Algorithms to Search Position Specific Scoring Matrices in


![LA over a Suffix Array In terms of suffix trees, skp[i] is the lexicographically LA over a Suffix Array In terms of suffix trees, skp[i] is the lexicographically](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-38.jpg)
![AC-Extension by LA n First position [C, 0. 1] 1 2 3 4 5 AC-Extension by LA n First position [C, 0. 1] 1 2 3 4 5](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-48.jpg)
![AC-Extension by LA n Second position [C, 0. 1] 1 2 3 4 5 AC-Extension by LA n Second position [C, 0. 1] 1 2 3 4 5](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-49.jpg)
![AC-Extension by LA n Third position [C, 0. 1] 1 2 3 4 5 AC-Extension by LA n Third position [C, 0. 1] 1 2 3 4 5](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-50.jpg)
![Example: M 0003, MSS=0. 85 [G, 18500] C. Pizzi, DEI – Univ. Of Padova Example: M 0003, MSS=0. 85 [G, 18500] C. Pizzi, DEI – Univ. Of Padova](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-59.jpg)
![Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] C. Pizzi, DEI – Univ. Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] C. Pizzi, DEI – Univ.](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-60.jpg)
![Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • GCC is Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • GCC is](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-61.jpg)
![Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • Save 5464 Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • Save 5464](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-62.jpg)




- Slides: 89

Algorithms to Search Position Specific Scoring Matrices in Biosequences Cinzia Pizzi Dipartimento di Ingegneria dell’Informazione Università degli Studi di Padova

Outline n n n Weighted patterns in Biology The problem of profile matching The look-ahead method n n n Suffix based Algorithms Aho-Corasick Extension (ACE) Look-ahead Filtration Algorithm (LFA) Superalphabet (NS) Some experimental results C. Pizzi, DEI – Univ. Of Padova (Italy) 2

What are Motifs? n n n Motifs are biologically significant elements that are responsible for common structures or functions Motifs are statistically significant substrings in bio-sequences Assumption: if two entities share same function or structure, common overrepresented elements might be responsible for observed similarity C. Pizzi, DEI – Univ. Of Padova (Italy) 3

Motif Discovery n n Take set of co-expressed genes Compare their promoter regions Common over-represented substrings are good candidates for TFBS Need counted/expected frequency Promoters of co-expressed genes C. Pizzi, DEI – Univ. Of Padova (Italy) 4

Motif Discovery n n TFBS, DNA motifs Motifs = binding sites = substrings Promoters of co-expressed genes n Intrinsic variability of biological sequences n Mismatches, indels, wildcards, superalphabets. . . C. Pizzi, DEI – Univ. Of Padova (Italy) 5

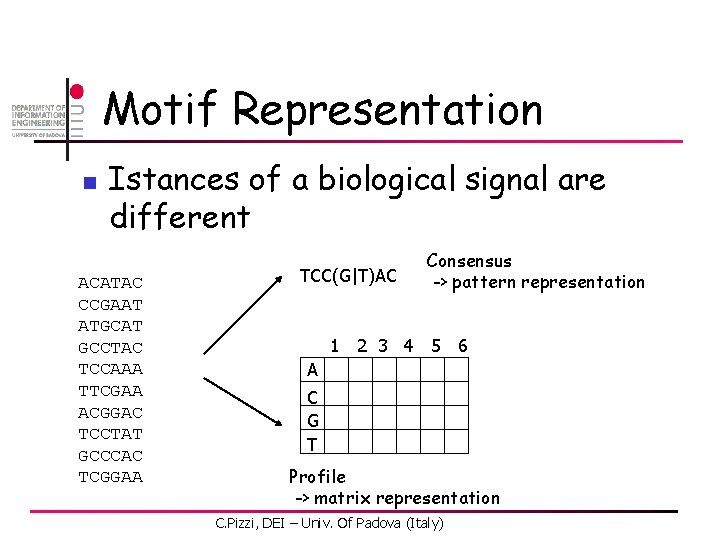
Motif Representation n Binding sites of the same factor are not exactly the same in all sequences ACATAC CCGAAT ATGCAT GCCTAC TCCAAA TTCGAA ACGGAC TCCTAT GCCCAC TCGGAA 1 2 3 4 5 6 A C G T Profile -> matrix representation C. Pizzi, DEI – Univ. Of Padova (Italy)

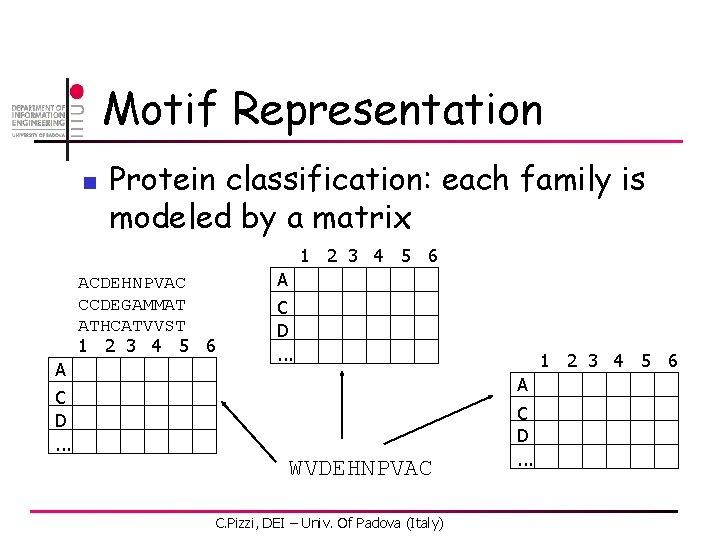
Motif Representation n Protein classification: each family is modeled by a matrix 1 2 3 4 5 6 ACDEHNPVAC CCDEGAMMAT ATHCATVVST 1 2 3 4 5 6 A A C D. . . 1 2 3 4 5 6 A C D. . . WVDEHNPVAC C. Pizzi, DEI – Univ. Of Padova (Italy) C D. . .

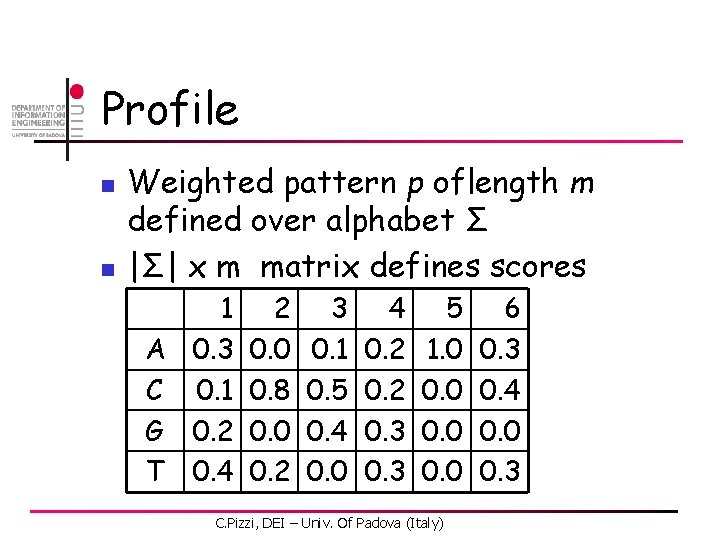
Profile n n Weighted pattern p oflength m defined over alphabet Σ |Σ| x m matrix defines scores 1 A 0. 3 C 0. 1 G 0. 2 T 0. 4 2 0. 0 0. 8 0. 0 0. 2 3 0. 1 0. 5 0. 4 0. 0 4 0. 2 0. 3 5 1. 0 0. 0 C. Pizzi, DEI – Univ. Of Padova (Italy) 6 0. 3 0. 4 0. 0 0. 3

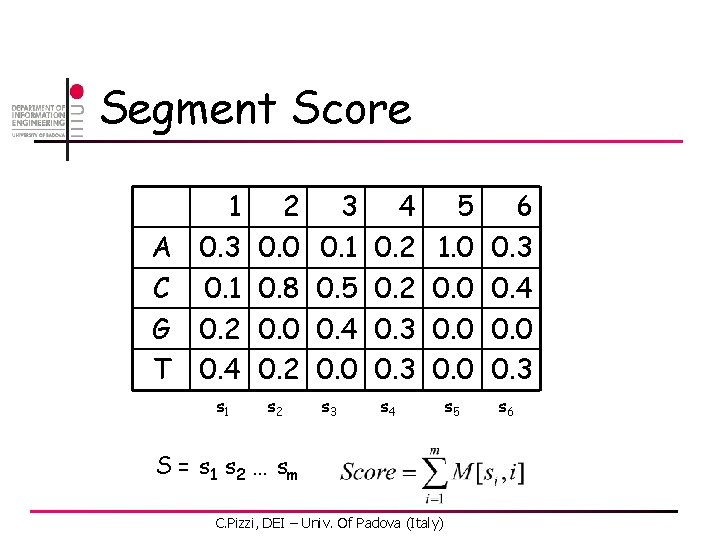
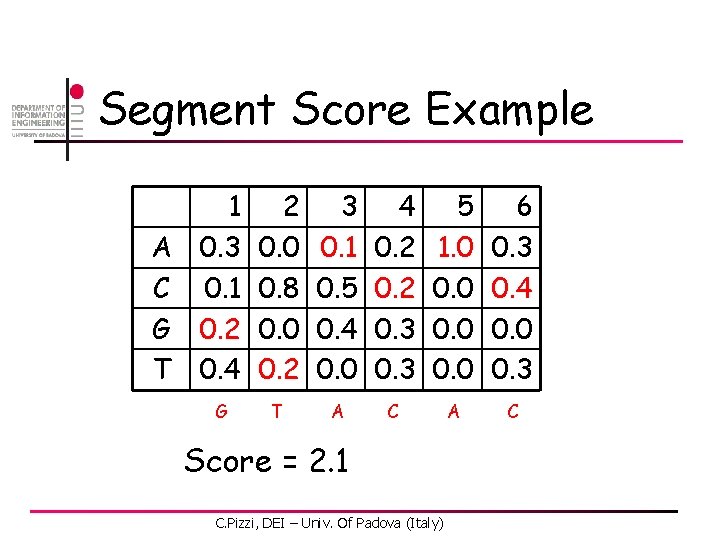
Segment Score A C G T 1 0. 3 0. 1 0. 2 0. 4 2 0. 0 0. 8 0. 0 0. 2 s 1 s 2 3 0. 1 0. 5 0. 4 0. 0 4 0. 2 0. 3 s 3 s 4 5 1. 0 0. 0 S = s 1 s 2 … s m C. Pizzi, DEI – Univ. Of Padova (Italy) s 5 6 0. 3 0. 4 0. 0 0. 3 s 6

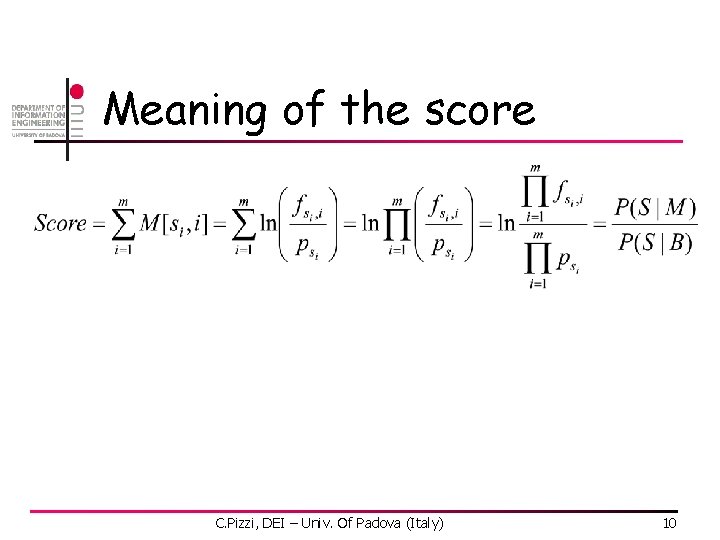
Meaning of the score C. Pizzi, DEI – Univ. Of Padova (Italy) 10

Segment Score Example A C G T 1 0. 3 0. 1 0. 2 0. 4 2 0. 0 0. 8 0. 0 0. 2 3 0. 1 0. 5 0. 4 0. 0 4 0. 2 0. 3 5 1. 0 0. 0 6 0. 3 0. 4 0. 0 0. 3 G T A C Score = 2. 1 C. Pizzi, DEI – Univ. Of Padova (Italy)

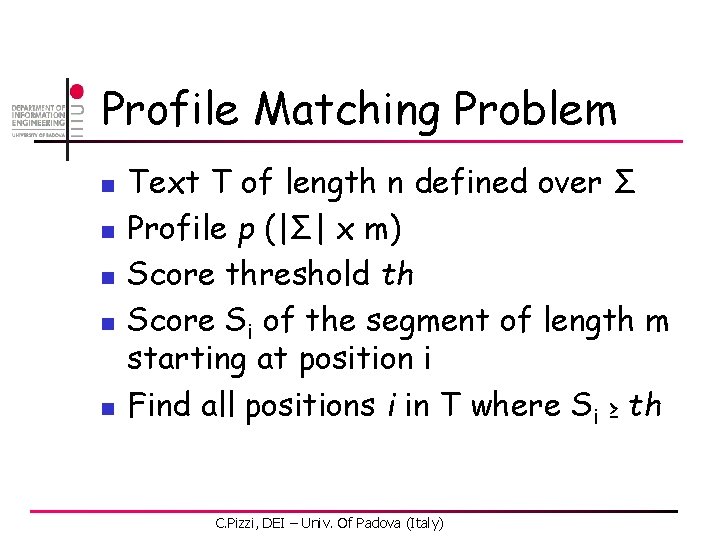
Profile Matching Problem n n n Text T of length n defined over Σ Profile p (|Σ| x m) Score threshold th Score Si of the segment of length m starting at position i Find all positions i in T where Si ≥ th C. Pizzi, DEI – Univ. Of Padova (Italy)

