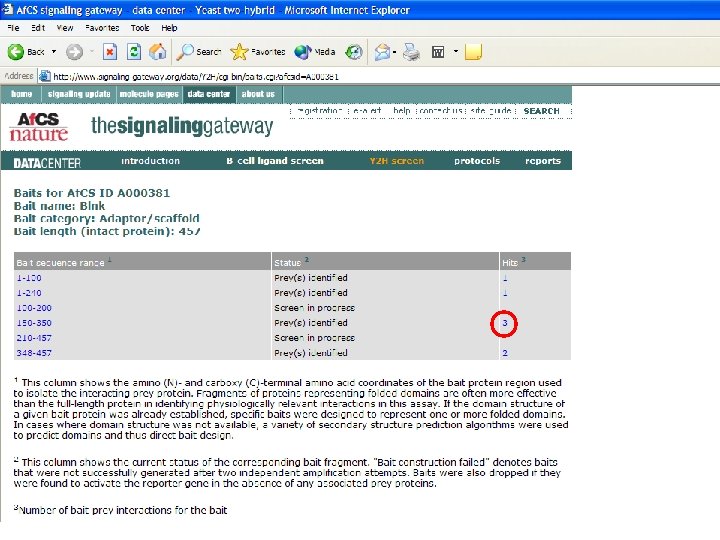
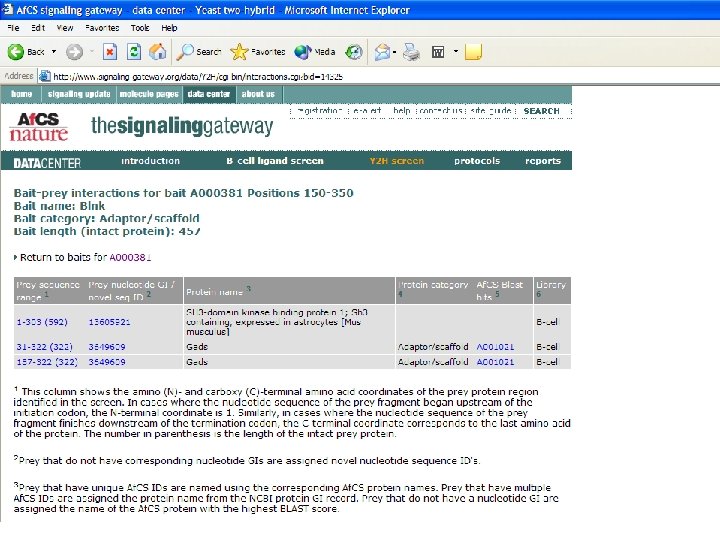
Bait Design • Fragments of proteins representing folded domains are often more effective than the full-length protein in identifying physiologically relevant interactions • If the domain structure of a given bait protein was already established, the specific baits were designed to represent one or more folded domains. • For cases in which domain structure was not available, a variety of secondary structure prediction algorithms were used to predict domains and thus direct bait design. • Baits were designed to cover the entire protein, with several overlapping fragments, as not all baits will work effectively.



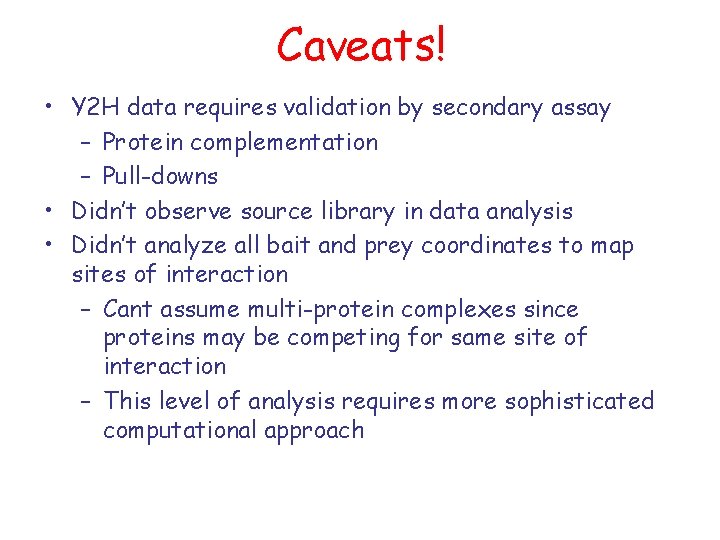
Caveats! • Y 2 H data requires validation by secondary assay – Protein complementation – Pull-downs • Didn’t observe source library in data analysis • Didn’t analyze all bait and prey coordinates to map sites of interaction – Cant assume multi-protein complexes since proteins may be competing for same site of interaction – This level of analysis requires more sophisticated computational approach


