Factor and Component Analysis esp Principal Component Analysis

























- Slides: 99

Factor and Component Analysis esp. Principal Component Analysis (PCA)

Why Factor or Component Analysis? • We study phenomena that can not be directly observed – ego, personality, intelligence in psychology – Underlying factors that govern the observed data • We want to identify and operate with underlying latent factors rather than the observed data – E. g. topics in news articles – Transcription factors in genomics • We want to discover and exploit hidden relationships – – “beautiful car” and “gorgeous automobile” are closely related So are “driver” and “automobile” But does your search engine know this? Reduces noise and error in results

Why Factor or Component Analysis? • We have too many observations and dimensions – – – To reason about or obtain insights from To visualize Too much noise in the data Need to “reduce” them to a smaller set of factors Better representation of data without losing much information Can build more effective data analyses on the reduceddimensional space: classification, clustering, pattern recognition • Combinations of observed variables may be more effective bases for insights, even if physical meaning is obscure

Factor or Component Analysis • Discover a new set of factors/dimensions/axes against which to represent, describe or evaluate the data – – – For more effective reasoning, insights, or better visualization Reduce noise in the data Typically a smaller set of factors: dimension reduction Better representation of data without losing much information Can build more effective data analyses on the reduceddimensional space: classification, clustering, pattern recognition • Factors are combinations of observed variables – May be more effective bases for insights, even if physical meaning is obscure – Observed data are described in terms of these factors rather than in terms of original variables/dimensions

Basic Concept • Areas of variance in data are where items can be best discriminated and key underlying phenomena observed – Areas of greatest “signal” in the data • If two items or dimensions are highly correlated or dependent – They are likely to represent highly related phenomena – If they tell us about the same underlying variance in the data, combining them to form a single measure is reasonable • Parsimony • Reduction in Error • So we want to combine related variables, and focus on uncorrelated or independent ones, especially those along which the observations have high variance • We want a smaller set of variables that explain most of the variance in the original data, in more compact and insightful form

Basic Concept • What if the dependences and correlations are not so strong or direct? • And suppose you have 3 variables, or 4, or 5, or 10000? • Look for the phenomena underlying the observed covariance/co-dependence in a set of variables – Once again, phenomena that are uncorrelated or independent, and especially those along which the data show high variance • These phenomena are called “factors” or “principal components” or “independent components, ” depending on the methods used – Factor analysis: based on variance/correlation – Independent Component Analysis: based on independence

Principal Component Analysis • Most common form of factor analysis • The new variables/dimensions – Are linear combinations of the original ones – Are uncorrelated with one another • Orthogonal in original dimension space – Capture as much of the original variance in the data as possible – Are called Principal Components

Some Simple Demos • http: //www. cs. mcgill. ca/~sqrt/dimre duction. html

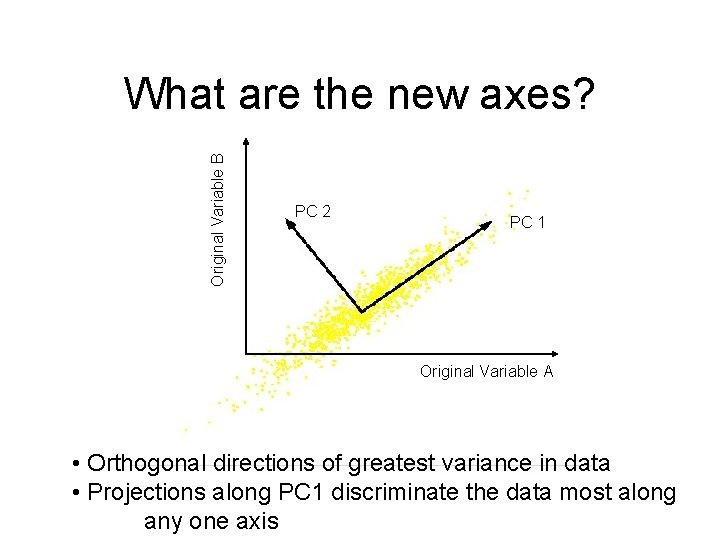
Original Variable B What are the new axes? PC 2 PC 1 Original Variable A • Orthogonal directions of greatest variance in data • Projections along PC 1 discriminate the data most along any one axis

Principal Components • First principal component is the direction of greatest variability (covariance) in the data • Second is the next orthogonal (uncorrelated) direction of greatest variability – So first remove all the variability along the first component, and then find the next direction of greatest variability • And so on …

Principal Components Analysis (PCA) • Principle – Linear projection method to reduce the number of parameters – Transfer a set of correlated variables into a new set of uncorrelated variables – Map the data into a space of lower dimensionality – Form of unsupervised learning • Properties – It can be viewed as a rotation of the existing axes to new positions in the space defined by original variables – New axes are orthogonal and represent the directions with maximum variability

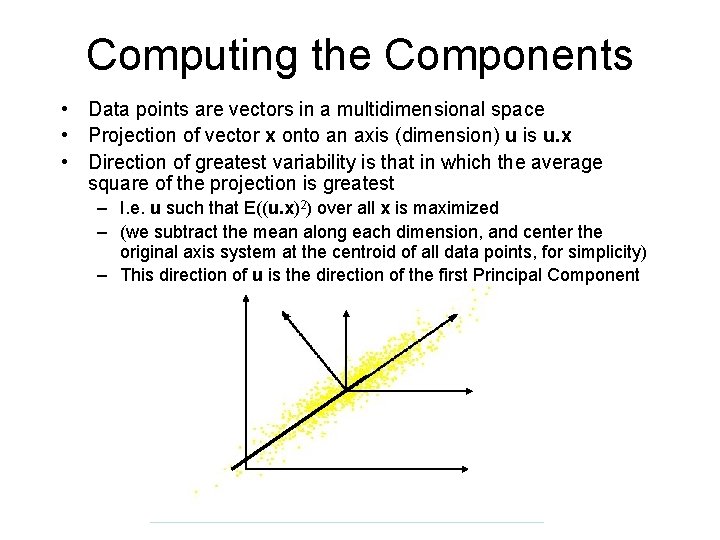
Computing the Components • Data points are vectors in a multidimensional space • Projection of vector x onto an axis (dimension) u is u. x • Direction of greatest variability is that in which the average square of the projection is greatest – I. e. u such that E((u. x)2) over all x is maximized – (we subtract the mean along each dimension, and center the original axis system at the centroid of all data points, for simplicity) – This direction of u is the direction of the first Principal Component

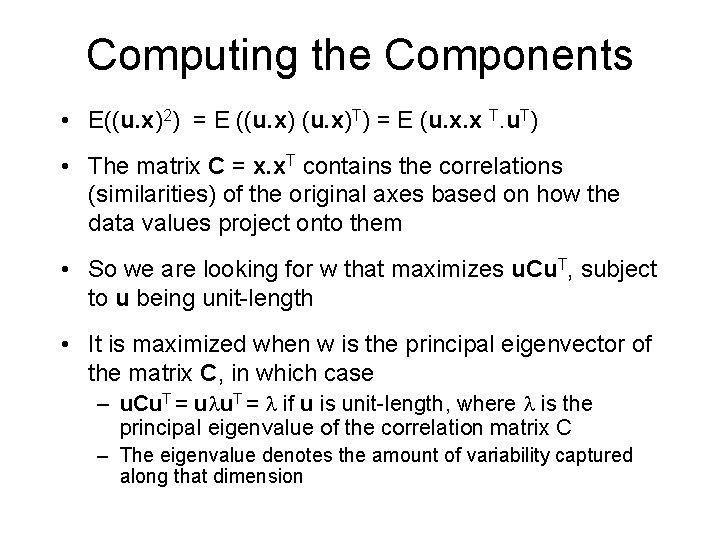
Computing the Components • E((u. x)2) = E ((u. x)T) = E (u. x. x T. u. T) • The matrix C = x. x. T contains the correlations (similarities) of the original axes based on how the data values project onto them • So we are looking for w that maximizes u. Cu. T, subject to u being unit-length • It is maximized when w is the principal eigenvector of the matrix C, in which case – u. Cu. T = ulu. T = l if u is unit-length, where l is the principal eigenvalue of the correlation matrix C – The eigenvalue denotes the amount of variability captured along that dimension

Why the Eigenvectors? Maximise u. Txx. Tu s. t u. Tu =1 Construct Langrangian u. Txx. Tu – λu. Tu Vector of partial derivatives set to zero xx. Tu – λu = (xx. T – λI) u = 0 As u ≠ 0 then u must be an eigenvector of xx. T with eigenvalue λ

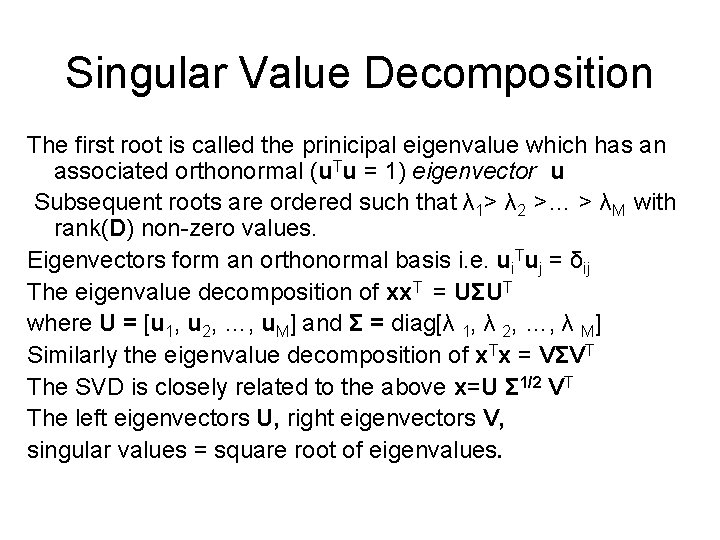
Singular Value Decomposition The first root is called the prinicipal eigenvalue which has an associated orthonormal (u. Tu = 1) eigenvector u Subsequent roots are ordered such that λ 1> λ 2 >… > λM with rank(D) non-zero values. Eigenvectors form an orthonormal basis i. e. ui. Tuj = δij The eigenvalue decomposition of xx. T = UΣUT where U = [u 1, u 2, …, u. M] and Σ = diag[λ 1, λ 2, …, λ M] Similarly the eigenvalue decomposition of x. Tx = VΣVT The SVD is closely related to the above x=U Σ 1/2 VT The left eigenvectors U, right eigenvectors V, singular values = square root of eigenvalues.

