Wellcome Trust Advanced Courses Genomic Epidemiology in Africa
















- Slides: 71

Wellcome Trust Advanced Courses; Genomic Epidemiology in Africa, 21 st – 26 th June 2015 Africa Centre for Health and Population Studies, University of Kwa. Zulu-Natal, Durban, South Africa Population genetics Dr Gavin Band

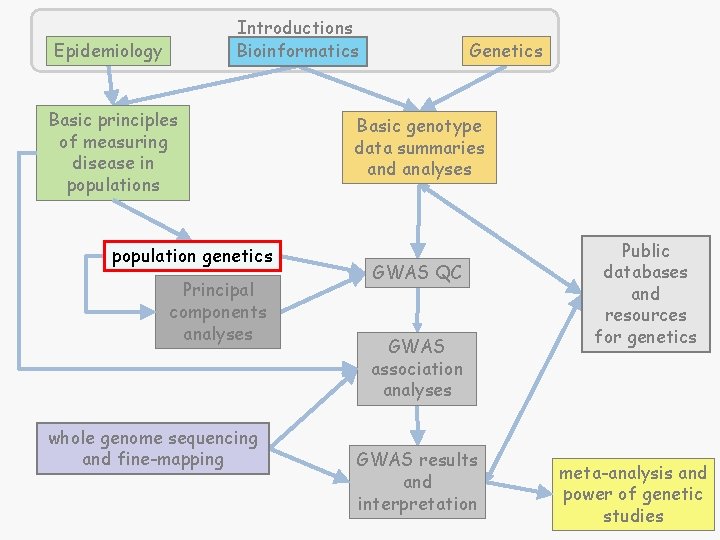
Introductions Bioinformatics Epidemiology Basic principles of measuring disease in populations population genetics Principal components analyses whole genome sequencing and fine-mapping Genetics Basic genotype data summaries and analyses GWAS QC GWAS association analyses GWAS results and interpretation Public databases and resources for genetics meta-analysis and power of genetic studies

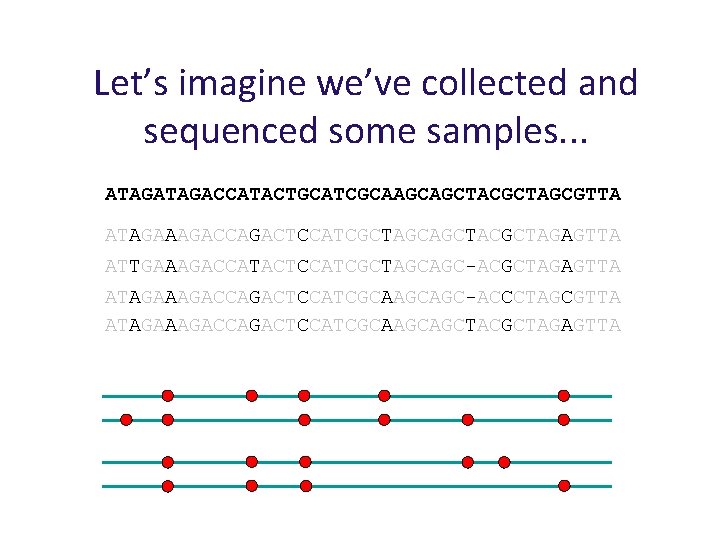
Let’s imagine we’ve collected and sequenced some samples. . . ATAGACCATACTGCATCGCAAGCAGCTACGCTAGCGTTA K samples ATAGAAAGACCAGACTCCATCGCTAGCAGCTACGCTAGAGTTA ATTGAAAGACCATACTCCATCGCTAGCAGC-ACGCTAGAGTTA ATAGAAAGACCAGACTCCATCGCAAGCAGC-ACCCTAGCGTTA ATAGAAAGACCAGACTCCATCGCAAGCAGCTACGCTAGAGTTA. . .

Let’s imagine we’ve collected and sequenced some samples. . . ATAGACCATACTGCATCGCAAGCAGCTACGCTAGCGTTA ATAGAAAGACCAGACTCCATCGCTAGCAGCTACGCTAGAGTTA ATTGAAAGACCATACTCCATCGCTAGCAGC-ACGCTAGAGTTA ATAGAAAGACCAGACTCCATCGCAAGCAGC-ACCCTAGCGTTA ATAGAAAGACCAGACTCCATCGCAAGCAGCTACGCTAGAGTTA SNPs Insertion / deletion polymorphism

Let’s imagine we’ve collected and sequenced some samples. . . ATAGACCATACTGCATCGCAAGCAGCTACGCTAGCGTTA ATAGAAAGACCAGACTCCATCGCTAGCAGCTACGCTAGAGTTA ATTGAAAGACCATACTCCATCGCTAGCAGC-ACGCTAGAGTTA ATAGAAAGACCAGACTCCATCGCAAGCAGC-ACCCTAGCGTTA ATAGAAAGACCAGACTCCATCGCAAGCAGCTACGCTAGAGTTA

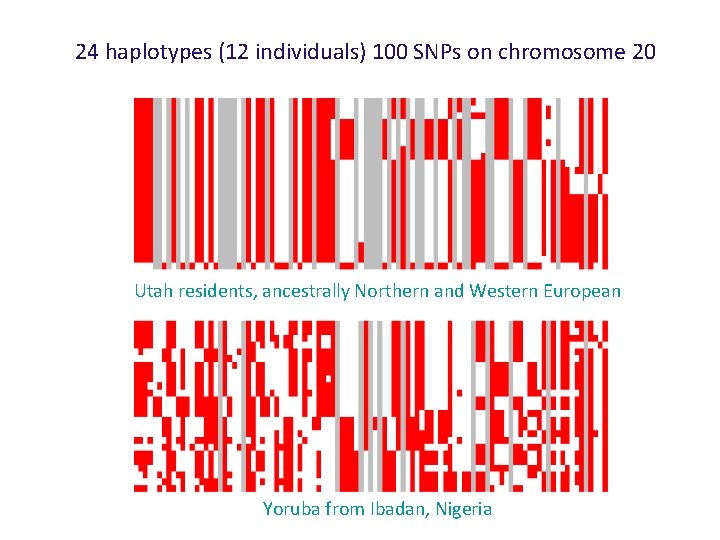
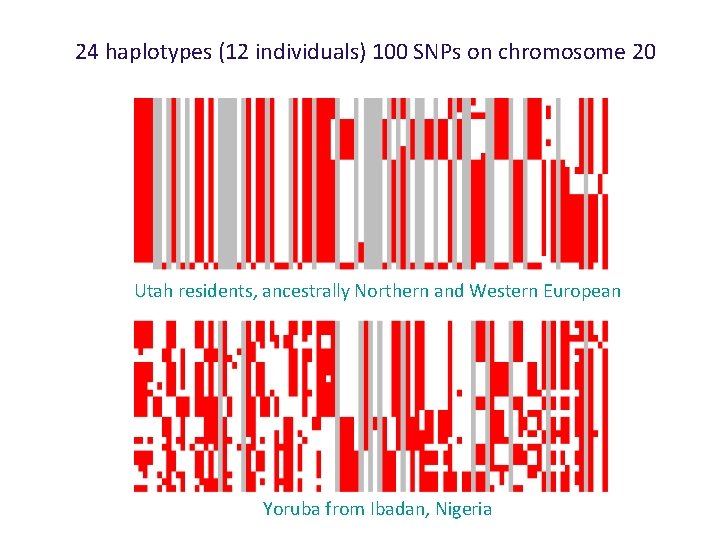
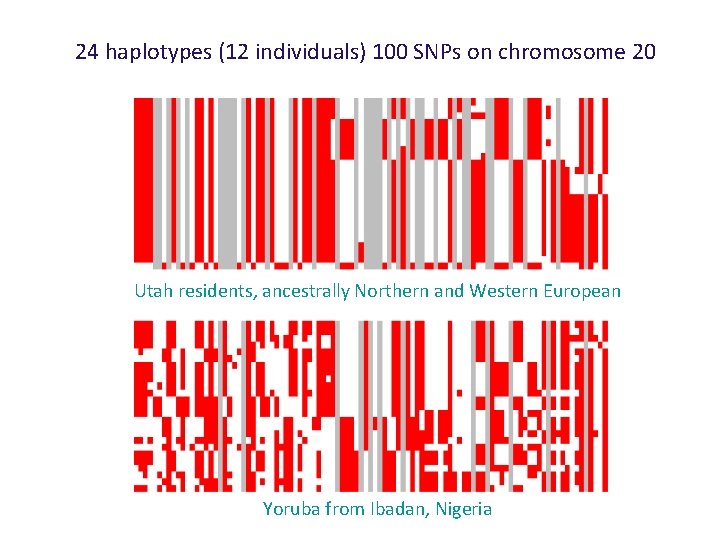
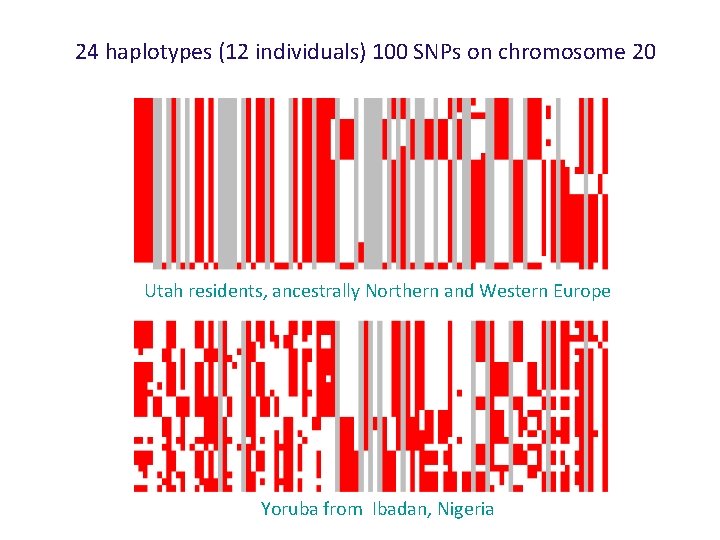
24 haplotypes (12 individuals) 100 SNPs on chromosome 20 Utah residents, ancestrally Northern and Western European Yoruba from Ibadan, Nigeria

Key questions • What should we expect to observe? • How can we interpret observed patterns? • What processes generated this data?