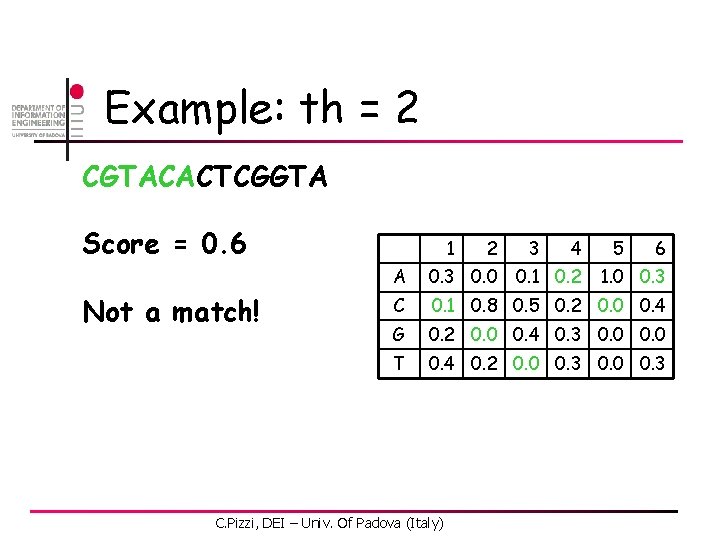
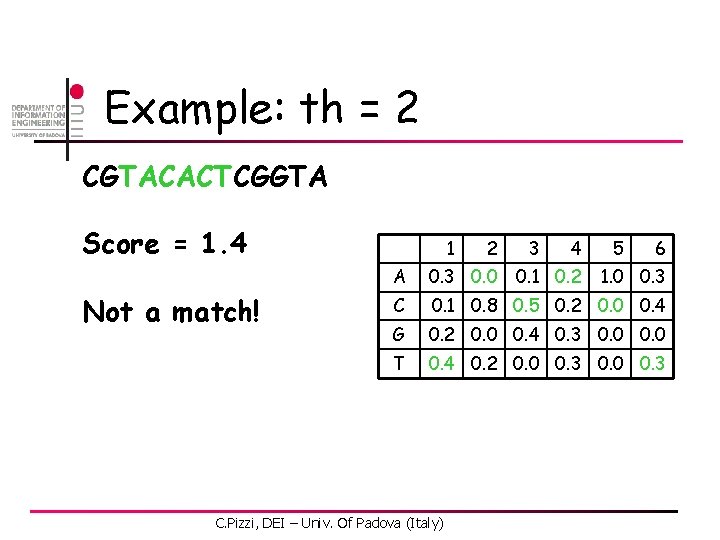
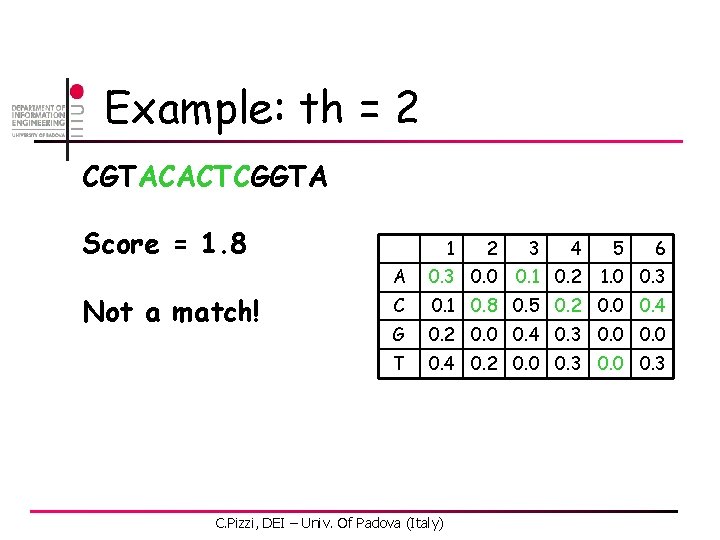
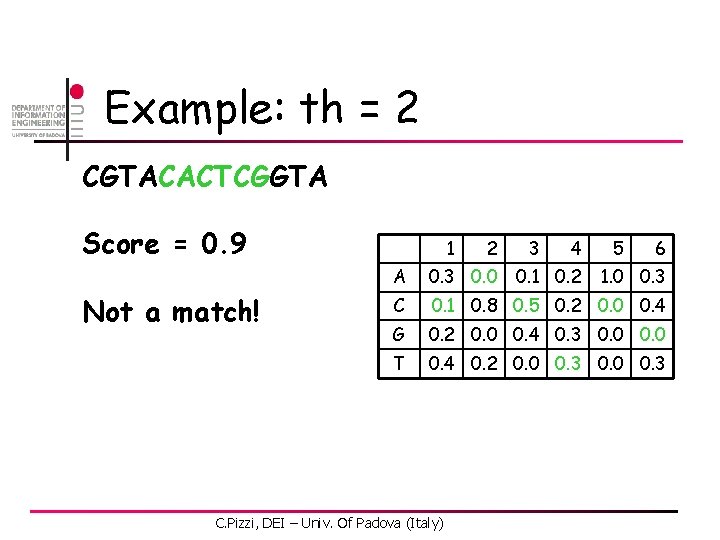
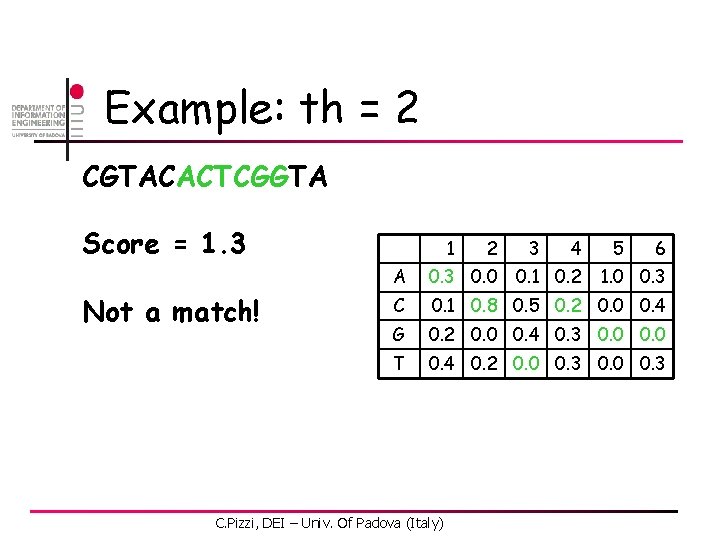
Example: th = 2 CGTACACTCGGTA Score = 0. 6 Not a match! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

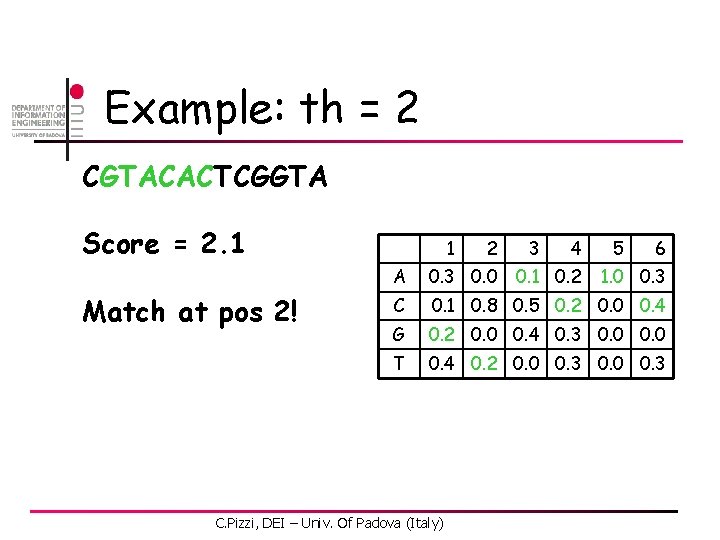
Example: th = 2 CGTACACTCGGTA Score = 2. 1 Match at pos 2! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Example: th = 2 CGTACACTCGGTA Score = 1. 4 Not a match! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Example: th = 2 CGTACACTCGGTA Score = 1. 8 Not a match! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Example: th = 2 CGTACACTCGGTA Score = 0. 9 Not a match! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Example: th = 2 CGTACACTCGGTA Score = 1. 3 Not a match! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Example: th = 2 CGTACACTCGGTA Score = 1. 4 Not a match! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Example: th = 2 CGTACACTCGGTA Score = 2. 2 Match at pos 8! 1 2 3 4 6 A 0. 3 0. 0 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) 0. 1 0. 2 5 1. 0 0. 3

Scenarios of applications n Online Algorithms (no indexing) n n n Database of profile matrices (e. g. TRANSFAC, JASPAR for TFBS) Input sequence to be searched Offline algorithms (indexing) n n Sequence or set of sequences Input matrix to search for matches C. Pizzi, DEI – Univ. Of Padova (Italy)

Summary of current methods n n Look-ahead method LA (Wu et al, 00) Offline methods based on LA: n n Suffix-tree (Dorohonceanu et al, 00) Suffix-array (Beckstette et al, 04, 06) Truncated Suffix Tree (Pizzi and Favaretto, 10) Online methods based on LA: n Aho-Corasick, Filtering(Pizzi et al. 07, 09) C. Pizzi, DEI – Univ. Of Padova (Italy)

Summary of current methods n Pattern Matching n n n Shift-Add (Salmela e Tarhio, 08) KMP (Liefoghee et al, 09) Matrix partitioning (Liefhooghe et al. , 06, Pizzi et al. , 07, 09) FFT based (Rajasekaran et al. , 02) Compression based(Freschi et al. , 05) C. Pizzi, DEI – Univ. Of Padova (Italy)

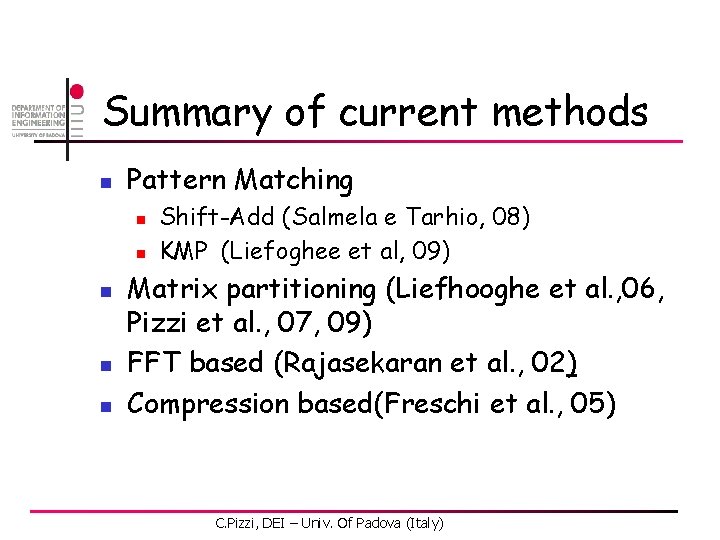
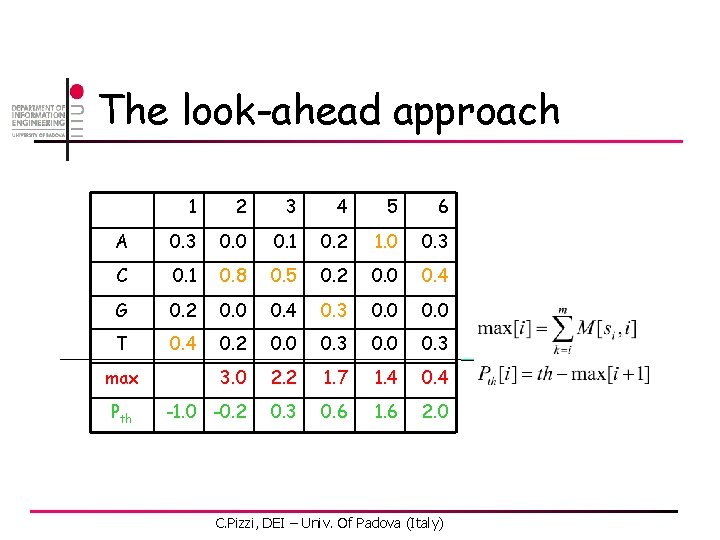
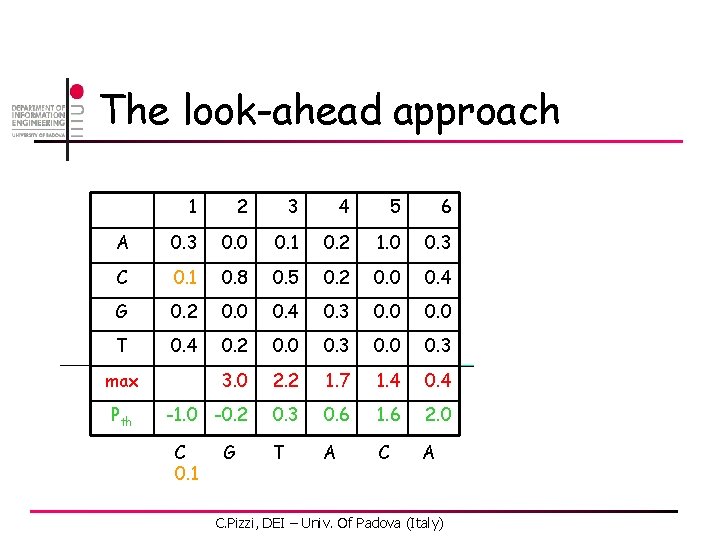
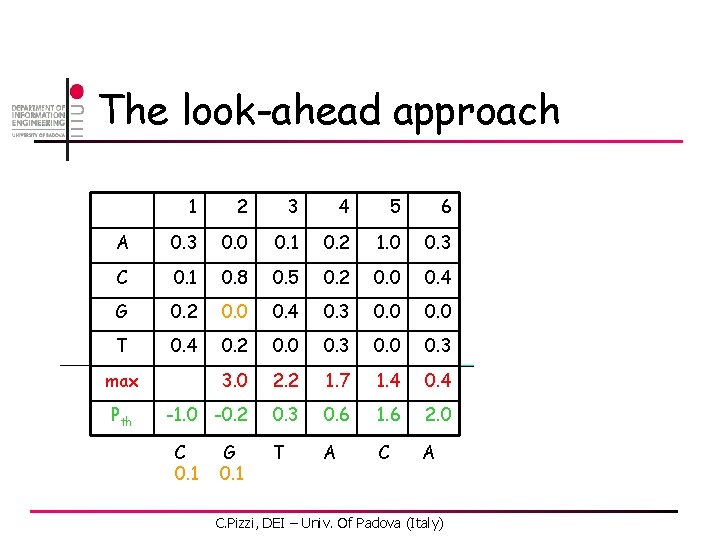
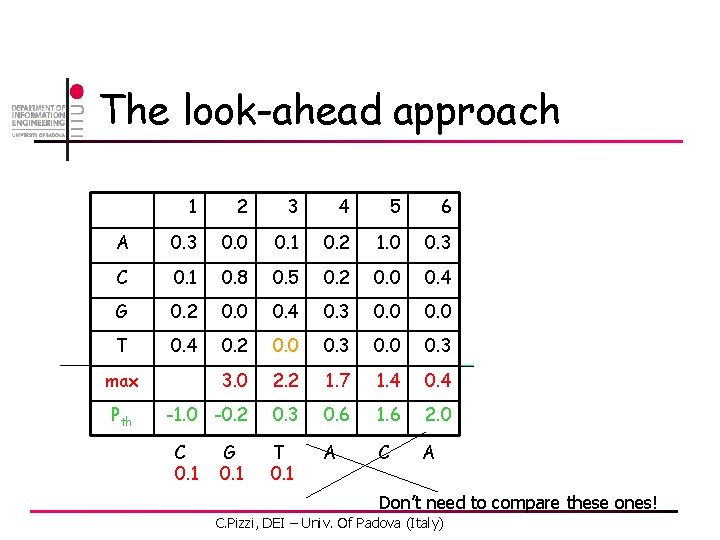
The look-ahead approach 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 3. 0 2. 2 1. 7 1. 4 0. 4 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 max Pth C. Pizzi, DEI – Univ. Of Padova (Italy)