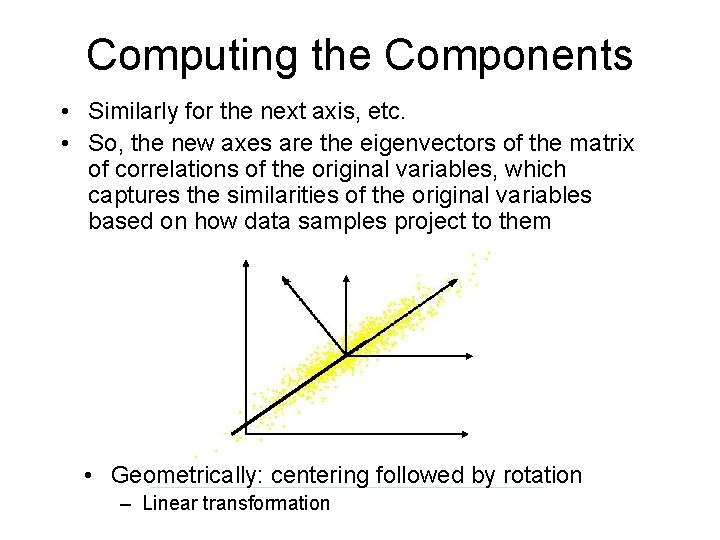
Computing the Components • Similarly for the next axis, etc. • So, the new axes are the eigenvectors of the matrix of correlations of the original variables, which captures the similarities of the original variables based on how data samples project to them • Geometrically: centering followed by rotation – Linear transformation

PCs, Variance and Least-Squares • The first PC retains the greatest amount of variation in the sample • The kth PC retains the kth greatest fraction of the variation in the sample • The kth largest eigenvalue of the correlation matrix C is the variance in the sample along the kth PC • The least-squares view: PCs are a series of linear least squares fits to a sample, each orthogonal to all previous ones

How Many PCs? • For n original dimensions, correlation matrix is nxn, and has up to n eigenvectors. So n PCs. • Where does dimensionality reduction come from?

Dimensionality Reduction Can ignore the components of lesser significance. You do lose some information, but if the eigenvalues are small, you don’t lose much – – n dimensions in original data calculate n eigenvectors and eigenvalues choose only the first p eigenvectors, based on their eigenvalues final data set has only p dimensions

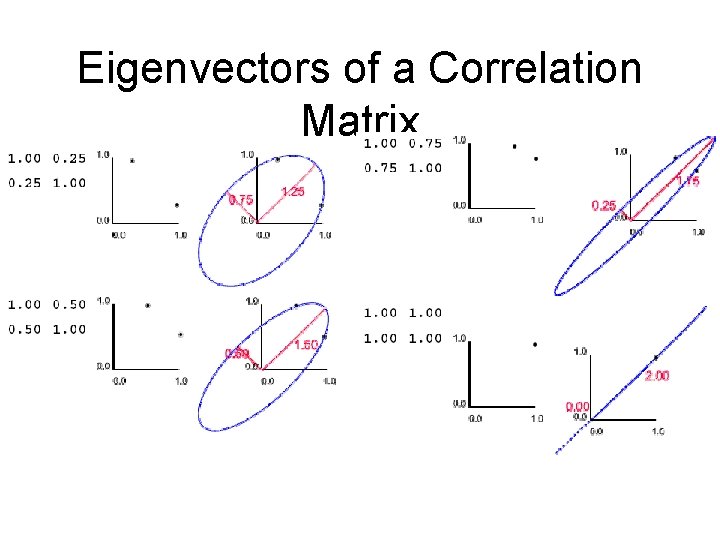
Eigenvectors of a Correlation Matrix

Computing and Using PCA: Text Search Example

PCA/FA • Principal Components Analysis – Extracts all the factor underlying a set of variables – The number of factors = the number of variables – Completely explains the variance in each variable • Parsimony? !? !? ! • Factor Analysis – Also known as Principal Axis Analysis – Analyses only the shared variance • NO ERROR!

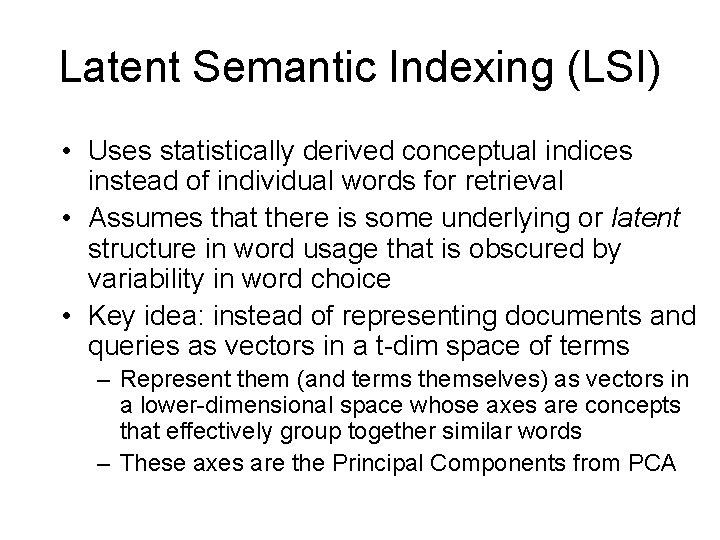
Vector Space Model for Documents • Documents are vectors in multi-dim Euclidean space – Each term is a dimension in the space: LOTS of them – Coordinate of doc d in dimension t is prop. to TF(d, t) • TF(d, t) is term frequency of t in document d • Term-document matrix, with entries being normalized TF values – t*d matrix, for t terms and d documents • Queries are also vectors in the same space – Treated just like documents • Rank documents based on similarity of their vectors with the query vector – Using cosine distance of vectors

Problems • Looks for literal term matches • Terms in queries (esp short ones) don’t always capture user’s information need well • Problems: – Synonymy: other words with the same meaning • Car and automobile – Polysemy: the same word having other meanings • Apple (fruit and company) • What if we could match against ‘concepts’, that represent related words, rather than words themselves

Example of Problems -- Relevant docs may not have the query terms but may have many “related” terms -- Irrelevant docs may have the query terms but may not have any “related” terms

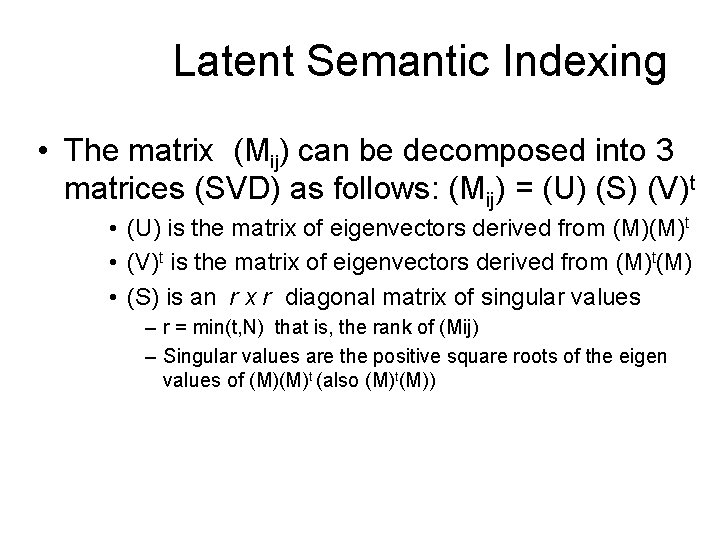
Latent Semantic Indexing (LSI) • Uses statistically derived conceptual indices instead of individual words for retrieval • Assumes that there is some underlying or latent structure in word usage that is obscured by variability in word choice • Key idea: instead of representing documents and queries as vectors in a t-dim space of terms – Represent them (and terms themselves) as vectors in a lower-dimensional space whose axes are concepts that effectively group together similar words – These axes are the Principal Components from PCA

Example • Suppose we have keywords – Car, automobile, driver, elephant • We want queries on car to also get docs about drivers and automobiles, but not about elephants – What if we could discover that the cars, automobiles and drivers axes are strongly correlated, but elephants is not – How? Via correlations observed through documents – If docs A & B don’t share any words with each other, but both share lots of words with doc C, then A & B will be considered similar – E. g A has cars and drivers, B has automobiles and drivers • When you scrunch down dimensions, small differences (noise) gets glossed over, and you get desired behavior

LSI and Our Earlier Example -- Relevant docs may not have the query terms but may have many “related” terms -- Irrelevant docs may have the query terms but may not have any “related” terms

LSI: Satisfying a query • Take the vector representation of the query in the original term space and transform it to new space – Turns out dq = xqt T S-1 – places the query pseudo-doc at the centroid of its corresponding terms’ locations in the new space • Similarity with existing docs computed by taking dot products with the rows of the matrix DS 2 Dt

Latent Semantic Indexing • The matrix (Mij) can be decomposed into 3 matrices (SVD) as follows: (Mij) = (U) (S) (V)t • (U) is the matrix of eigenvectors derived from (M)(M)t • (V)t is the matrix of eigenvectors derived from (M)t(M) • (S) is an r x r diagonal matrix of singular values – r = min(t, N) that is, the rank of (Mij) – Singular values are the positive square roots of the eigen values of (M)(M)t (also (M)t(M))

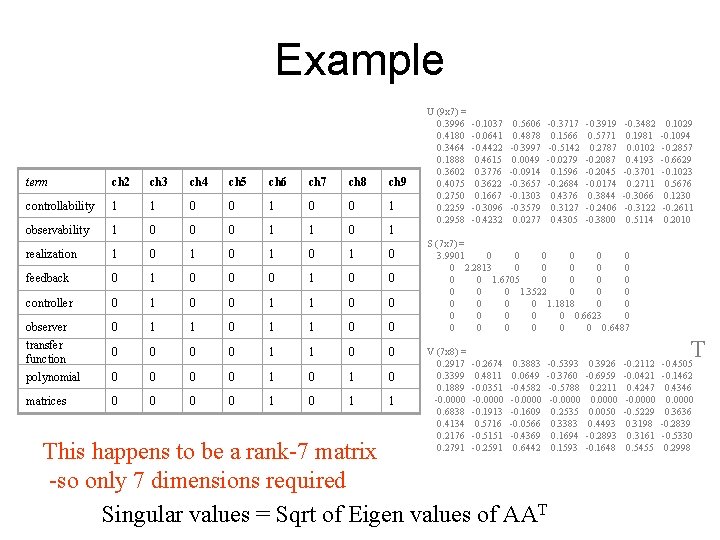
Example term ch 2 ch 3 ch 4 ch 5 ch 6 ch 7 ch 8 ch 9 controllability 1 1 0 0 1 0 0 1 observability 1 0 0 0 1 1 0 1 realization 1 0 1 0 feedback 0 1 0 0 controller 0 1 0 0 1 1 0 0 observer transfer function polynomial 0 1 1 0 1 1 0 0 0 0 1 1 0 0 1 0 matrices 0 0 1 0 1 1 U (9 x 7) = 0. 3996 -0. 1037 0. 5606 -0. 3717 -0. 3919 -0. 3482 0. 1029 0. 4180 -0. 0641 0. 4878 0. 1566 0. 5771 0. 1981 -0. 1094 0. 3464 -0. 4422 -0. 3997 -0. 5142 0. 2787 0. 0102 -0. 2857 0. 1888 0. 4615 0. 0049 -0. 0279 -0. 2087 0. 4193 -0. 6629 0. 3602 0. 3776 -0. 0914 0. 1596 -0. 2045 -0. 3701 -0. 1023 0. 4075 0. 3622 -0. 3657 -0. 2684 -0. 0174 0. 2711 0. 5676 0. 2750 0. 1667 -0. 1303 0. 4376 0. 3844 -0. 3066 0. 1230 0. 2259 -0. 3096 -0. 3579 0. 3127 -0. 2406 -0. 3122 -0. 2611 0. 2958 -0. 4232 0. 0277 0. 4305 -0. 3800 0. 5114 0. 2010 S (7 x 7) = 3. 9901 0 0 0 0 2. 2813 0 0 0 0 1. 6705 0 0 1. 3522 0 0 1. 1818 0 0 0 0 0. 6623 0 0 0 0 0. 6487 T V (7 x 8) = 0. 2917 -0. 2674 0. 3883 -0. 5393 0. 3926 -0. 2112 -0. 4505 0. 3399 0. 4811 0. 0649 -0. 3760 -0. 6959 -0. 0421 -0. 1462 0. 1889 -0. 0351 -0. 4582 -0. 5788 0. 2211 0. 4247 0. 4346 -0. 0000 0. 0000 0. 6838 -0. 1913 -0. 1609 0. 2535 0. 0050 -0. 5229 0. 3636 0. 4134 0. 5716 -0. 0566 0. 3383 0. 4493 0. 3198 -0. 2839 0. 2176 -0. 5151 -0. 4369 0. 1694 -0. 2893 0. 3161 -0. 5330 0. 2791 -0. 2591 0. 6442 0. 1593 -0. 1648 0. 5455 0. 2998 This happens to be a rank-7 matrix -so only 7 dimensions required Singular values = Sqrt of Eigen values of AAT