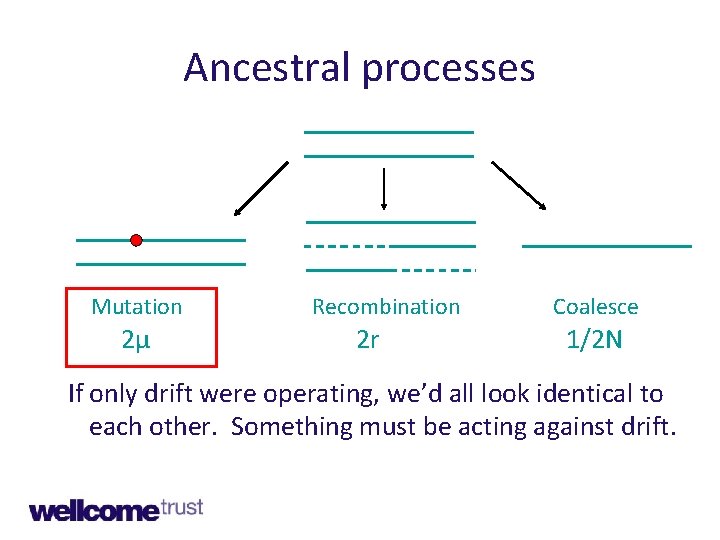
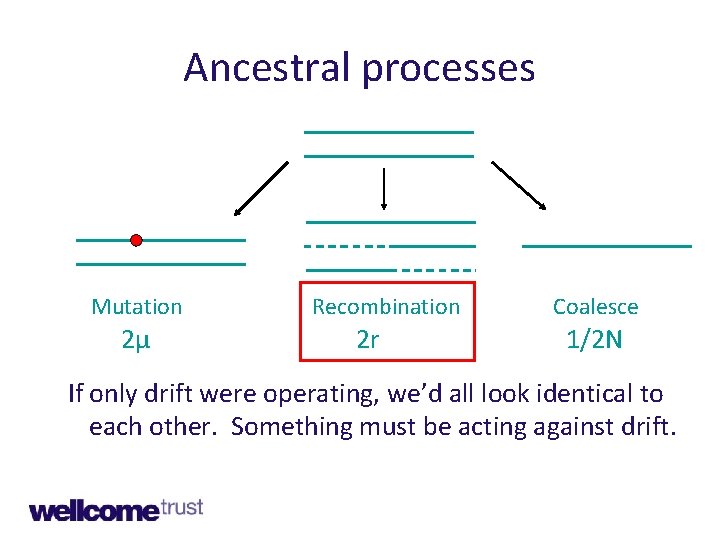
Key ancestral processes • Genetic drift • Mutation • Recombination • (and selection)

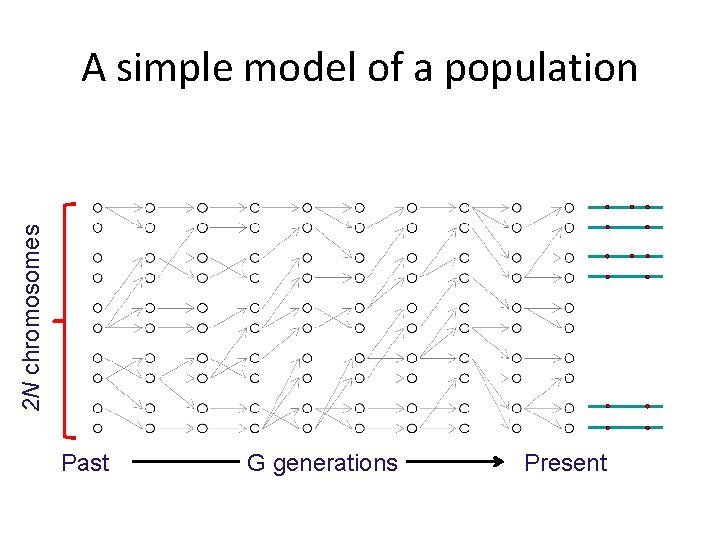
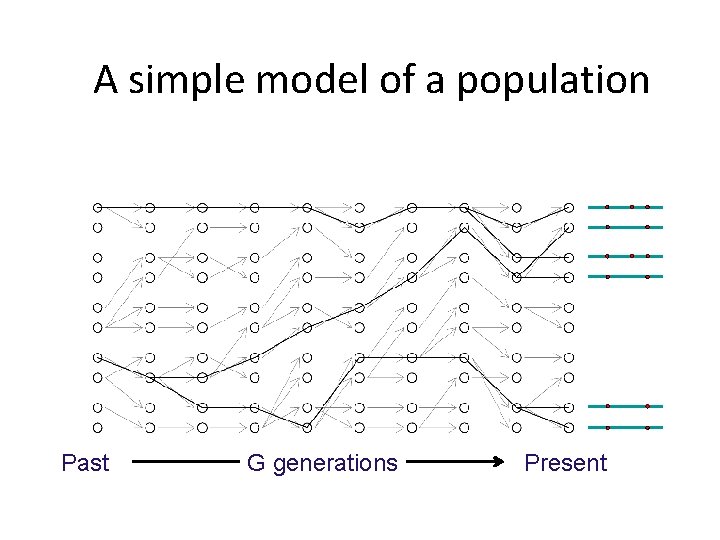
2 N chromosomes A simple model of a population Past G generations Present

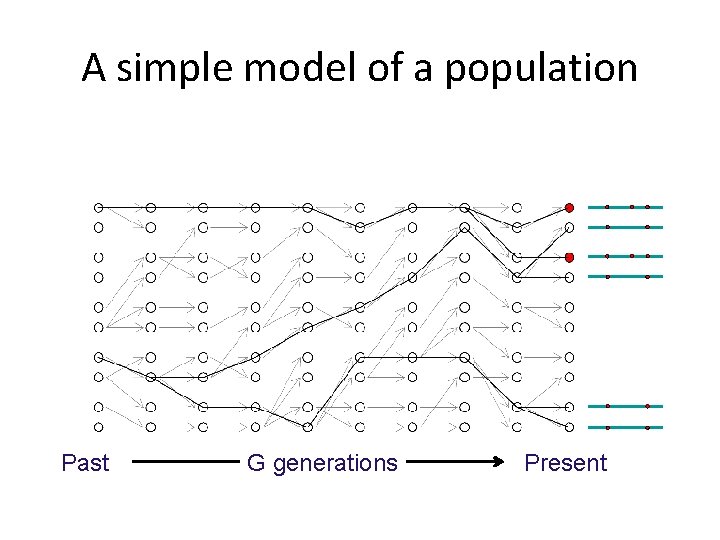
A simple model of a population Past G generations Present