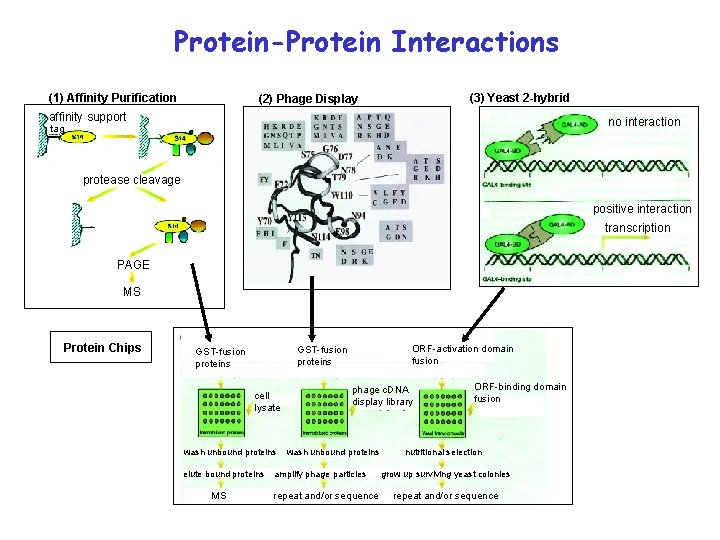
Protein-Protein Interactions (1) Affinity Purification (3) Yeast 2 -hybrid (2) Phage Display affinity support no interaction tag protease cleavage positive interaction transcription PAGE MS Protein Chips cell lysate wash unbound proteins elute bound proteins MS ORF-activation domain fusion GST-fusion proteins phage c. DNA display library wash unbound proteins amplify phage particles repeat and/or sequence ORF-binding domain fusion nutritional selection grow up surviving yeast colonies repeat and/or sequence

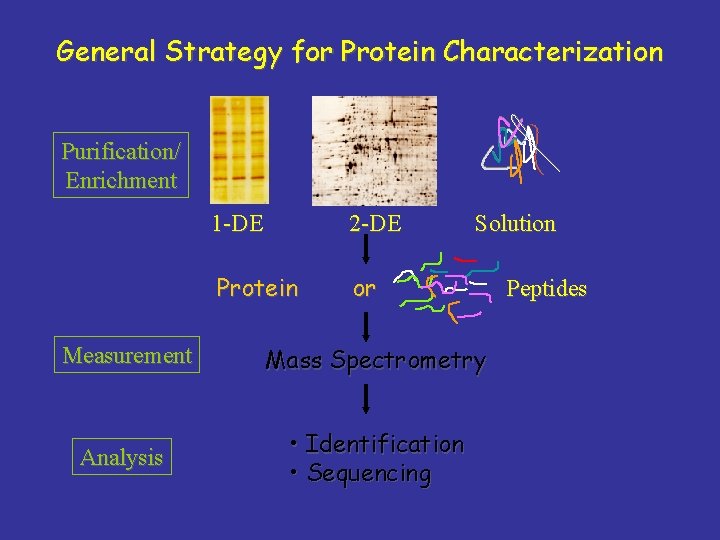
General Strategy for Protein Characterization Purification/ Enrichment 1 -DE 2 -DE Protein or Solution Measurement Mass Spectrometry Analysis • Identification • Sequencing Peptides

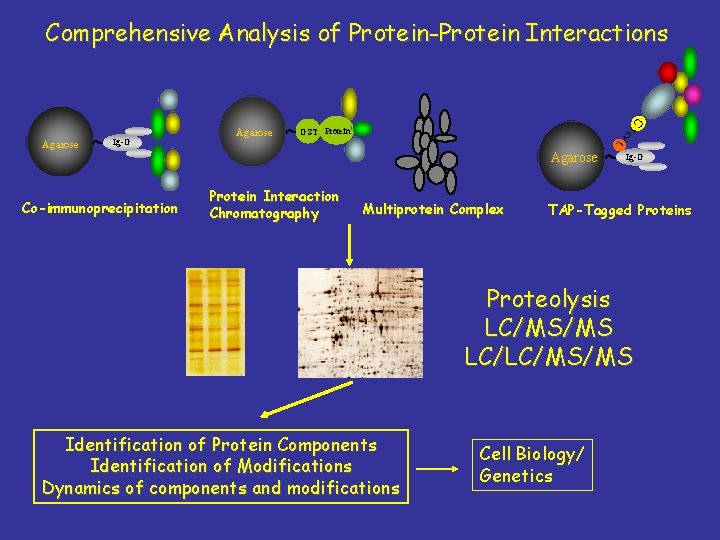
Comprehensive Analysis of Protein-Protein Interactions C GST Protein L Co-immunoprecipitation Agarose V Ig-G TE Agarose Protein Interaction Chromatography Multiprotein Complex Ig-G TAP-Tagged Proteins Proteolysis LC/MS/MS LC/LC/MS/MS Identification of Protein Components Identification of Modifications Dynamics of components and modifications Cell Biology/ Genetics