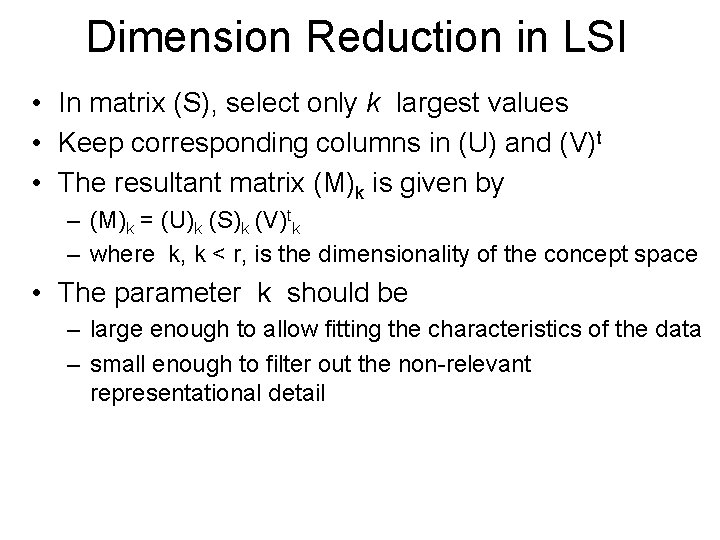
Dimension Reduction in LSI • The key idea is to map documents and queries into a lower dimensional space (i. e. , composed of higher level concepts which are in fewer number than the index terms) • Retrieval in this reduced concept space might be superior to retrieval in the space of index terms

Dimension Reduction in LSI • In matrix (S), select only k largest values • Keep corresponding columns in (U) and (V)t • The resultant matrix (M)k is given by – (M)k = (U)k (S)k (V)tk – where k, k < r, is the dimensionality of the concept space • The parameter k should be – large enough to allow fitting the characteristics of the data – small enough to filter out the non-relevant representational detail

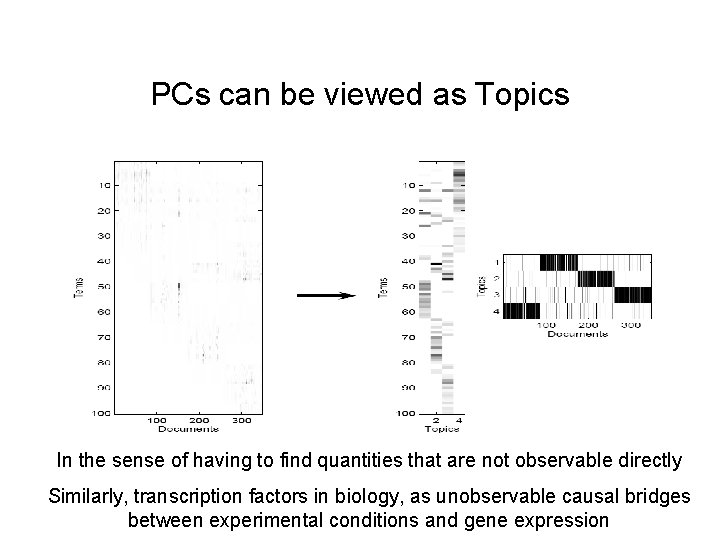
PCs can be viewed as Topics In the sense of having to find quantities that are not observable directly Similarly, transcription factors in biology, as unobservable causal bridges between experimental conditions and gene expression

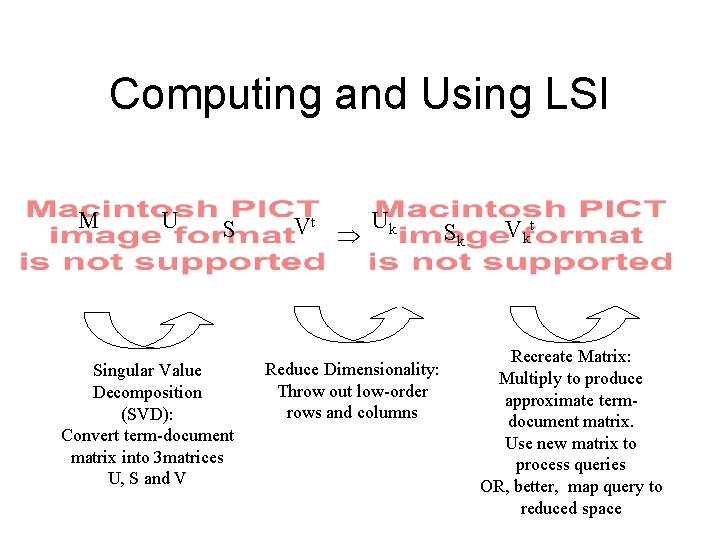
Computing and Using LSI M U S Singular Value Decomposition (SVD): Convert term-document matrix into 3 matrices U, S and V Vt Þ Uk Reduce Dimensionality: Throw out low-order rows and columns Sk V kt Recreate Matrix: Multiply to produce approximate termdocument matrix. Use new matrix to process queries OR, better, map query to reduced space

Following the Example term ch 2 ch 3 ch 4 ch 5 ch 6 ch 7 ch 8 ch 9 controllability 1 1 0 0 1 0 0 1 observability 1 0 0 0 1 1 0 1 realization 1 0 1 0 feedback 0 1 0 0 controller 0 1 0 0 1 1 0 0 observer transfer function polynomial 0 1 1 0 1 1 0 0 0 0 1 1 0 0 1 0 matrices 0 0 1 0 1 1 U (9 x 7) = 0. 3996 -0. 1037 0. 5606 -0. 3717 -0. 3919 -0. 3482 0. 1029 0. 4180 -0. 0641 0. 4878 0. 1566 0. 5771 0. 1981 -0. 1094 0. 3464 -0. 4422 -0. 3997 -0. 5142 0. 2787 0. 0102 -0. 2857 0. 1888 0. 4615 0. 0049 -0. 0279 -0. 2087 0. 4193 -0. 6629 0. 3602 0. 3776 -0. 0914 0. 1596 -0. 2045 -0. 3701 -0. 1023 0. 4075 0. 3622 -0. 3657 -0. 2684 -0. 0174 0. 2711 0. 5676 0. 2750 0. 1667 -0. 1303 0. 4376 0. 3844 -0. 3066 0. 1230 0. 2259 -0. 3096 -0. 3579 0. 3127 -0. 2406 -0. 3122 -0. 2611 0. 2958 -0. 4232 0. 0277 0. 4305 -0. 3800 0. 5114 0. 2010 S (7 x 7) = 3. 9901 0 0 0 0 2. 2813 0 0 0 0 1. 6705 0 0 1. 3522 0 0 1. 1818 0 0 0 0 0. 6623 0 0 0 0 0. 6487 T V (7 x 8) = 0. 2917 -0. 2674 0. 3883 -0. 5393 0. 3926 -0. 2112 -0. 4505 0. 3399 0. 4811 0. 0649 -0. 3760 -0. 6959 -0. 0421 -0. 1462 0. 1889 -0. 0351 -0. 4582 -0. 5788 0. 2211 0. 4247 0. 4346 -0. 0000 0. 0000 0. 6838 -0. 1913 -0. 1609 0. 2535 0. 0050 -0. 5229 0. 3636 0. 4134 0. 5716 -0. 0566 0. 3383 0. 4493 0. 3198 -0. 2839 0. 2176 -0. 5151 -0. 4369 0. 1694 -0. 2893 0. 3161 -0. 5330 0. 2791 -0. 2591 0. 6442 0. 1593 -0. 1648 0. 5455 0. 2998 This happens to be a rank-7 matrix -so only 7 dimensions required Singular values = Sqrt of Eigen values of AAT

Formally, this will be the rank-k (2) matrix that is closest to M in the U (9 x 7) = matrix norm sense 0. 3996 -0. 1037 0. 5606 -0. 3717 -0. 3919 -0. 3482 0. 1029 0. 4180 -0. 0641 0. 4878 0. 1566 0. 5771 0. 1981 -0. 1094 0. 3464 -0. 4422 -0. 3997 -0. 5142 0. 2787 0. 0102 -0. 2857 0. 1888 0. 4615 0. 0049 -0. 0279 -0. 2087 0. 4193 -0. 6629 0. 3602 0. 3776 -0. 0914 0. 1596 -0. 2045 -0. 3701 -0. 1023 0. 4075 0. 3622 -0. 3657 -0. 2684 -0. 0174 0. 2711 0. 5676 0. 2750 0. 1667 -0. 1303 0. 4376 0. 3844 -0. 3066 0. 1230 0. 2259 -0. 3096 -0. 3579 0. 3127 -0. 2406 -0. 3122 -0. 2611 0. 2958 -0. 4232 0. 0277 0. 4305 -0. 3800 0. 5114 0. 2010 S (7 x 7) = 3. 9901 0 0 0 0 2. 2813 0 0 0 0 1. 6705 0 0 1. 3522 0 0 1. 1818 0 0 0 0 0. 6623 0 0 0 0 0. 6487 V (7 x 8) = 0. 2917 -0. 2674 0. 3883 -0. 5393 0. 3926 -0. 2112 -0. 4505 0. 3399 0. 4811 0. 0649 -0. 3760 -0. 6959 -0. 0421 -0. 1462 0. 1889 -0. 0351 -0. 4582 -0. 5788 0. 2211 0. 4247 0. 4346 -0. 0000 0. 0000 0. 6838 -0. 1913 -0. 1609 0. 2535 0. 0050 -0. 5229 0. 3636 0. 4134 0. 5716 -0. 0566 0. 3383 0. 4493 0. 3198 -0. 2839 0. 2176 -0. 5151 -0. 4369 0. 1694 -0. 2893 0. 3161 -0. 5330 0. 2791 -0. 2591 0. 6442 0. 1593 -0. 1648 0. 5455 0. 2998 U 2 (9 x 2) = 0. 3996 -0. 1037 0. 4180 -0. 0641 0. 3464 -0. 4422 0. 1888 0. 4615 0. 3602 0. 3776 0. 4075 0. 3622 0. 2750 0. 1667 0. 2259 -0. 3096 0. 2958 -0. 4232 S 2 (2 x 2) = 3. 9901 0 0 2. 2813 V 2 (8 x 2) = 0. 2917 -0. 2674 T 0. 3399 0. 4811 0. 1889 -0. 0351 -0. 0000 0. 6838 -0. 1913 0. 4134 0. 5716 0. 2176 -0. 5151 0. 2791 -0. 2591 U 2*S 2*V 2 will be a 9 x 8 matrix That approximates original matrix