A simple model of a population Past G generations Present

A simple model of a population Past G generations Present

A simple model of a population Past G generations Present

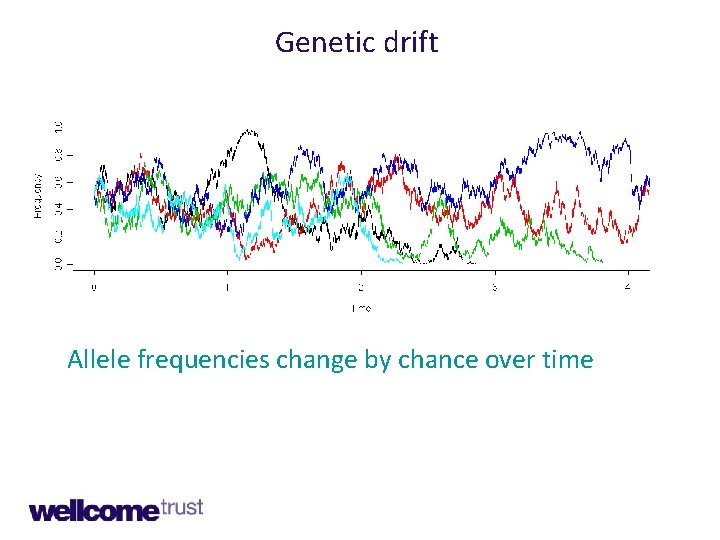
Genetic drift

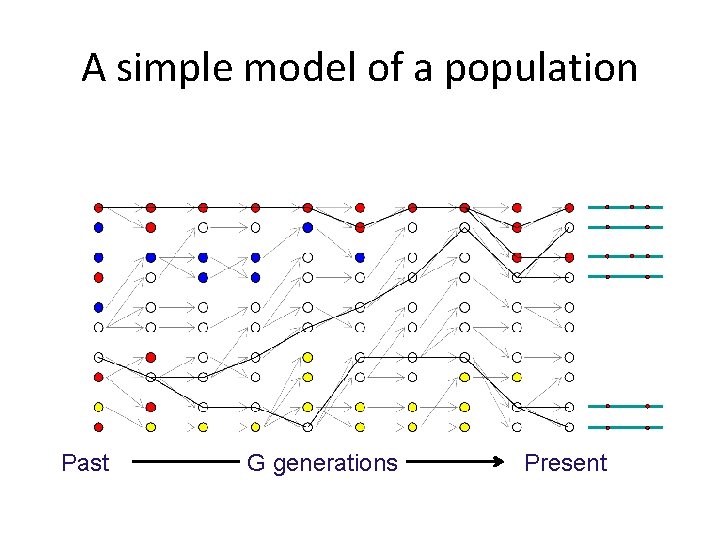
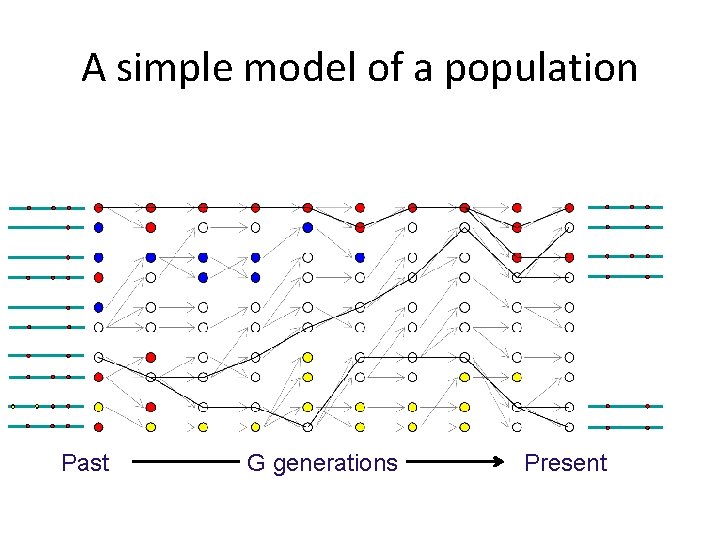
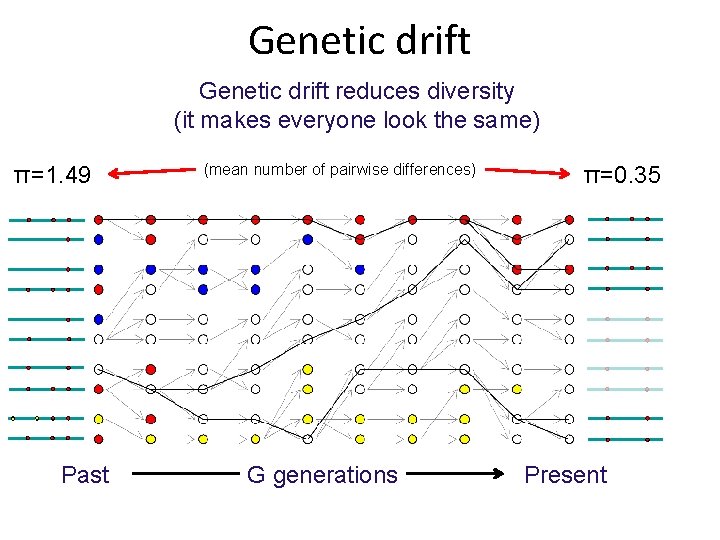
Genetic drift reduces diversity (it makes everyone look the same) π=1. 49 Past (mean number of pairwise differences) G generations π=0. 35 Present

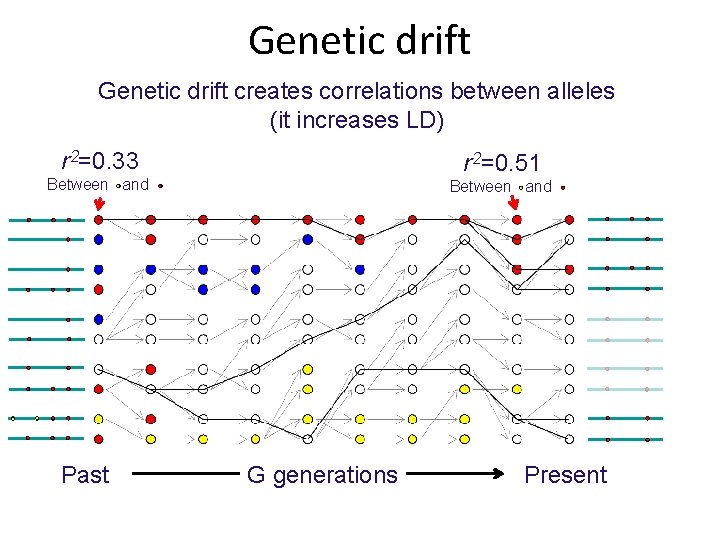
Genetic drift creates correlations between alleles (it increases LD) r 2=0. 33 r 2=0. 51 Between and Past Between and G generations Present

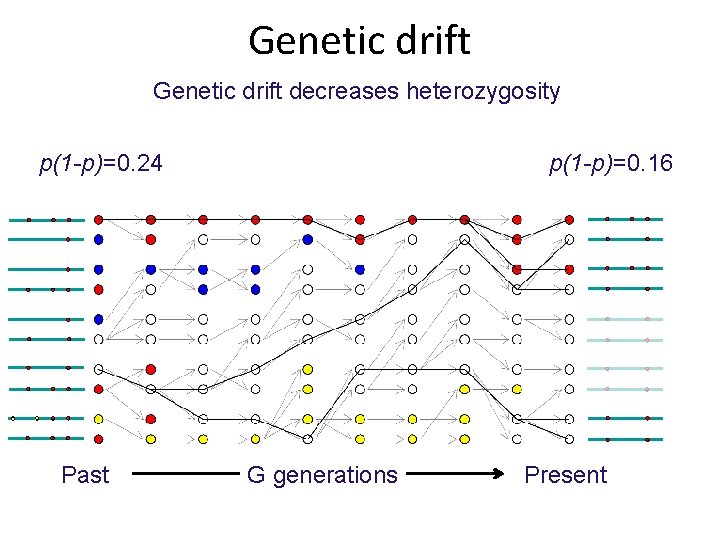
Genetic drift decreases heterozygosity p(1 -p)=0. 24 Past p(1 -p)=0. 16 G generations Present

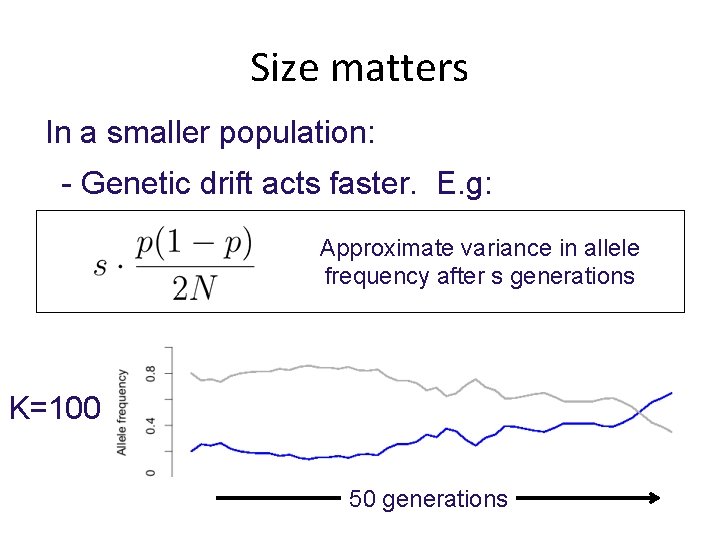
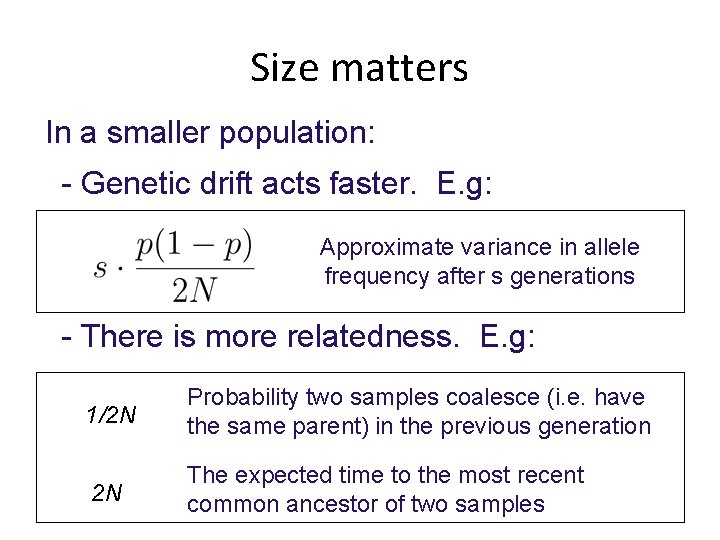
Size matters In a smaller population: - Genetic drift acts faster. E. g: Approximate variance in allele frequency after s generations K=100 50 generations

Size matters In a smaller population: - Genetic drift acts faster. E. g: Approximate variance in allele frequency after s generations - There is more relatedness. E. g: 1/2 N Probability two samples coalesce (i. e. have the same parent) in the previous generation 2 N The expected time to the most recent common ancestor of two samples

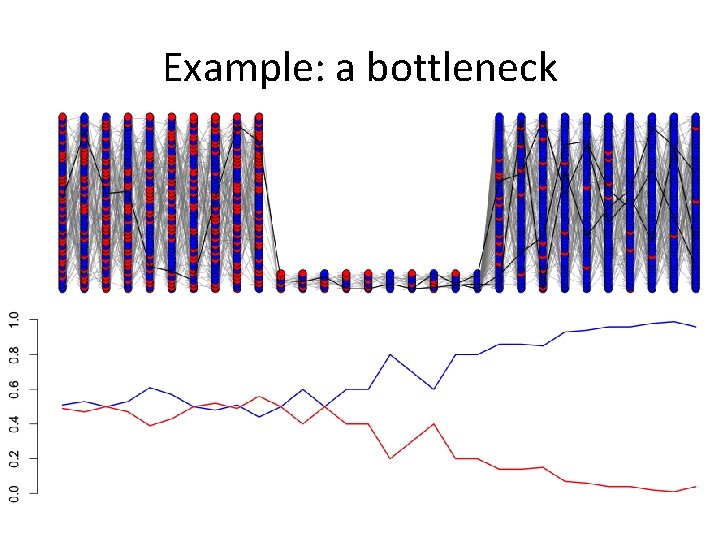
Example: a bottleneck

24 haplotypes (12 individuals) 100 SNPs on chromosome 20 Utah residents, ancestrally Northern and Western European Yoruba from Ibadan, Nigeria

