Contents Introduction of protein Principle and methods for


![Ion Exchange Separation Example [salt] UV Ion Exchange Separation Example [salt] UV](https://slidetodoc.com/presentation_image_h/7cc2bf28cf5b64eafdf413a6b0d7f413/image-19.jpg)









- Slides: 74

Contents • Introduction of protein • Principle and methods for protein characterization • Principle and methods for protein purification • Principle and methods for detection of protein-mediated interactions • Principle and methods for structural determination of proteins

Methods and Principle for Protein Purification Yong Chen SIBCB

Proteins in a cell • In a cell, the protein concentration is very high (~300 mg/ml) • Other components – Membranes, DNA, RNA, sugars, structural elements • Goal: Isolating a specific protein from a cell for further study Ellis and Minton, Nature 425, 27 - 28 (04 Sep 2003) 3 Medalia, O. et al. Science 298, 1209– 1213 (2002)

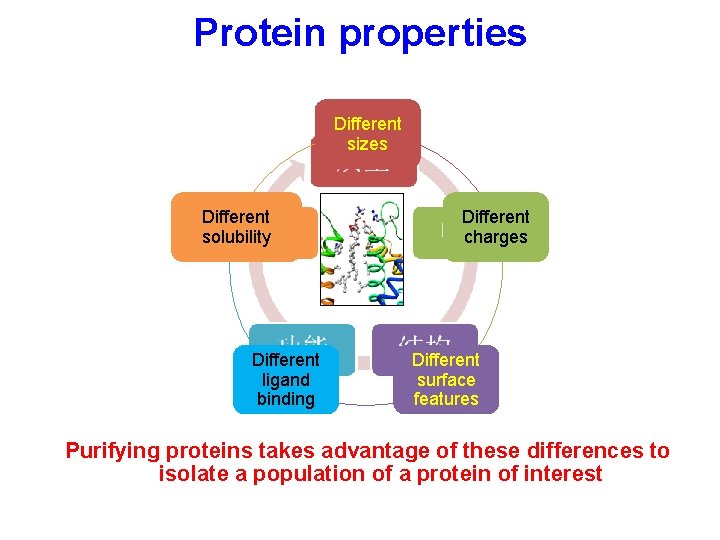
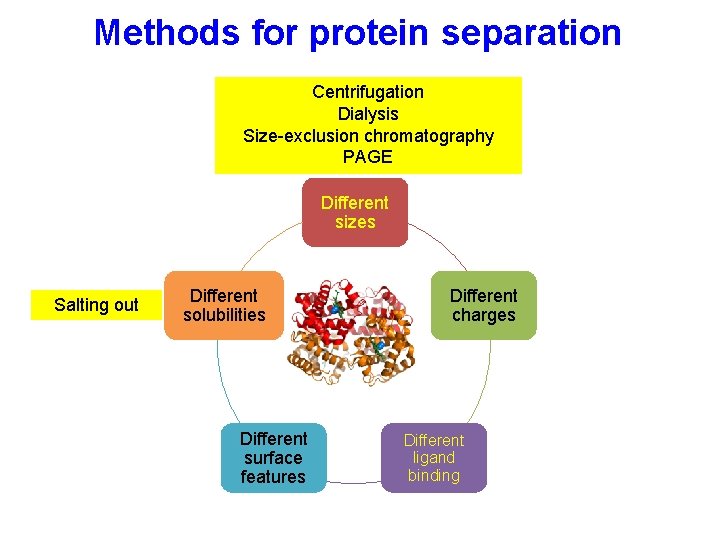
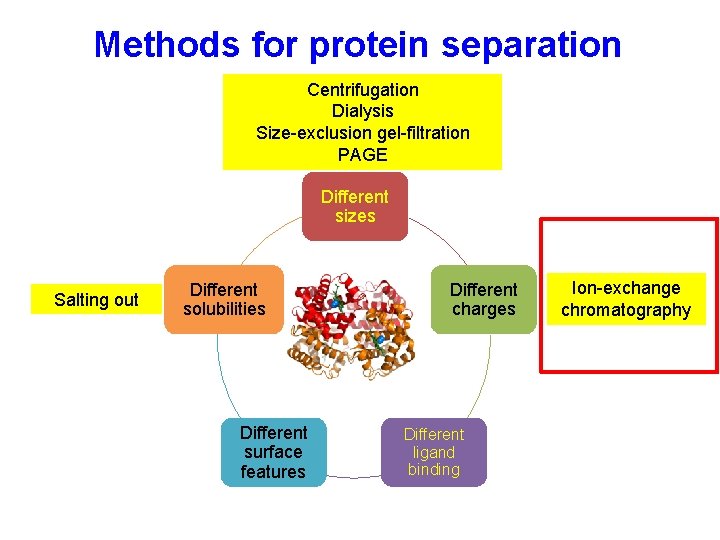
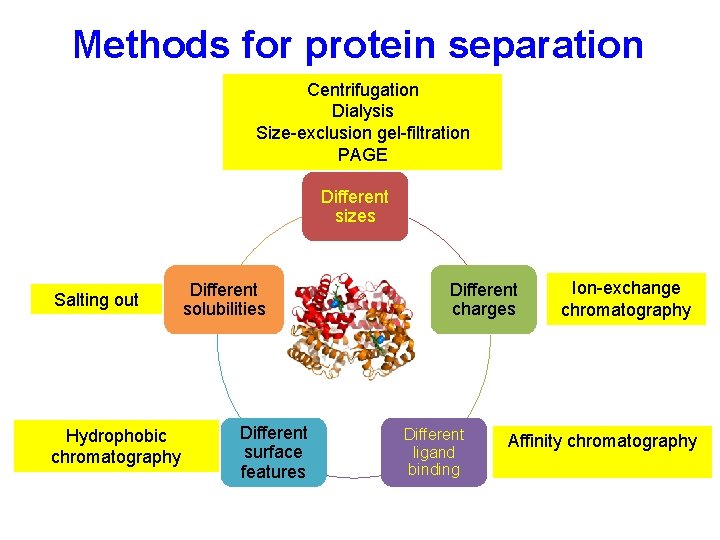
Protein properties Different sizes Different solubility Different ligand binding Different charges Different surface features Purifying proteins takes advantage of these differences to isolate a population of a protein of interest

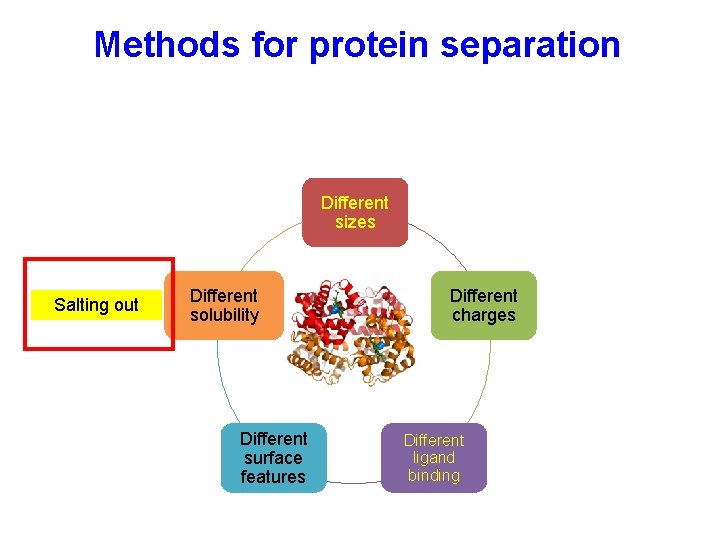
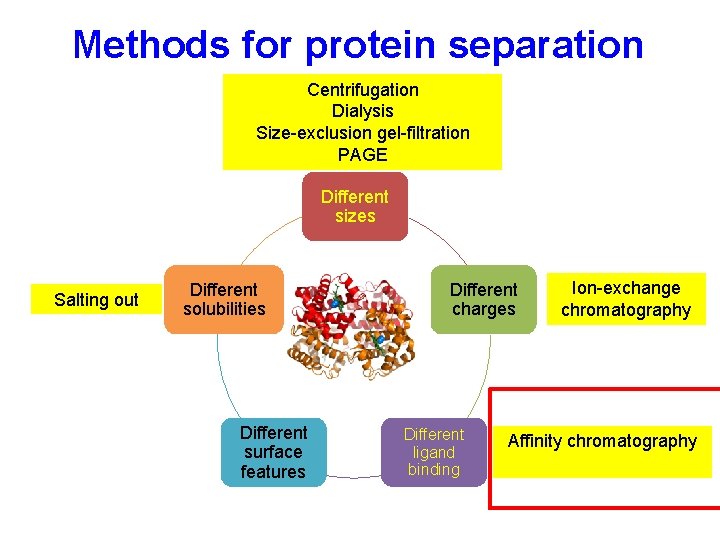
Methods for protein separation Different sizes Salting out Different solubility Different surface features Different charges Different ligand binding

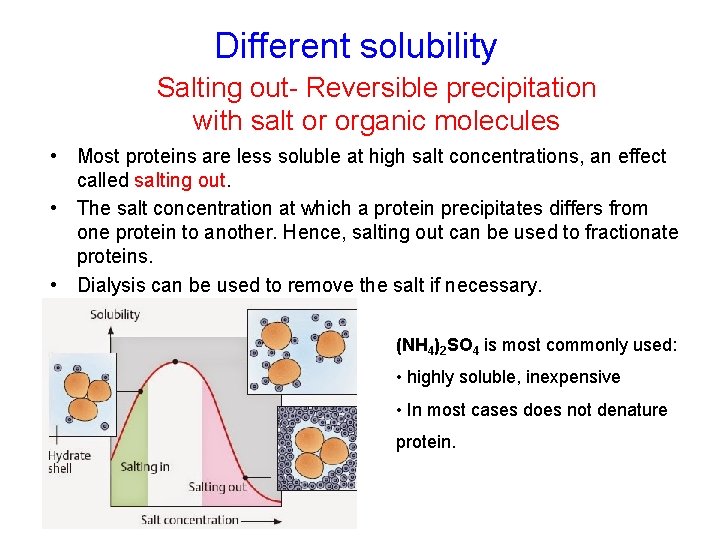
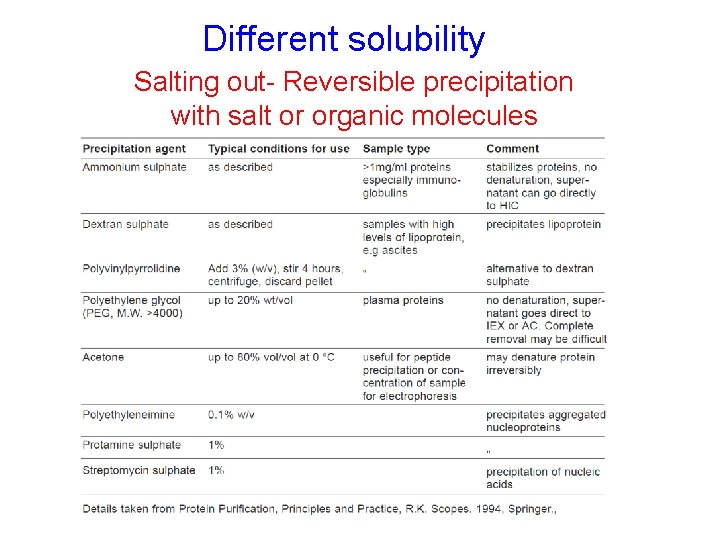
Different solubility Salting out- Reversible precipitation with salt or organic molecules • Most proteins are less soluble at high salt concentrations, an effect called salting out. • The salt concentration at which a protein precipitates differs from one protein to another. Hence, salting out can be used to fractionate proteins. • Dialysis can be used to remove the salt if necessary. (NH 4)2 SO 4 is most commonly used: • highly soluble, inexpensive • In most cases does not denature protein.