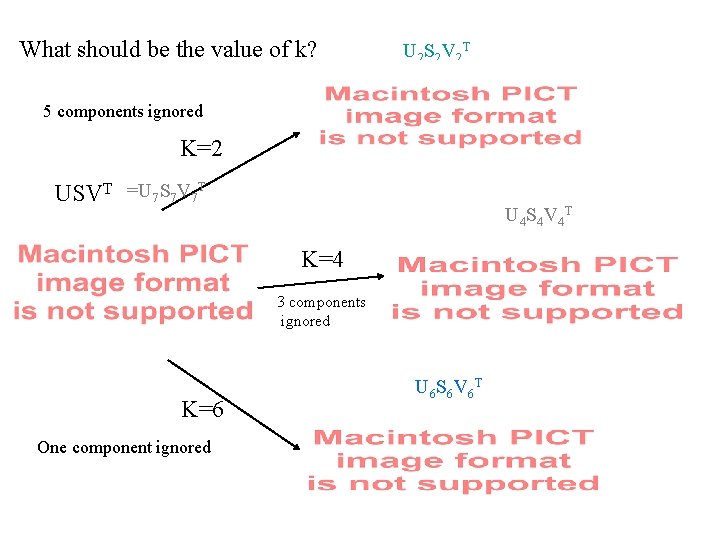
What should be the value of k? U 2 S 2 V 2 T 5 components ignored K=2 USVT =U 7 S 7 V 7 T U 4 S 4 V 4 T K=4 3 components ignored K=6 One component ignored U 6 S 6 V 6 T

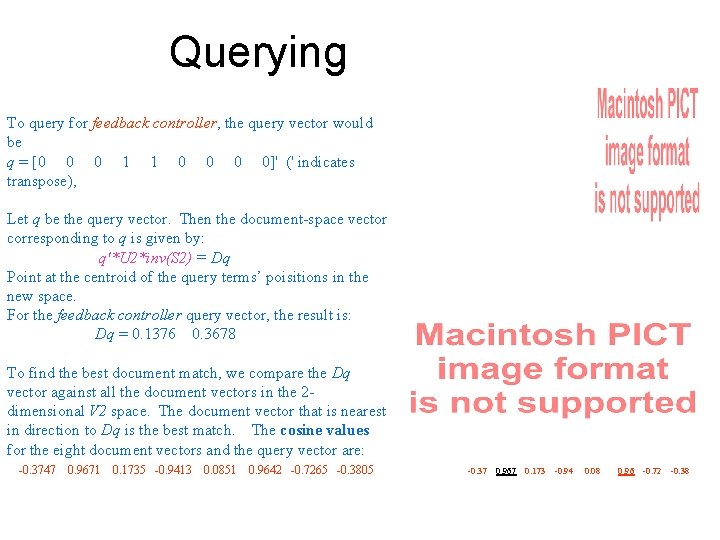
Querying To query for feedback controller, the query vector would be q = [0 0 0 1 1 0 0]' (' indicates transpose), Let q be the query vector. Then the document-space vector corresponding to q is given by: q'*U 2*inv(S 2) = Dq Point at the centroid of the query terms’ poisitions in the new space. For the feedback controller query vector, the result is: Dq = 0. 1376 0. 3678 To find the best document match, we compare the Dq vector against all the document vectors in the 2 dimensional V 2 space. The document vector that is nearest in direction to Dq is the best match. The cosine values for the eight document vectors and the query vector are: -0. 3747 0. 9671 0. 1735 -0. 9413 0. 0851 0. 9642 -0. 7265 -0. 3805 -0. 37 0. 967 0. 173 -0. 94 0. 08 0. 96 -0. 72 -0. 38

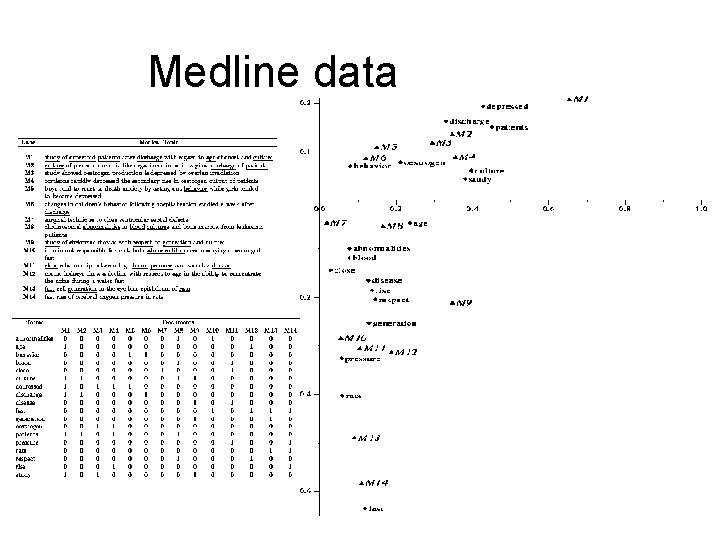
Medline data

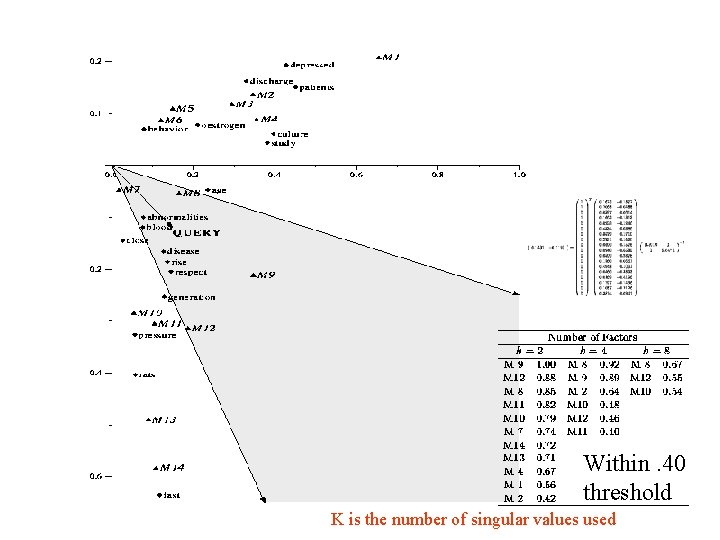
Within. 40 threshold K is the number of singular values used

What LSI can do • LSI analysis effectively does – – Dimensionality reduction Noise reduction Exploitation of redundant data Correlation analysis and Query expansion (with related words) • Some of the individual effects can be achieved with simpler techniques (e. g. thesaurus construction). LSI does them together. • LSI handles synonymy well, not so much polysemy • Challenge: SVD is complex to compute (O(n 3)) – Needs to be updated as new documents are found/updated

SVD Properties • There is an implicit assumption that the observed data distribution is multivariate Gaussian • Can consider as a probabilistic generative model – latent variables are Gaussian – sub-optimal in likelihood terms for non-Gaussian distribution • Employed in signal processing for noise filtering – dominant subspace contains majority of information bearing part of signal • Similar rationale when applying SVD to LSI

LSI Conclusions – SVD defined basis provide P/R improvements over term matching • • Interpretation difficult Optimal dimension – open question Variable performance on LARGE collections Supercomputing muscle required – Probabilistic approaches provide improvements over SVD • Clear interpretation of decomposition • Optimal dimension – open question • High variability of results due to nonlinear optimisation over HUGE parameter space – Improvements marginal in relation to cost

Factor Analysis (e. g. PCA) is not the most sophisticated dimensionality reduction technique • Dimensionality reduction is a useful technique for any classification/regression problem – Text retrieval can be seen as a classification problem • There are many other dimensionality reduction techniques – Neural nets, support vector machines etc. • Compared to them, LSI is limited in the sense that it is a “linear” technique – It cannot capture non-linear dependencies between original dimensions (e. g. data that are circularly distributed).

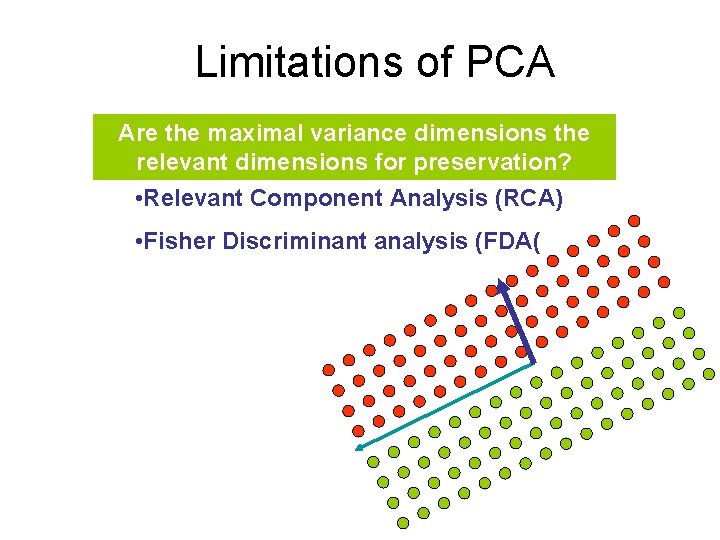
Limitations of PCA Are the maximal variance dimensions the relevant dimensions for preservation? • Relevant Component Analysis (RCA) • Fisher Discriminant analysis (FDA(