The look-ahead approach 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 3. 0 2. 2 1. 7 1. 4 0. 4 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 T A C A max Pth C 0. 1 G C. Pizzi, DEI – Univ. Of Padova (Italy)

The look-ahead approach 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 3. 0 2. 2 1. 7 1. 4 0. 4 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 T A C A max Pth C 0. 1 G 0. 1 C. Pizzi, DEI – Univ. Of Padova (Italy)

The look-ahead approach 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 3. 0 2. 2 1. 7 1. 4 0. 4 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 T 0. 1 A C A max Pth C 0. 1 G 0. 1 Don’t need to compare these ones! C. Pizzi, DEI – Univ. Of Padova (Italy)

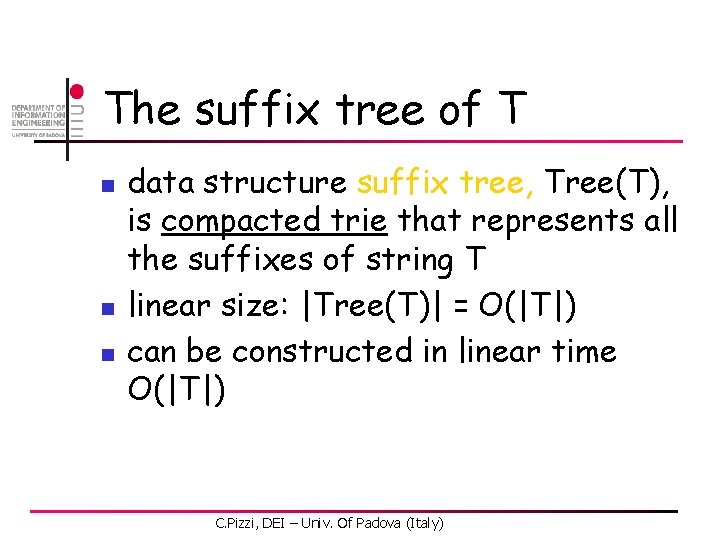
The suffix tree of T n n n data structure suffix tree, Tree(T), is compacted trie that represents all the suffixes of string T linear size: |Tree(T)| = O(|T|) can be constructed in linear time O(|T|) C. Pizzi, DEI – Univ. Of Padova (Italy)

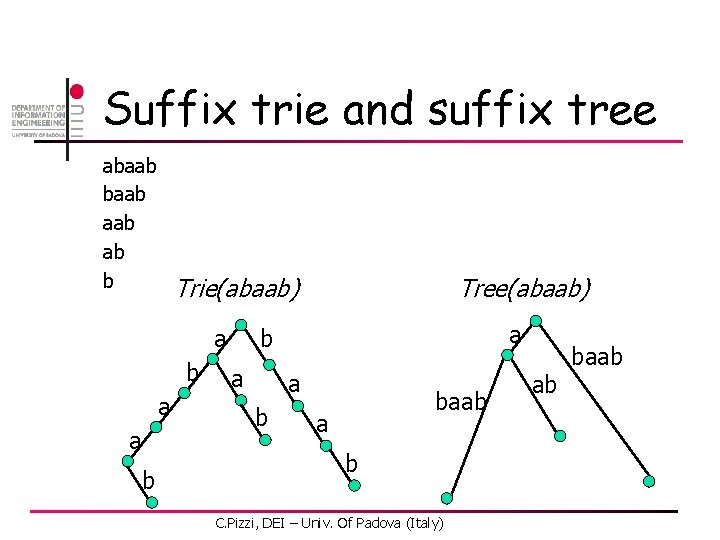
Suffix trie and suffix tree abaab aab ab b Trie(abaab) a b a a b Tree(abaab) a b a a b baab a b C. Pizzi, DEI – Univ. Of Padova (Italy) ab baab

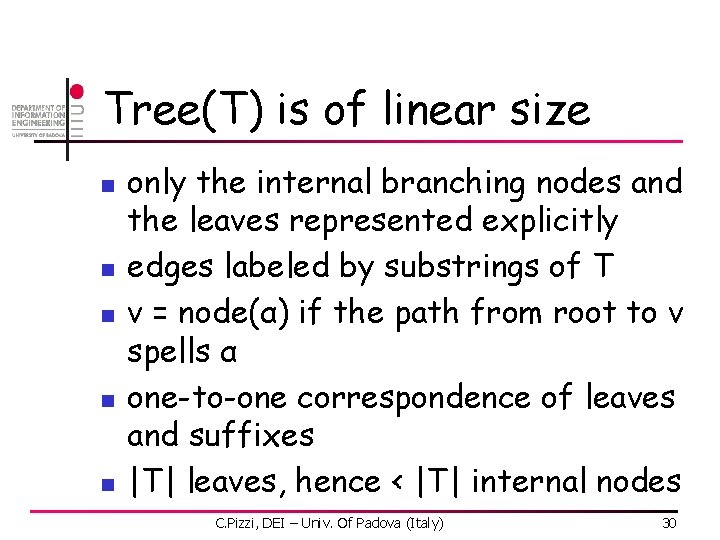
Tree(T) is of linear size n n n only the internal branching nodes and the leaves represented explicitly edges labeled by substrings of T v = node(α) if the path from root to v spells α one-to-one correspondence of leaves and suffixes |T| leaves, hence < |T| internal nodes C. Pizzi, DEI – Univ. Of Padova (Italy) 30

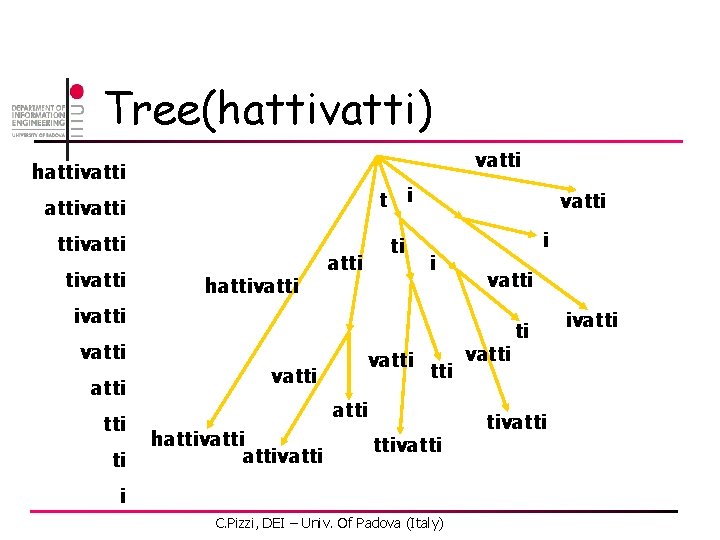
Tree(hattivatti) vatti hattivatti i t attivatti hattivatti ti atti vatti ivatti ti vatti tti atti hattivatti i C. Pizzi, DEI – Univ. Of Padova (Italy) vatti ti tivatti

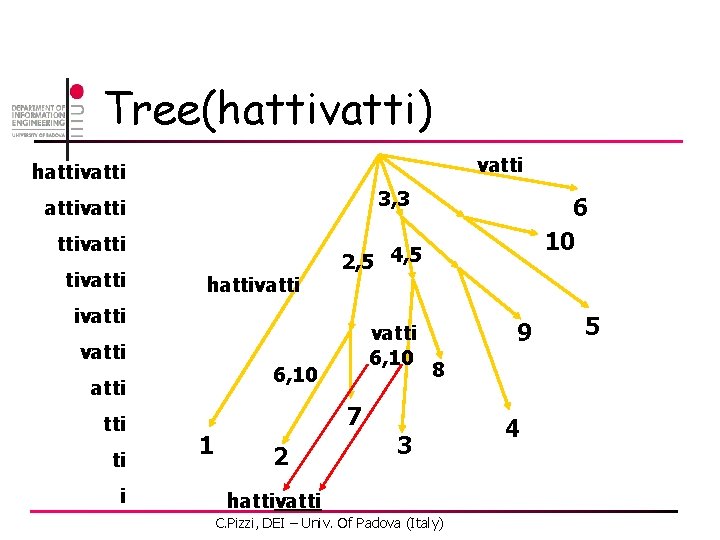
Tree(hattivatti) vatti hattivatti 3, 3 attivatti hattivatti 2, 5 4, 5 ivatti tti ti i vatti 6, 10 atti 1 7 2 6 10 9 8 3 hattivatti C. Pizzi, DEI – Univ. Of Padova (Italy) 4 5

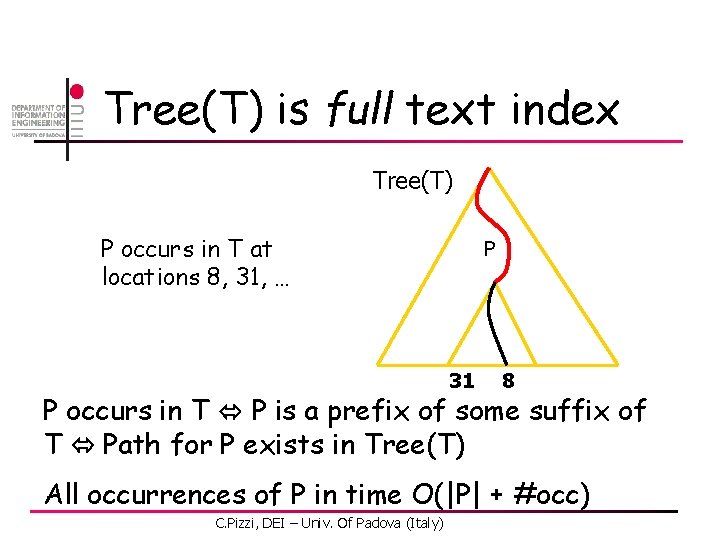
Tree(T) is full text index Tree(T) P occurs in T at locations 8, 31, … P 31 8 P occurs in T P is a prefix of some suffix of T Path for P exists in Tree(T) All occurrences of P in time O(|P| + #occ) C. Pizzi, DEI – Univ. Of Padova (Italy)

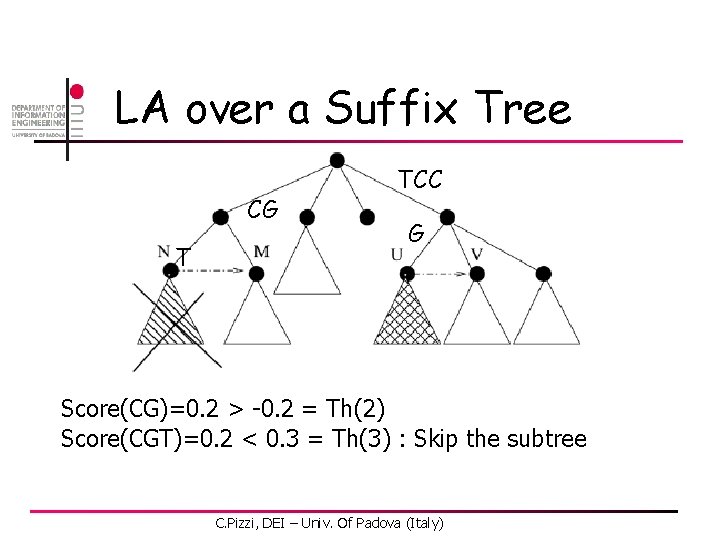
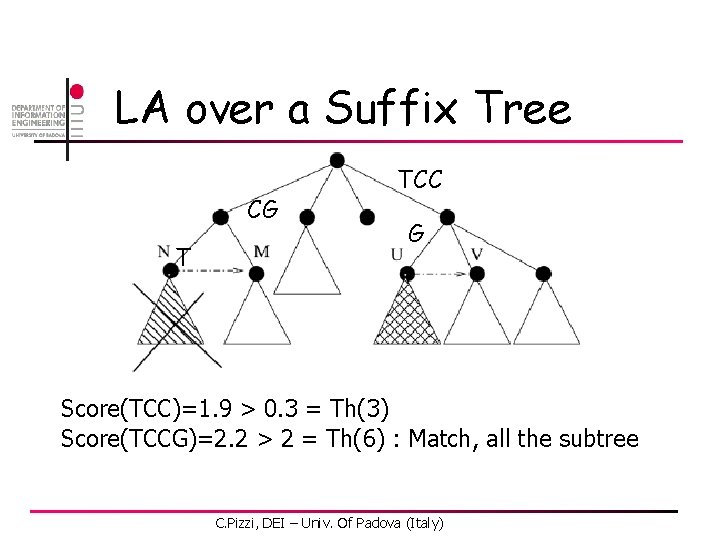
LA over a Suffix Tree CG T TCC G Score(CG)=0. 2 > -0. 2 = Th(2) Score(CGT)=0. 2 < 0. 3 = Th(3) : Skip the subtree C. Pizzi, DEI – Univ. Of Padova (Italy)