Different solubility Salting out- Reversible precipitation with salt or organic molecules

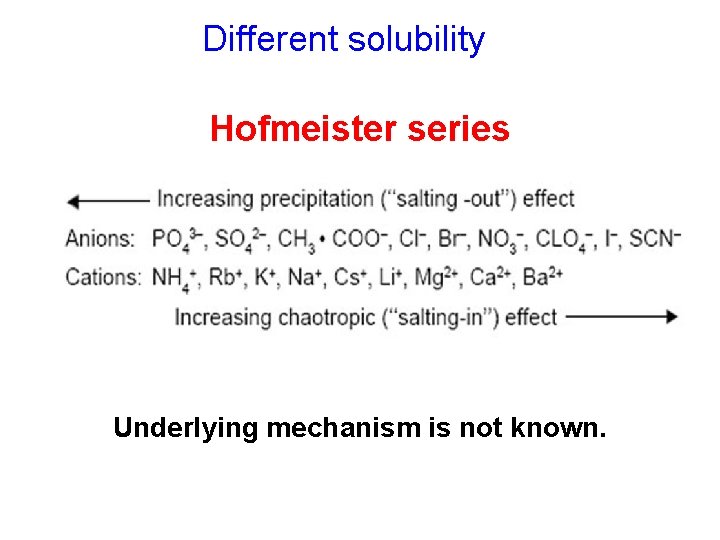
Different solubility Hofmeister series Underlying mechanism is not known.

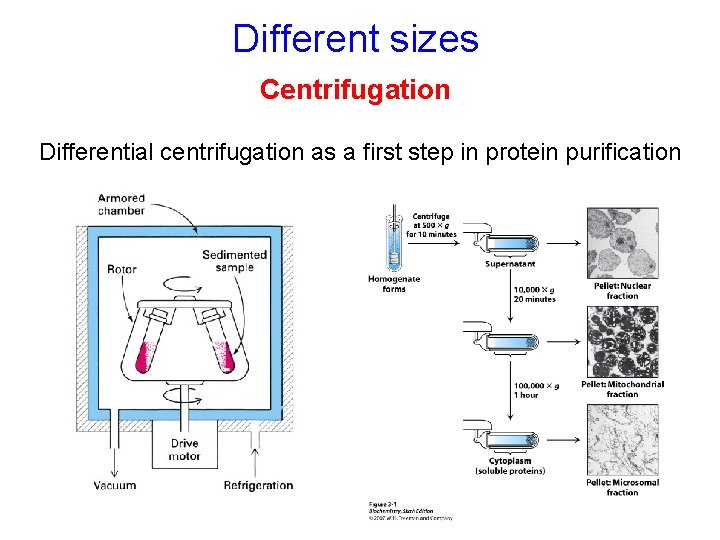
Methods for protein separation Centrifugation Dialysis Size-exclusion chromatography PAGE Different sizes Salting out Different solubilities Different surface features Different charges Different ligand binding

Different sizes Centrifugation Differential centrifugation as a first step in protein purification

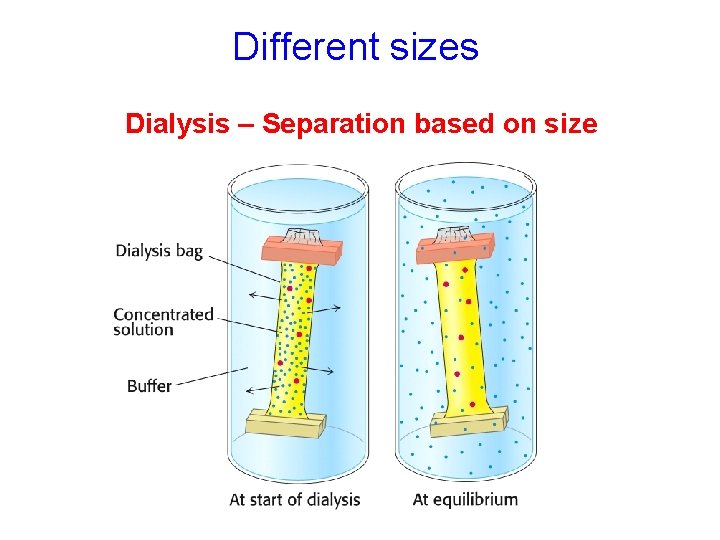
Different sizes Dialysis – Separation based on size

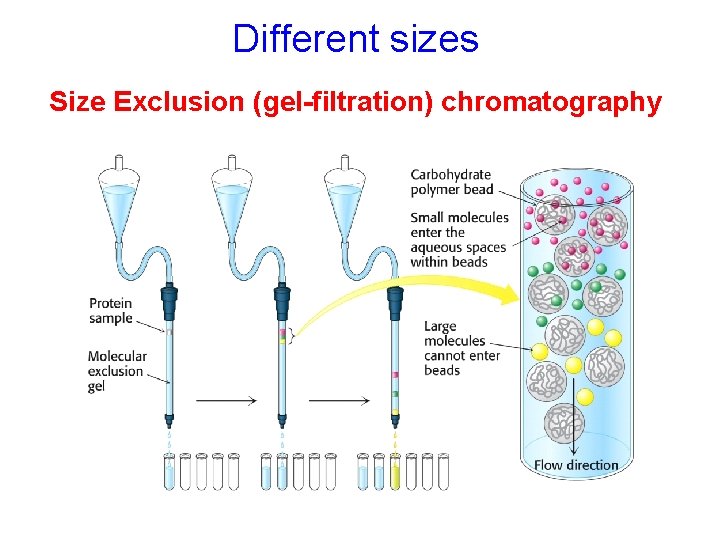
Different sizes Size Exclusion (gel-filtration) chromatography

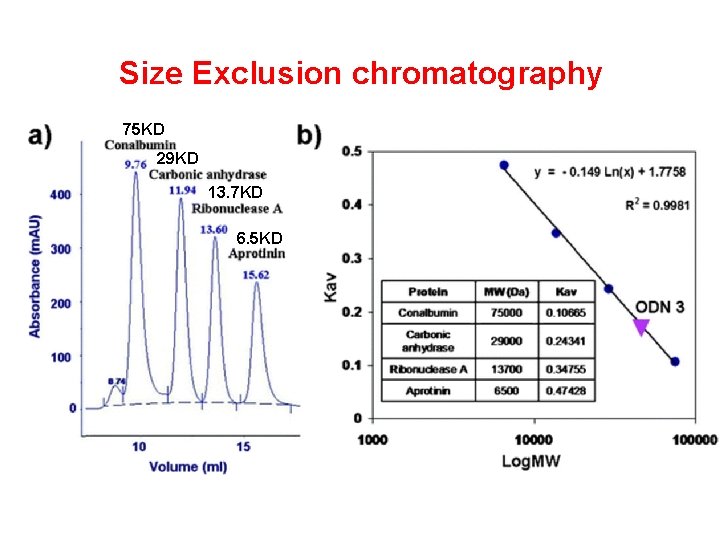
Size Exclusion chromatography 75 KD 29 KD 13. 7 KD 6. 5 KD

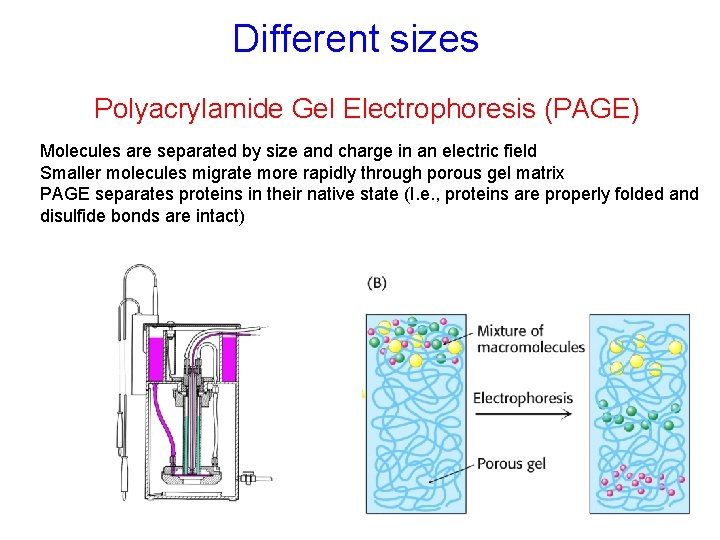
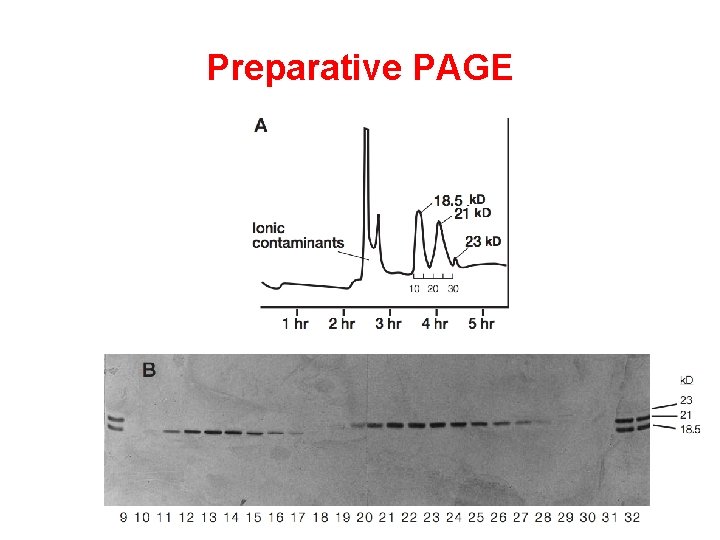
Different sizes Polyacrylamide Gel Electrophoresis (PAGE) Molecules are separated by size and charge in an electric field Smaller molecules migrate more rapidly through porous gel matrix PAGE separates proteins in their native state (I. e. , proteins are properly folded and disulfide bonds are intact)

Preparative PAGE

Methods for protein separation Centrifugation Dialysis Size-exclusion gel-filtration PAGE Different sizes Salting out Different solubilities Different surface features Different charges Different ligand binding Ion-exchange chromatography