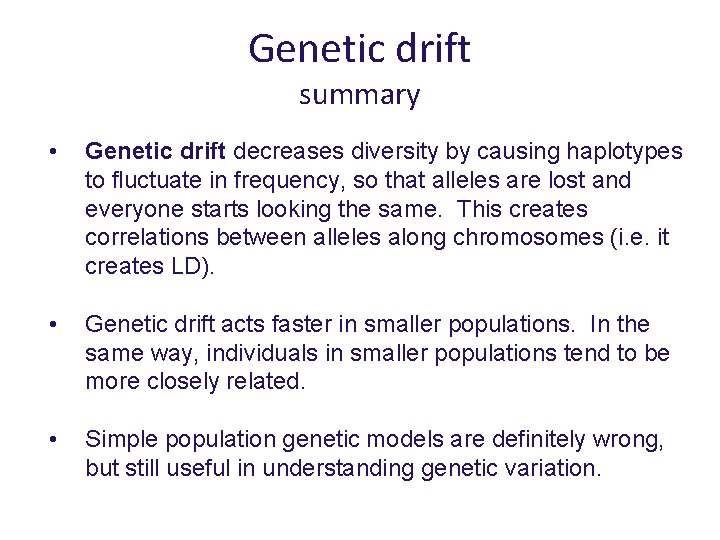
Genetic drift summary • Genetic drift decreases diversity by causing haplotypes to fluctuate in frequency, so that alleles are lost and everyone starts looking the same. This creates correlations between alleles along chromosomes (i. e. it creates LD). • Genetic drift acts faster in smaller populations. In the same way, individuals in smaller populations tend to be more closely related. • Simple population genetic models are definitely wrong, but still useful in understanding genetic variation.

An acknowledgement To make these slides I’ve used modified version of code originally written by Graham Coop. I’ll make this code available on the course materials site, but the original code is here: https: //github. com/cooplab/popgen-notes/ Graham’s group website www. gcbias. org is also a good place to look for information on population genetics topics.

Ancestral processes Mutation 2μ Recombination 2 r Coalesce 1/2 N If only drift were operating, we’d all look identical to each other. Something must be acting against drift.

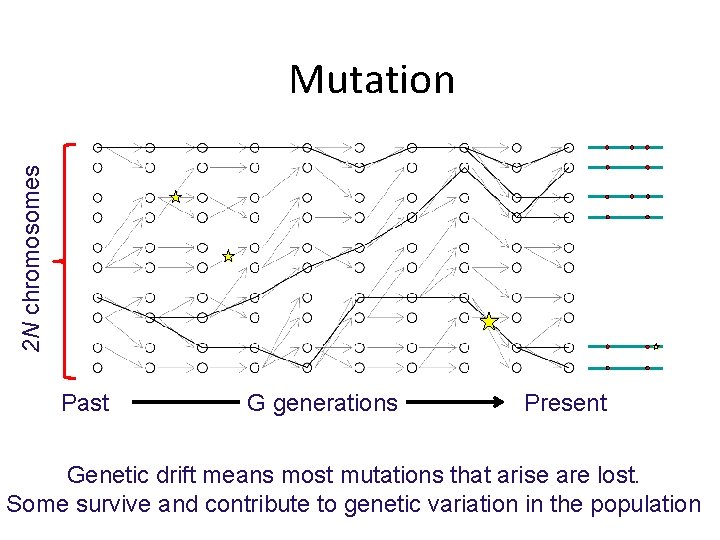
2 N chromosomes Mutation Past G generations Present Genetic drift means most mutations that arise are lost. Some survive and contribute to genetic variation in the population

Ancestral processes Mutation 2μ Recombination 2 r Coalesce 1/2 N If only drift were operating, we’d all look identical to each other. Something must be acting against drift.

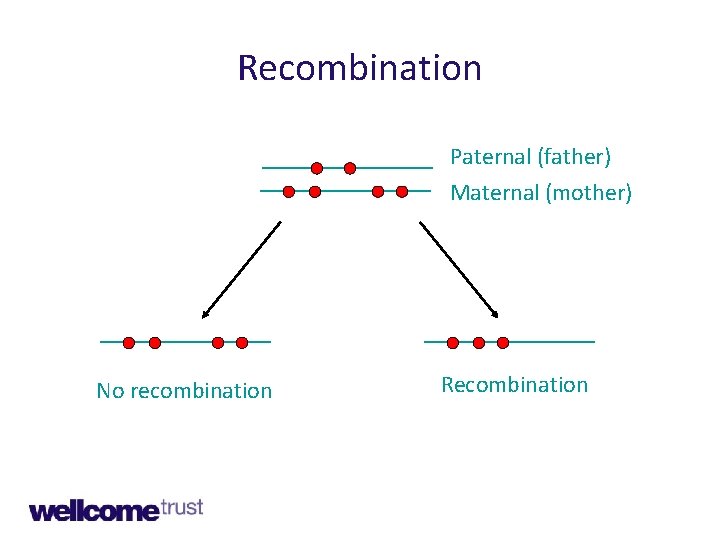
Recombination Paternal (father) Maternal (mother) No recombination Recombination

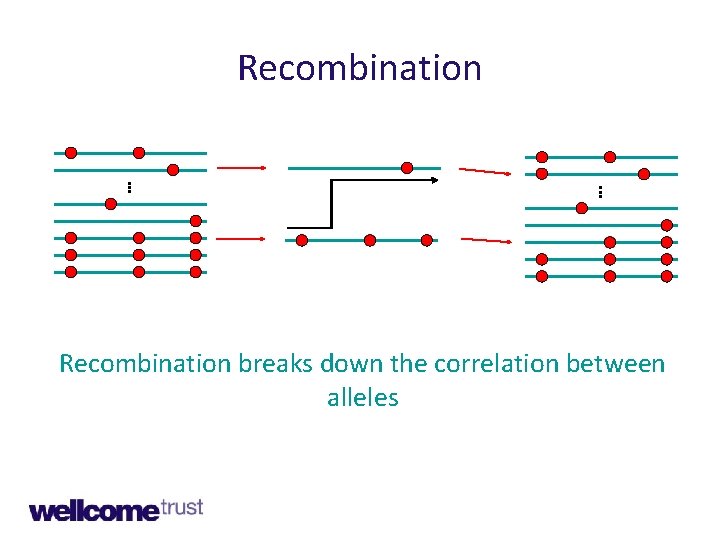
Recombination. . . Recombination breaks down the correlation between alleles

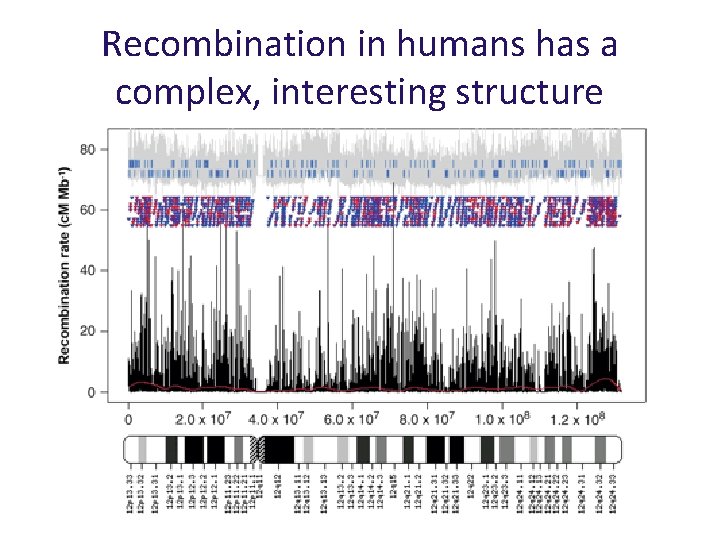
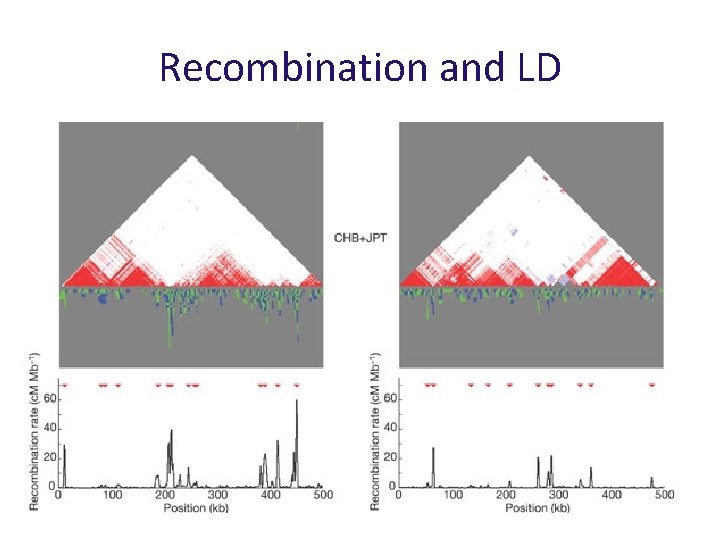
Recombination in humans has a complex, interesting structure