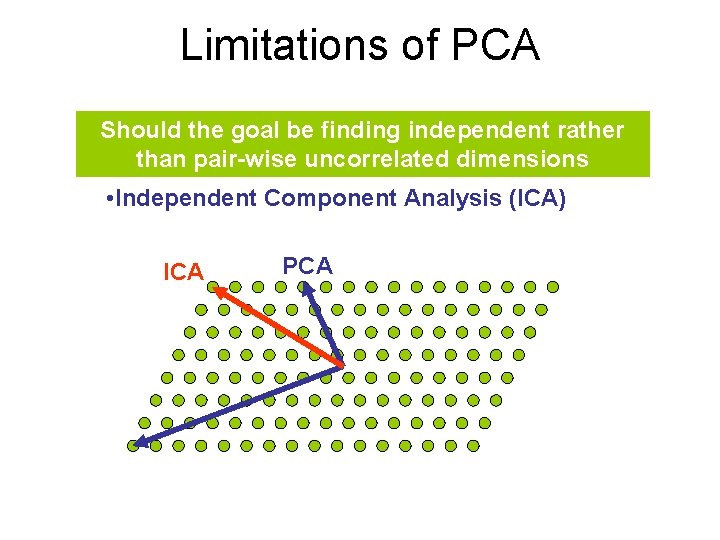
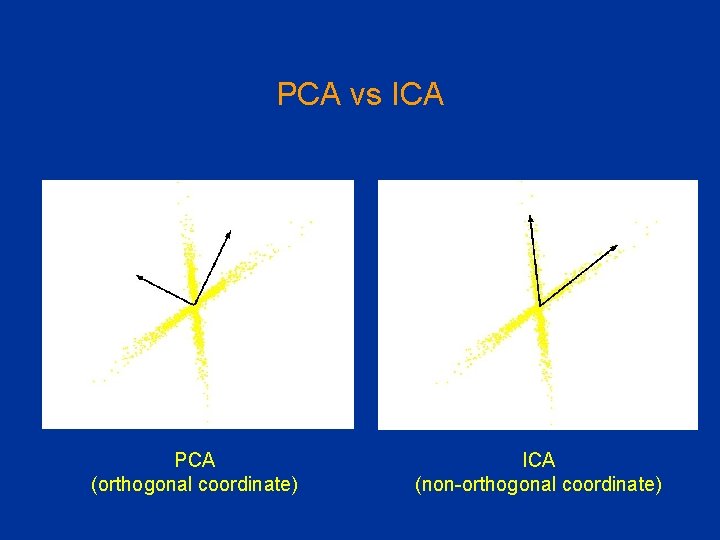
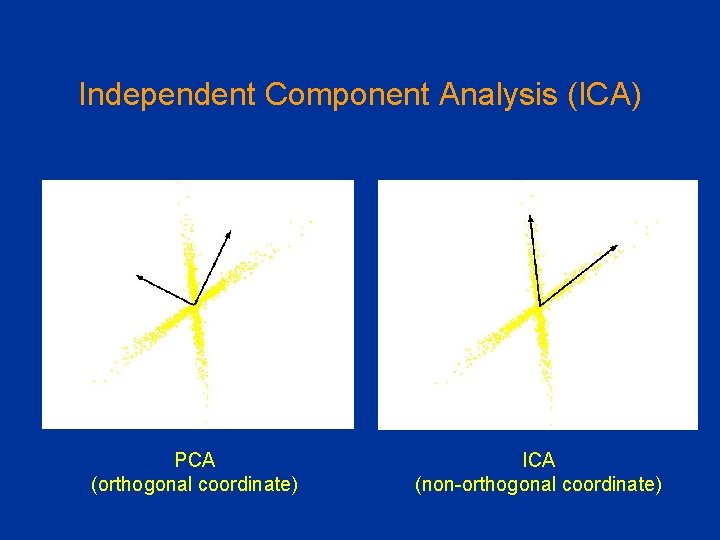
Limitations of PCA Should the goal be finding independent rather than pair-wise uncorrelated dimensions • Independent Component Analysis (ICA) ICA PCA

PCA vs ICA PCA (orthogonal coordinate) ICA (non-orthogonal coordinate)

Limitations of PCA • The reduction of dimensions for complex distributions may need non linear processing • Curvilenear Component Analysis (CCA) – Non linear extension of PCA – Preserves the proximity between the points in the input space i. e. local topology of the distribution – Enables to unfold some varieties in the input data – Keep the local topology

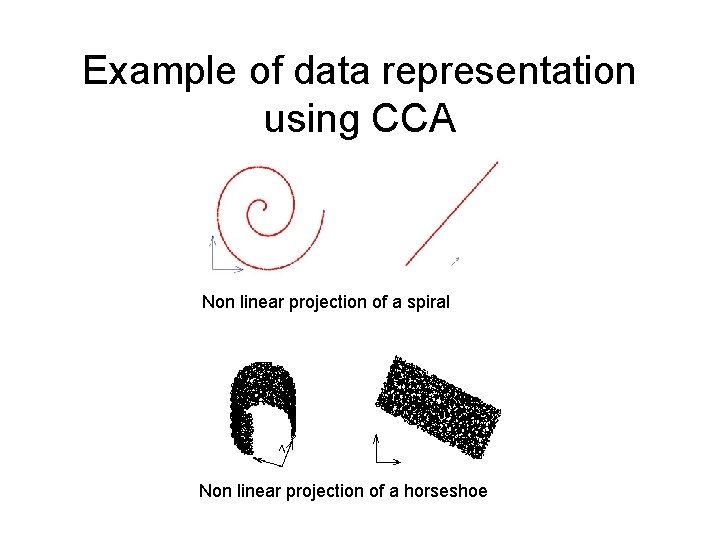
Example of data representation using CCA Non linear projection of a spiral Non linear projection of a horseshoe

PCA applications -Eigenfaces • Eigenfaces are the eigenvectors of the covariance matrix of the probability distribution of the vector space of human faces • Eigenfaces are the ‘standardized face ingredients’ derived from the statistical analysis of many pictures of human faces • A human face may be considered to be a combination of these standard faces

PCA applications -Eigenfaces To generate a set of eigenfaces: 1. 2. 3. 4. Large set of digitized images of human faces is taken under the same lighting conditions. The images are normalized to line up the eyes and mouths. The eigenvectors of the covariance matrix of the statistical distribution of face image vectors are then extracted. These eigenvectors are called eigenfaces.

PCA applications -Eigenfaces • the principal eigenface looks like a bland androgynous average human face http: //en. wikipedia. org/wiki/Image: Eigenfaces. png

Eigenfaces – Face Recognition • When properly weighted, eigenfaces can be summed together to create an approximate grayscale rendering of a human face. • Remarkably few eigenvector terms are needed to give a fair likeness of most people's faces • Hence eigenfaces provide a means of applying data compression to faces for identification purposes. • Similarly, Expert Object Recognition in Video

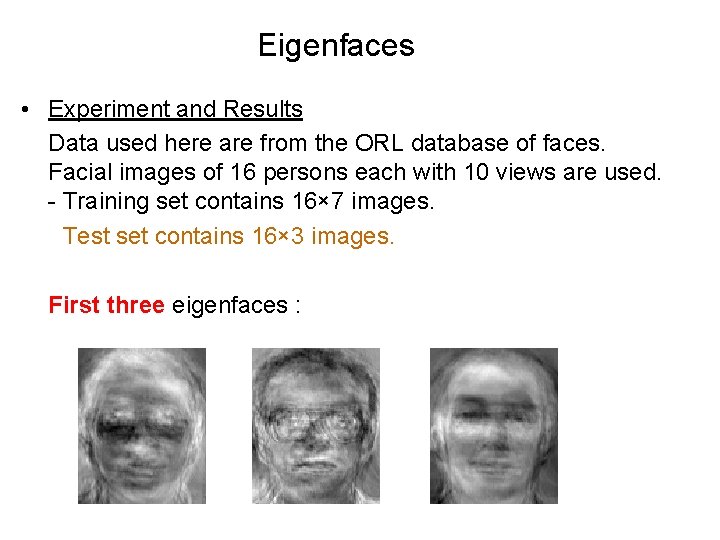
Eigenfaces • Experiment and Results Data used here are from the ORL database of faces. Facial images of 16 persons each with 10 views are used. - Training set contains 16× 7 images. - Test set contains 16× 3 images. First three eigenfaces :

Classification Using Nearest Neighbor • Save average coefficients for each person. Classify new face as the person with the closest average. • Recognition accuracy increases with number of eigenfaces till 15. Later eigenfaces do not help much with recognition. Best recognition rates Training set 99% Test set 89%

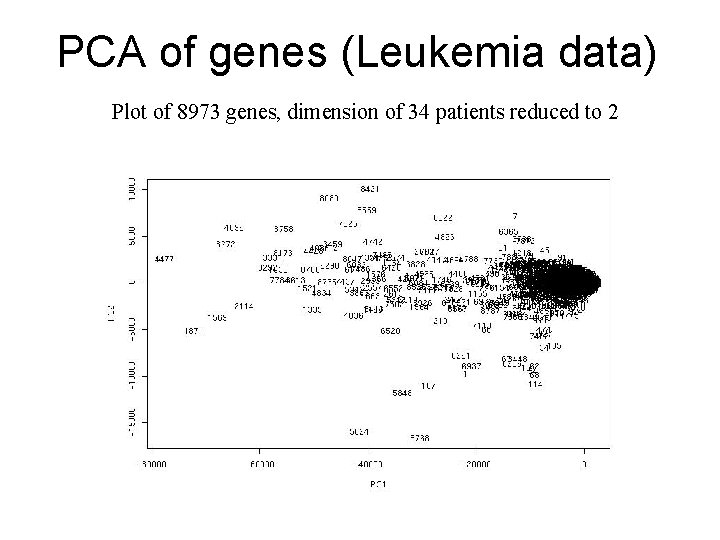
PCA of Genes (Leukemia data, precursor B and T cells) • 34 patients, dimension of 8973 genes reduced to 2

PCA of genes (Leukemia data) Plot of 8973 genes, dimension of 34 patients reduced to 2

Leukemia data - clustering of patients on original gene dimensions

Leukemia data - clustering of patients on top 100 eigen-genes

Leukemia data - clustering of genes

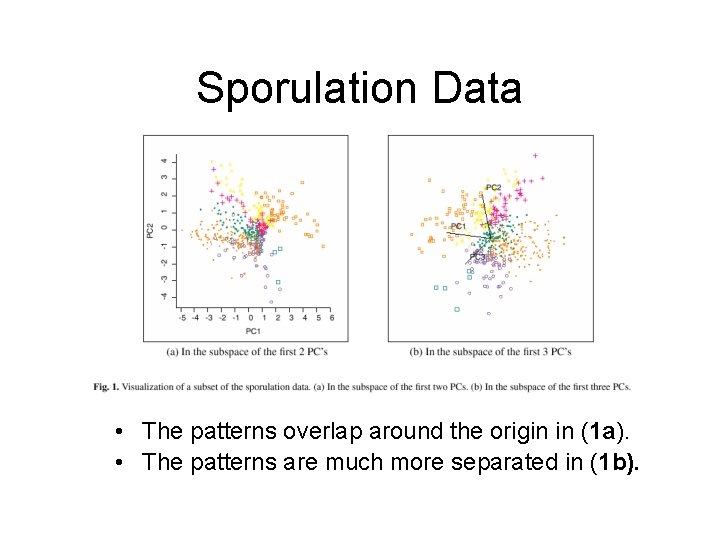
Sporulation Data Example • Data: A subset of sporulation data (477 genes) were classified into seven temporal patterns (Chu et al. , 1998) • The first 2 PCs contains 85. 9% of the variation in the data. (Figure 1 a) • The first 3 PCs contains 93. 2% of the variation in the data. (Figure 1 b)

Sporulation Data • The patterns overlap around the origin in (1 a). • The patterns are much more separated in (1 b).

PCA for Process Monitoring Variation in processes: • Chance • Natural variation inherent in a process. Cumulative effect of many small, unavoidable causes. • Assignable • Variations in raw material, machine tools, mechanical failure and human error. These are accountable circumstances and are normally larger.