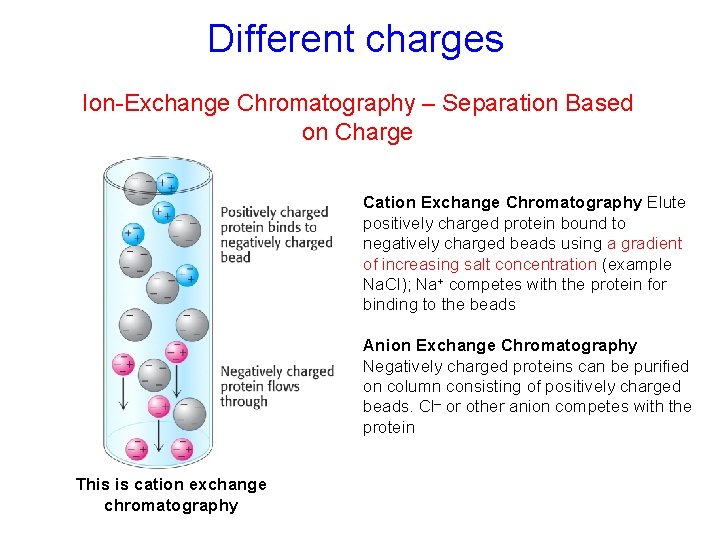
Different charges Ion-Exchange Chromatography – Separation Based on Charge Cation Exchange Chromatography Elute positively charged protein bound to negatively charged beads using a gradient of increasing salt concentration (example Na. Cl); Na+ competes with the protein for binding to the beads Anion Exchange Chromatography Negatively charged proteins can be purified on column consisting of positively charged beads. Cl– or other anion competes with the protein This is cation exchange chromatography

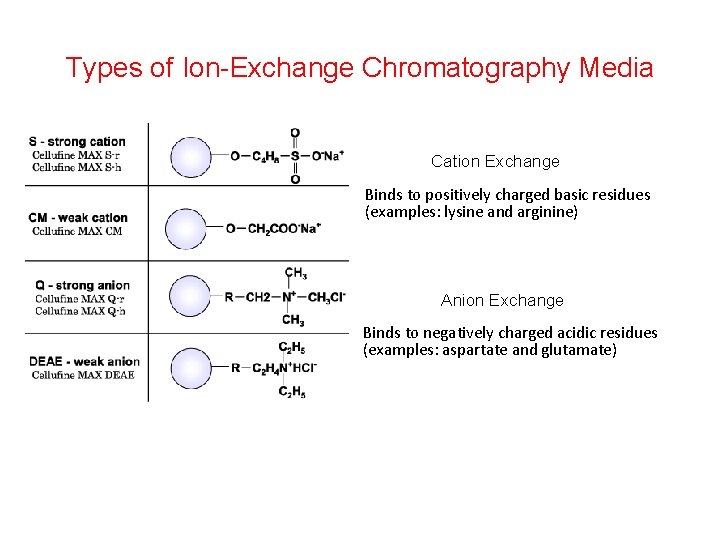
Types of Ion-Exchange Chromatography Media Cation Exchange Binds to positively charged basic residues (examples: lysine and arginine) Anion Exchange Binds to negatively charged acidic residues (examples: aspartate and glutamate)
![Ion Exchange Separation Example salt UV Ion Exchange Separation Example [salt] UV](https://slidetodoc.com/presentation_image_h/7cc2bf28cf5b64eafdf413a6b0d7f413/image-19.jpg)
Ion Exchange Separation Example [salt] UV

Methods for protein separation Centrifugation Dialysis Size-exclusion gel-filtration PAGE Different sizes Salting out Different solubilities Different surface features Different charges Different ligand binding Ion-exchange chromatography Affinity chromatography

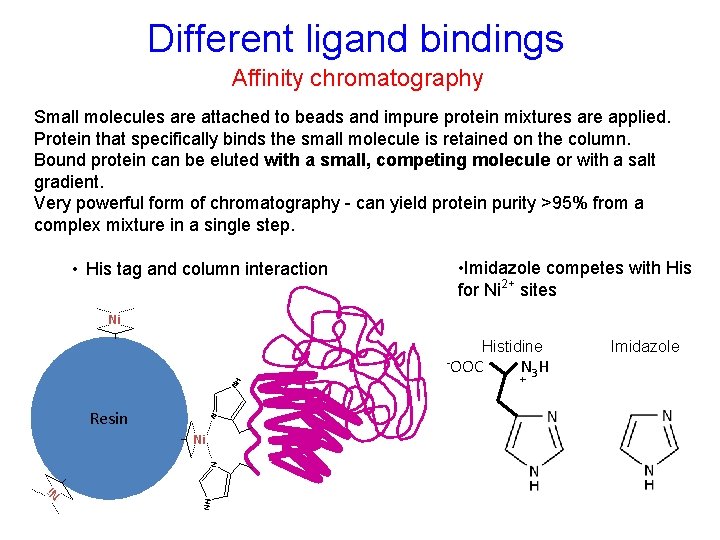
Different ligand bindings Affinity chromatography Small molecules are attached to beads and impure protein mixtures are applied. Protein that specifically binds the small molecule is retained on the column. Bound protein can be eluted with a small, competing molecule or with a salt gradient. Very powerful form of chromatography - can yield protein purity >95% from a complex mixture in a single step. • His tag and column interaction • Imidazole competes with His for Ni 2+ sites Ni Histidine Resin N NH -OOC NH N Ni N 3 H + Imidazole Ni

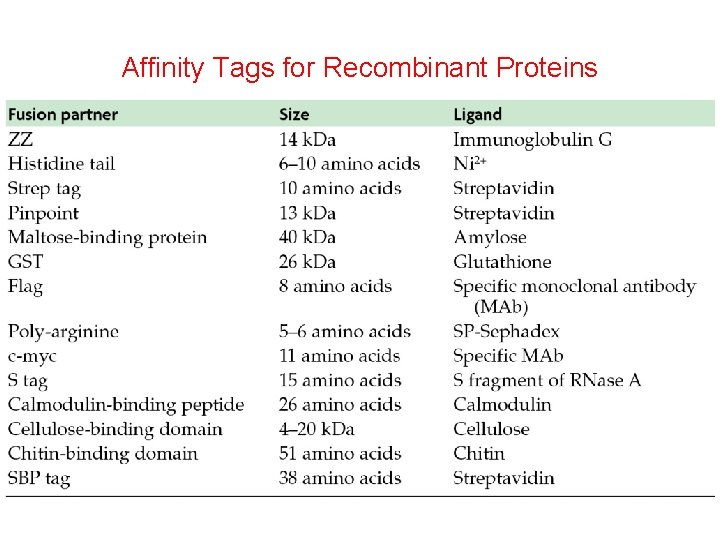
Affinity Tags for Recombinant Proteins

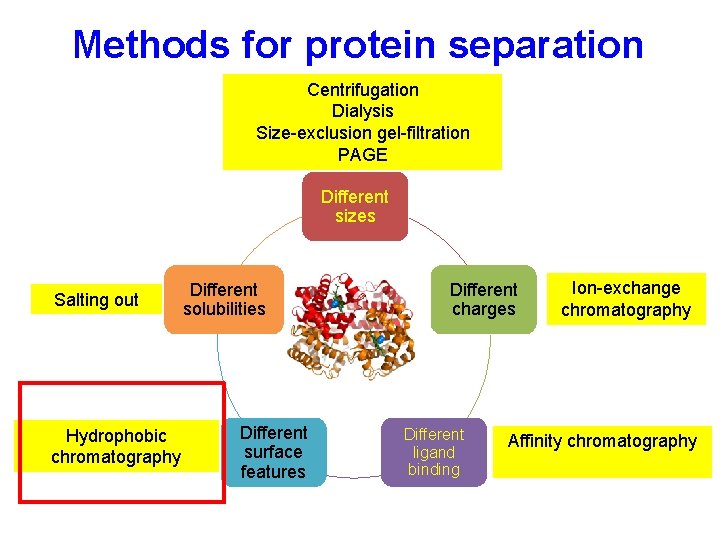
Methods for protein separation Centrifugation Dialysis Size-exclusion gel-filtration PAGE Different sizes Salting out Hydrophobic chromatography Different solubilities Different surface features Different charges Different ligand binding Ion-exchange chromatography Affinity chromatography

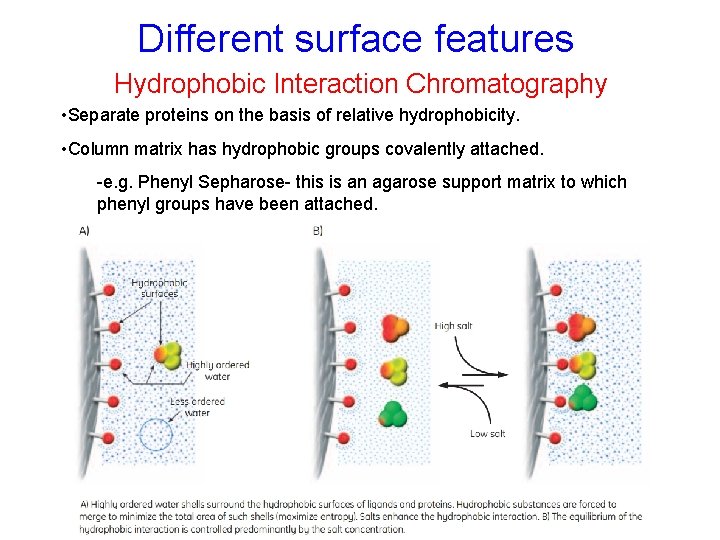
Different surface features Hydrophobic Interaction Chromatography • Separate proteins on the basis of relative hydrophobicity. • Column matrix has hydrophobic groups covalently attached. -e. g. Phenyl Sepharose- this is an agarose support matrix to which phenyl groups have been attached. • Proteins bind to an HIC column under high salt concentrations and are eluted in low salt concentrations.