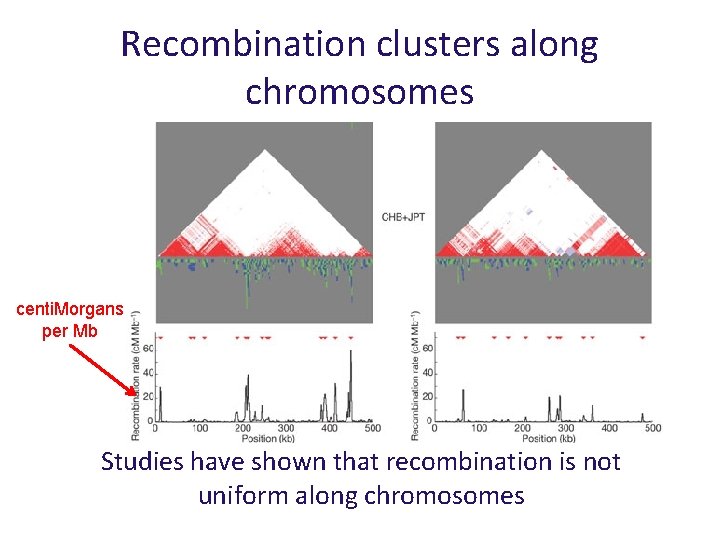
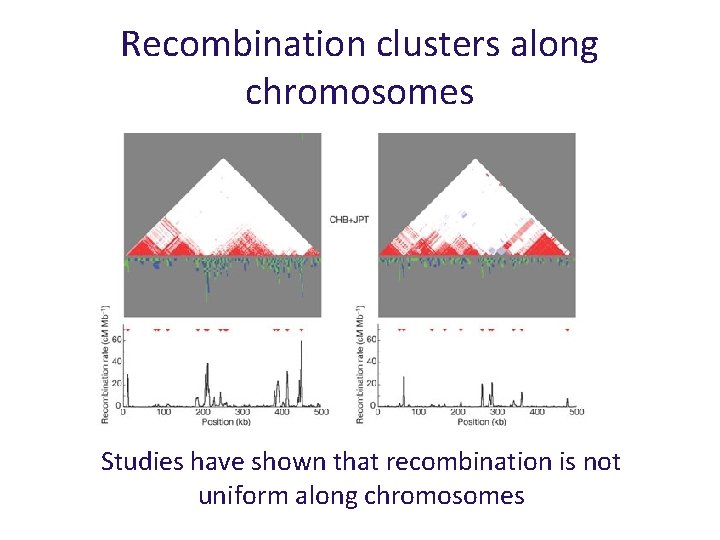
Recombination clusters along chromosomes centi. Morgans per Mb Studies have shown that recombination is not uniform along chromosomes

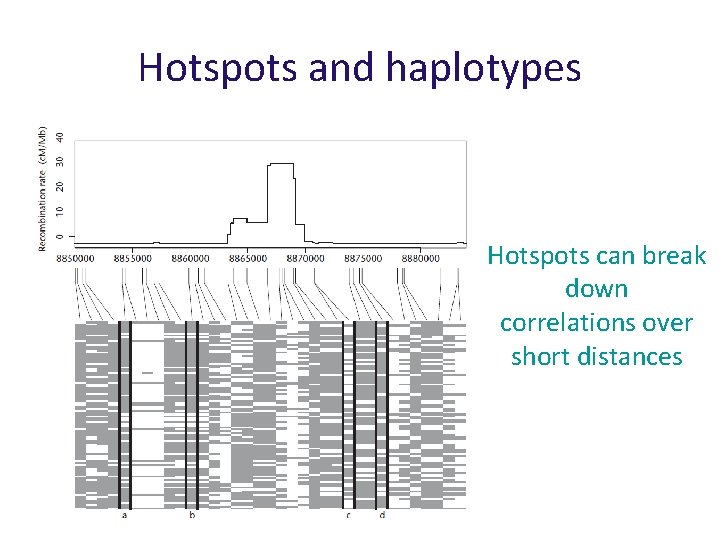
Hotspots and haplotypes Hotspots can break down correlations over short distances

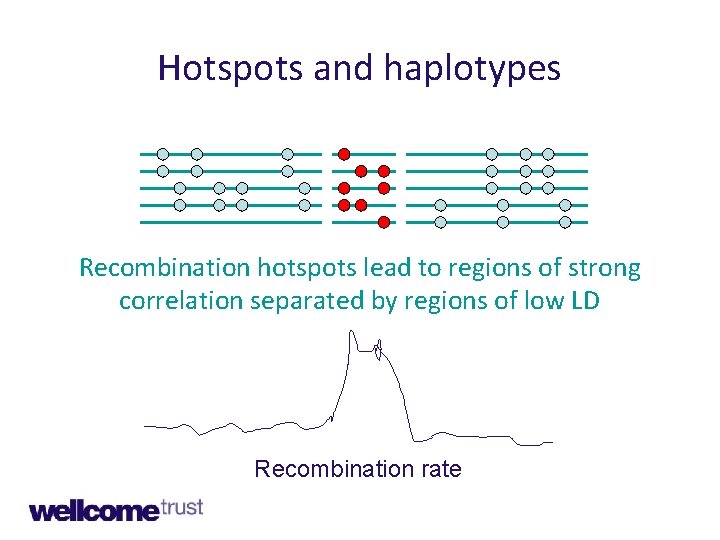
Hotspots and haplotypes Recombination hotspots lead to regions of strong correlation separated by regions of low LD Recombination rate

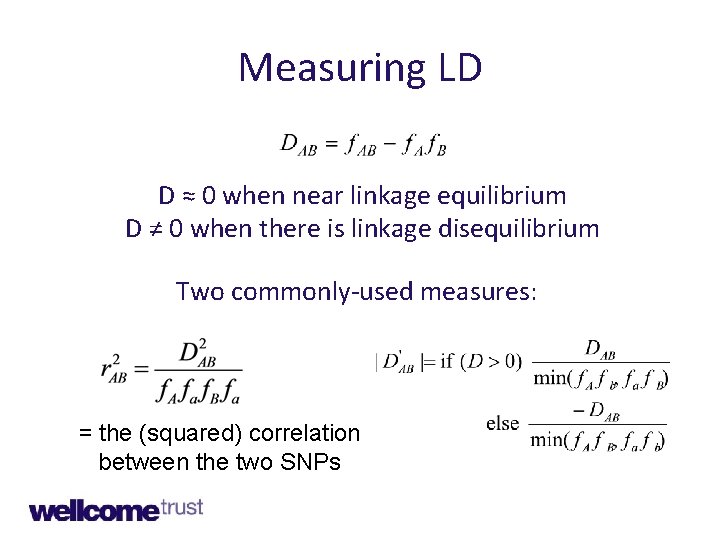
Measuring correlations • In genetics correlation between alleles is called linkage disequilibrium (LD) • There are several measures of LD • Understanding LD in natural populations is important for genomic epidemiology

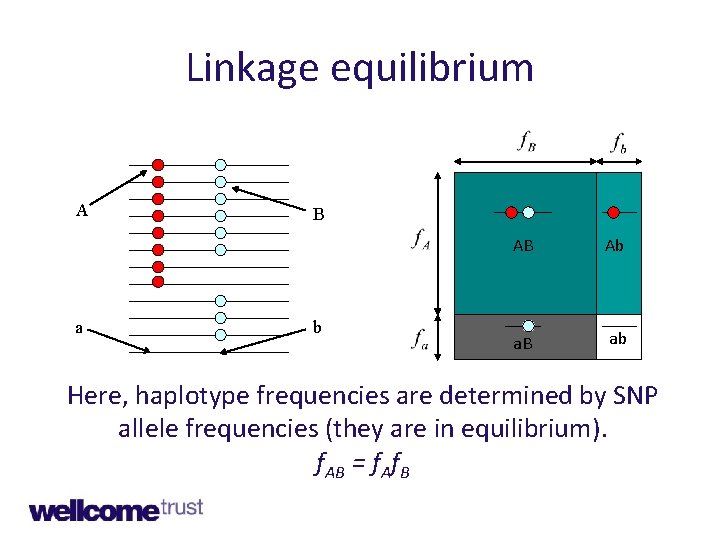
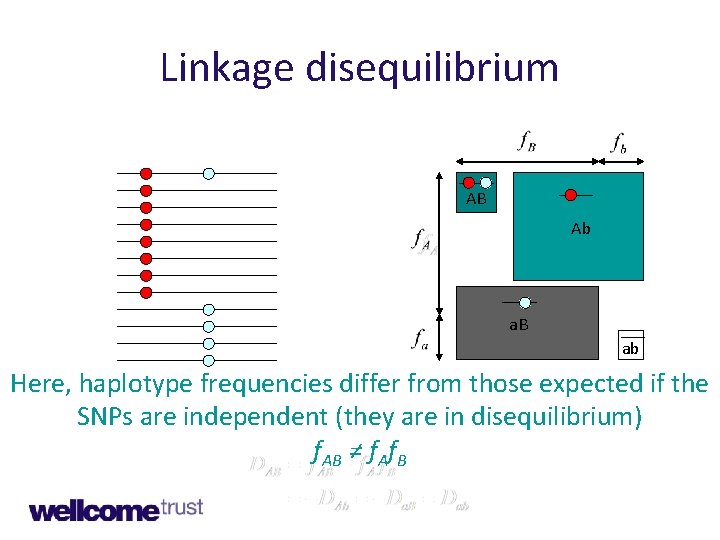
Linkage equilibrium A a B b AB Ab a. B ab Here, haplotype frequencies are determined by SNP allele frequencies (they are in equilibrium). f. AB = f. Af. B