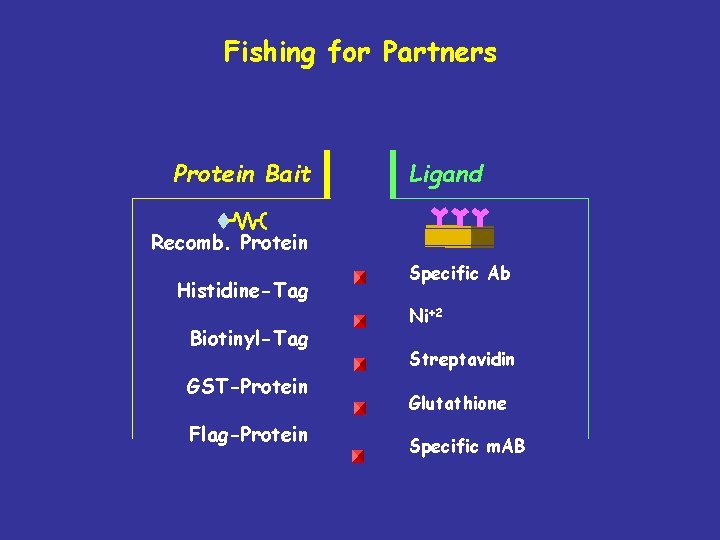
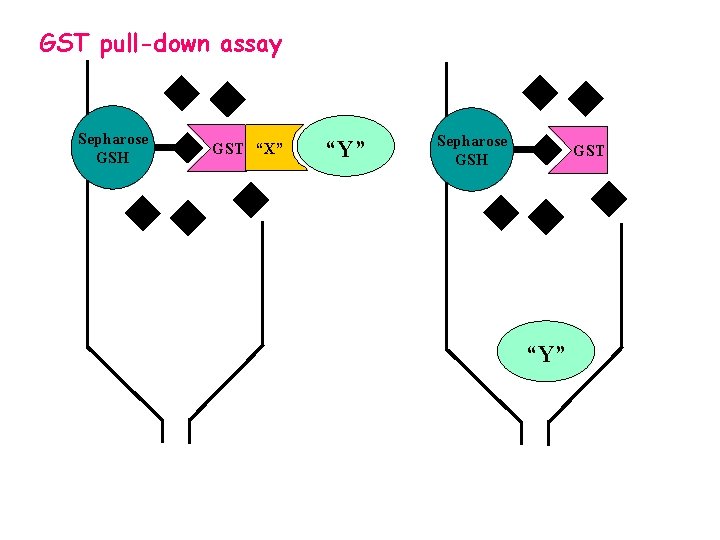
Fishing for Partners Protein Bait Ligand Recomb. Protein Histidine-Tag Biotinyl-Tag GST-Protein Flag-Protein Specific Ab Ni+2 Streptavidin Glutathione Specific m. AB


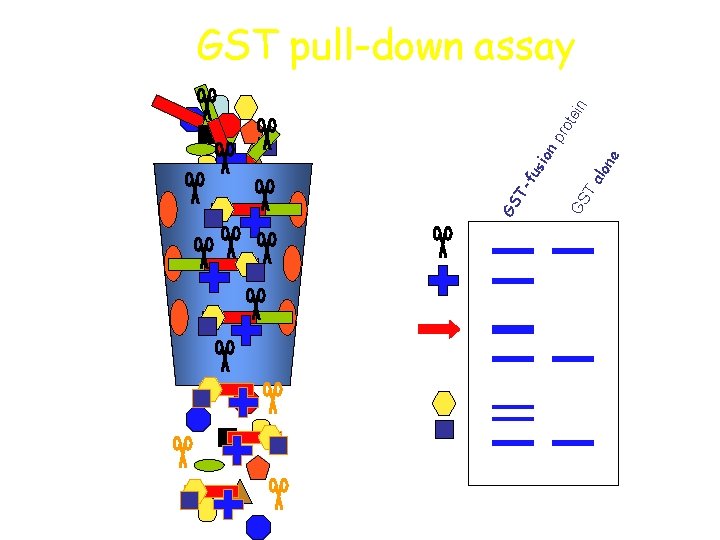
GST pull-down assay Sepharose GSH GST “X” “Y” Sepharose GSH GST “Y”

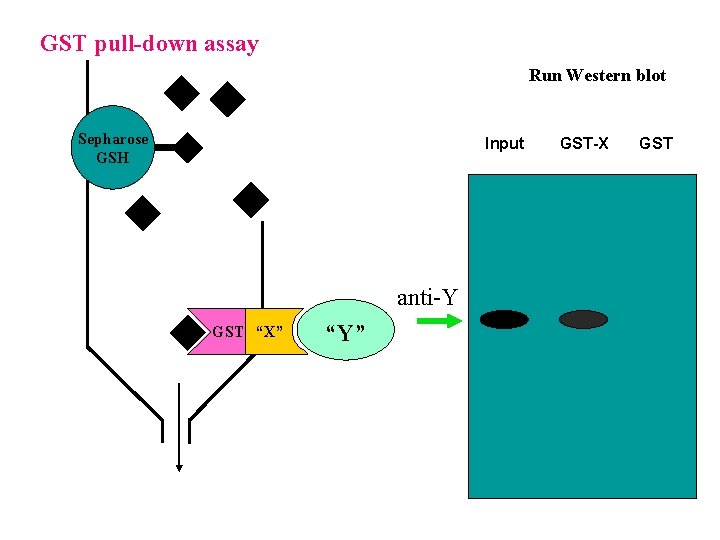
GST pull-down assay Run Western blot Sepharose GSH Input anti-Y GST “X” “Y” GST-X GST