Methods for protein separation Centrifugation Dialysis Size-exclusion gel-filtration PAGE Different sizes Salting out Hydrophobic chromatography Different solubilities Different surface features Different charges Different ligand binding Ion-exchange chromatography Affinity chromatography

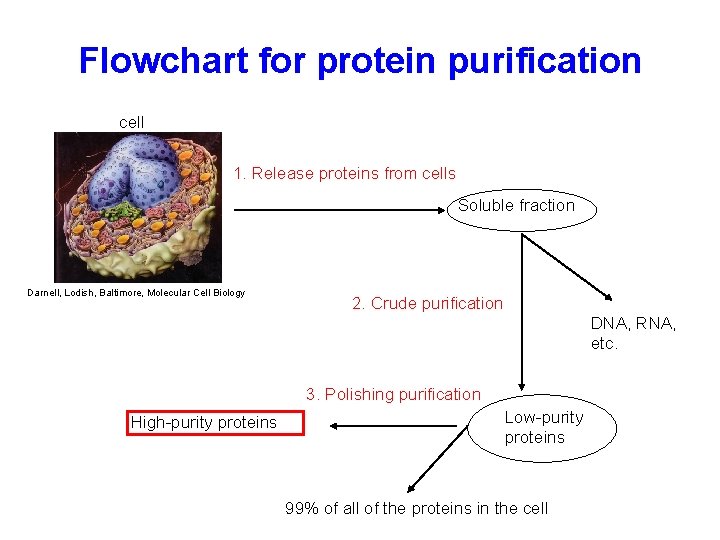
Flowchart for protein purification cell 1. Release proteins from cells Soluble fraction Darnell, Lodish, Baltimore, Molecular Cell Biology 2. Crude purification DNA, RNA, etc. 3. Polishing purification High-purity proteins Low-purity proteins 99% of all of the proteins in the cell

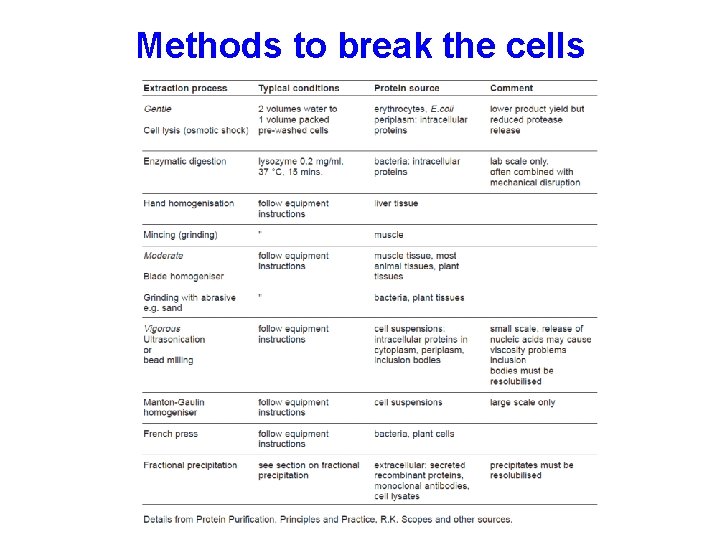
Releasing proteins from the cell • Physical methods – Shearing open the cells using pressure or strong sound waves or mechanical means • Chemical methods – Osmotic shock, detergents, or enzymatic digestion

Methods to break the cells

The purification strategy KEEP IT SIMPLE! • Define objectives for purity, activity and quantity required of final product to avoid over or under developing a method

Define objectives for your purification Goal: To set minimum objectives for purity and quantity, maintenance of biological activity and economy in terms of money and time. Define purity requirements according to the final use of the product.

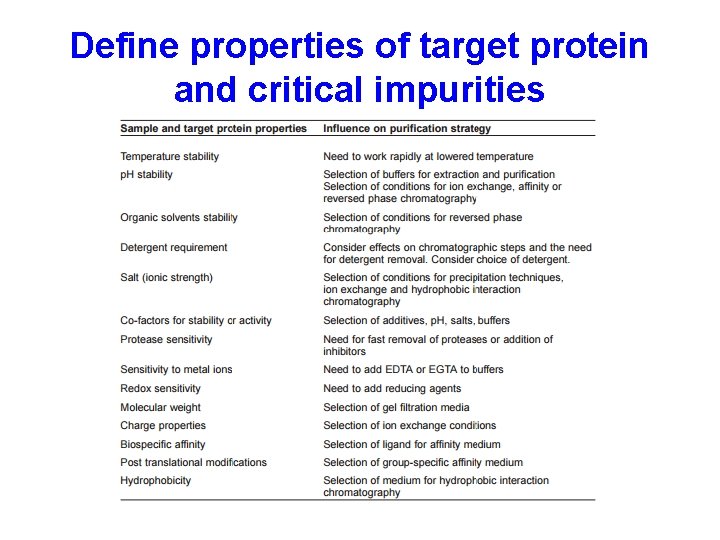
The purification strategy KEEP IT SIMPLE! • Define objectives for purity, activity and quantity required of final product to avoid over or under developing a method • Define properties of target protein and critical impurities to simplify technique selection and optimization

Define properties of target protein and critical impurities

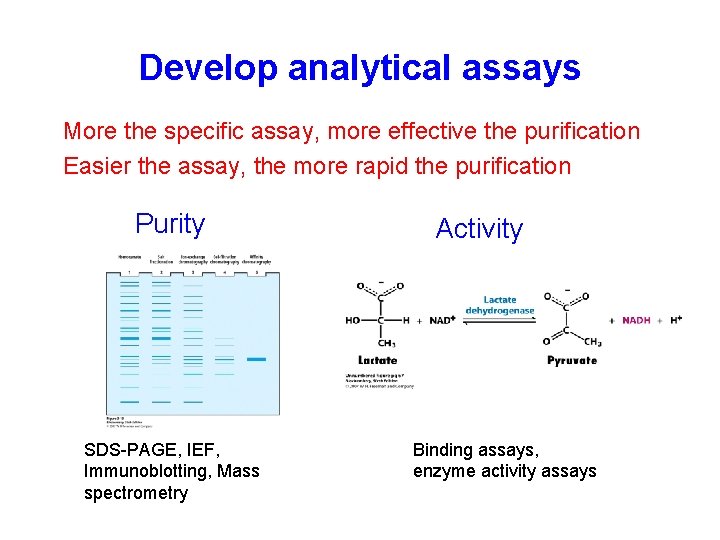
The purification strategy KEEP IT SIMPLE! • Define objectives for purity, activity and quantity required of final product to avoid over or under developing a method • Define properties of target protein and critical impurities to simplify technique selection and optimization • Develop analytical assays for fast detection of protein activity/recovery and critical contaminants

Develop analytical assays More the specific assay, more effective the purification Easier the assay, the more rapid the purification Purity SDS-PAGE, IEF, Immunoblotting, Mass spectrometry Activity Binding assays, enzyme activity assays

The purification strategy KEEP IT SIMPLE! • Define objectives for purity, activity and quantity required of final product to avoid over or under developing a method • Define properties of target protein and critical impurities to simplify technique selection and optimisation • Develop analytical assays for fast detection of protein activity/recovery and critical contaminants • Remove damaging contaminants early, e. g. proteases • Use a different technique at each step to take advantage of sample characteristics which can be used for separation (size, charge, hydrophobicity, ligand specificity) • Minimize number of steps and sample handling at every stage to avoid lengthy procedures which risk losing activity/reducing recovery

Summary of protein purification • Proteins have different physical properties. • These properties can be used to isolate populations of specific proteins from one another. • The target protein should be purified efficiently, economically and in sufficient purity and quantity, by the combination of different purification steps. (多、快、好、省)

Contents • Introduction of protein • Principle and methods for protein characterization • Principle and methods for protein purification • Principle and methods for detection of protein-mediated interactions • Principle and methods for structural determination of proteins

Principle and methods for detection of proteinmediated interactions Yong Chen SIBCB

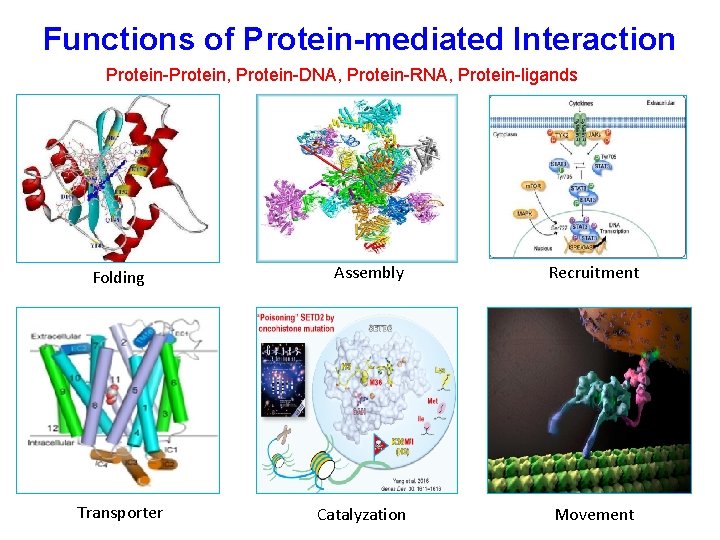
Functions of Protein-mediated Interaction Protein-Protein, Protein-DNA, Protein-RNA, Protein-ligands Folding Transporter Assembly Recruitment Catalyzation Movement

Methods for detecting proteinmediated interactions • Identifying the protein-interaction partners 1. Immunoprecipitation-Mass Spectrometry (IP-MS) 2. Yeast-two-hybrid (Y 2 H) 3. Phage display • Verifying protein-mediated interactions

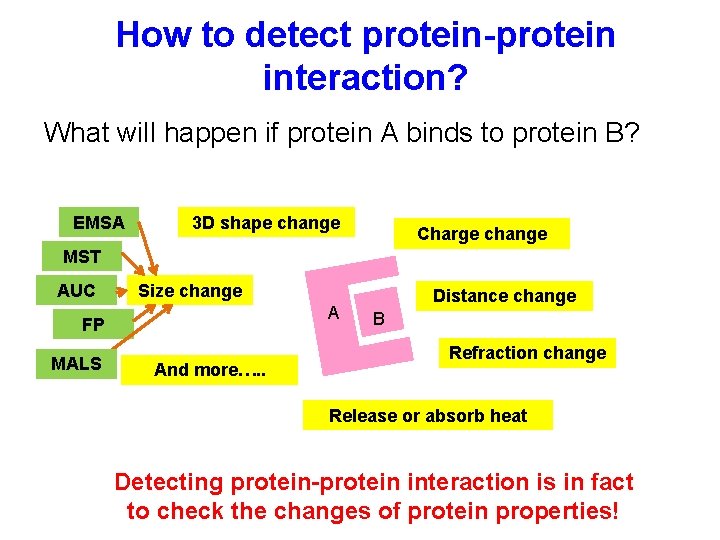
How to detect protein-protein interaction? What will happen if protein A binds to protein B? EMSA 3 D shape change Charge change MST AUC Size change A FP MALS And more…. . Distance change B Refraction change Release or absorb heat Detecting protein-protein interaction is in fact to check the changes of protein properties!