LA over a Suffix Tree CG T TCC G Score(TCC)=1. 9 > 0. 3 = Th(3) Score(TCCG)=2. 2 > 2 = Th(6) : Match, all the subtree C. Pizzi, DEI – Univ. Of Padova (Italy)

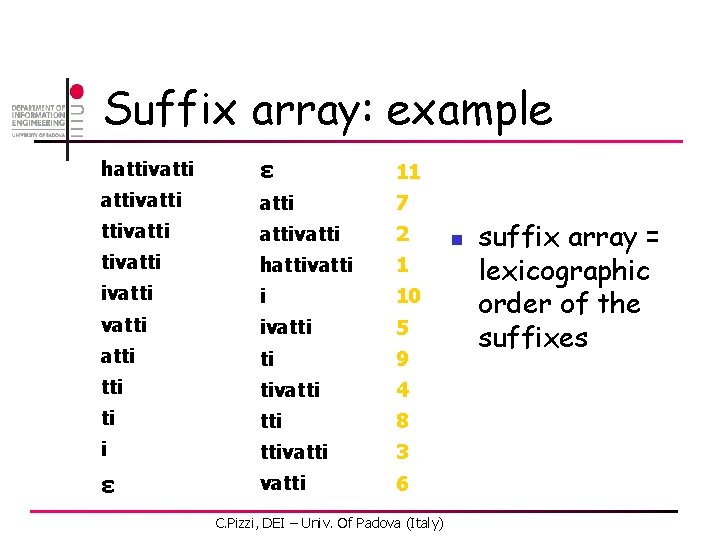
Suffix array: example hattivatti ε 11 attivatti 7 ttivattivatti 2 tivatti hattivatti 1 ivatti i 10 vatti ivatti 5 atti ti 9 tti tivatti 4 ti tti 8 i ttivatti 3 ε vatti 6 C. Pizzi, DEI – Univ. Of Padova (Italy) n suffix array = lexicographic order of the suffixes

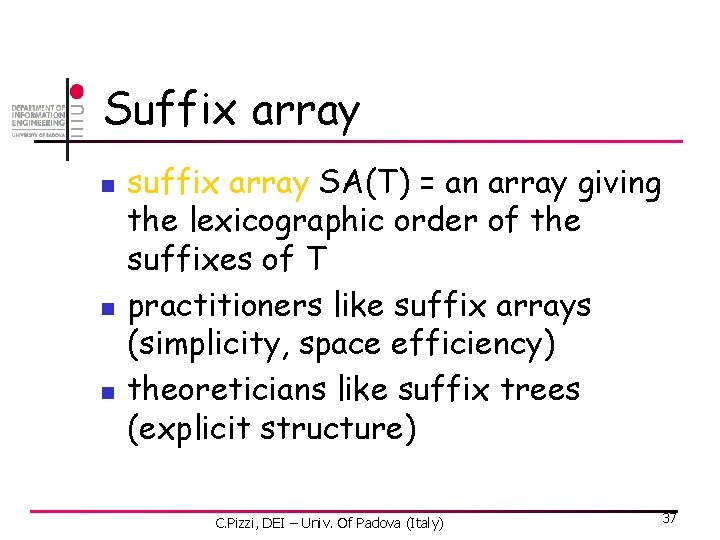
Suffix array n n n suffix array SA(T) = an array giving the lexicographic order of the suffixes of T practitioners like suffix arrays (simplicity, space efficiency) theoreticians like suffix trees (explicit structure) C. Pizzi, DEI – Univ. Of Padova (Italy) 37
![LA over a Suffix Array In terms of suffix trees skpi is the lexicographically LA over a Suffix Array In terms of suffix trees, skp[i] is the lexicographically](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-38.jpg)
LA over a Suffix Array In terms of suffix trees, skp[i] is the lexicographically next leaf that does not occur in the subtree below the branching node corresponding to the longest common prefix of Ssuf[i-1] and Ssuf[i]. skp[i] = min({n + 1} U [ j in [i + 1; n] | lcp[i] > lcp[j]) C. Pizzi, DEI – Univ. Of Padova (Italy)

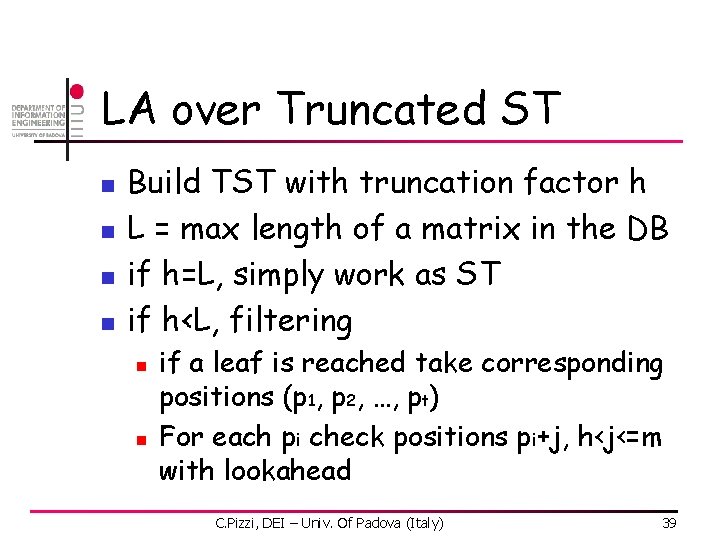
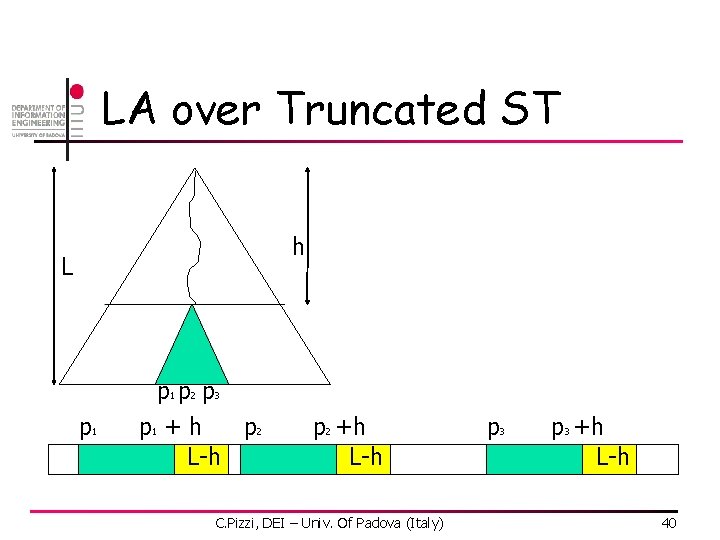
LA over Truncated ST n n Build TST with truncation factor h L = max length of a matrix in the DB if h=L, simply work as ST if h<L, filtering n n if a leaf is reached take corresponding positions (p 1, p 2, …, pt) For each pi check positions pi+j, h<j<=m with lookahead C. Pizzi, DEI – Univ. Of Padova (Italy) 39

LA over Truncated ST h L p 1 p 2 p 3 p 2 p 1 + h L-h p 2 +h L-h C. Pizzi, DEI – Univ. Of Padova (Italy) p 3 +h L-h 40

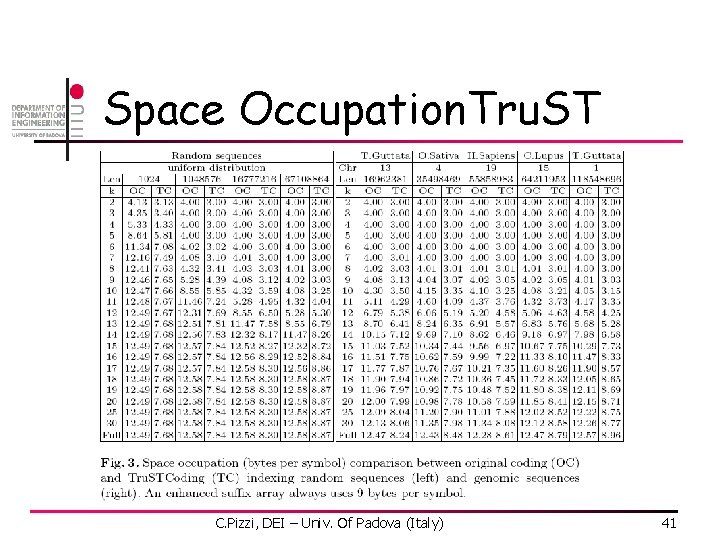
Space Occupation. Tru. ST C. Pizzi, DEI – Univ. Of Padova (Italy) 41

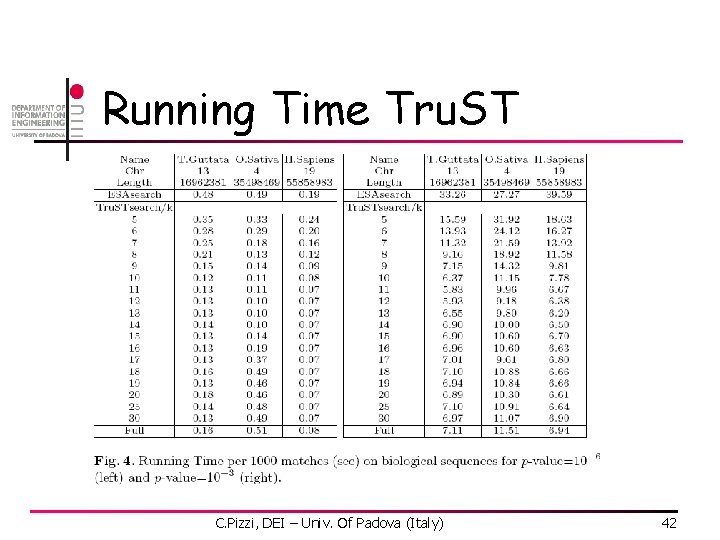
Running Time Tru. ST C. Pizzi, DEI – Univ. Of Padova (Italy) 42

Online Profile Matching n Aho-Corasick Expansion (ACE) n n Lookahead Filtration Algorithm(LFA) n n Pattern matching + LA Score for fixed length prefix as a filter + LA Naive Superalphabet (NS) n Encode k-mers in superalphabet symbol C. Pizzi, DEI – Univ. Of Padova (Italy)

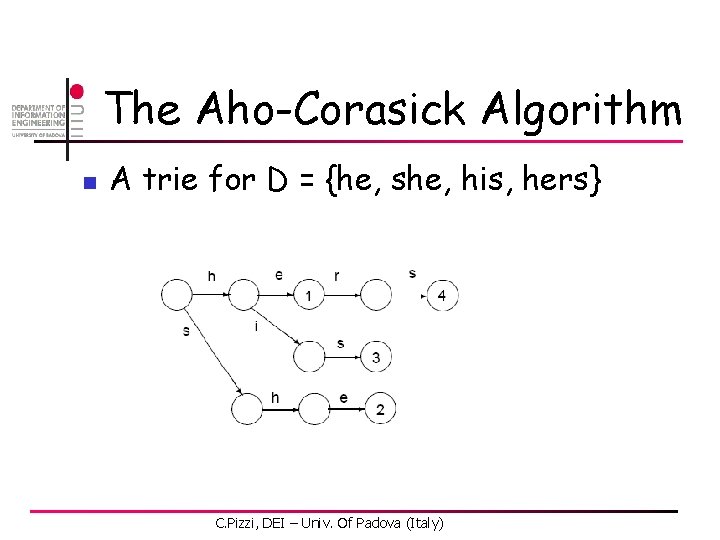
The Aho-Corasick Algorithm n A trie for D = {he, she, his, hers} C. Pizzi, DEI – Univ. Of Padova (Italy)

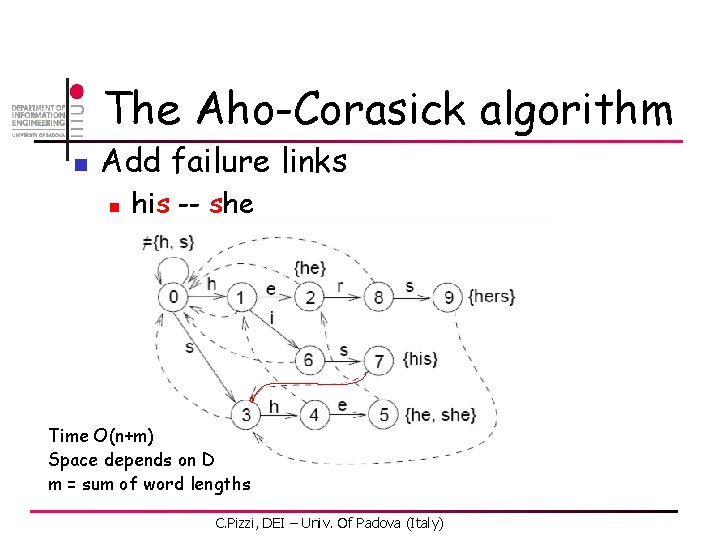
The Aho-Corasick algorithm n Add failure links n his -- she Time O(n+m) Space depends on D m = sum of word lengths C. Pizzi, DEI – Univ. Of Padova (Italy)