Linkage disequilibrium AB Ab a. B ab Here, haplotype frequencies differ from those expected if the SNPs are independent (they are in disequilibrium) f. AB ≠ f. Af. B

Measuring LD D ≈ 0 when near linkage equilibrium D ≠ 0 when there is linkage disequilibrium Two commonly-used measures: = the (squared) correlation between the two SNPs

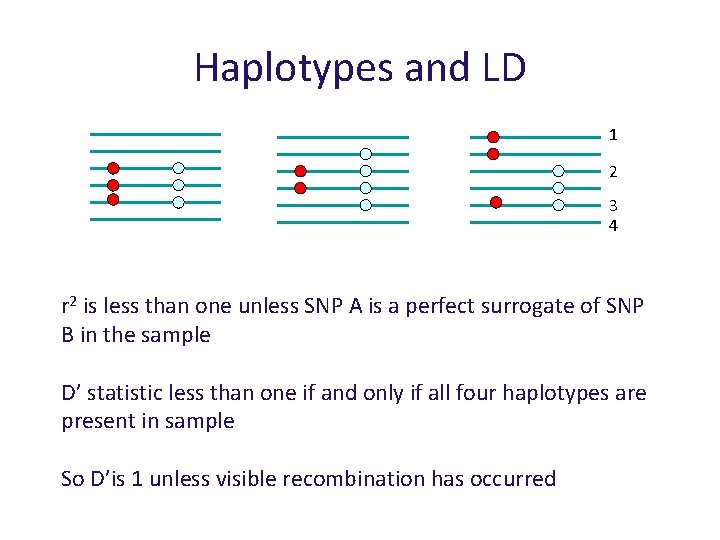
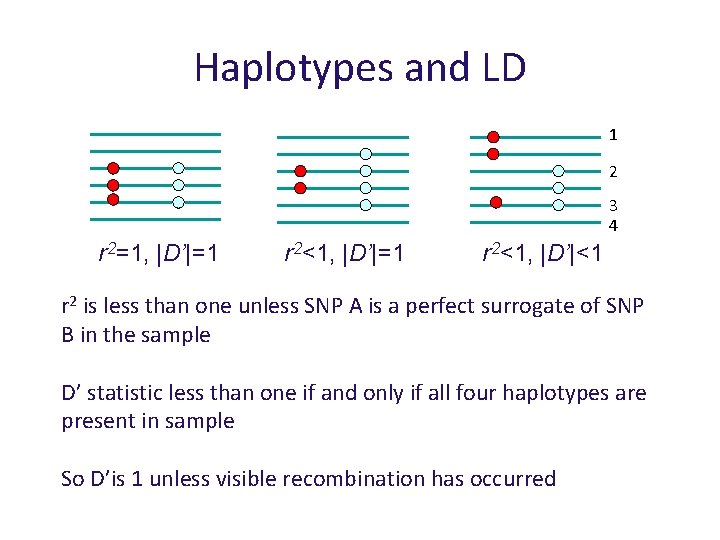
Haplotypes and LD 1 2 3 4 r 2 is less than one unless SNP A is a perfect surrogate of SNP B in the sample D’ statistic less than one if and only if all four haplotypes are present in sample So D’is 1 unless visible recombination has occurred

Haplotypes and LD 1 2 3 4 r 2=1, |D’|=1 r 2<1, |D’|<1 r 2 is less than one unless SNP A is a perfect surrogate of SNP B in the sample D’ statistic less than one if and only if all four haplotypes are present in sample So D’is 1 unless visible recombination has occurred

Recombination and LD

Population genetic processes summary • Genetic drift decreases diversity and heterozygosity, and increases levels of LD. It acts faster in smaller populations. • Mutations occur at about 60 mutations per diploid genome per generation. But most are lost due to drift. • Recombination breaks down correlations between alleles. It occurs in a highly nonuniform manner, clustered into recombination hotspots.

Population size matters • We’ve seen that in larger populations we have to go further back in time to find the common ancestor • Consequently there is more opportunity for – Mutation, increasing genetic diversity – Recombination, decreasing correlation between alleles

The power of population genetic inference from a large genome The human genome is very large, and broken up into essentially independent chunks by recombination. This gives us many observations of the ancestral process, and considerable power to understand ancestry. Will give two examples.

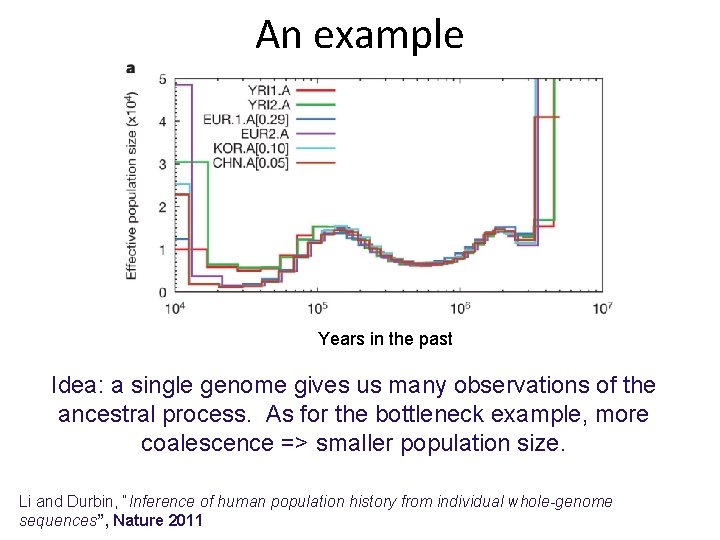
An example Years in the past Idea: a single genome gives us many observations of the ancestral process. As for the bottleneck example, more coalescence => smaller population size. Li and Durbin, “Inference of human population history from individual whole-genome sequences”, Nature 2011

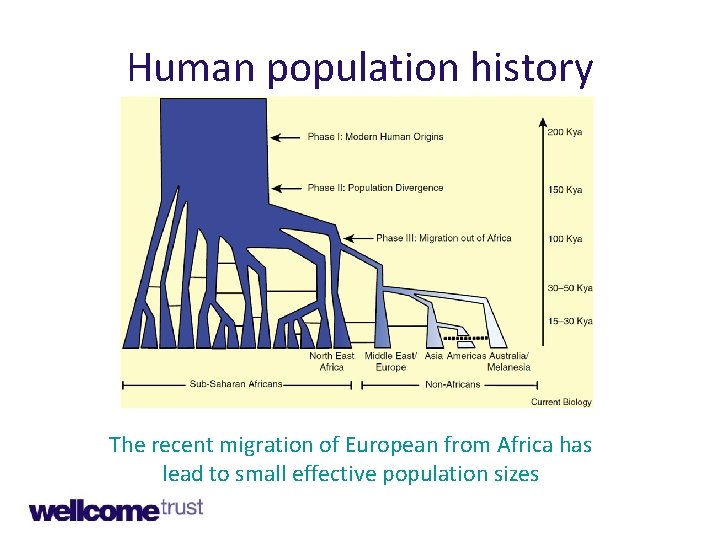
Human population history The recent migration of European from Africa has lead to small effective population sizes

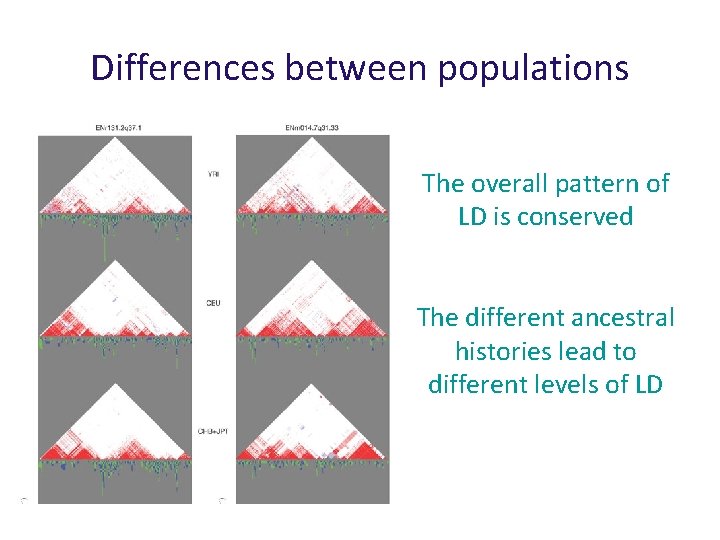
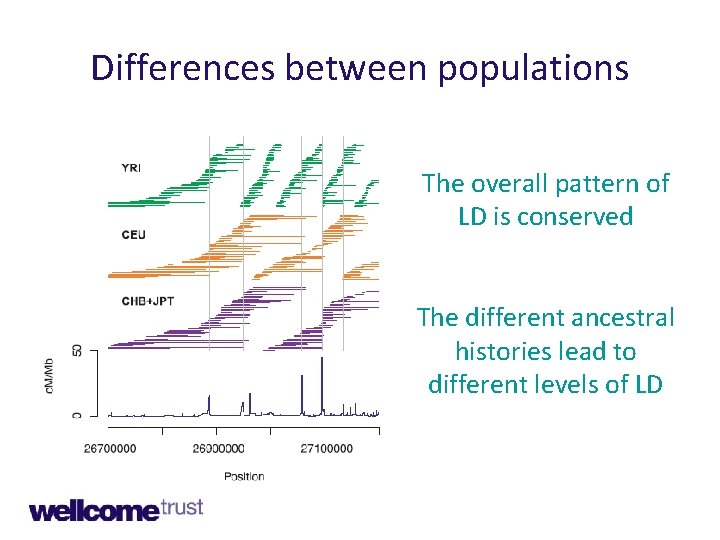
Differences between populations The overall pattern of LD is conserved The different ancestral histories lead to different levels of LD