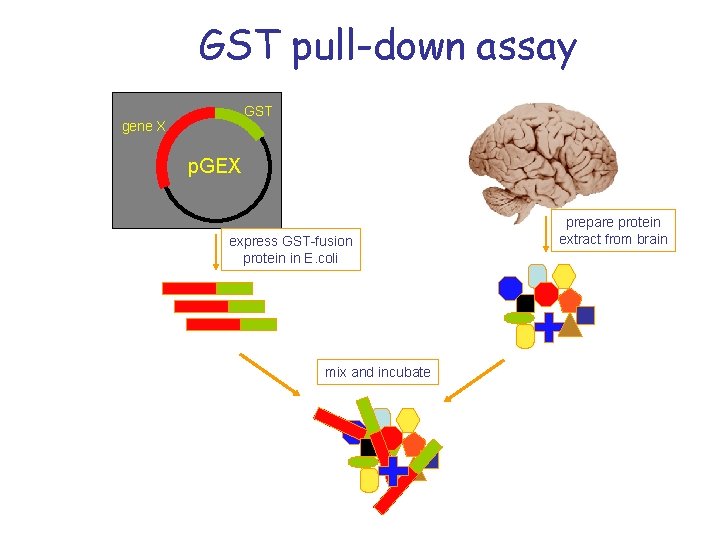
GST pull-down assay GST gene X p. GEX express GST-fusion protein in E. coli mix and incubate prepare protein extract from brain

us ion Ta lon e GS Tf GS n tei pro GST pull-down assay

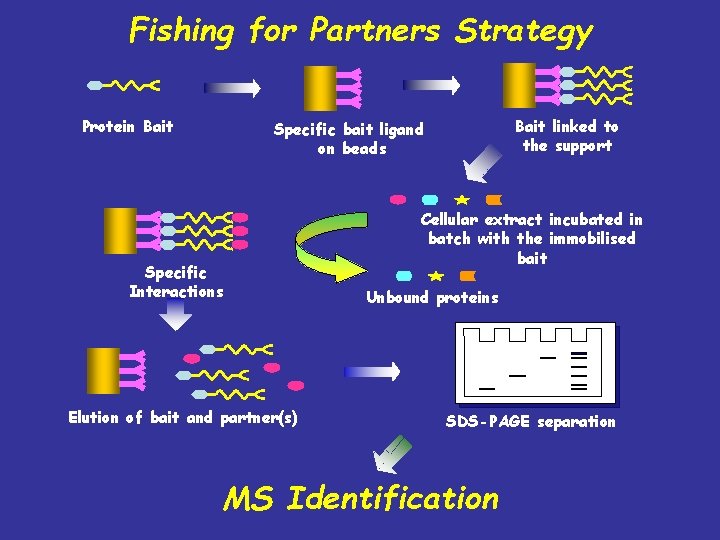
Fishing for Partners Strategy Protein Bait linked to the support Specific bait ligand on beads Specific Interactions Elution of bait and partner(s) Cellular extract incubated in batch with the immobilised bait Unbound proteins SDS-PAGE separation MS Identification

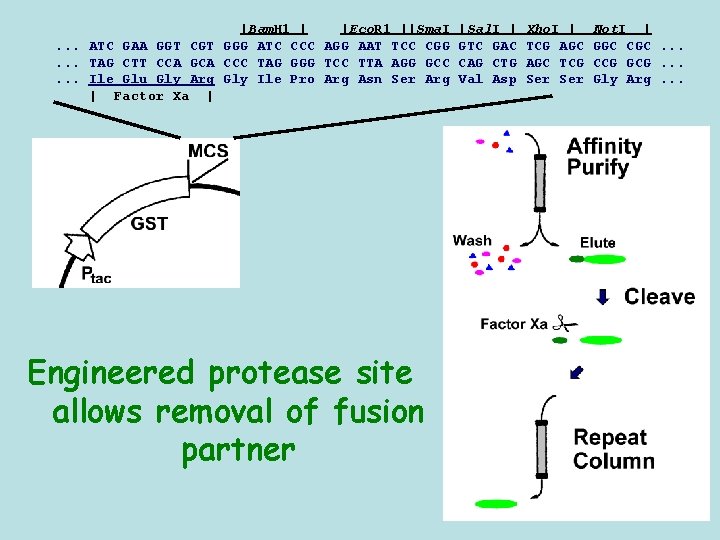
|Bam. H 1 | |Eco. R 1 ||Sma. I. . . ATC GAA GGT CGT GGG ATC CCC AGG AAT TCC CGG. . . TAG CTT CCA GCA CCC TAG GGG TCC TTA AGG GCC. . . Ile Glu Gly Arg Gly Ile Pro Arg Asn Ser Arg | Factor Xa | Engineered protease site allows removal of fusion partner |Sal. I | GTC GAC CAG CTG Val Asp Xho. I | TCG AGC TCG Ser Not. I | GGC CGC. . . CCG GCG. . . Gly Arg. . .