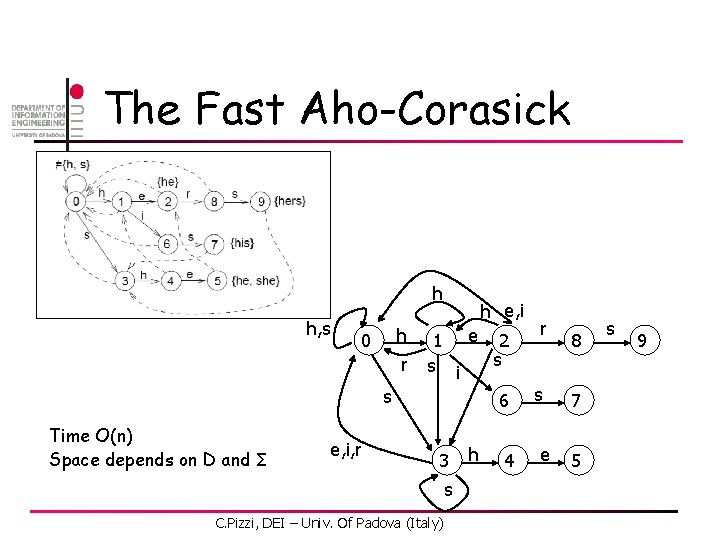
The Fast Aho-Corasick h h, s h 0 r h e, i 1 e 2 s s i s Time O(n) Space depends on D and Σ e, i, r 6 3 s C. Pizzi, DEI – Univ. Of Padova (Italy) h 4 r s e 8 7 5 s 9

AC and profile matching n Build AC automaton for all the words that are a match for the matrix n n LA partial threshold limits the number of words to those that actually match O(|D||Σ|m + m|Σ|) pre-processing |D|≤|Σ|m depends on matrix and threshold Search the text with AC automaton n O(n) search C. Pizzi, DEI – Univ. Of Padova (Italy)
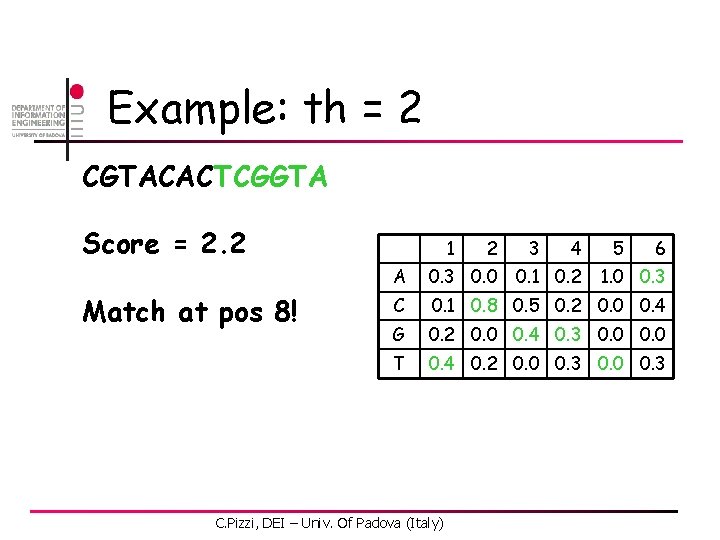
![ACExtension by LA n First position C 0 1 1 2 3 4 5 AC-Extension by LA n First position [C, 0. 1] 1 2 3 4 5](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-48.jpg)
AC-Extension by LA n First position [C, 0. 1] 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 Pth C. Pizzi, DEI – Univ. Of Padova (Italy) [G, 0. 2] [T, 0. 4] [A, 0. 3]
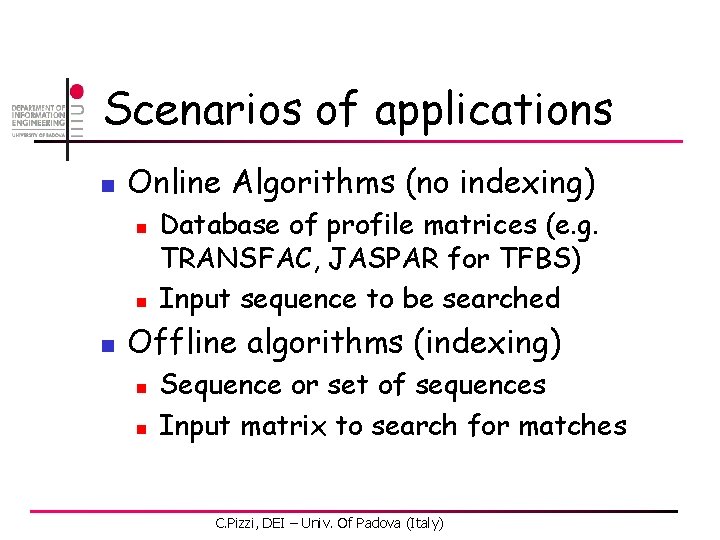
![ACExtension by LA n Second position C 0 1 1 2 3 4 5 AC-Extension by LA n Second position [C, 0. 1] 1 2 3 4 5](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-49.jpg)
AC-Extension by LA n Second position [C, 0. 1] 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 Pth [G, 0. 2] [T, 0. 4] [A, 0. 3] [A, 0. 1] [G, 0. 1] C. Pizzi, DEI – Univ. Of Padova (Italy) [T, 0. 3] [C, 0. 9]
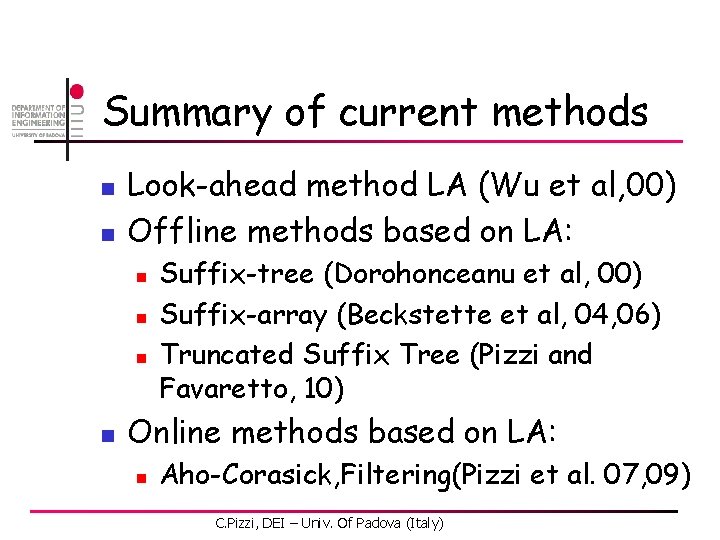
![ACExtension by LA n Third position C 0 1 1 2 3 4 5 AC-Extension by LA n Third position [C, 0. 1] 1 2 3 4 5](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-50.jpg)
AC-Extension by LA n Third position [C, 0. 1] 1 2 3 4 5 6 A 0. 3 0. 0 0. 1 0. 2 1. 0 0. 3 C 0. 1 0. 8 0. 5 0. 2 0. 0 0. 4 G 0. 2 0. 0 0. 4 0. 3 0. 0 T 0. 4 0. 2 0. 0 0. 3 -1. 0 -0. 2 0. 3 0. 6 1. 6 2. 0 Pth [G, 0. 2] [T, 0. 4] [A, 0. 3] [A, 0. 1] [G, 0. 5] [C, 0. 6] C. Pizzi, DEI – Univ. Of Padova (Italy) [T, 0. 3] [C, 0. 9]

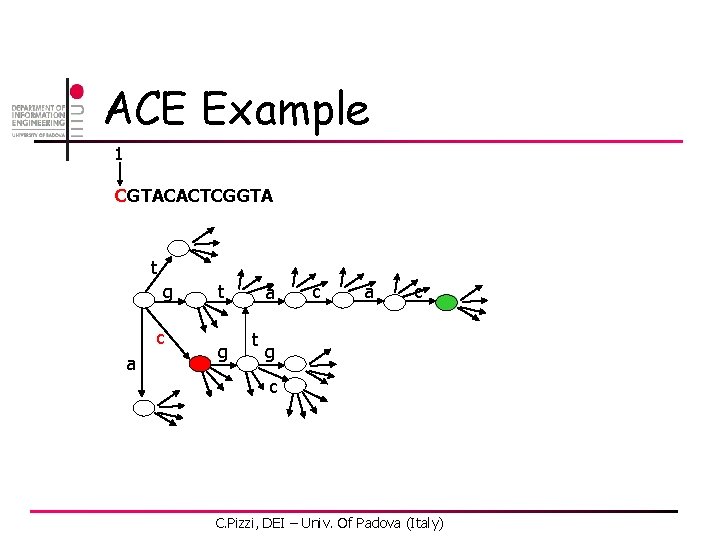
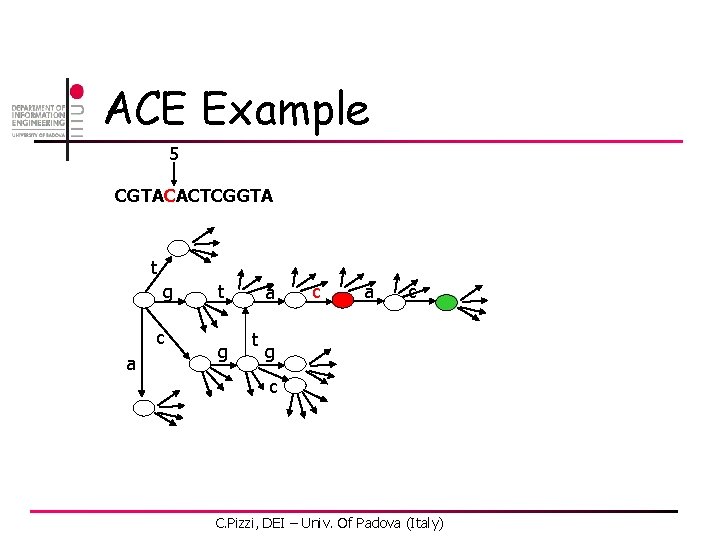
ACE Example 1 CGTACACTCGGTA t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

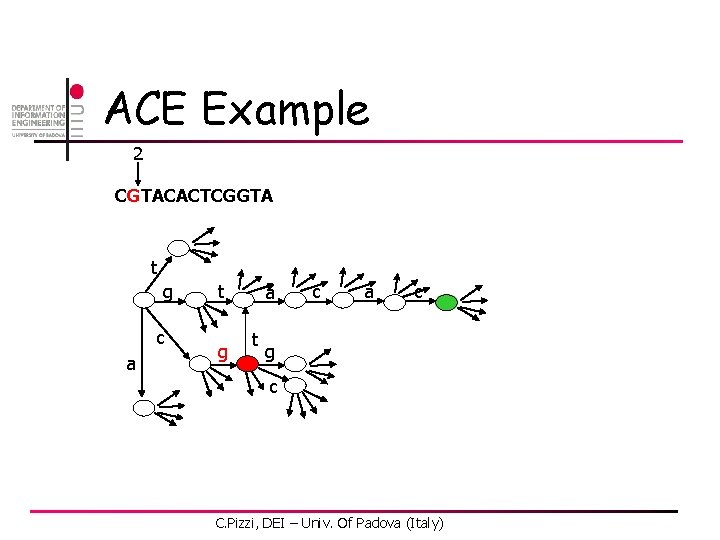
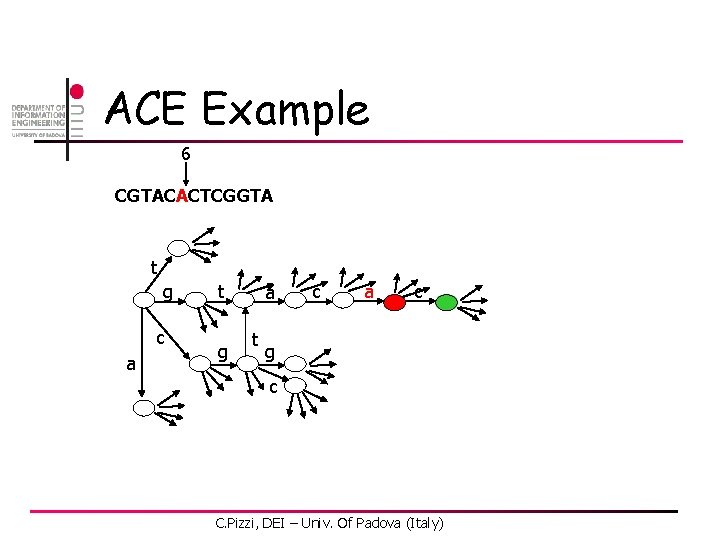
ACE Example 2 CGTACACTCGGTA t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

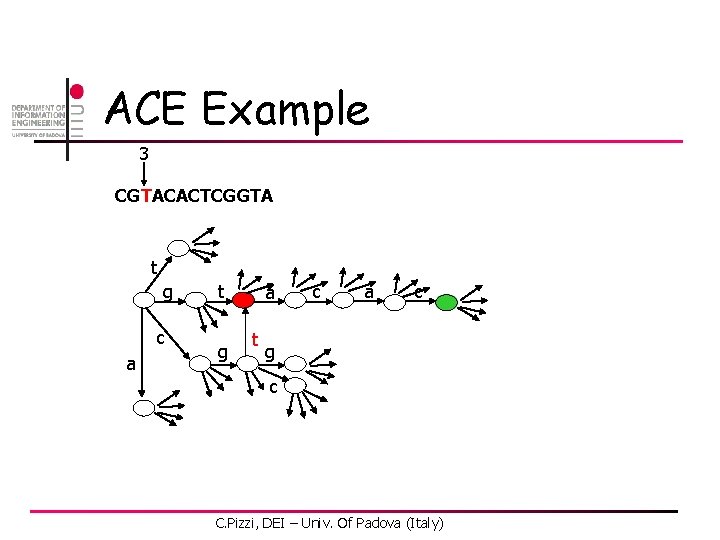
ACE Example 3 CGTACACTCGGTA t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