Methods to characterize protein-mediated interactions Qualitative methods Yeast-two-hybrid, Bimolecular fluorescence complementation, Fluorescence Resonance Energy Transfer , Pull-down (or Co-IP), Size-exclusion chromatography, Light scattering, Crosslinking Quantitative methods Analytic ultracentrifugation, Electrophoretic mobility shift assay, Microscale Thermophoresis, Fluorescence Polarization, Isothermal Titration Calorimetry, Surface plasmon resonance, Bio-layer Interferometry

Introduction of Qualitative Methods

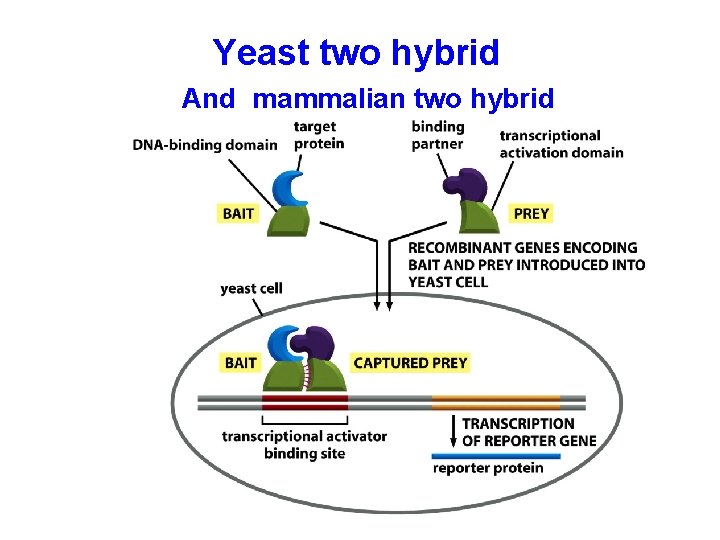
Yeast two hybrid And mammalian two hybrid

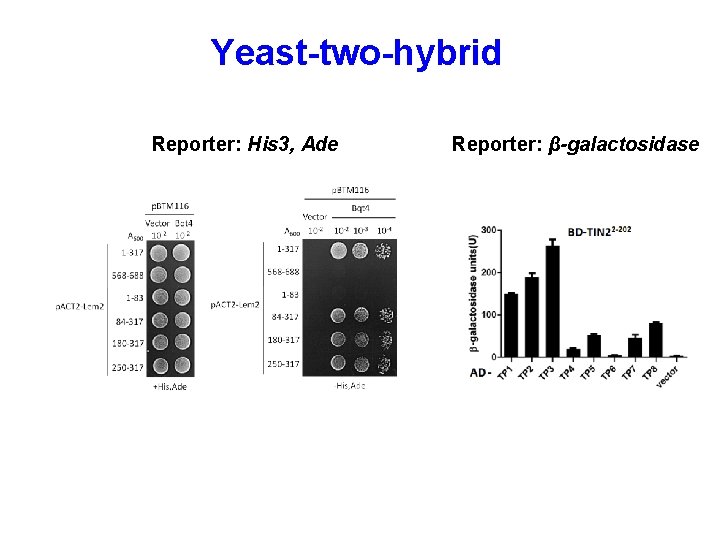
Yeast-two-hybrid Reporter: His 3, Ade Reporter: β-galactosidase

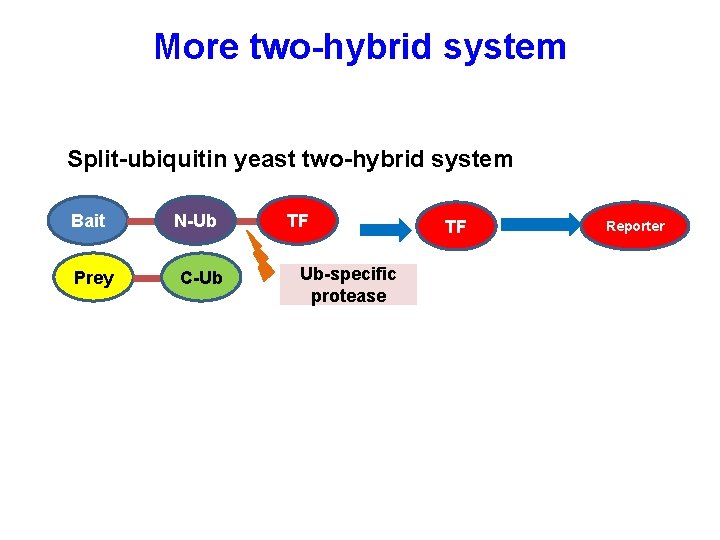
More two-hybrid system Split-ubiquitin yeast two-hybrid system Bait Prey N-Ub C-Ub TF Ub-specific protease TF Reporter

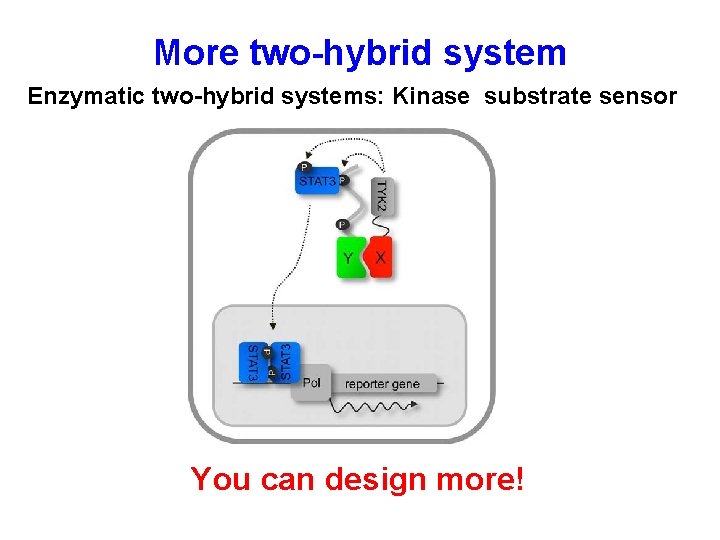
More two-hybrid system Enzymatic two-hybrid systems: Kinase substrate sensor You can design more!

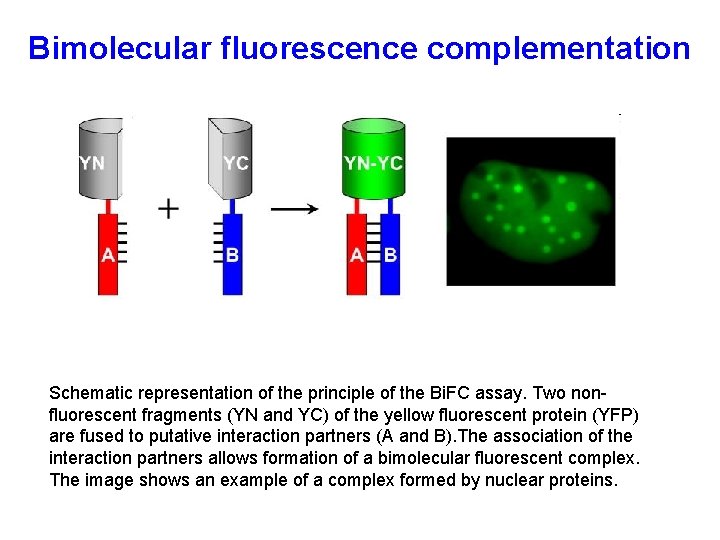
Bimolecular fluorescence complementation Schematic representation of the principle of the Bi. FC assay. Two nonfluorescent fragments (YN and YC) of the yellow fluorescent protein (YFP) are fused to putative interaction partners (A and B). The association of the interaction partners allows formation of a bimolecular fluorescent complex. The image shows an example of a complex formed by nuclear proteins.

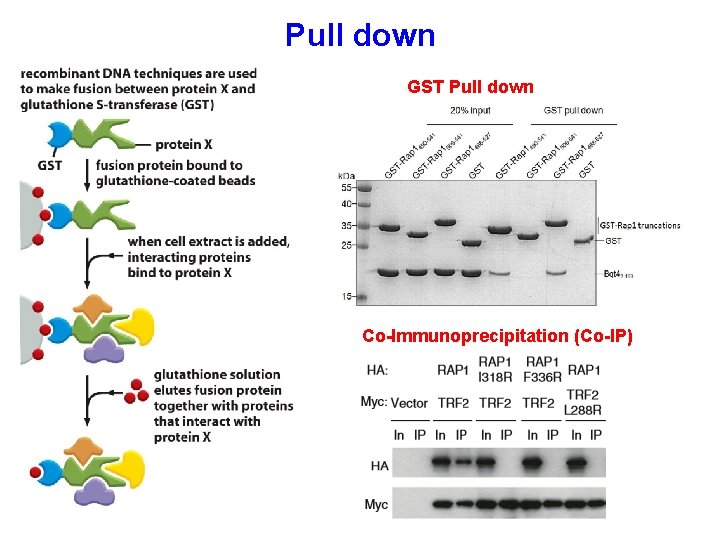
Pull down GST Pull down Co-Immunoprecipitation (Co-IP)

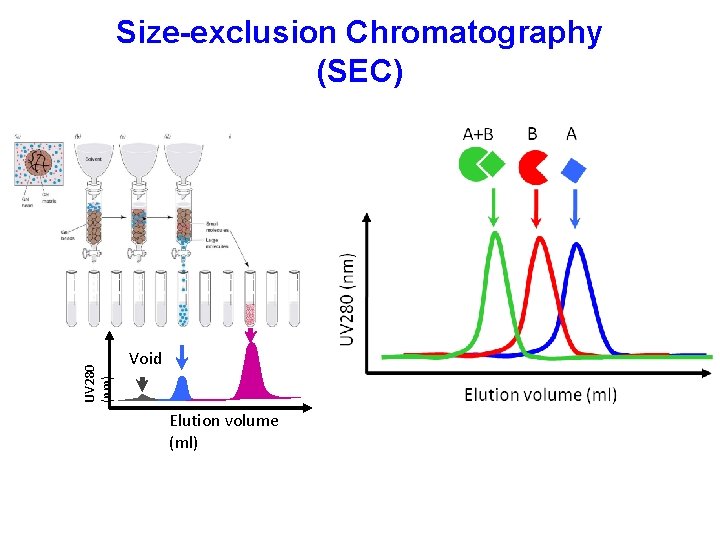
UV 280 (nm) Size-exclusion Chromatography (SEC) Void Elution volume (ml)

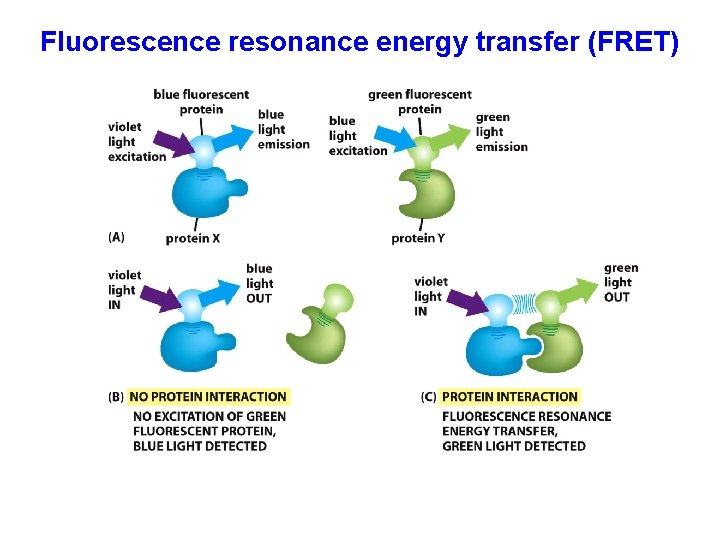
Fluorescence resonance energy transfer (FRET)

