Scoring Matrices Scoring matrices PSSMs and HMMs Reading








- Slides: 23

Scoring Matrices Scoring matrices, PSSMs, and HMMs Reading: Ch 6. 1 BIO 520 Bioinformatics Jim Lund

Alignment scoring matrix • DNA matrix: A C G T 5 -4 -4 5 -4 -4 5

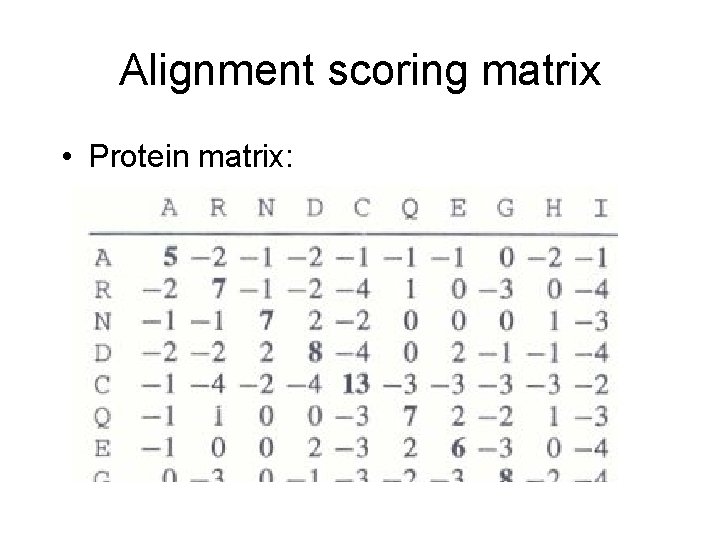
Alignment scoring matrix • Protein matrix:

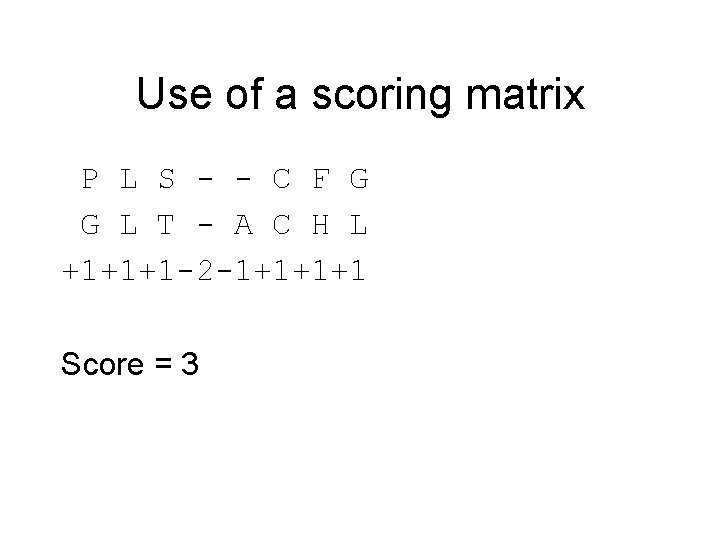
Use of a scoring matrix P L S - - C F G G L T - A C H L +1+1+1 -2 -1+1+1+1 Score = 3

Consensus sequences Different ways to describe a consensus, from crude to refined: • • Consensus site Sequence logos Position Specific Score Matrix (PSSM) Hidden Markov Model (HMM)

Consensus sequences and sequence logos GTMGFGLPAAIGAKLARPDRRVVAIDGDGSFQMTVQELST Consensus sequence Sequence logo

Constructing (and using) a consensus sequence 1. Collect sequences 2. Align sequences (consensus sites are descriptions of the alignment) 3. Condense the set of sequences into a consensus (to a consensus, PSSM, HMM). 4. Apply the scoring matrix in alignments/searches.

Position Specific Score Matrix (PSSM) • A position specific scoring matrix (PSSM) is a matrix based on the amino acid frequencies (or nucleic acid frequencies) at every position of a multiple alignment. • From these frequencies, the PSSM that will be calculated will result in a matrix that will assign superior scores to residues that appear more often than by chance at a certain position.