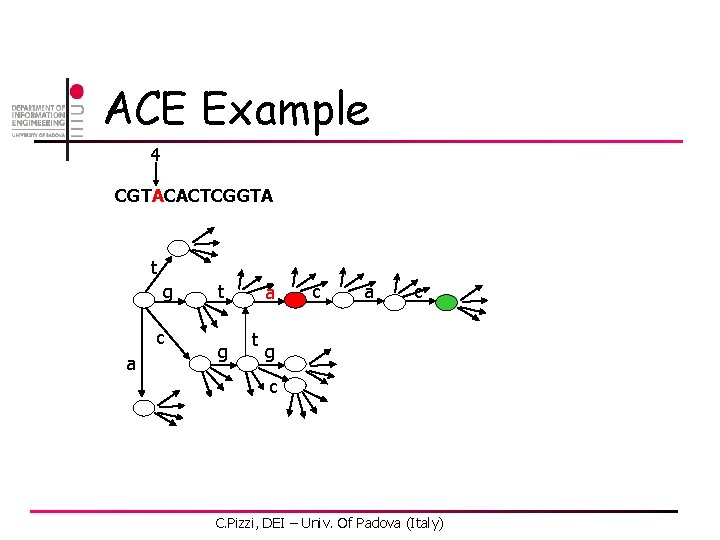
ACE Example 4 CGTACACTCGGTA t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

ACE Example 5 CGTACACTCGGTA t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

ACE Example 6 CGTACACTCGGTA t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

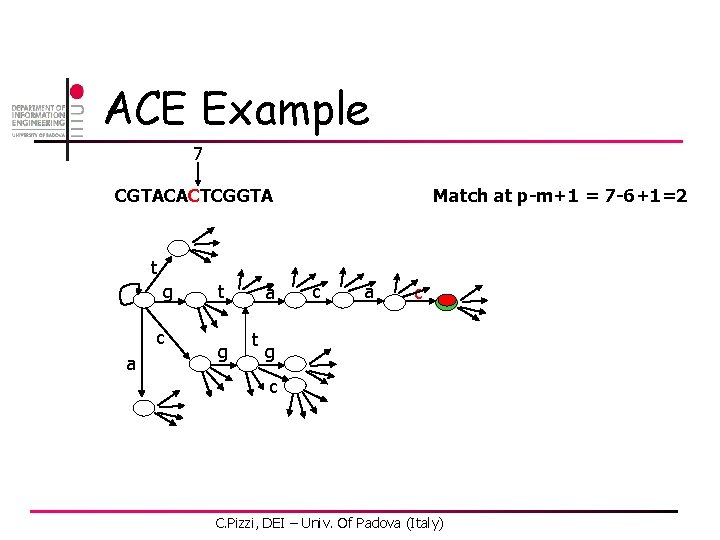
ACE Example 7 CGTACACTCGGTA Match at p-m+1 = 7 -6+1=2 t g c a t g a t c a c g c C. Pizzi, DEI – Univ. Of Padova (Italy)

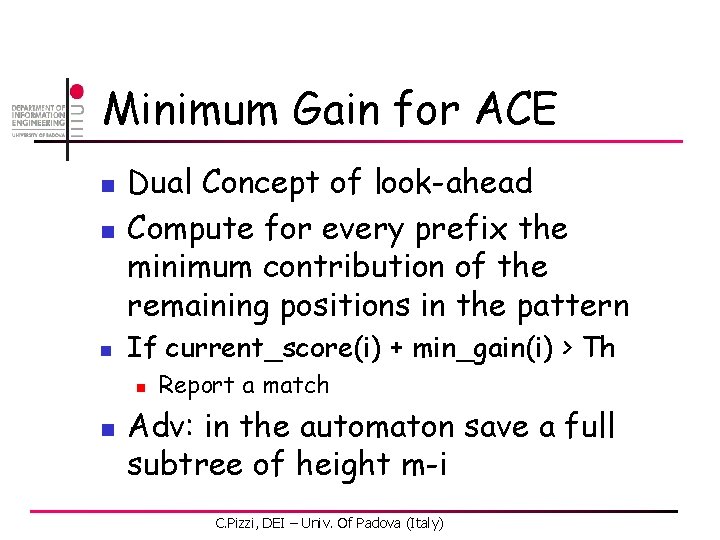
Minimum Gain for ACE n n n Dual Concept of look-ahead Compute for every prefix the minimum contribution of the remaining positions in the pattern If current_score(i) + min_gain(i) > Th n n Report a match Adv: in the automaton save a full subtree of height m-i C. Pizzi, DEI – Univ. Of Padova (Italy)
![Example M 0003 MSS0 85 G 18500 C Pizzi DEI Univ Of Padova Example: M 0003, MSS=0. 85 [G, 18500] C. Pizzi, DEI – Univ. Of Padova](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-59.jpg)
Example: M 0003, MSS=0. 85 [G, 18500] C. Pizzi, DEI – Univ. Of Padova (Italy)
![Example M 0003 MSS0 85 G 18500 C 37000 C Pizzi DEI Univ Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] C. Pizzi, DEI – Univ.](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-60.jpg)
Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] C. Pizzi, DEI – Univ. Of Padova (Italy)
![Example M 0003 MSS0 85 G 18500 C 37000 C 55500 GCC is Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • GCC is](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-61.jpg)
Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • GCC is sufficient to detect a match C. Pizzi, DEI – Univ. Of Padova (Italy) h=3
![Example M 0003 MSS0 85 G 18500 C 37000 C 55500 Save 5464 Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • Save 5464](https://slidetodoc.com/presentation_image/2ad59afc40105dc688c8712cd7b7a9d0/image-62.jpg)
Example: M 0003, MSS=0. 85 [G, 18500] [C, 37000] [C, 55500] • Save 5464 nodes out of 5468 C. Pizzi, DEI – Univ. Of Padova (Italy) h=3

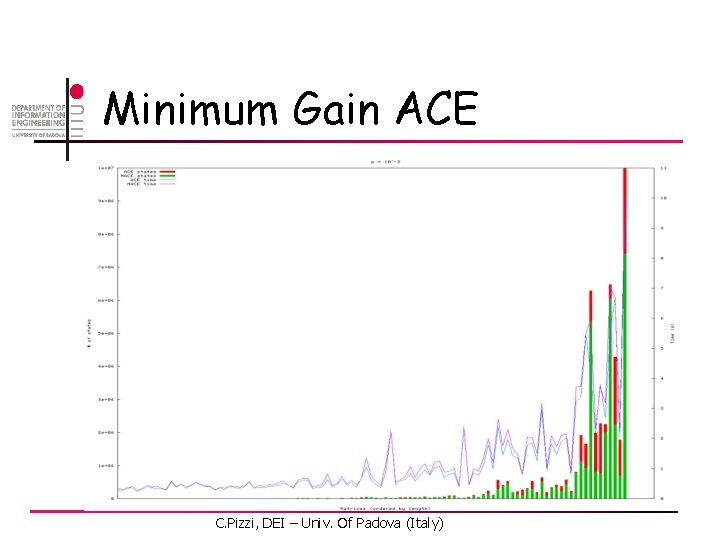
Minimum Gain ACE C. Pizzi, DEI – Univ. Of Padova (Italy)

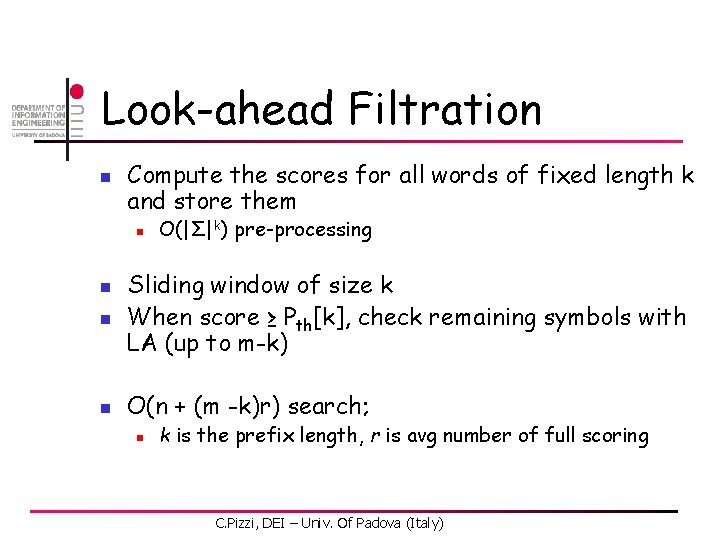
Look-ahead Filtration n Compute the scores for all words of fixed length k and store them n n O(|Σ|k) pre-processing Sliding window of size k When score ≥ Pth[k], check remaining symbols with LA (up to m-k) O(n + (m -k)r) search; n k is the prefix length, r is avg number of full scoring C. Pizzi, DEI – Univ. Of Padova (Italy)

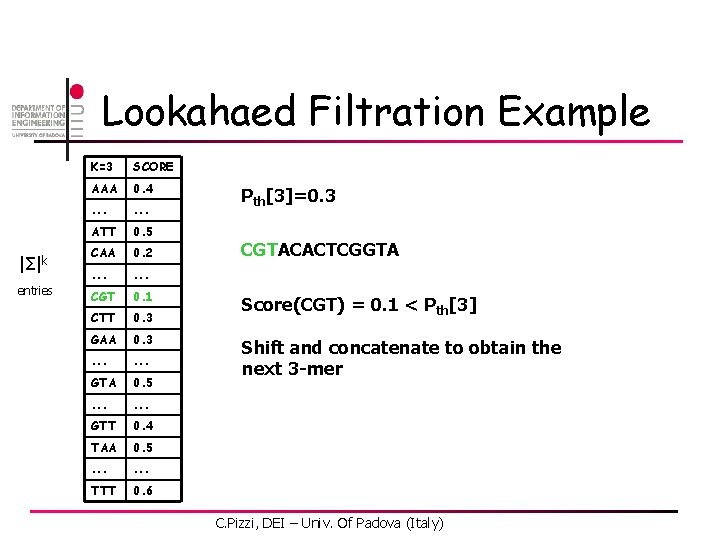
Lookahaed Filtration Example |Σ|k entries K=3 SCORE AAA 0. 4 . . . ATT 0. 5 CAA 0. 2 . . . CGT 0. 1 CTT 0. 3 GAA 0. 3 . . . GTA 0. 5 . . . GTT 0. 4 TAA 0. 5 . . . TTT 0. 6 Pth[3]=0. 3 CGTACACTCGGTA Score(CGT) = 0. 1 < Pth[3] Shift and concatenate to obtain the next 3 -mer C. Pizzi, DEI – Univ. Of Padova (Italy)

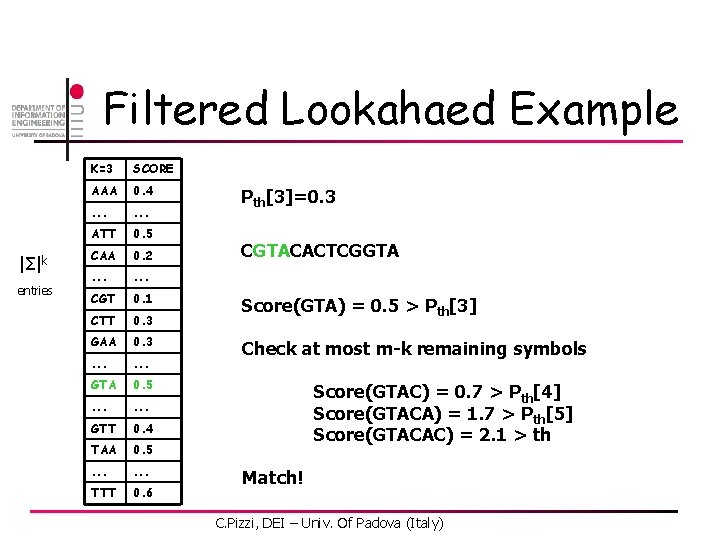
Filtered Lookahaed Example |Σ|k entries K=3 SCORE AAA 0. 4 . . . ATT 0. 5 CAA 0. 2 . . . CGT 0. 1 CTT 0. 3 GAA 0. 3 . . . GTA 0. 5 . . . GTT 0. 4 TAA 0. 5 . . . TTT 0. 6 Pth[3]=0. 3 CGTACACTCGGTA Score(GTA) = 0. 5 > Pth[3] Check at most m-k remaining symbols Score(GTAC) = 0. 7 > Pth[4] Score(GTACA) = 1. 7 > Pth[5] Score(GTACAC) = 2. 1 > th Match! C. Pizzi, DEI – Univ. Of Padova (Italy)

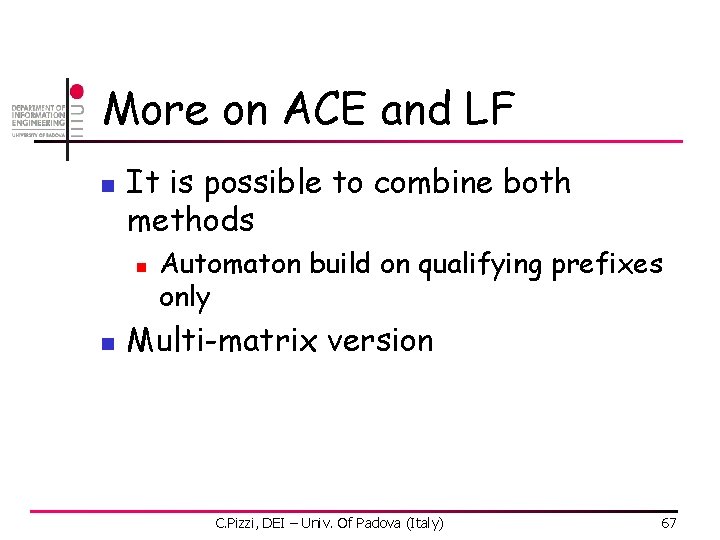
More on ACE and LF n It is possible to combine both methods n n Automaton build on qualifying prefixes only Multi-matrix version C. Pizzi, DEI – Univ. Of Padova (Italy) 67