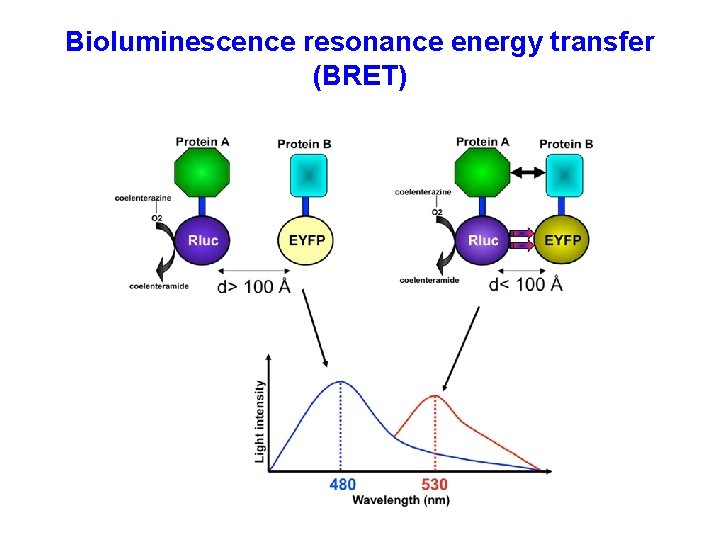
Bioluminescence resonance energy transfer (BRET)

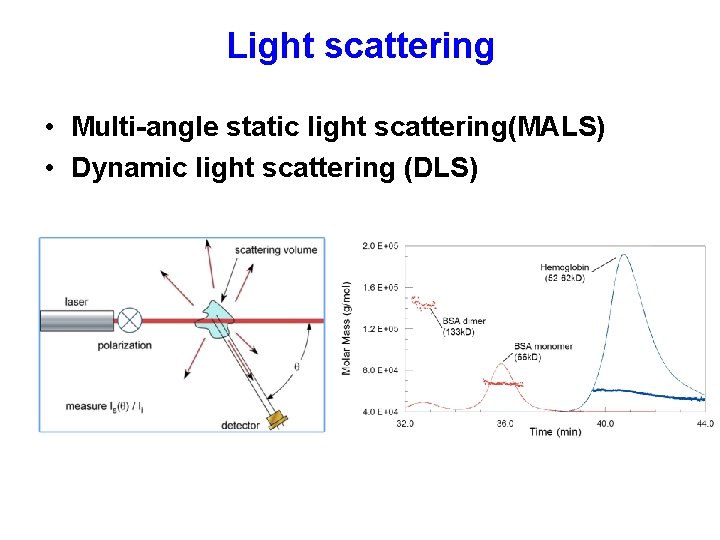
Light scattering • Multi-angle static light scattering(MALS) • Dynamic light scattering (DLS)

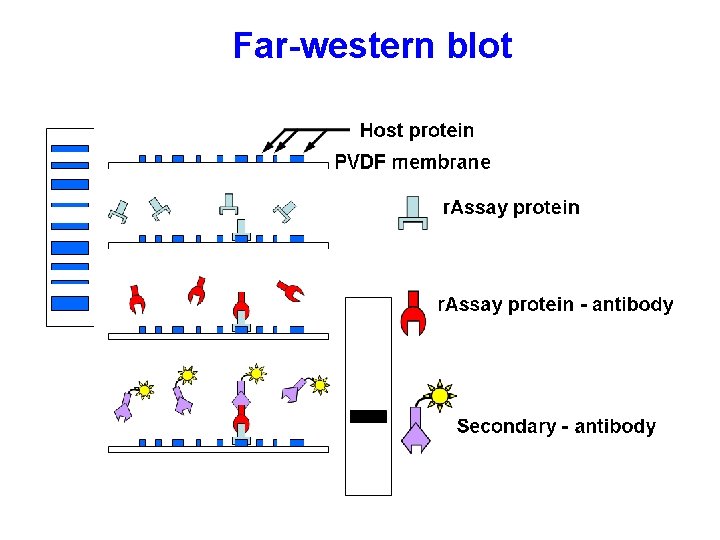
Far-western blot

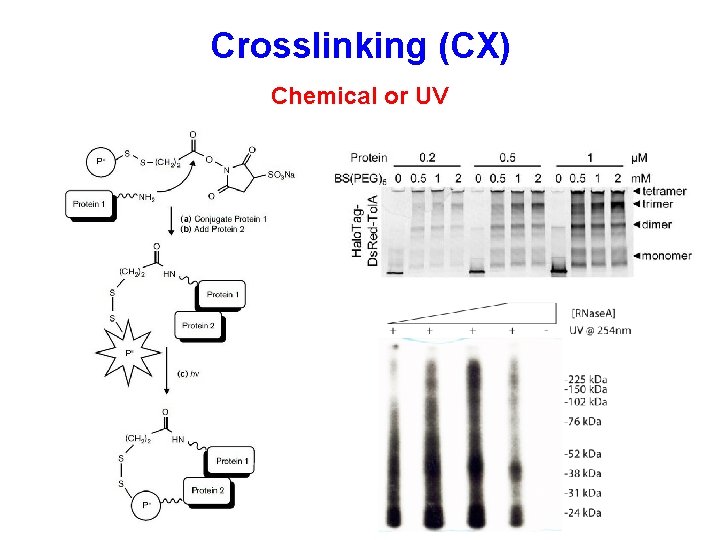
Crosslinking (CX) Chemical or UV

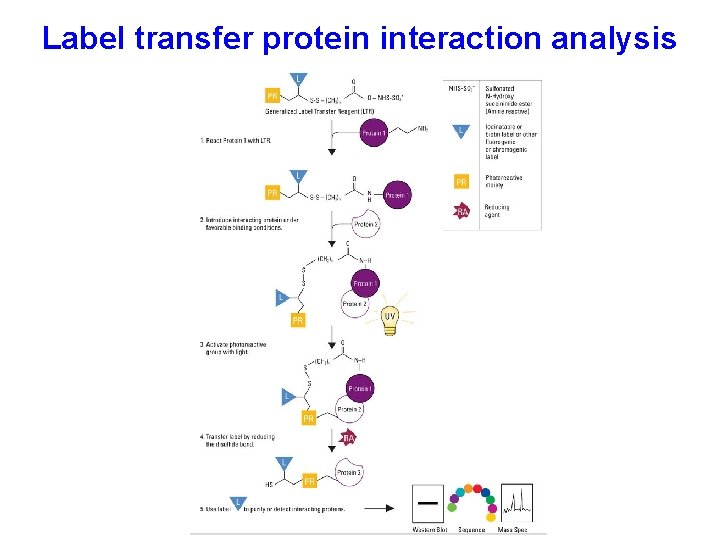
Label transfer protein interaction analysis

Introduction of Quantitative Methods

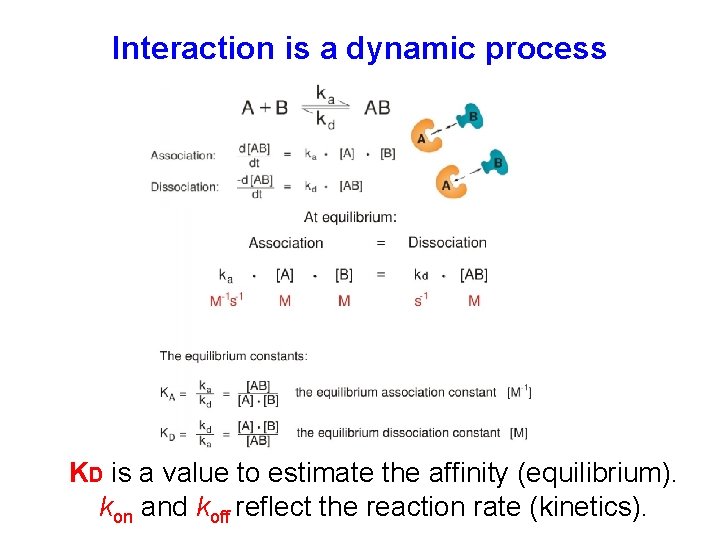
Interaction is a dynamic process KD is a value to estimate the affinity (equilibrium). kon and koff reflect the reaction rate (kinetics).

Analytical Ultracentrifugation (AUC)

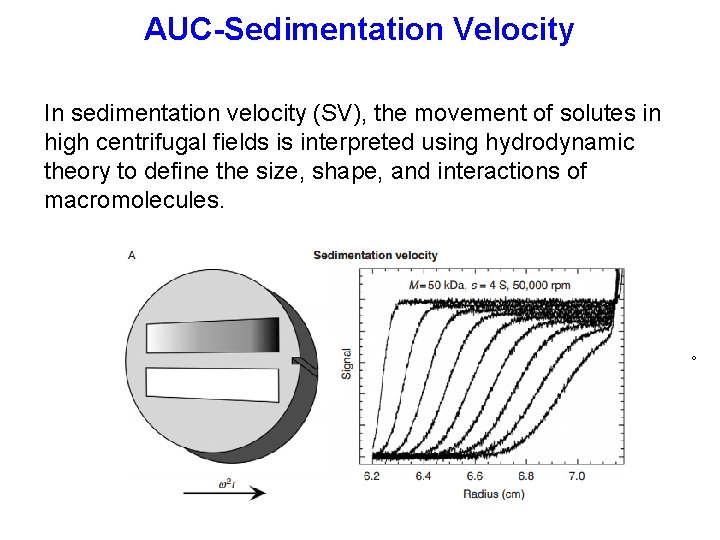
AUC-Sedimentation Velocity In sedimentation velocity (SV), the movement of solutes in high centrifugal fields is interpreted using hydrodynamic theory to define the size, shape, and interactions of macromolecules. 。

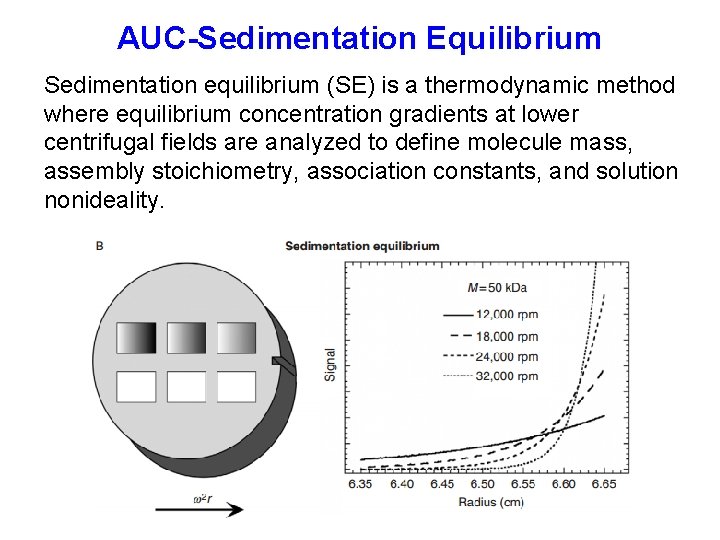
AUC-Sedimentation Equilibrium Sedimentation equilibrium (SE) is a thermodynamic method where equilibrium concentration gradients at lower centrifugal fields are analyzed to define molecule mass, assembly stoichiometry, association constants, and solution nonideality.