Population genetics • Genetic drift generates correlations between alleles • Recombination breaks them down • The ancestral population size and history determines the amount of diversity and how it is structured • Natural selection can generate strong differences between populations

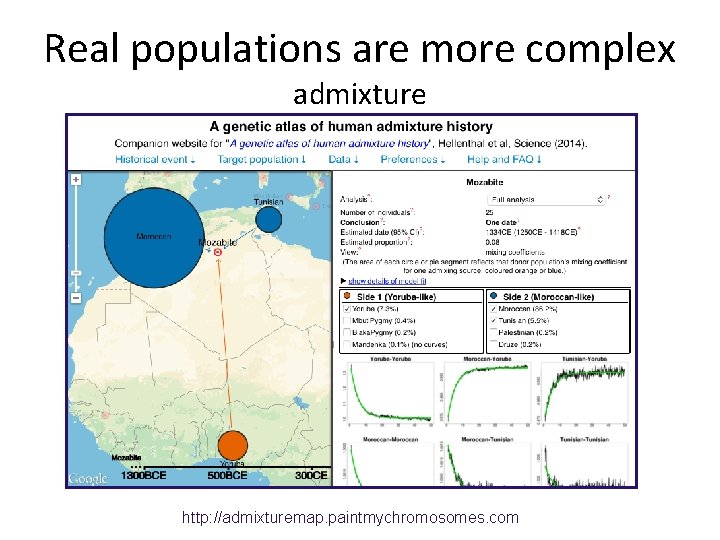
Real populations are more complex admixture http: //admixturemap. paintmychromosomes. com

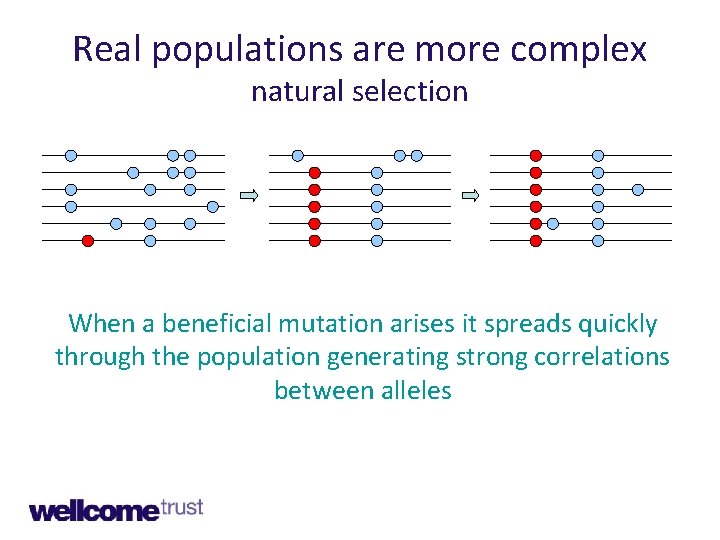
Real populations are more complex natural selection When a beneficial mutation arises it spreads quickly through the population generating strong correlations between alleles

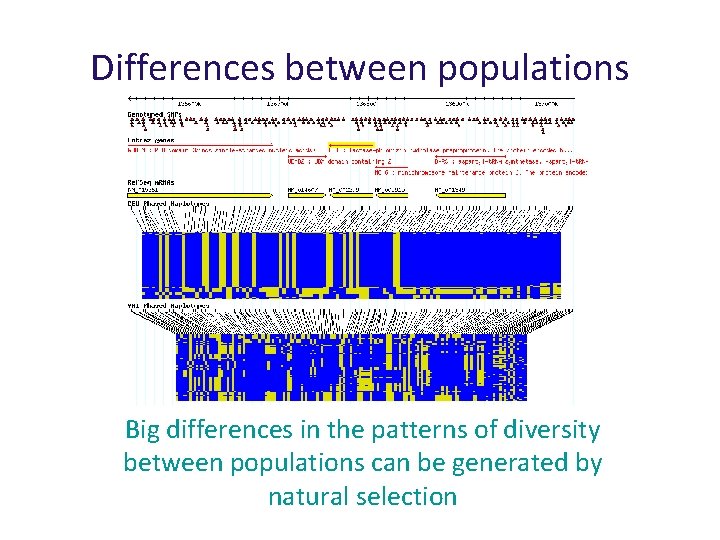
Natural Selection Big differences in the patterns of diversity between populations can be generated by natural selection

Differences between populations Big differences in the patterns of diversity between populations can be generated by natural selection

24 haplotypes (12 individuals) 100 SNPs on chromosome 20 Utah residents, ancestrally Northern and Western European Yoruba from Ibadan, Nigeria

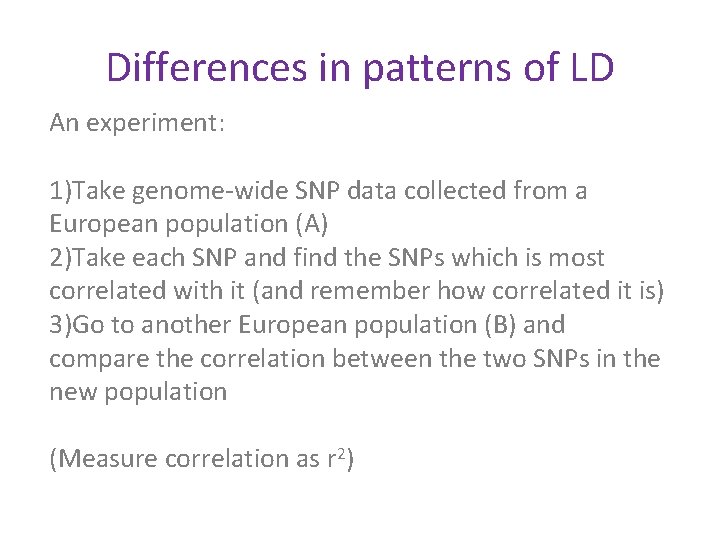
Differences in patterns of LD An experiment: 1)Take genome-wide SNP data collected from a European population (A) 2)Take each SNP and find the SNPs which is most correlated with it (and remember how correlated it is) 3)Go to another European population (B) and compare the correlation between the two SNPs in the new population (Measure correlation as r 2)

Differences in patterns of LD Across Europe Within Kenya We will look at this in the practical

Thanks!

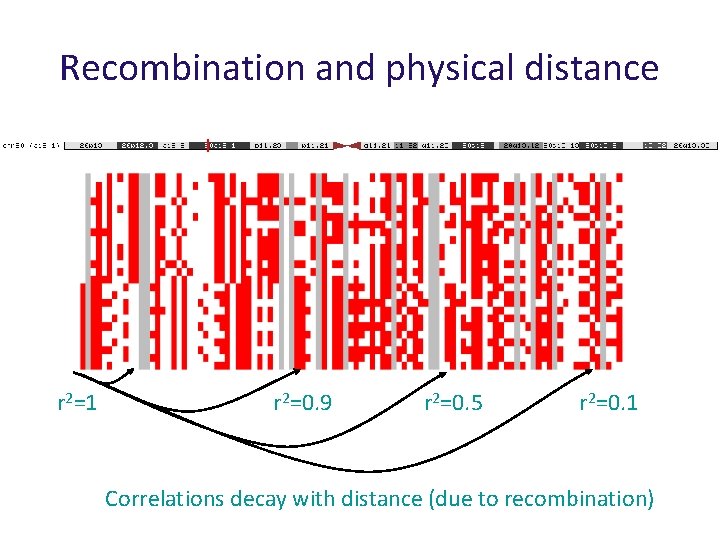
Recombination and physical distance r 2=1 r 2=0. 9 r 2=0. 5 r 2=0. 1 Correlations decay with distance (due to recombination)

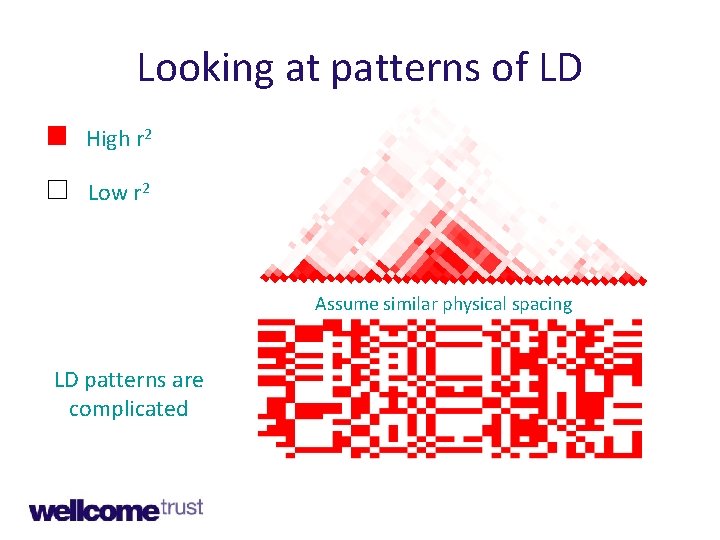
Looking at patterns of LD High r 2 Low r 2 Assume similar physical spacing LD patterns are complicated

Recombination clusters along chromosomes Studies have shown that recombination is not uniform along chromosomes

The power of population genetic inference from a large genome

24 haplotypes (12 individuals) 100 SNPs on chromosome 20 Utah residents, ancestrally Northern and Western Europe Yoruba from Ibadan, Nigeria

LD and Recombination • There are lots of ways to measure LD • Recombination is not uniform along chromosomes • Much of the recombination happens in hotspots and these demark breakdown in correlations • Correlations do persist across hot spots