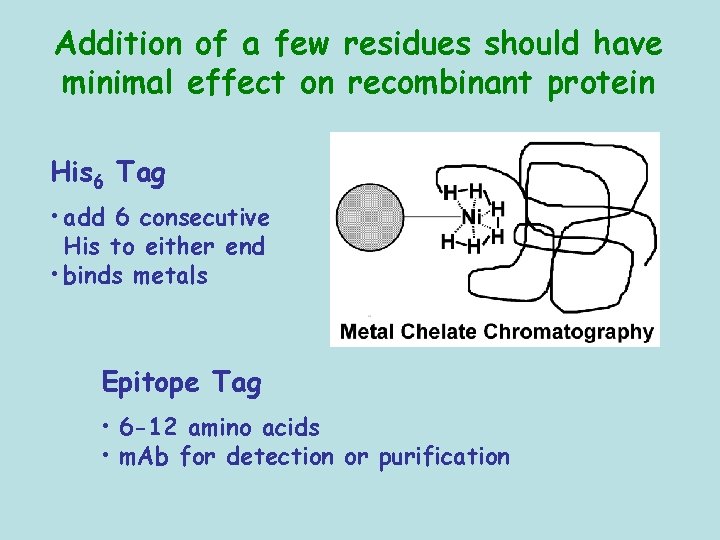
Addition of a few residues should have minimal effect on recombinant protein His 6 Tag • add 6 consecutive His to either end • binds metals Epitope Tag • 6 -12 amino acids • m. Ab for detection or purification

Immunoprecipitation • affinity purification based on isolation of Ag-Ab complexes • analyze by gel electrophoresis • initially based on centrifugation of large supramolecular complexes • [high] and equal amounts • isolation of Ag-Ab complexes • protein A-agarose • protein G-agarose Bacterial proteins that bind Ig. G (Fc): • protein A (Staphylococcus aureus) • protein G (Streptococcus) • binds more species and subclasses

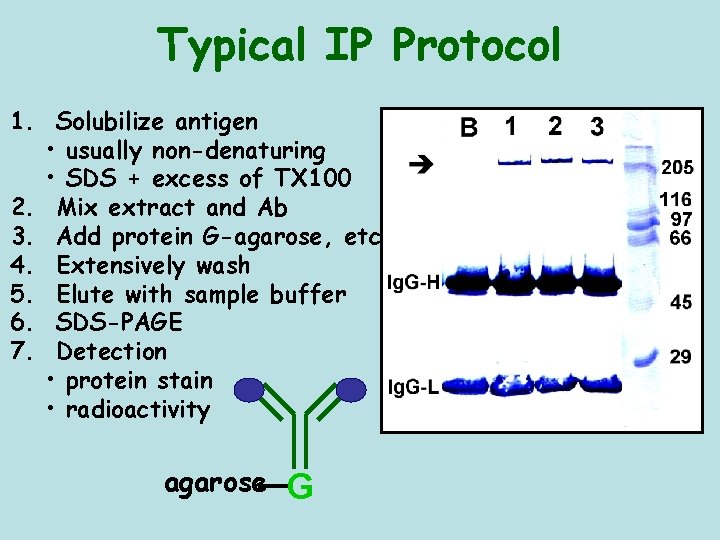
Typical IP Protocol 1. Solubilize antigen • usually non-denaturing • SDS + excess of TX 100 2. Mix extract and Ab 3. Add protein G-agarose, etc 4. Extensively wash 5. Elute with sample buffer 6. SDS-PAGE 7. Detection • protein stain • radioactivity agarose G