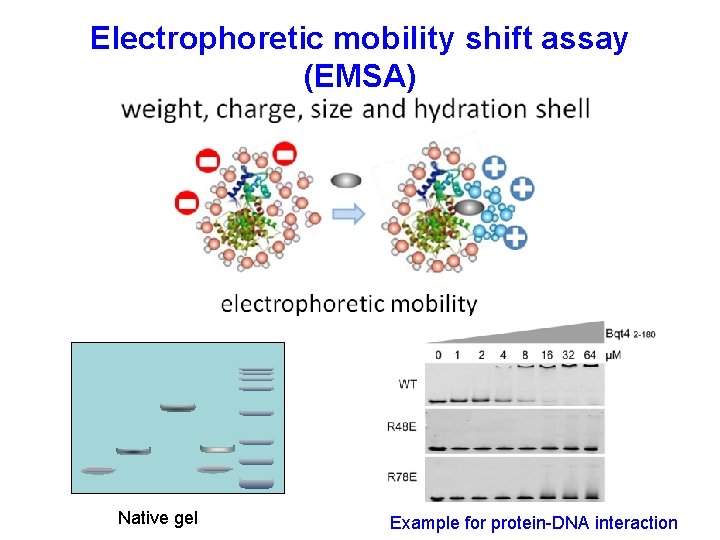
Electrophoretic mobility shift assay (EMSA) Native gel Example for protein-DNA interaction

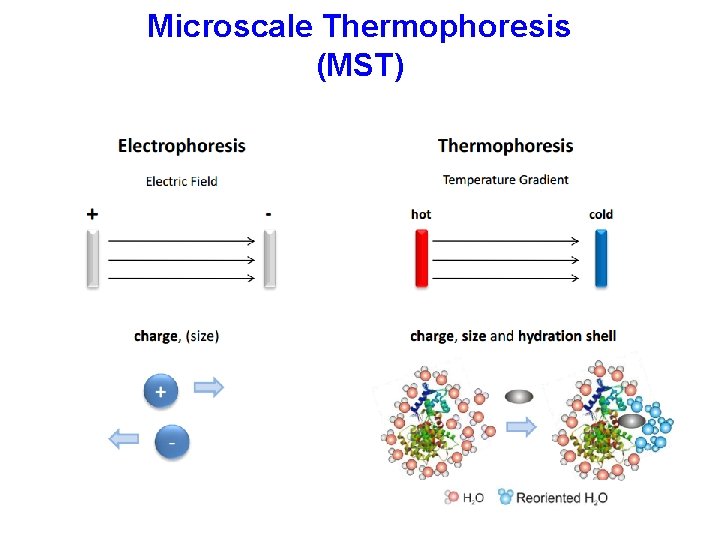
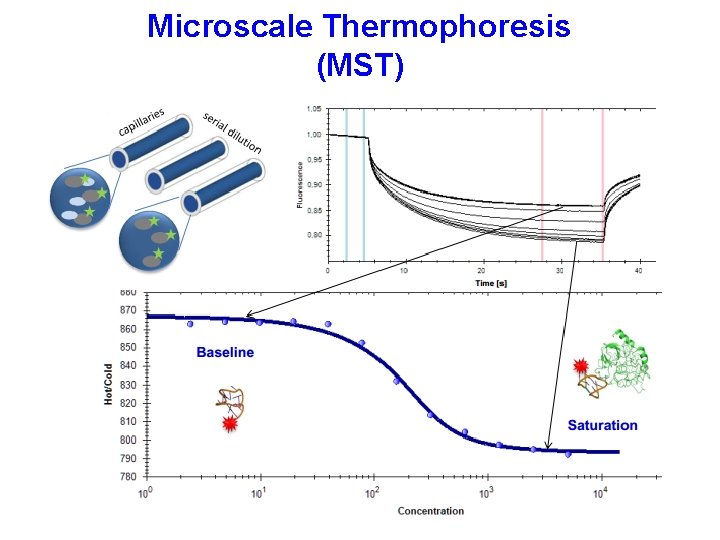
Microscale Thermophoresis (MST)

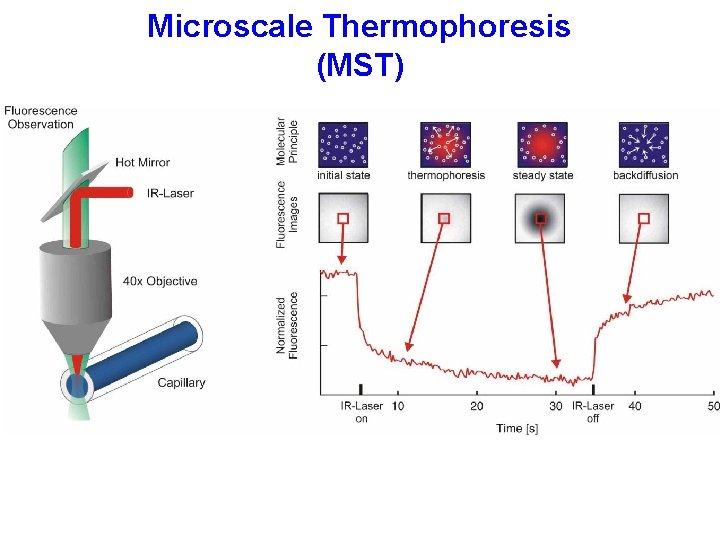
Microscale Thermophoresis (MST)

Microscale Thermophoresis (MST)

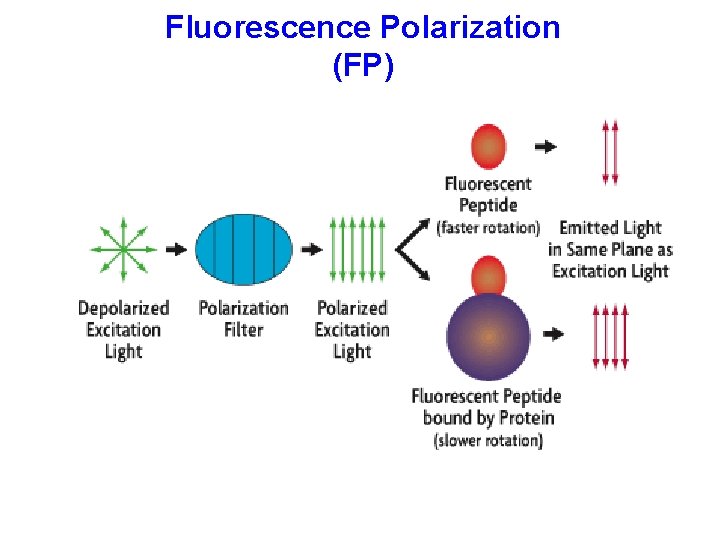
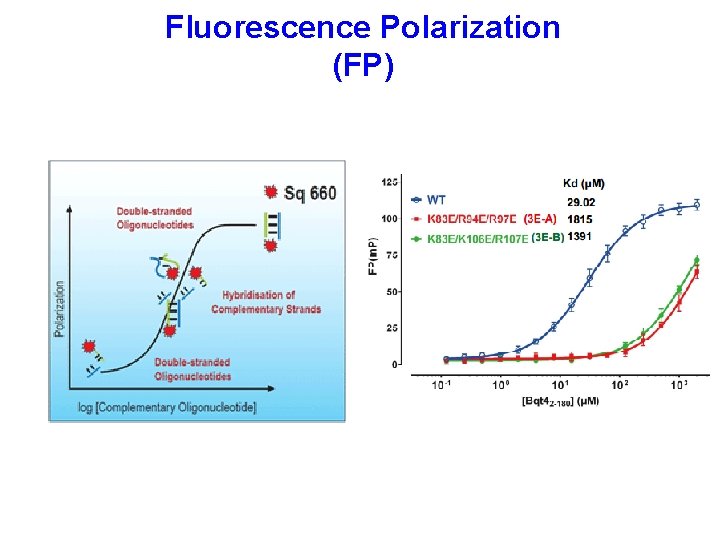
Fluorescence Polarization (FP)

Fluorescence Polarization (FP)

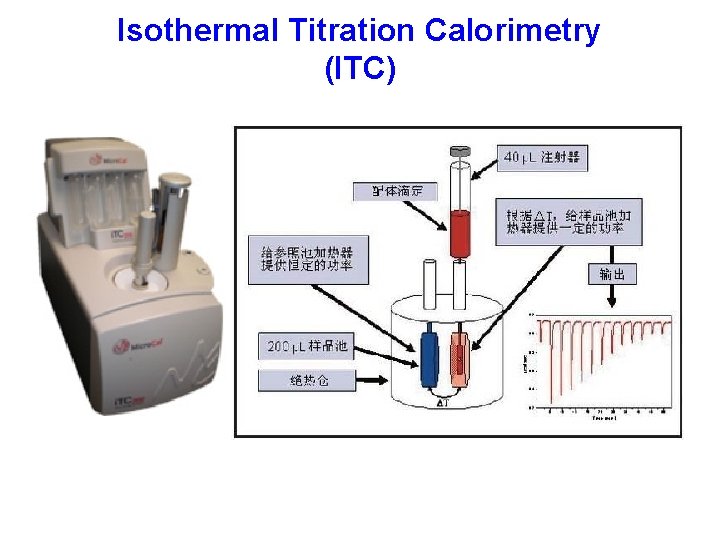
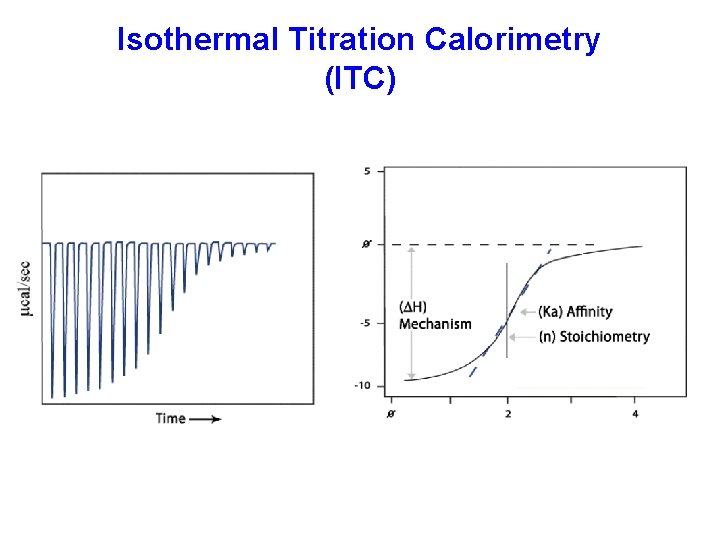
Isothermal Titration Calorimetry (ITC)

Isothermal Titration Calorimetry (ITC)