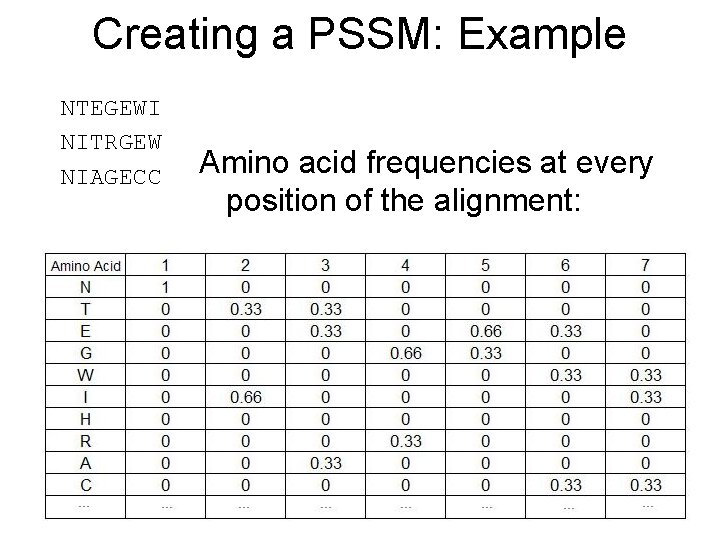
Creating a PSSM: Example NTEGEWI NITRGEW NIAGECC Amino acid frequencies at every position of the alignment:

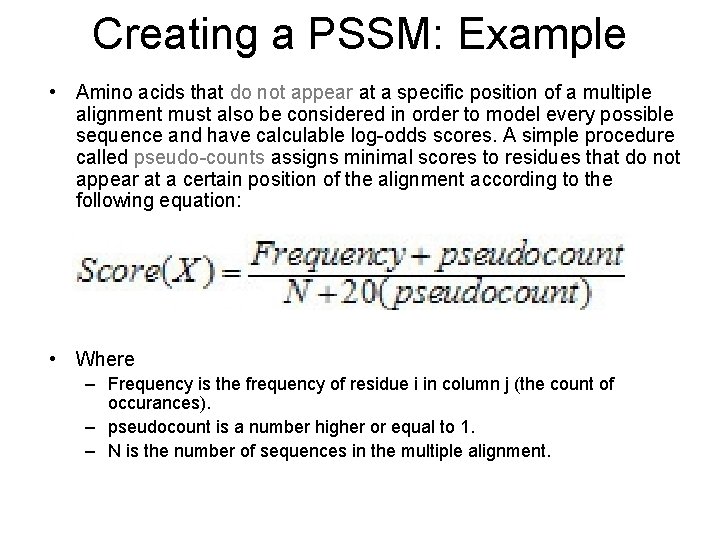
Creating a PSSM: Example • Amino acids that do not appear at a specific position of a multiple alignment must also be considered in order to model every possible sequence and have calculable log-odds scores. A simple procedure called pseudo-counts assigns minimal scores to residues that do not appear at a certain position of the alignment according to the following equation: • Where – Frequency is the frequency of residue i in column j (the count of occurances). – pseudocount is a number higher or equal to 1. – N is the number of sequences in the multiple alignment.