Protein identification In situ digestion Peptide extraction S -M M I D AL

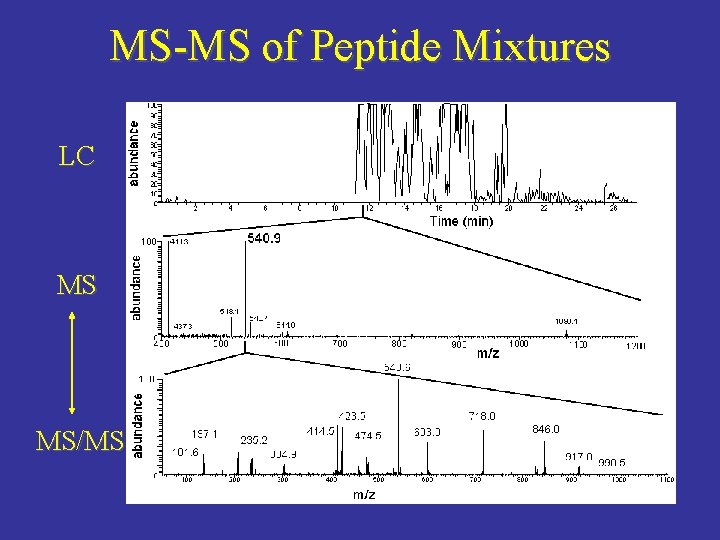
MS-MS of Peptide Mixtures LC MS MS/MS

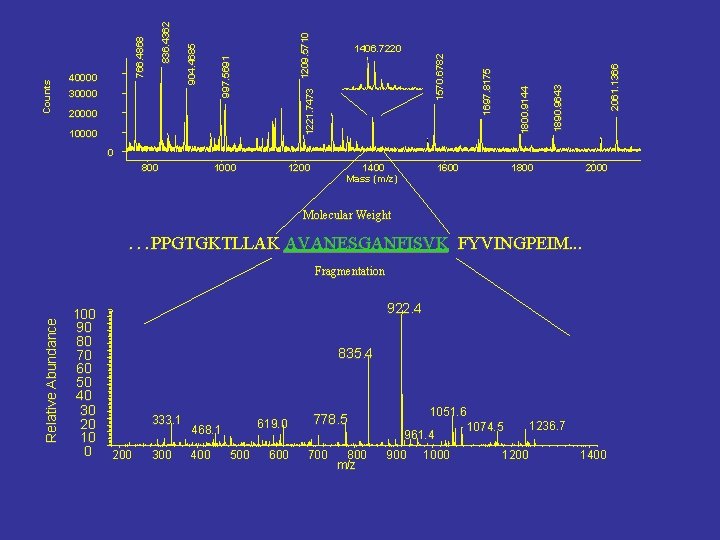
2061. 1366 1570. 6782 1890. 9643 10000 1800. 9144 20000 1697. 8175 1221. 7473 1209. 5710 30000 997. 5691 836. 4362 766. 4868 904. 4685 Counts 40000 1406. 7220 0 800 1000 1200 1400 Mass (m/z) 1600 1800 2000 Molecular Weight …PPGTGKTLLAK AVANESGANFISVK FYVINGPEIM. . . Relative Abundance Fragmentation 100 90 80 70 60 50 40 30 20 10 0 922. 4 835. 4 333. 1 200 300 619. 0 468. 1 400 500 600 778. 5 700 800 m/z 1051. 6 1074. 5 961. 4 900 1000 1236. 7 1200 1400

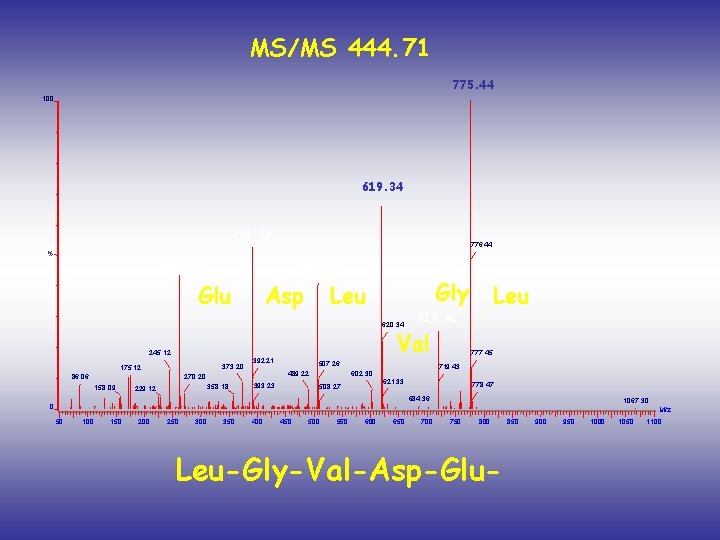
MS/MS 444. 71 775. 44 100 619. 34 391. 22 % 776. 44 262. 16 506. 24 Glu Asp Gly Leu 620. 34 373. 20 175. 12 86. 06 392. 21 158. 09 358. 18 229. 12 507. 26 489. 22 270. 20 718. 42 Val 245. 12 393. 23 777. 45 719. 43 602. 30 621. 33 508. 27 Leu 778. 47 684. 36 1067. 30 0 M/z 50 100 150 200 250 300 350 400 450 500 550 600 650 700 750 800 Leu-Gly-Val-Asp-Glu- 850 900 950 1000 1050 1100

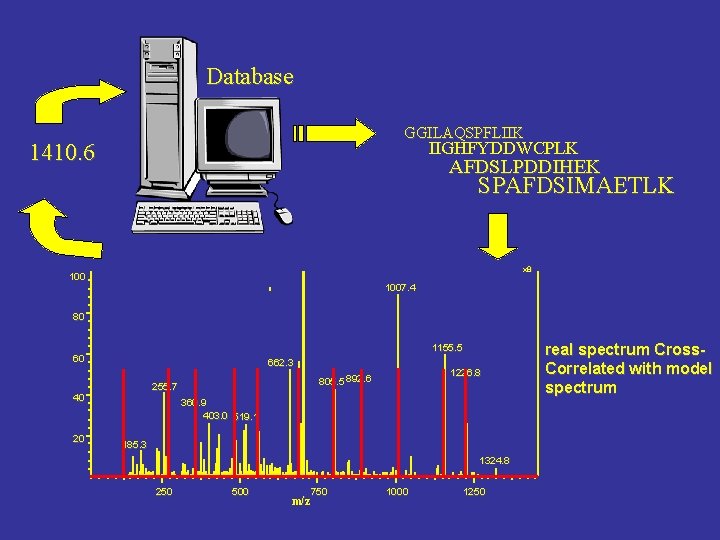
Database GGILAQSPFLIIK IIGHFYDDWCPLK 1410. 6 AFDSLPDDIHEK SPAFDSIMAETLK x 8 1007. 4 80 1155. 5 60 662. 3 40 20 1226. 8 805. 5 892. 6 255. 7 360. 9 403. 0 519. 1 185. 3 1324. 8 250 500 750 m/z 1000 1250 real spectrum Cross. Correlated with model spectrum