PCA for process monitoring • Latent variables can sometimes be interpreted as measures of physical characteristics of a process i. e. , temp, pressure. • Variable reduction can increase the sensitivity of a control scheme to assignable causes • Application of PCA to monitoring is increasing – Start with a reference set defining normal operation conditions, look for assignable causes – Generate a set of indicator variables that best describe the dynamics of the process – PCA is sensitive to data types

Independent Component Analysis (ICA) PCA (orthogonal coordinate) ICA (non-orthogonal coordinate)

Cocktail-party Problem • Multiple sound sources in room (independent) • Multiple sensors receiving signals which are mixture of original signals • Estimate original source signals from mixture of received signals • Can be viewed as Blind-Source Separation as mixing parameters are not known

DEMO: BLIND SOURCE SEPARATION http: //www. cis. hut. fi/projects/ica/cocktail_en. cgi

BSS and ICA • Cocktail party or Blind Source Separation (BSS) problem – Ill posed problem, unless assumptions are made! • Most common assumption is that source signals are statistically independent. This means knowing value of one of them gives no information about the other. • Methods based on this assumption are called Independent Component Analysis methods – statistical techniques for decomposing a complex data set into independent parts. • It can be shown that under some reasonable conditions, if the ICA assumption holds, then the source signals can be recovered up to permutation and scaling.

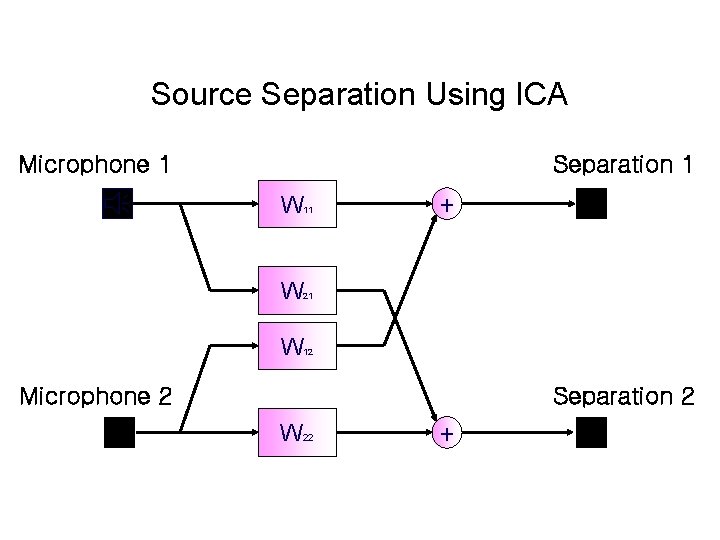
Source Separation Using ICA Microphone 1 Separation 1 W 11 W 21 W 12 + Microphone 2 Separation 2 W 22 +

Original signals (hidden sources) s 1(t), s 2(t), s 3(t), s 4(t), t=1: T

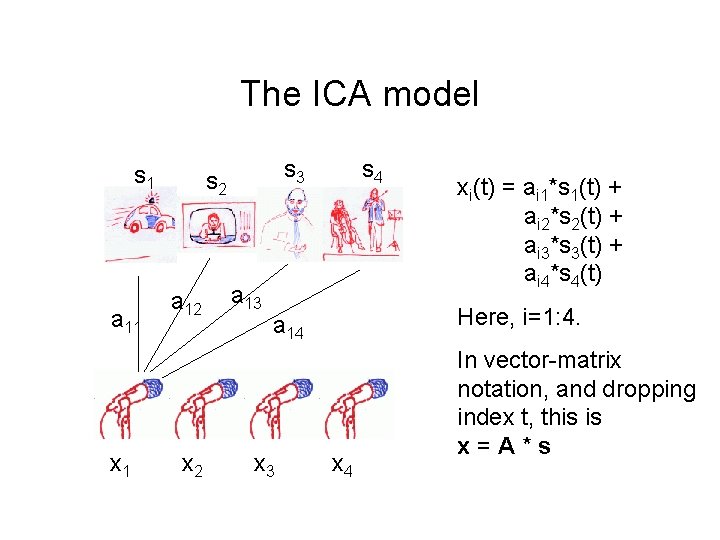
The ICA model s 1 a 11 x 1 s 3 s 2 a 12 x 2 a 13 s 4 Here, i=1: 4. a 14 x 3 xi(t) = ai 1*s 1(t) + ai 2*s 2(t) + ai 3*s 3(t) + ai 4*s 4(t) x 4 In vector-matrix notation, and dropping index t, this is x=A*s

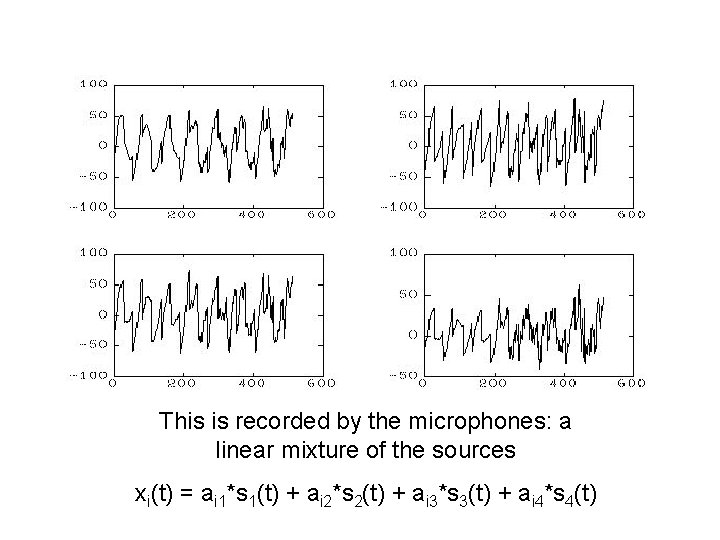
This is recorded by the microphones: a linear mixture of the sources xi(t) = ai 1*s 1(t) + ai 2*s 2(t) + ai 3*s 3(t) + ai 4*s 4(t)

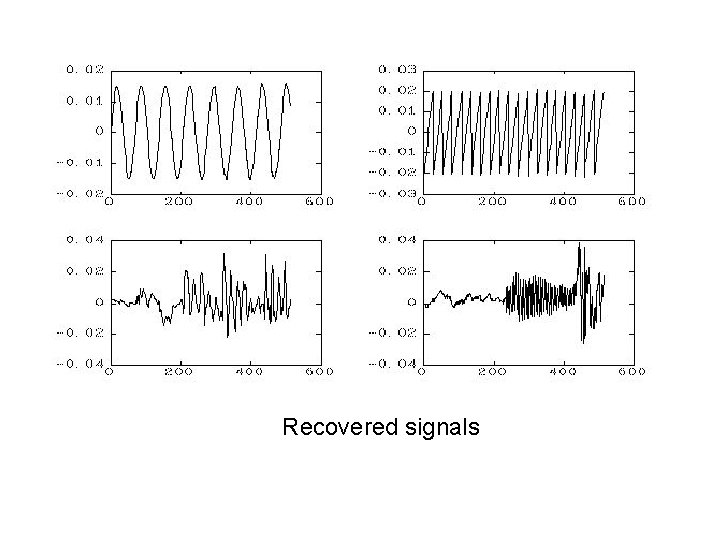
Recovered signals

BSS Problem: Determine the source signals s, given only the mixtures x. • If we knew the mixing parameters aij then we would just need to solve a linear system of equations. • We know neither aij nor si. • ICA was initially developed to deal with problems closely related to the cocktail party problem • Later it became evident that ICA has many other applications – e. g. from electrical recordings of brain activity from different locations of the scalp (EEG signals) recover underlying components of brain activity

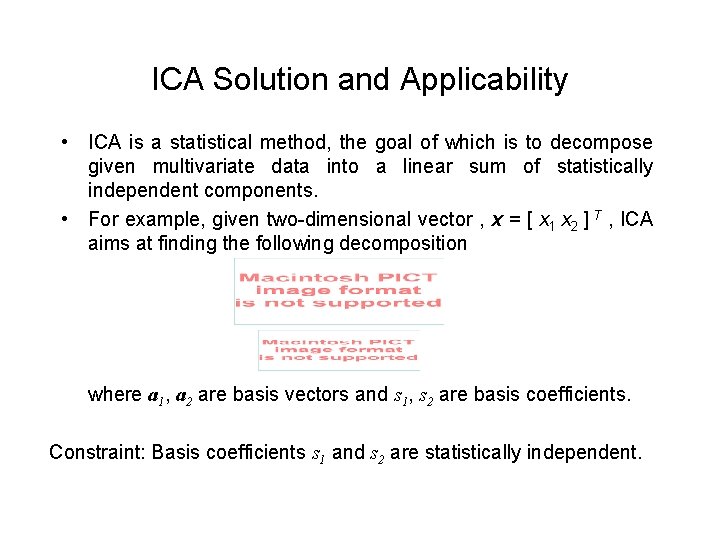
ICA Solution and Applicability • ICA is a statistical method, the goal of which is to decompose given multivariate data into a linear sum of statistically independent components. • For example, given two-dimensional vector , x = [ x 1 x 2 ] T , ICA aims at finding the following decomposition where a 1, a 2 are basis vectors and s 1, s 2 are basis coefficients. Constraint: Basis coefficients s 1 and s 2 are statistically independent.

Application domains of ICA • • • Blind source separation Image denoising Medical signal processing – f. MRI, ECG, EEG Modelling of the hippocampus and visual cortex Feature extraction, face recognition Compression, redundancy reduction Watermarking Clustering Time series analysis (stock market, microarray data) Topic extraction Econometrics: Finding hidden factors in financial data

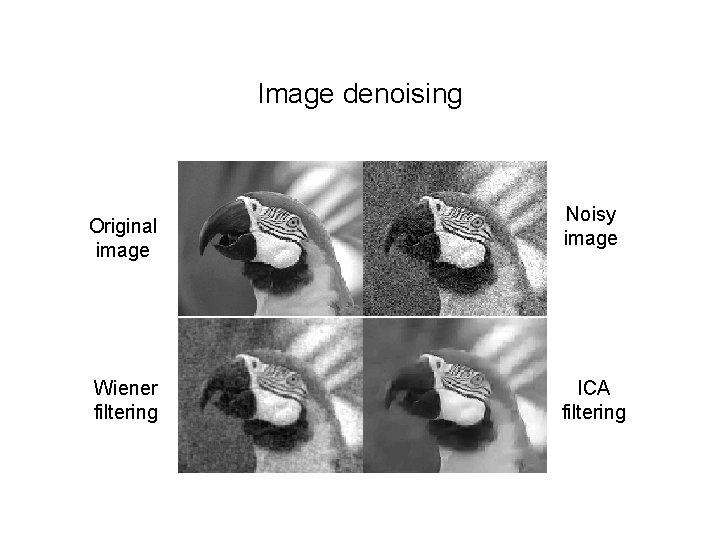
Image denoising Original image Wiener filtering Noisy image ICA filtering

Blind Separation of Information from Galaxy Spectra

Approaches to ICA • Unsupervised Learning – Factorial coding (Minimum entropy coding, Redundancy reduction) – Maximum likelihood learning – Nonlinear information maximization (entropy maximization) – Negentropy maximization – Bayesian learning • Statistical Signal Processing – Higher-order moments or cumulants – Joint approximate diagonalization – Maximum likelihood estimation