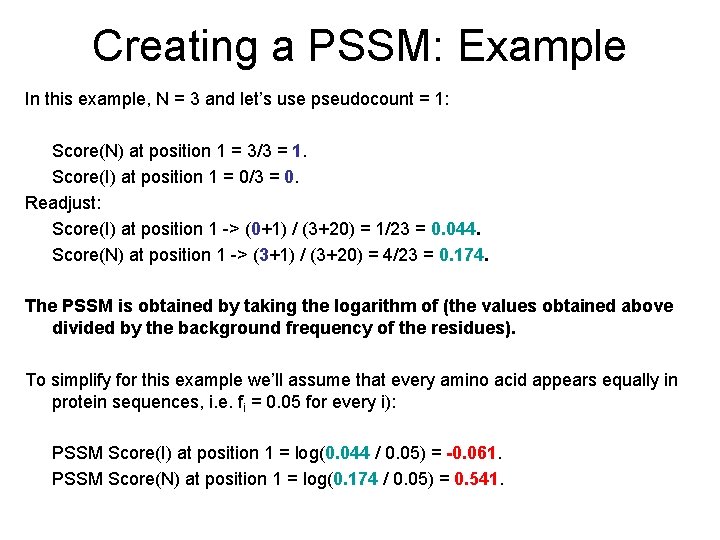
Creating a PSSM: Example In this example, N = 3 and let’s use pseudocount = 1: Score(N) at position 1 = 3/3 = 1. Score(I) at position 1 = 0/3 = 0. Readjust: Score(I) at position 1 -> (0+1) / (3+20) = 1/23 = 0. 044. Score(N) at position 1 -> (3+1) / (3+20) = 4/23 = 0. 174. The PSSM is obtained by taking the logarithm of (the values obtained above divided by the background frequency of the residues). To simplify for this example we’ll assume that every amino acid appears equally in protein sequences, i. e. fi = 0. 05 for every i): PSSM Score(I) at position 1 = log(0. 044 / 0. 05) = -0. 061. PSSM Score(N) at position 1 = log(0. 174 / 0. 05) = 0. 541.

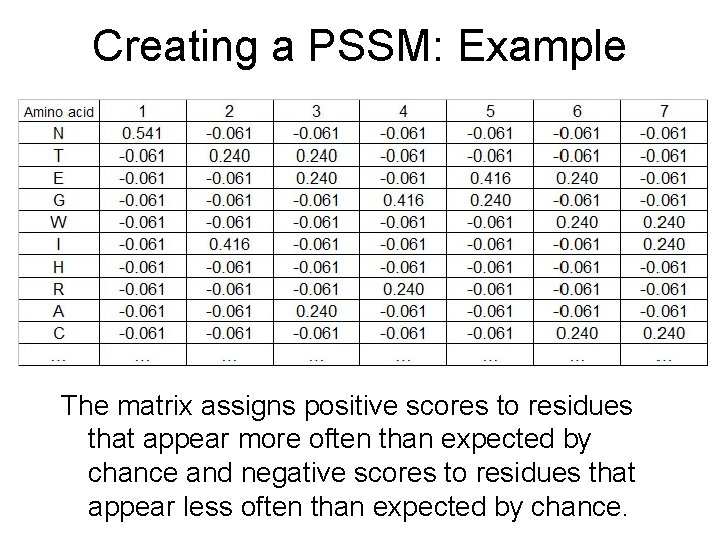
Creating a PSSM: Example The matrix assigns positive scores to residues that appear more often than expected by chance and negative scores to residues that appear less often than expected by chance.

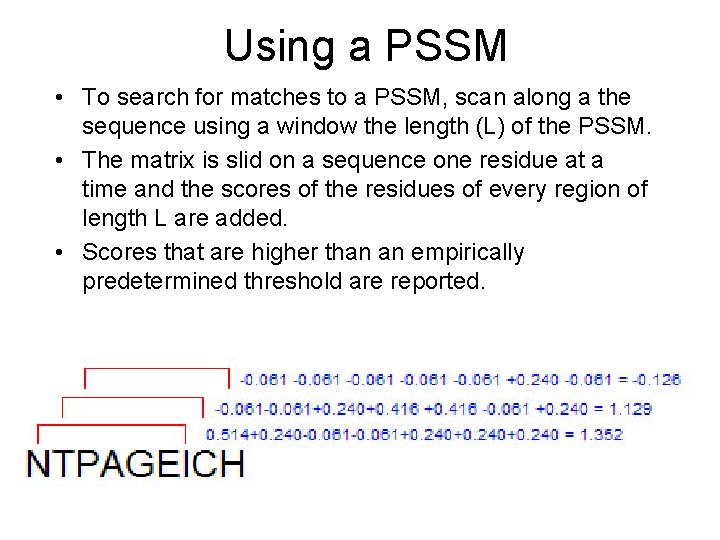
Using a PSSM • To search for matches to a PSSM, scan along a the sequence using a window the length (L) of the PSSM. • The matrix is slid on a sequence one residue at a time and the scores of the residues of every region of length L are added. • Scores that are higher than an empirically predetermined threshold are reported.