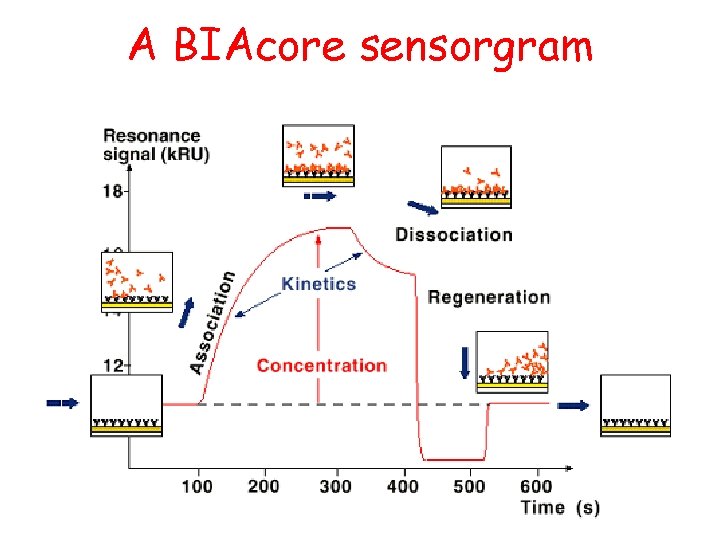
What can Biacore do for you? • SPR (surface plasmon resonance) technology enables real-time detection and monitoring of biomolecular events and provides quantitative information on: 1. 2. 3. 4. 5. Specificity – how specific is the binding between two moelcules? Concentration – how much of a given molecule is present and active? Kinetics – what is the rate of association/dissociation? Affinity – how strong is the binding? Binding partners - provide identification of binding targets by linking SPR to MS

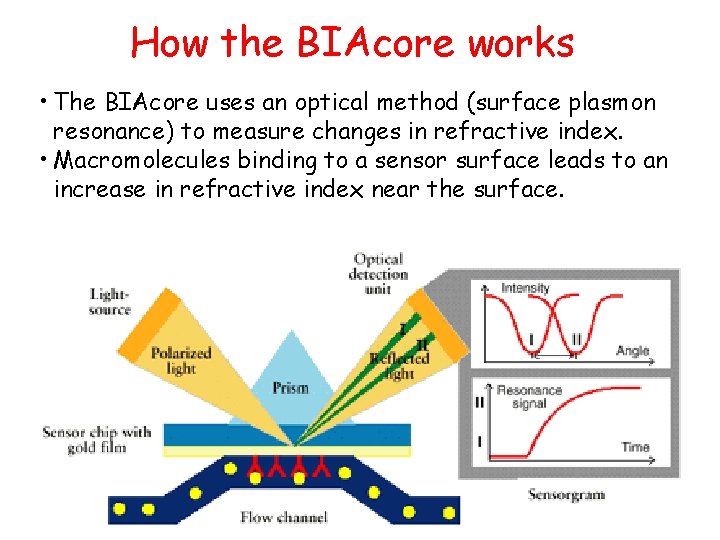
How the BIAcore works • The BIAcore uses an optical method (surface plasmon resonance) to measure changes in refractive index. • Macromolecules binding to a sensor surface leads to an increase in refractive index near the surface.

A BIAcore sensorgram

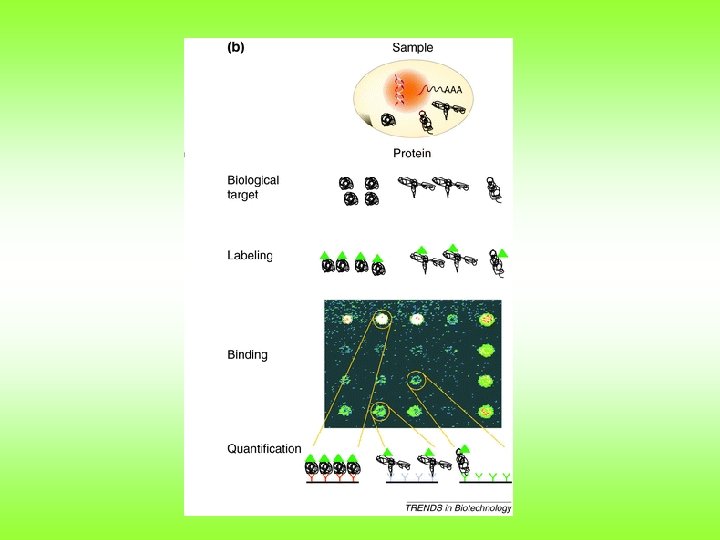
Micro. Array • Analysis of thousands of proteins at one time. • Many different types – Antibody arrayed detect many proteins – Proteins arrayed detect interacting small molecules – Etc.