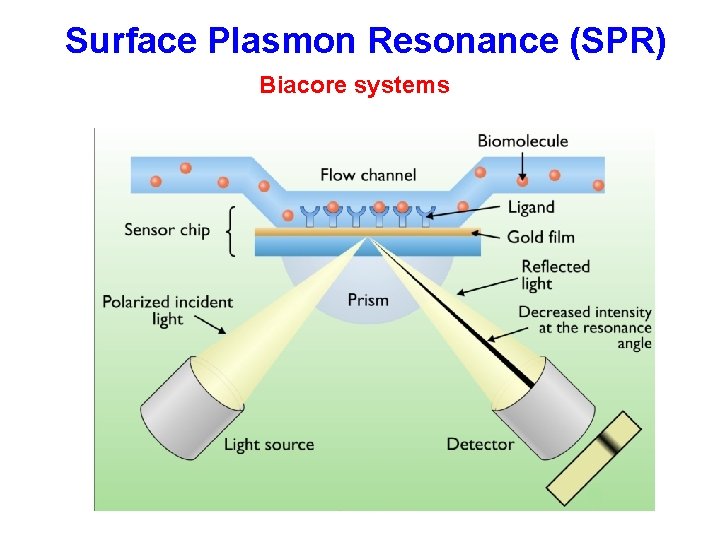
Surface Plasmon Resonance (SPR) Biacore systems

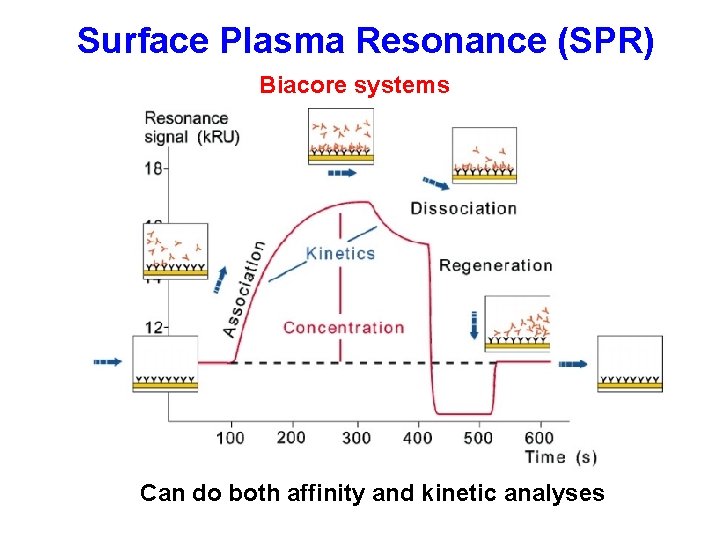
Surface Plasma Resonance (SPR) Biacore systems Can do both affinity and kinetic analyses

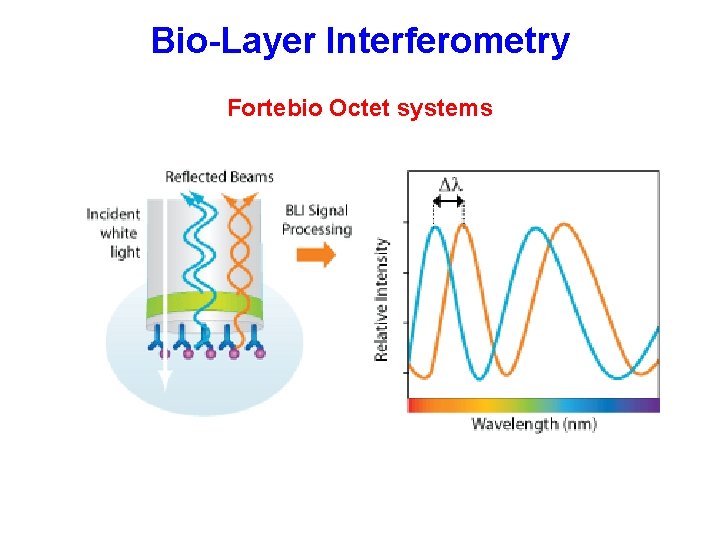
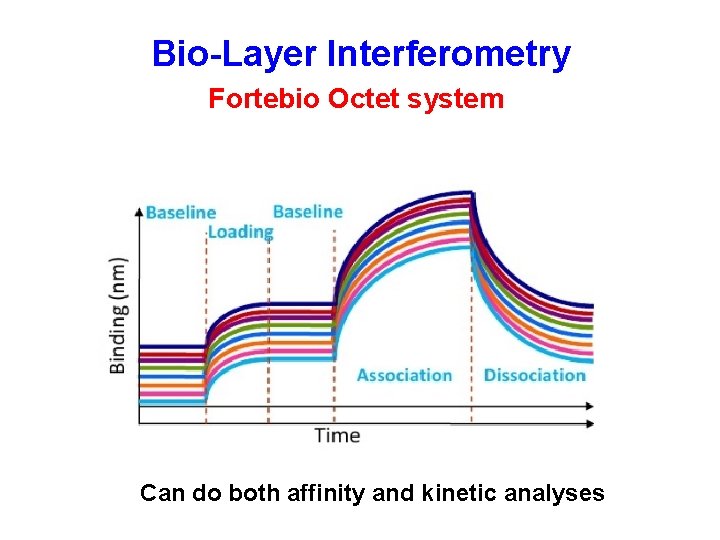
Bio-Layer Interferometry Fortebio Octet systems

Bio-Layer Interferometry Fortebio Octet system Can do both affinity and kinetic analyses

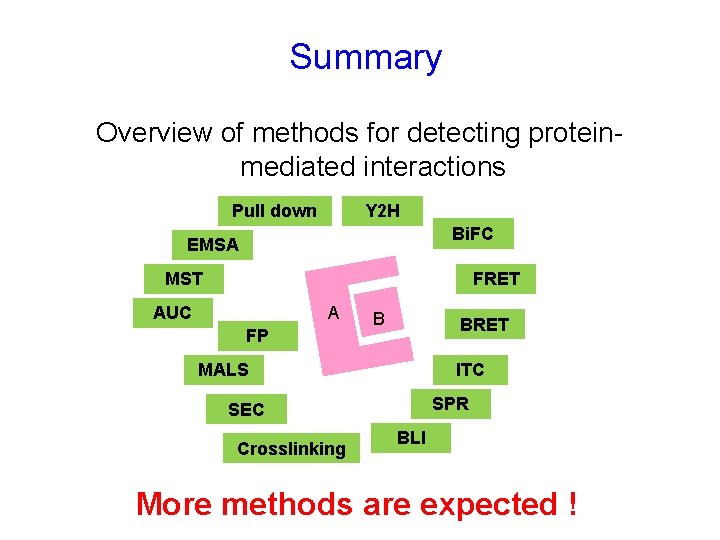
Summary Overview of methods for detecting proteinmediated interactions Pull down Y 2 H Bi. FC EMSA MST FRET AUC A FP B BRET MALS ITC SPR SEC Crosslinking BLI More methods are expected !
 Channel vs carrier proteins
Channel vs carrier proteins Protein-protein docking
Protein-protein docking Protein characterization techniques
Protein characterization techniques Contents introduction
Contents introduction Fast protein liquid chromatography principle
Fast protein liquid chromatography principle Indirect wax pattern
Indirect wax pattern What is proteins
What is proteins Introduction to proteins
Introduction to proteins Fspos
Fspos Typiska drag för en novell
Typiska drag för en novell Tack för att ni lyssnade bild
Tack för att ni lyssnade bild Ekologiskt fotavtryck
Ekologiskt fotavtryck Shingelfrisyren
Shingelfrisyren En lathund för arbete med kontinuitetshantering
En lathund för arbete med kontinuitetshantering Personalliggare bygg undantag
Personalliggare bygg undantag Tidbok
Tidbok Sura för anatom
Sura för anatom Densitet vatten
Densitet vatten Datorkunskap för nybörjare
Datorkunskap för nybörjare Tack för att ni lyssnade bild
Tack för att ni lyssnade bild Debatt artikel mall
Debatt artikel mall Delegerande ledarstil
Delegerande ledarstil Nyckelkompetenser för livslångt lärande
Nyckelkompetenser för livslångt lärande Påbyggnader för flakfordon
Påbyggnader för flakfordon Lufttryck formel
Lufttryck formel Svenskt ramverk för digital samverkan
Svenskt ramverk för digital samverkan Jag har gått inunder stjärnor text
Jag har gått inunder stjärnor text Presentera för publik crossboss
Presentera för publik crossboss Argument för teckenspråk som minoritetsspråk
Argument för teckenspråk som minoritetsspråk Kanaans land
Kanaans land Treserva lathund
Treserva lathund Mjälthilus
Mjälthilus Bästa kameran för astrofoto
Bästa kameran för astrofoto Cks
Cks Verifikationsplan
Verifikationsplan Mat för idrottare
Mat för idrottare Verktyg för automatisering av utbetalningar
Verktyg för automatisering av utbetalningar Rutin för avvikelsehantering
Rutin för avvikelsehantering Smärtskolan kunskap för livet
Smärtskolan kunskap för livet Ministerstyre för och nackdelar
Ministerstyre för och nackdelar Tack för att ni har lyssnat
Tack för att ni har lyssnat Referatmarkering
Referatmarkering Redogör för vad psykologi är
Redogör för vad psykologi är Matematisk modellering eksempel
Matematisk modellering eksempel Tack för att ni har lyssnat
Tack för att ni har lyssnat Borra hål för knoppar
Borra hål för knoppar Vilken grundregel finns det för tronföljden i sverige?
Vilken grundregel finns det för tronföljden i sverige? Hur räknar man standardavvikelse
Hur räknar man standardavvikelse Tack för att ni har lyssnat
Tack för att ni har lyssnat Steg för steg rita
Steg för steg rita Vad är verksamhetsanalys
Vad är verksamhetsanalys Tobinskatten för och nackdelar
Tobinskatten för och nackdelar Blomman för dagen drog
Blomman för dagen drog Datumr
Datumr Egg för emanuel
Egg för emanuel Elektronik för barn
Elektronik för barn Klädsel i rom
Klädsel i rom Strategi för svensk viltförvaltning
Strategi för svensk viltförvaltning Kung dog 1611
Kung dog 1611 Ellika andolf
Ellika andolf Romarriket tidslinje
Romarriket tidslinje Tack för att ni lyssnade
Tack för att ni lyssnade Matte större än tecken
Matte större än tecken Dikter som rimmar
Dikter som rimmar Inköpsprocessen steg för steg
Inköpsprocessen steg för steg Rådet för byggkompetens
Rådet för byggkompetens Etik och ledarskap etisk kod för chefer
Etik och ledarskap etisk kod för chefer Skivepiteldysplasi
Skivepiteldysplasi Myndigheten för delaktighet
Myndigheten för delaktighet Trög för kemist
Trög för kemist Sju principer för tillitsbaserad styrning
Sju principer för tillitsbaserad styrning Läkarutlåtande för livränta
Läkarutlåtande för livränta Kraftledning karttecken
Kraftledning karttecken Gumman cirkel
Gumman cirkel Shaktismen
Shaktismen