Advantages of PSSM • Weights sequence according to observed diversity specific to the family of interest • Minimal assumptions • Easy to compute • Can be used in comprehensive evaluations.

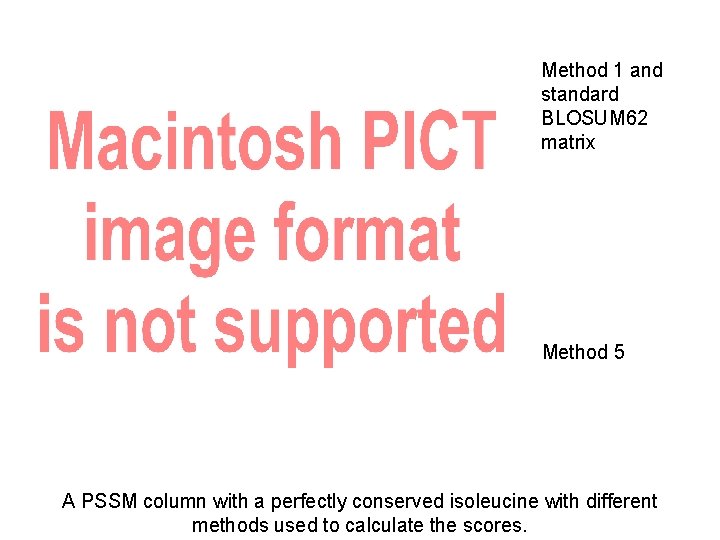
More sophisticated PSSMs From less to more complicated 1. PSSM with pseudocounts. 2. Giving pseudocounts less weight when more alignment data is available. 3. Weight pseudocount amino acids by their frequency of occurrence in proteins. 4. Instead of giving pseudocounts all the same value, weight them by their similarity to the consensus (like BLOSUM 62 does) at each position. (PSI-BLAST method). 5. Combine 2 & 4 (Dirichlet mixture method).

Method 1 and standard BLOSUM 62 matrix Method 5 A PSSM column with a perfectly conserved isoleucine with different methods used to calculate the scores.

Using Hidden Markov models to describe sequence alignment profiles • A profile HMM can represent a sequence alignment profile similar to how a PSSM does. • A profile HMM includes information on the amino acid consensus at each position in the alignment like a PSSM. • A profile HMM also has position-specific scores for gap insertion and extensions.

Background: Creating HMMs To create an HMM to model data we need to determine two things: • The structure/topology of the HMM—states and transitions • The values of the parameters—emission and transition probabilities. • Determining the parameters is called “training”.