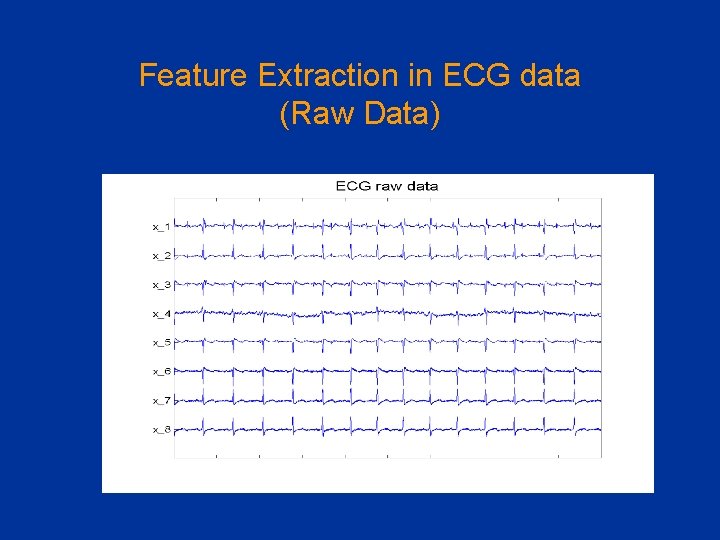
Feature Extraction in ECG data (Raw Data)

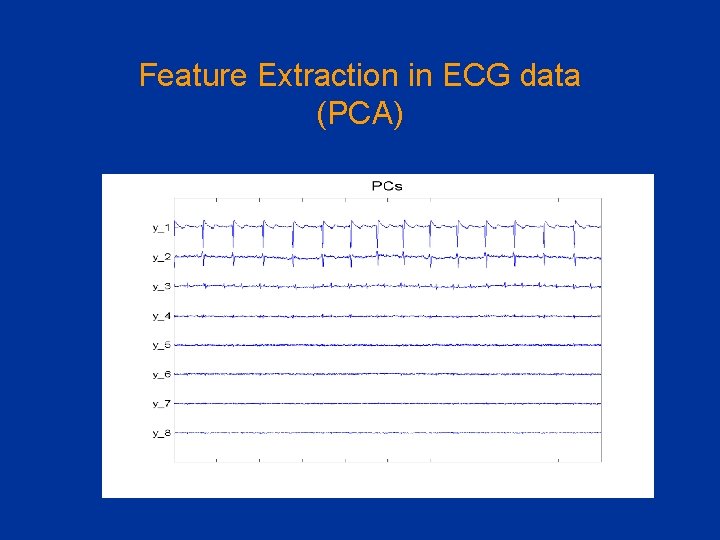
Feature Extraction in ECG data (PCA)

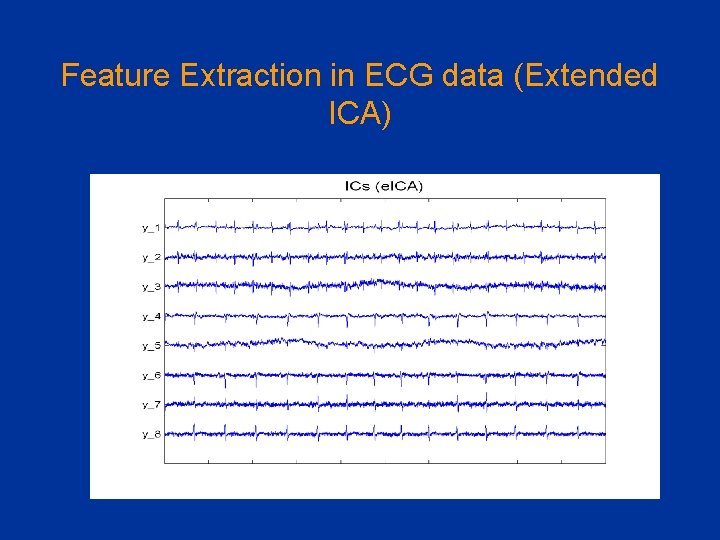
Feature Extraction in ECG data (Extended ICA)

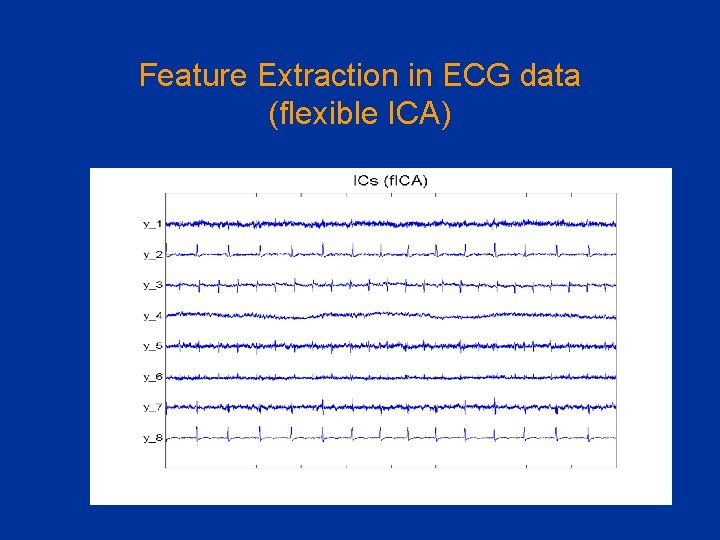
Feature Extraction in ECG data (flexible ICA)

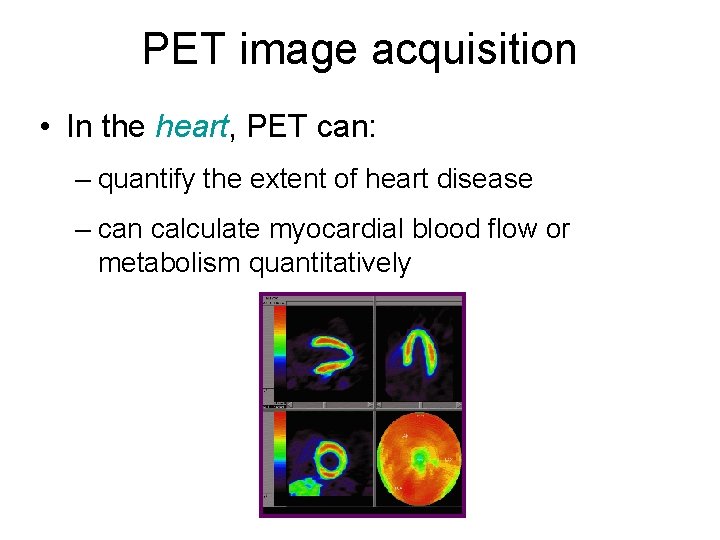
PET image acquisition • In the heart, PET can: – quantify the extent of heart disease – can calculate myocardial blood flow or metabolism quantitatively

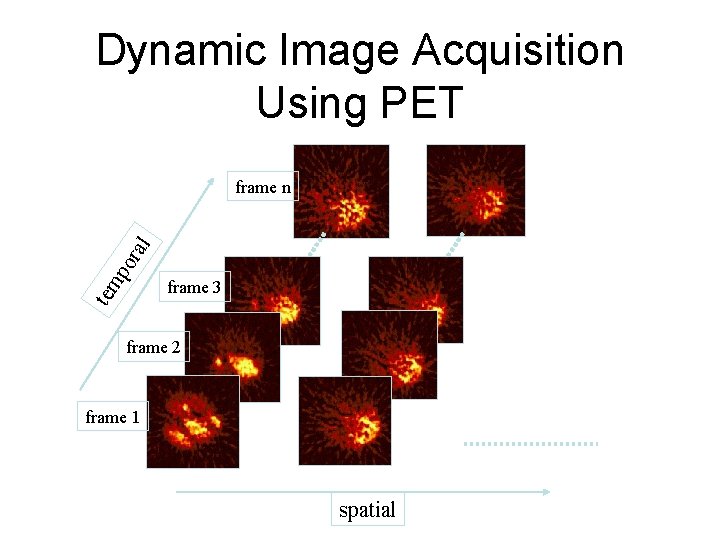
PET image acquisition • Static image – accumulating the data during the acquisition • Dynamic image – for the quantitative analysis (ex. blood flow, metabolism) – acquiring the data sequentially with a time interval

Dynamic Image Acquisition Using PET tem po ral frame n frame 3 frame 2 frame 1 spatial

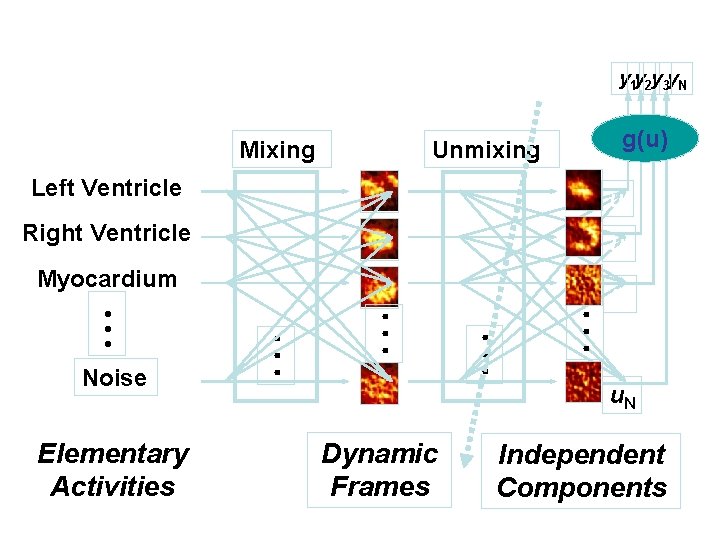
y 1 y 2 y 3 y. N Mixing g(u) Unmixing Left Ventricle u 1 Right Ventricle u 2 Elementary Activities • • • Noise • • • Myocardium u 3 u. N Dynamic Frames Independent Components

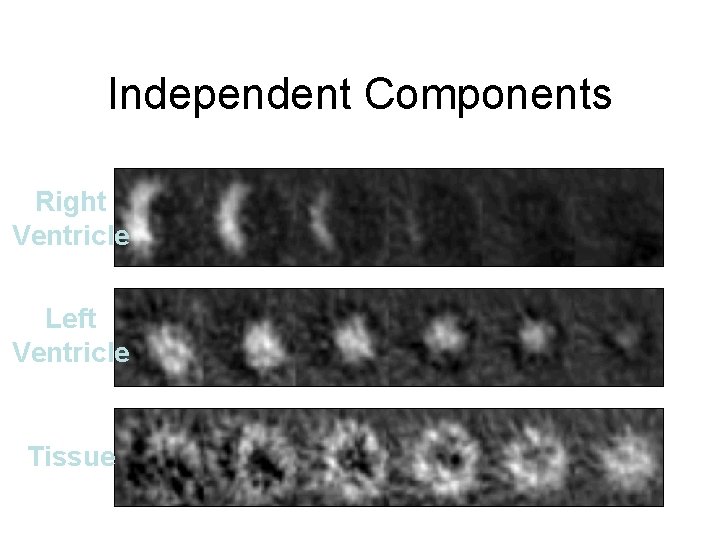
Independent Components Right Ventricle Left Ventricle Tissue

Other PET applications • In cancer, PET can: • distinguish benign from malignant tumors • stage cancer by showing metastases anywhere in body • In the brain, PET can: • calculate cerebral blood flow or metabolism quantitatively. • positively diagnose Alzheimer's disease for early intervention. • locate tumors in the brain and distinguish tumor from scar tissue. • locate the focus of seizures for some patients with epilepsy. • find regions related with specific task like memory, behavior etc.