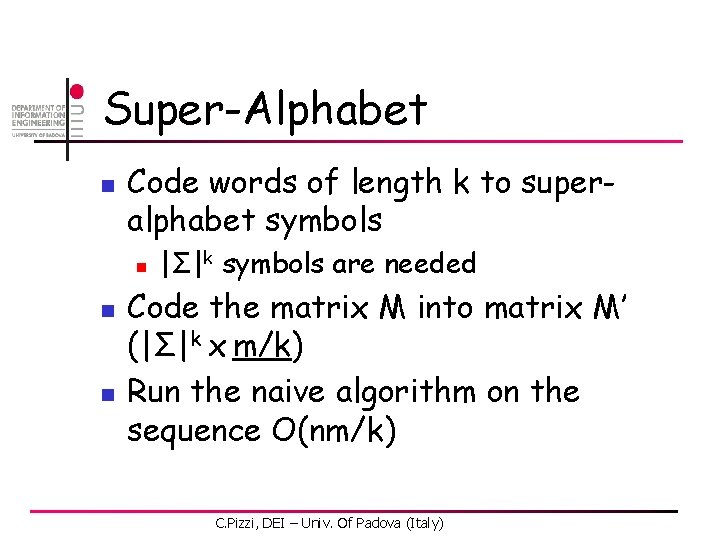
Super-Alphabet n Code words of length k to superalphabet symbols n n n |Σ|k symbols are needed Code the matrix M into matrix M’ (|Σ|k x m/k) Run the naive algorithm on the sequence O(nm/k) C. Pizzi, DEI – Univ. Of Padova (Italy)

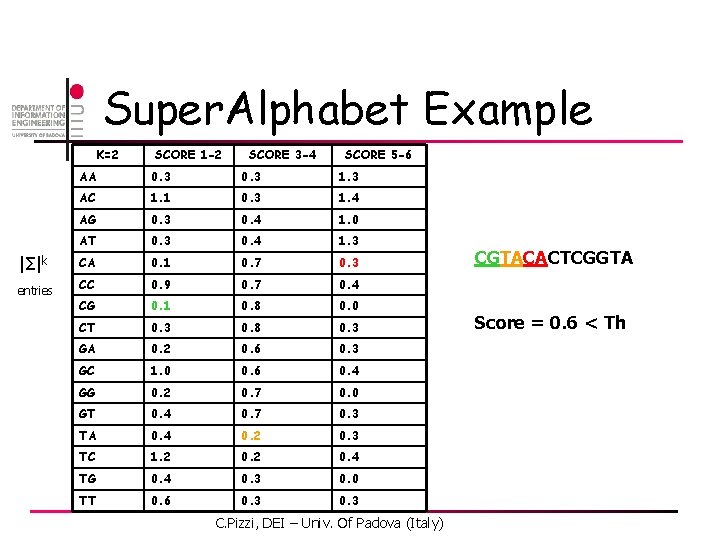
Super. Alphabet Example K=2 SCORE 1 -2 SCORE 3 -4 SCORE 5 -6 AA 0. 3 1. 3 AC 1. 1 0. 3 1. 4 AG 0. 3 0. 4 1. 0 AT 0. 3 0. 4 1. 3 |Σ|k CA 0. 1 0. 7 0. 3 entries CC 0. 9 0. 7 0. 4 CG 0. 1 0. 8 0. 0 CT 0. 3 0. 8 0. 3 GA 0. 2 0. 6 0. 3 GC 1. 0 0. 6 0. 4 GG 0. 2 0. 7 0. 0 GT 0. 4 0. 7 0. 3 TA 0. 4 0. 2 0. 3 TC 1. 2 0. 4 TG 0. 4 0. 3 0. 0 TT 0. 6 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) CGTACACTCGGTA Score = 0. 6 < Th

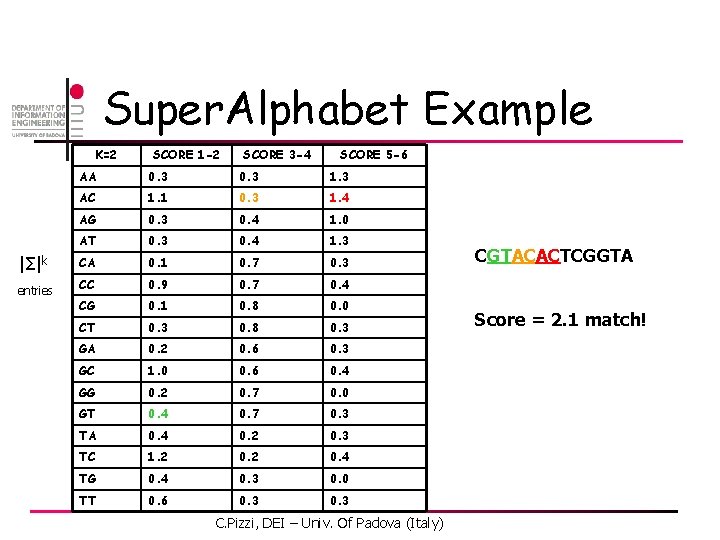
Super. Alphabet Example K=2 SCORE 1 -2 SCORE 3 -4 SCORE 5 -6 AA 0. 3 1. 3 AC 1. 1 0. 3 1. 4 AG 0. 3 0. 4 1. 0 AT 0. 3 0. 4 1. 3 |Σ|k CA 0. 1 0. 7 0. 3 entries CC 0. 9 0. 7 0. 4 CG 0. 1 0. 8 0. 0 CT 0. 3 0. 8 0. 3 GA 0. 2 0. 6 0. 3 GC 1. 0 0. 6 0. 4 GG 0. 2 0. 7 0. 0 GT 0. 4 0. 7 0. 3 TA 0. 4 0. 2 0. 3 TC 1. 2 0. 4 TG 0. 4 0. 3 0. 0 TT 0. 6 0. 3 C. Pizzi, DEI – Univ. Of Padova (Italy) CGTACACTCGGTA Score = 2. 1 match!

Experiments n n Jaspar Database: 123 TFBS matrices (DNA), PRINTS database (proteins) Test sequence about 50 M bases P-value defines threshold 3 GHz Intel Pentium IV processor with 2 gigabytes of main memory, running under Linux. C. Pizzi, DEI – Univ. Of Padova (Italy)

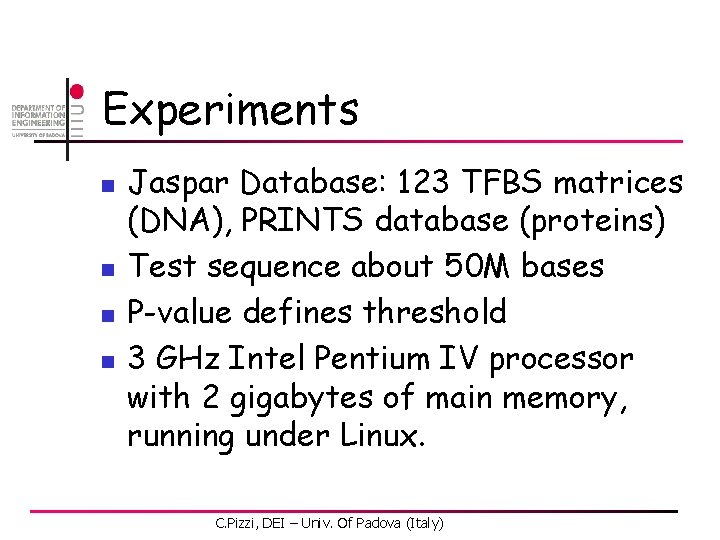
DNA – avg running times per matrix C. Pizzi, DEI – Univ. Of Padova (Italy) 72

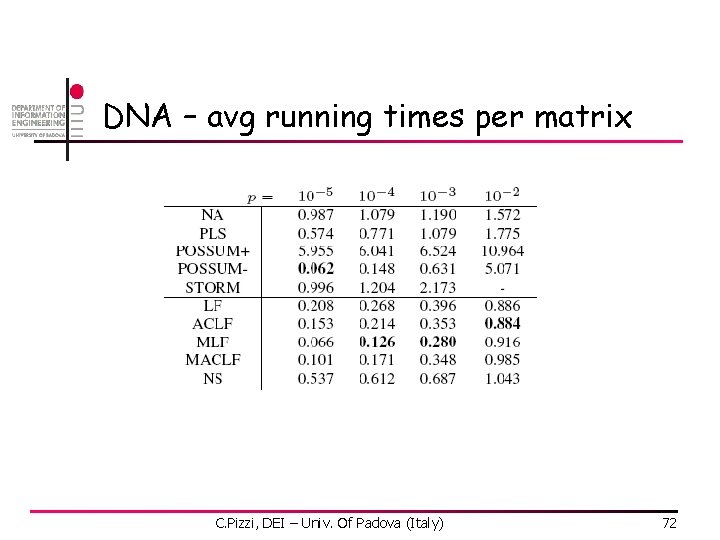
DNA- matrix length C. Pizzi, DEI – Univ. Of Padova (Italy) 73

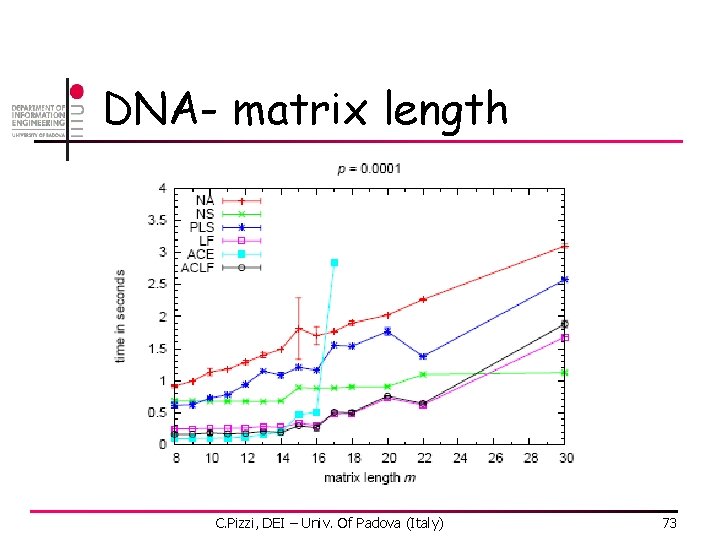
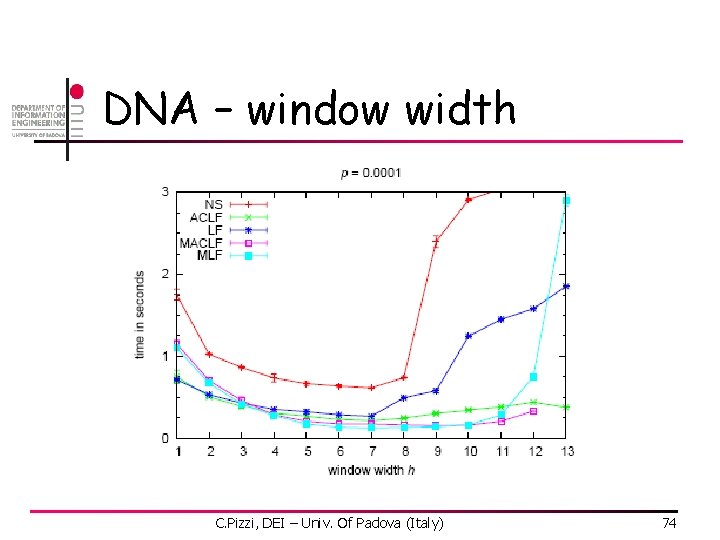
DNA – window width C. Pizzi, DEI – Univ. Of Padova (Italy) 74

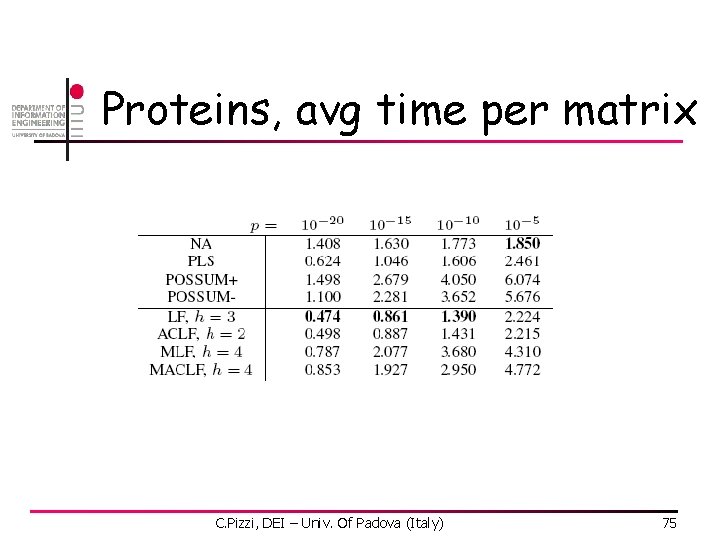
Proteins, avg time per matrix C. Pizzi, DEI – Univ. Of Padova (Italy) 75

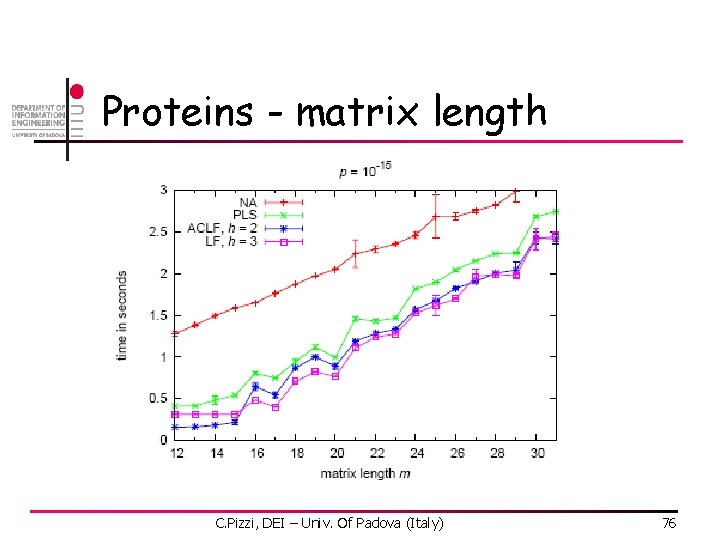
Proteins - matrix length C. Pizzi, DEI – Univ. Of Padova (Italy) 76