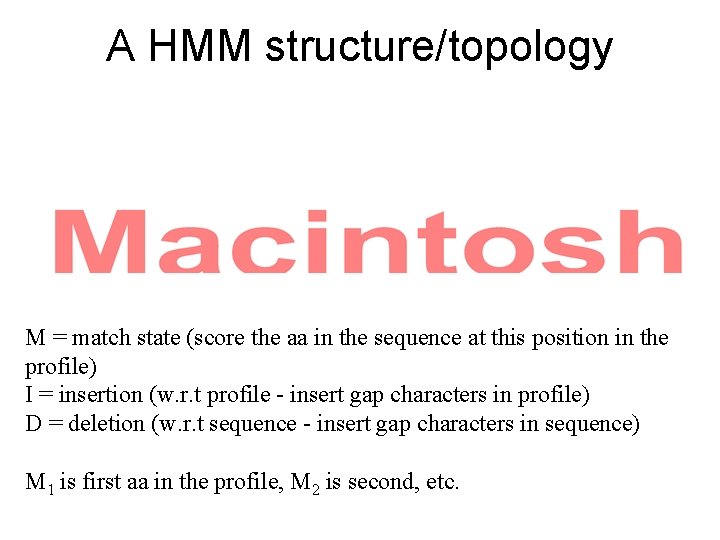
A HMM structure/topology M = match state (score the aa in the sequence at this position in the profile) I = insertion (w. r. t profile - insert gap characters in profile) D = deletion (w. r. t sequence - insert gap characters in sequence) M 1 is first aa in the profile, M 2 is second, etc.

Example HMMER parameters NULE 595 -1558 85 338 -294 453 -1158 (. . . ) -21 -313 45 531 201 384 HMM A C D E F G H (. . . ) m->m m->i m->d i->m i->i d->m d->d b->m m->e 1 -1084 390 -8597 -8255 -5793 -8424 -8268 (. . . ) 1 - -149 -500 233 43 -381 399 106 (. . . ) C -1 -11642 -12684 -894 -1115 -701 -1378 -16 * 2 -2140 -3785 -6293 -2251 3226 -2495 -727 (. . . ) 2 - -149 -500 233 43 -381 399 106 (. . . ) C -1 -11642 -12684 -894 -1115 -701 -1378 * * (. . . ) 76 -2255 -5128 -302 363 -784 -2353 1398 (. . . ) 103 - -149 -500 233 43 -381 399 106 (. . . ) E -1 -11642 -12684 -894 -1115 -701 -1378 * * 77 -633 879 -2198 -5620 -1457 -5498 -4367 (. . . ) 104 - * * * * (. . . ) C * * * * 0 //

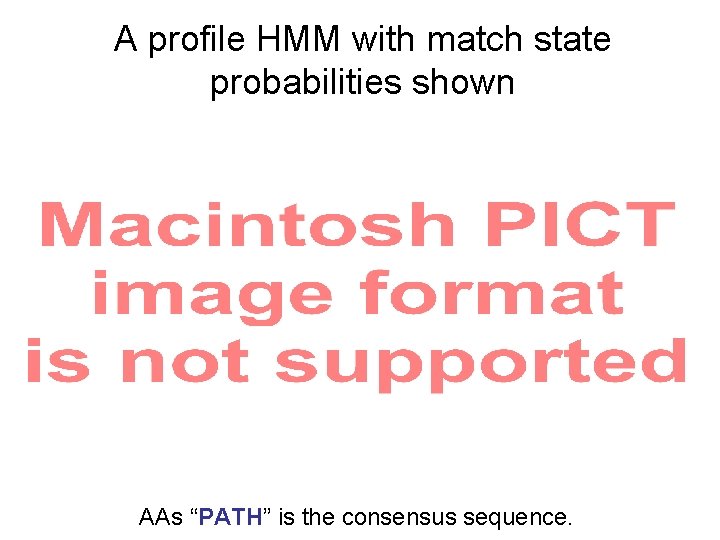
A profile HMM with match state probabilities shown AAs “PATH” is the consensus sequence.

Building a profile HMM • Pick a HMM structure/topology. • Estimate initial parameters. • Train the HMM by running sequences through it. • Transitions that get used are given higher probabilities, those rarely used are given lower probabilities.

Protein profile HMMs • Better (in theory) representations than PSSMs. – More complicated. – Not hand-tuned by curators. • Used in some protein profile databases: – Pfam (http: //pfam. sanger. ac. uk/) – SMART (http: //smart. embl-heidelberg. de/) • Difficult to describe in human readable formats. Schuster-Böckler et al. , 2004 (http: //www. biomedcentral. com/1471 -2105/5/7)