Differences between populations The overall pattern of LD is conserved The different ancestral histories lead to different levels of LD

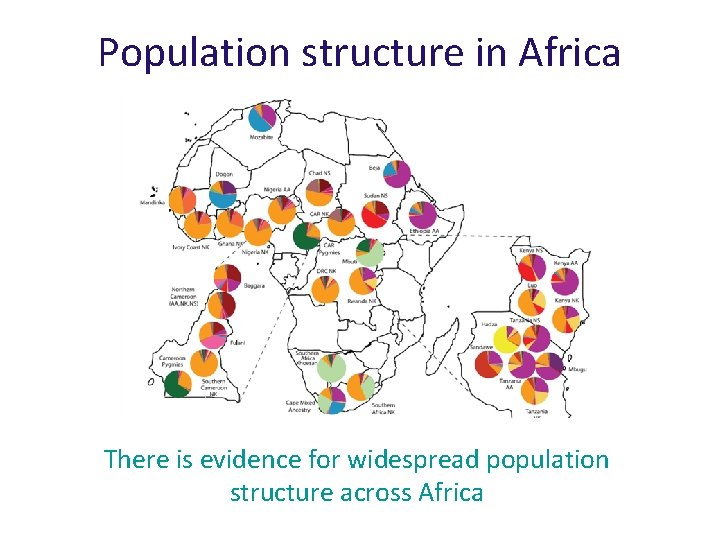
Population structure in Africa There is evidence for widespread population structure across Africa

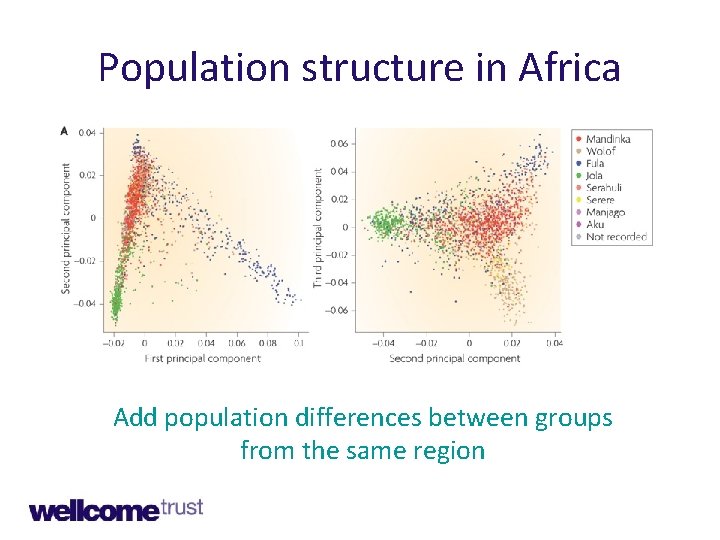
Population structure in Africa Add population differences between groups from the same region

24 haplotypes (12 individuals) 100 SNPs on chromosome 20 Luhya in Webuye, Kenya Maasai in Kinyawa, Kenya


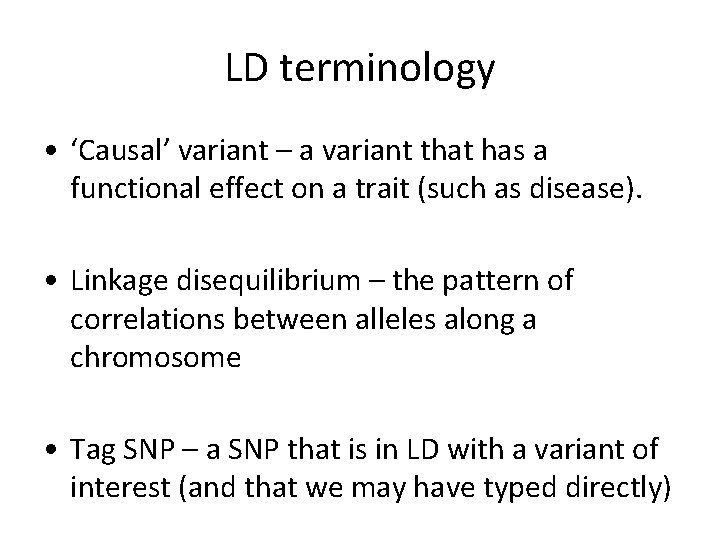
LD terminology • ‘Causal’ variant – a variant that has a functional effect on a trait (such as disease). • Linkage disequilibrium – the pattern of correlations between alleles along a chromosome • Tag SNP – a SNP that is in LD with a variant of interest (and that we may have typed directly)

Summary • Different ancestral histories have led to different patterns of diversity • Natural selection can generate strong differences in haplotype patterns • Population structure across Africa, and between groups in Africa, will lead to differences in the structure of LD



Genetic drift Allele frequencies change by chance over time

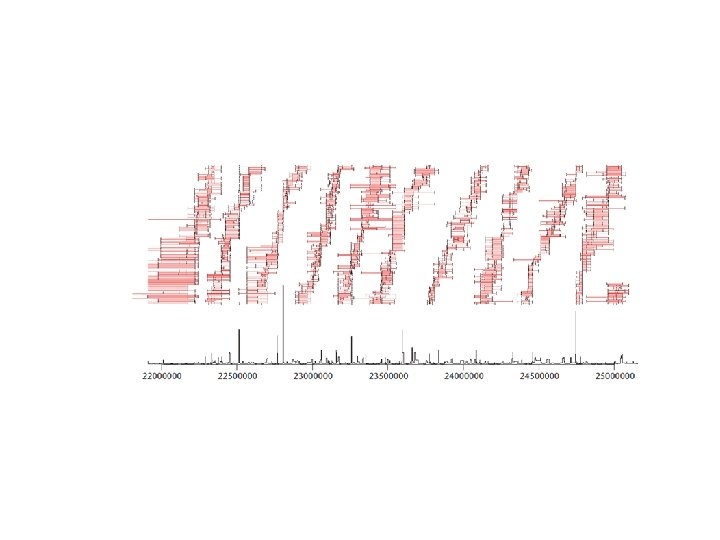
Genetic diversity 180 haplotypes (90 individuals) from Luhya in Webuye, Kenya typed at 6856 SNPs in 10 Mb region on chromosome 20
 Wellcome trust courses
Wellcome trust courses Wellcome trust courses
Wellcome trust courses Wellcome trust
Wellcome trust Advanced placement courses
Advanced placement courses Genomic instability
Genomic instability Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic england
Genomic england Genomic
Genomic Genomic equivalence definition
Genomic equivalence definition Genomic england
Genomic england Genomic imprinting definition
Genomic imprinting definition Anneke seller
Anneke seller Genomic signal processing
Genomic signal processing Principle of genomic equivalence
Principle of genomic equivalence Bending moment formula pdf
Bending moment formula pdf Gif welcome images
Gif welcome images Waterlow classification
Waterlow classification Wellcome to my presentation
Wellcome to my presentation Wellcome
Wellcome Wellcome to my presentation
Wellcome to my presentation Wellcome classification
Wellcome classification Clasificacion wellcome
Clasificacion wellcome Wellcome to our class
Wellcome to our class Sir henry wellcome postdoctoral fellowship
Sir henry wellcome postdoctoral fellowship Henry wellcome fellowship
Henry wellcome fellowship Definition of protein-energy malnutrition
Definition of protein-energy malnutrition Wellcome classification
Wellcome classification Charitable work
Charitable work Consortium for advanced research training in africa
Consortium for advanced research training in africa Nutrition epidemiology definition
Nutrition epidemiology definition Epidemiology triangle
Epidemiology triangle Ramboman analysis
Ramboman analysis How dr. wafaa elsadr epidemiology professor
How dr. wafaa elsadr epidemiology professor Certification board of infection control and epidemiology
Certification board of infection control and epidemiology Formula for attack rate
Formula for attack rate Attack rate formula
Attack rate formula How dr. wafaa elsadr epidemiology professor
How dr. wafaa elsadr epidemiology professor Distribution in epidemiology
Distribution in epidemiology Spurious association
Spurious association Descriptive vs analytical epidemiology
Descriptive vs analytical epidemiology Logistic regression epidemiology
Logistic regression epidemiology John snow epidemiology
John snow epidemiology Define epidemiology
Define epidemiology Epi
Epi Gordon nichols
Gordon nichols Defination of epidemiology
Defination of epidemiology Gate frame epidemiology
Gate frame epidemiology Thesourceagents
Thesourceagents Epidemiology definition
Epidemiology definition Effect modification vs confounding
Effect modification vs confounding Field epidemiology ppt
Field epidemiology ppt Bibliography of epidemiology
Bibliography of epidemiology Measures of association in epidemiology
Measures of association in epidemiology Prevalence calculation example
Prevalence calculation example Epidemiology made easy
Epidemiology made easy Mp under microscope
Mp under microscope Epidemiology kept simple
Epidemiology kept simple Wheel of causation example
Wheel of causation example Scope of epidemiology
Scope of epidemiology Epidemiology person place time
Epidemiology person place time Seven uses of epidemiology
Seven uses of epidemiology Distribution in epidemiology
Distribution in epidemiology Prevalence
Prevalence Aims of epidemiology
Aims of epidemiology Temporal relationship epidemiology example
Temporal relationship epidemiology example Cross sectional study advantages and disadvantages
Cross sectional study advantages and disadvantages Attack rate epidemiology
Attack rate epidemiology Diabetic ketoacidosis epidemiology
Diabetic ketoacidosis epidemiology Defination of proportion
Defination of proportion What is descriptive study in epidemiology
What is descriptive study in epidemiology Difference between descriptive and analytical epidemiology
Difference between descriptive and analytical epidemiology Dual enrollment valencia requirements
Dual enrollment valencia requirements