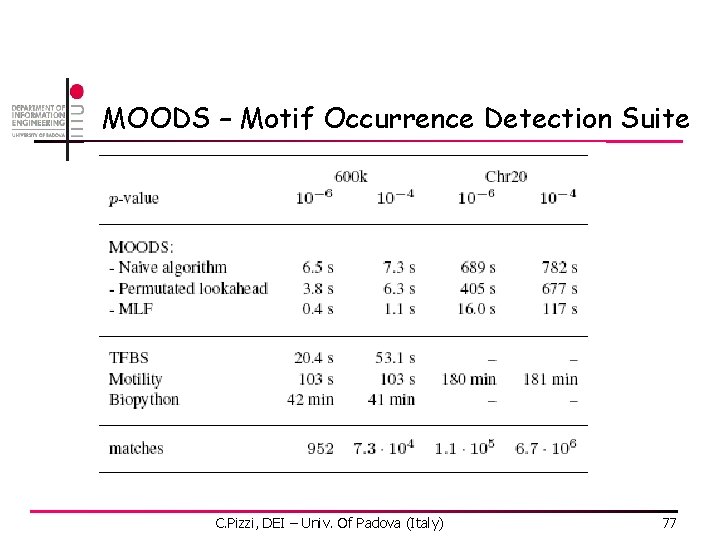
MOODS – Motif Occurrence Detection Suite C. Pizzi, DEI – Univ. Of Padova (Italy) 77

Conclusions n n n Searching matrix is a core step for many bioinformatics applications (searching, discovery, classification…) Several approaches have been developed in recent years Online methods based on filtering are currently the most efficient C. Pizzi, DEI – Univ. Of Padova (Italy) 78

References n n C. Pizzi, P. Rastas, E. Ukkonen Fast Search Algorithms for Position Specific Scoring Matrices In Proc. of the 1 st Conference on Bioinformatics Research and Development (BIRD 07), Berlin, Germany, March 2007, LNCS/LCBI 4414 pp 239 --250 C. Pizzi, E. Ukkonen Fast Profile Matching Algorithms - a survey Theoretical Computer Science, 395(2 -3), 2008, pp 137 --157, Special Issue SAIL: String Algorithms, Information and Learning C. Pizzi, P. Rastas, E. Ukkonen Finding significant matches of position weight matrices in linear time Accepted for publication by IEEE Transaction on Computational Biology and Bioinformatics, 2009 J. Korhonen, P. Martinmaki, C. Pizzi, P. Rastas, E. Ukkonen MOODS: fast search for position weight matrix matches in DNA sequences Bioinformatics 2009 25(23): 3181 -3182 C. Pizzi, DEI – Univ. Of Padova (Italy) 79

Thanks C. Pizzi, DEI – Univ. Of Padova (Italy) 80

Acknowledgements n Esko Ukkonen, Pasi Rastas, Janne Korhonen, P. Martinmaki n n n Academy of Finland grant “From Data to knowledge” EU Project “Regulatory Networks” Premio di Ricerca `Avere Trent’Anni’ n Univ. Padova, Parco Scientifico Galileo, Il Mattino, Giovani Confindustria, Scuola Galileiana di Studi Superiori C. Pizzi, DEI – Univ. Of Padova (Italy)

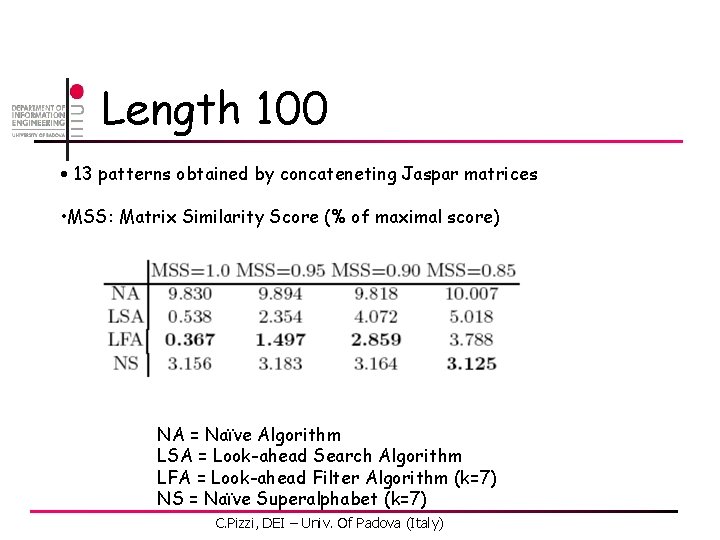
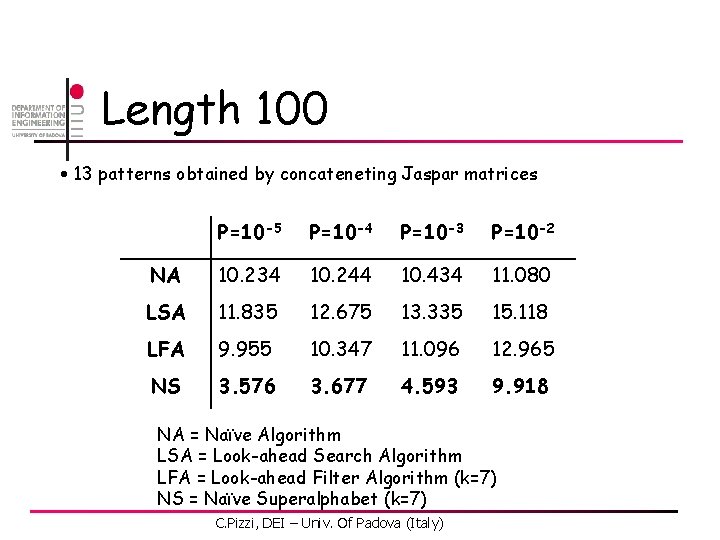
Length 100 • 13 patterns obtained by concateneting Jaspar matrices • MSS: Matrix Similarity Score (% of maximal score) NA = Naïve Algorithm LSA = Look-ahead Search Algorithm LFA = Look-ahead Filter Algorithm (k=7) NS = Naïve Superalphabet (k=7) C. Pizzi, DEI – Univ. Of Padova (Italy)

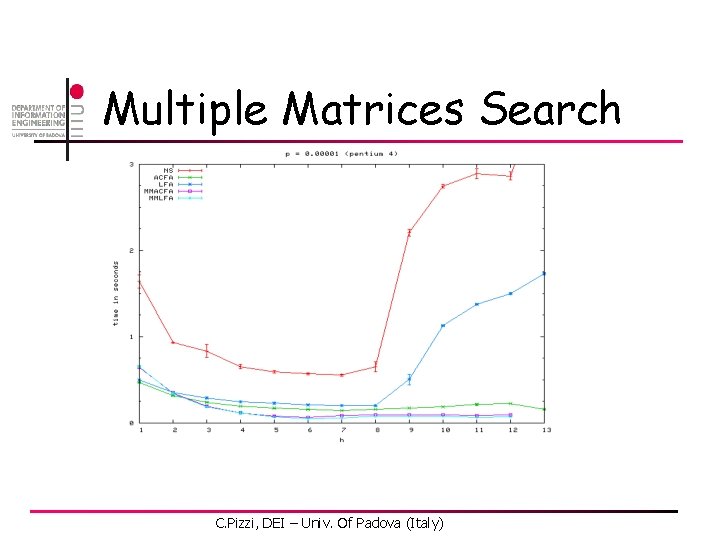
Multiple Matrices Search C. Pizzi, DEI – Univ. Of Padova (Italy)

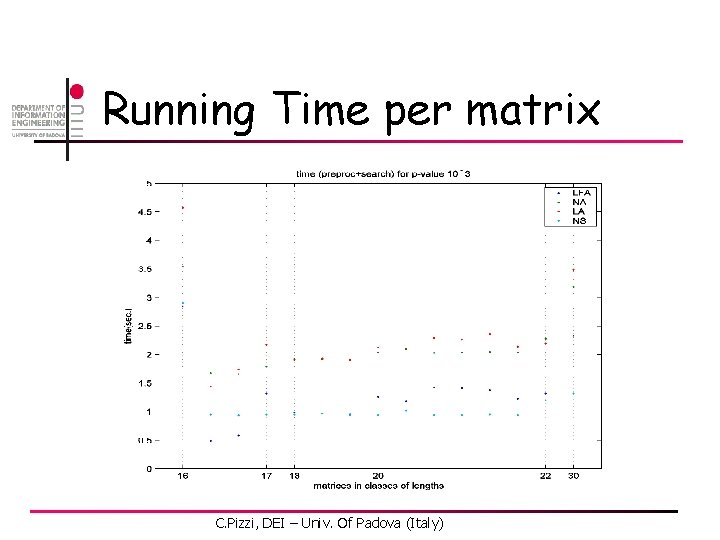
Running Time per matrix C. Pizzi, DEI – Univ. Of Padova (Italy)

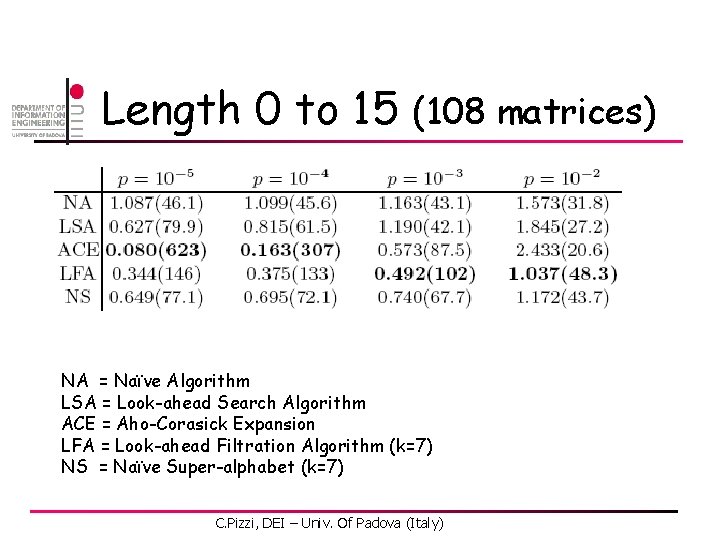
Length 0 to 15 (108 matrices) NA = Naïve Algorithm LSA = Look-ahead Search Algorithm ACE = Aho-Corasick Expansion LFA = Look-ahead Filtration Algorithm (k=7) NS = Naïve Super-alphabet (k=7) C. Pizzi, DEI – Univ. Of Padova (Italy)

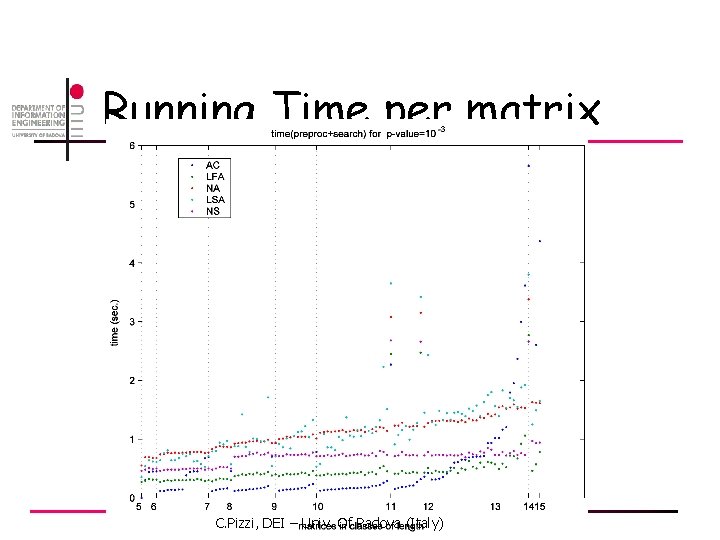
Running Time per matrix C. Pizzi, DEI – Univ. Of Padova (Italy)

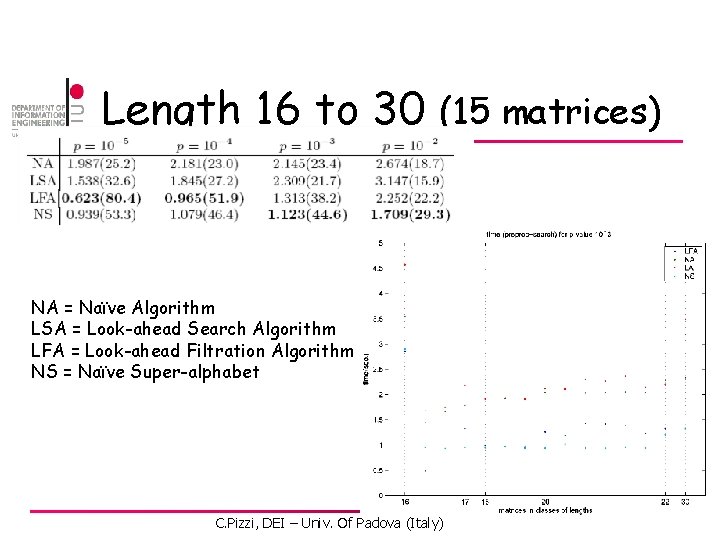
Length 16 to 30 (15 matrices) NA = Naïve Algorithm LSA = Look-ahead Search Algorithm LFA = Look-ahead Filtration Algorithm NS = Naïve Super-alphabet C. Pizzi, DEI – Univ. Of Padova (Italy)

Length 100 • 13 patterns obtained by concateneting Jaspar matrices P=10 -5 P=10 -4 P=10 -3 P=10 -2 NA 10. 234 10. 244 10. 434 11. 080 LSA 11. 835 12. 675 13. 335 15. 118 LFA 9. 955 10. 347 11. 096 12. 965 NS 3. 576 3. 677 4. 593 9. 918 NA = Naïve Algorithm LSA = Look-ahead Search Algorithm LFA = Look-ahead Filter Algorithm (k=7) NS = Naïve Superalphabet (k=7) C. Pizzi, DEI – Univ. Of Padova (Italy)

Motif Representation n Istances of a biological signal are different ACATAC CCGAAT ATGCAT GCCTAC TCCAAA TTCGAA ACGGAC TCCTAT GCCCAC TCGGAA TCC(G|T)AC Consensus -> pattern representation 1 2 3 4 5 6 A C G T Profile -> matrix representation C. Pizzi, DEI – Univ. Of Padova (Italy)