Protein: protein interactions


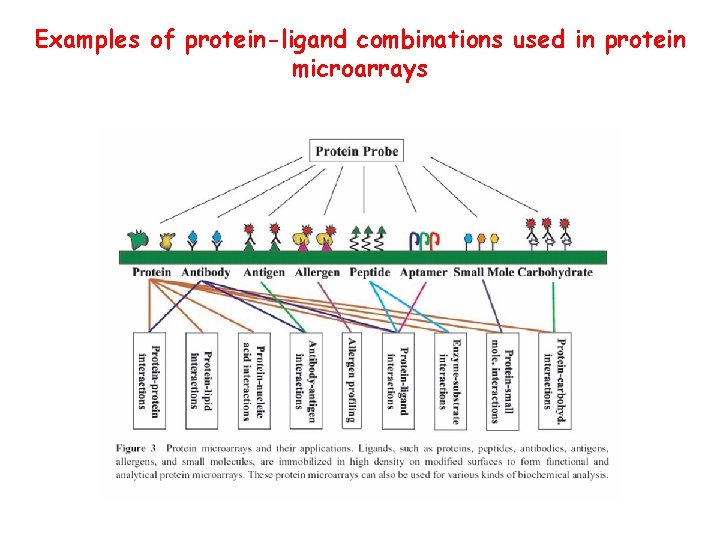
Examples of protein-ligand combinations used in protein microarrays


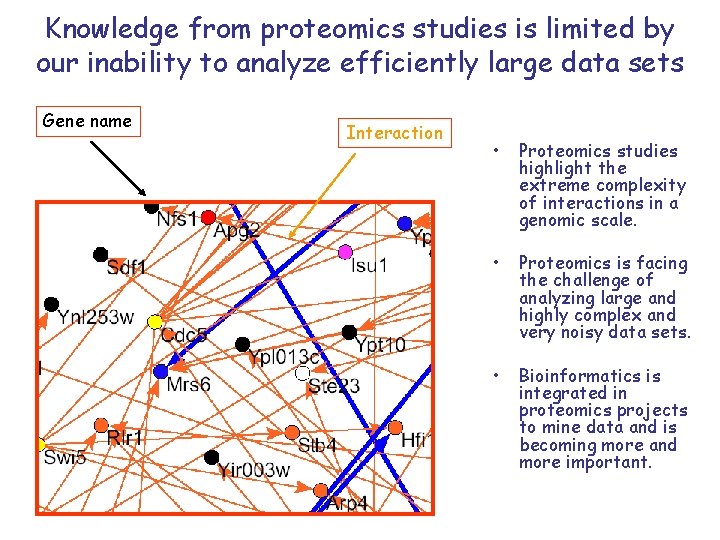
Knowledge from proteomics studies is limited by our inability to analyze efficiently large data sets Gene name Interaction • Proteomics studies highlight the extreme complexity of interactions in a genomic scale. • Proteomics is facing the challenge of analyzing large and highly complex and very noisy data sets. • Bioinformatics is integrated in proteomics projects to mine data and is becoming more and more important.

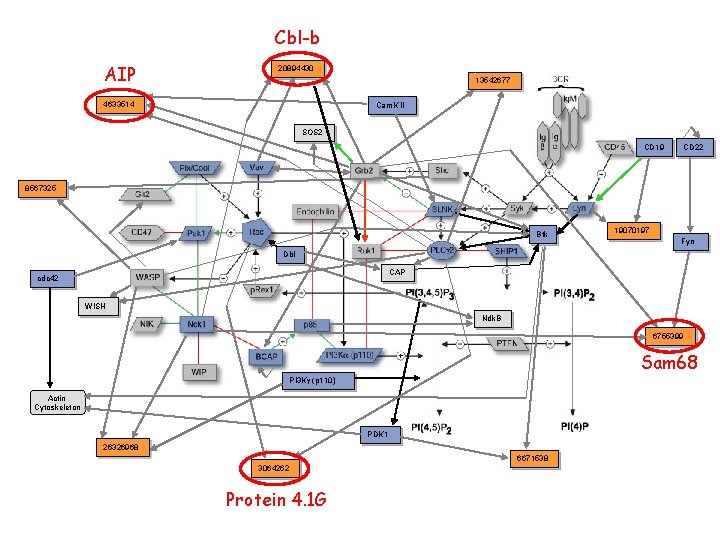
Cam. K II SOS 2 CD 19 Btk Dbl CAP cdc 42 WISH Ndk. B PI 3 Kg (p 110) Actin Cytoskeleton PDK 1 CD 22 Fyn

Cbl-b AIP 20894430 13542677 4633514 Cam. K II SOS 2 CD 19 CD 22 8567325 Btk 19070197 Fyn Dbl CAP cdc 42 WISH Ndk. B 6755399 Sam 68 PI 3 Kg (p 110) Actin Cytoskeleton PDK 1 26326968 6671538 3064262 Protein 4. 1 G




Protein Interactions • Databases of experimental protein interaction data – SPIN-PP: http: //honiglab. cpmc. columbia. edu/SPIN/main. html (existing protein-protein interfaces in the PDB) – MIPS: http: //mips. gsf. de/proj/yeast/CYGD/interaction/ (protein-protein interactions in saccharomyces cerevisae) – Inter. Act: http: //www. ebi. ac. uk/intact/index. html (protein interactions from literature curation) – DIP: http: //dip. doe-mbi. ucla. edu/ – BIND: http: //bind. ca/

Protein Interactomics