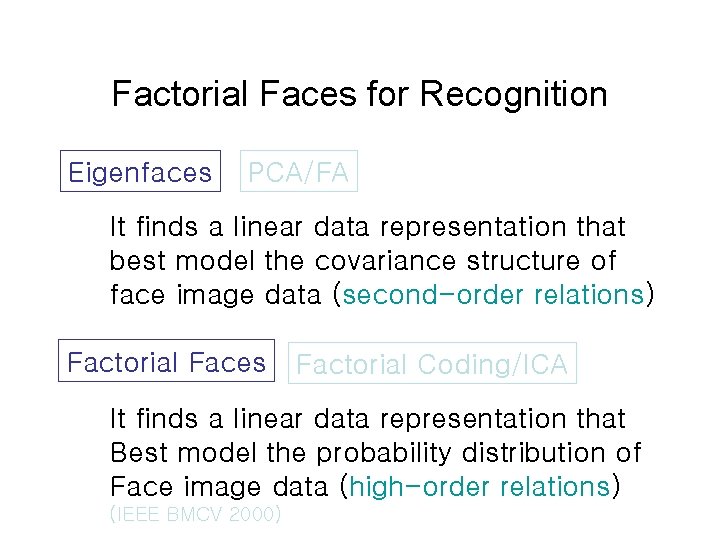
Factorial Faces for Recognition Eigenfaces PCA/FA It finds a linear data representation that best model the covariance structure of face image data (second-order relations) Factorial Faces Factorial Coding/ICA It finds a linear data representation that Best model the probability distribution of Face image data (high-order relations) (IEEE BMCV 2000)

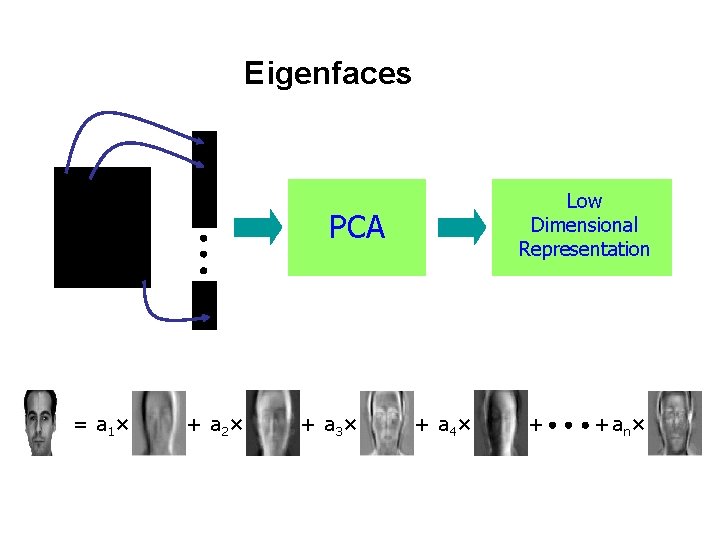
Eigenfaces Low Dimensional Representation PCA = a 1 × + a 2 × + a 3 × + a 4 × + + an×

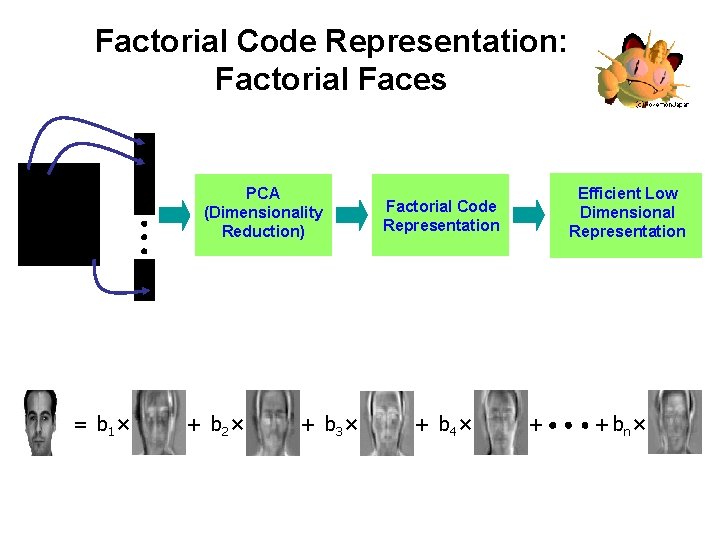
Factorial Code Representation: Factorial Faces PCA (Dimensionality Reduction) = b 1 × + b 2 × + b 3 × Efficient Low Dimensional Representation Factorial Code Representation + b 4 × + + bn ×

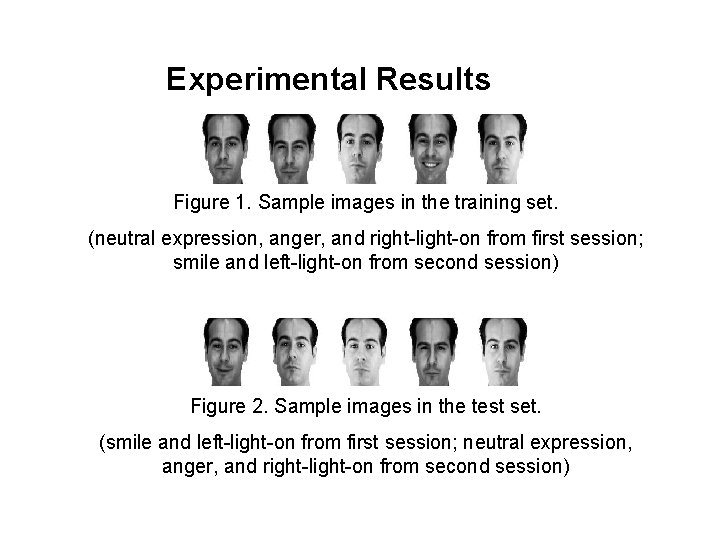
Experimental Results Figure 1. Sample images in the training set. (neutral expression, anger, and right-light-on from first session; smile and left-light-on from second session) Figure 2. Sample images in the test set. (smile and left-light-on from first session; neutral expression, anger, and right-light-on from second session)

Eigenfaces vs Factorial Faces (a) (b) Figure 3. First 20 basis images: (a) in eigenface method; (b) factorial code. They are ordered by column, then, by row.

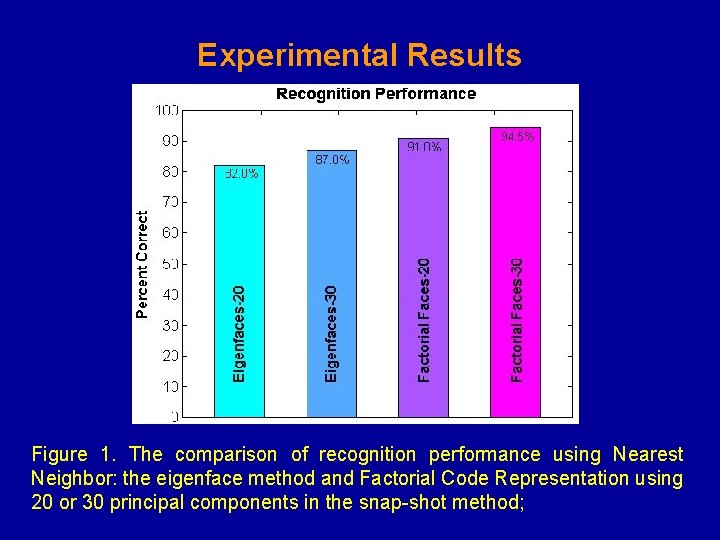
Experimental Results Figure 1. The comparison of recognition performance using Nearest Neighbor: the eigenface method and Factorial Code Representation using 20 or 30 principal components in the snap-shot method;

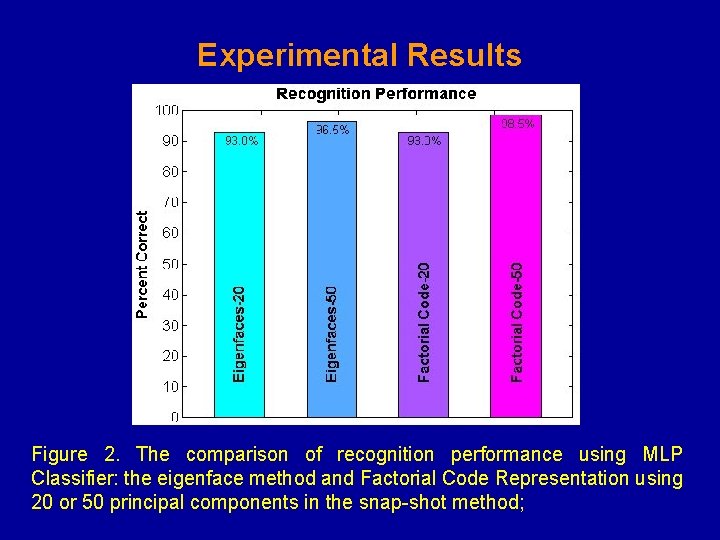
Experimental Results Figure 2. The comparison of recognition performance using MLP Classifier: the eigenface method and Factorial Code Representation using 20 or 50 principal components in the snap-shot method;

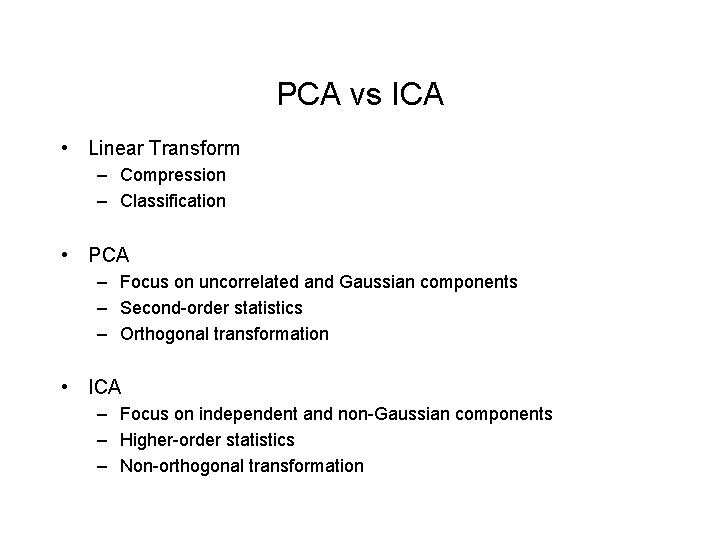
PCA vs ICA • Linear Transform – Compression – Classification • PCA – Focus on uncorrelated and Gaussian components – Second-order statistics – Orthogonal transformation • ICA – Focus on independent and non-Gaussian components – Higher-order statistics – Non-orthogonal transformation

Gaussians and ICA • If some components are gaussian and some are non-gaussian. – Can estimate all non-gaussian components – Linear combination of gaussian components can be estimated. – If only one gaussian component, model can be estimated • ICA sometimes viewed as non-Gaussian factor analysis