Genomic Signal Processing Dr C Q Chang Dept








- Slides: 22

Genomic Signal Processing Dr. C. Q. Chang Dept. of EEE

Outline • • • Basic Genomics Signal Processing for Genomic Sequences Signal Processing for Gene Expression Resources and Co-operations Challenges and Future Work

Basic Genomics

Genome • Every human cell contains 6 feet of double stranded (ds) DNA • This DNA has 3, 000, 000 base pairs representing 50, 000100, 000 genes • This DNA contains our complete genetic code or genome • DNA regulates all cell functions including response to disease, aging and development • Gene expression pattern: snapshot of DNA in a cell • Gene expression profile: DNA mutation or polymorphism over time • Genetic pathways: changes in genetic code accompanying metabolic and functional changes, e. g. disease or aging.


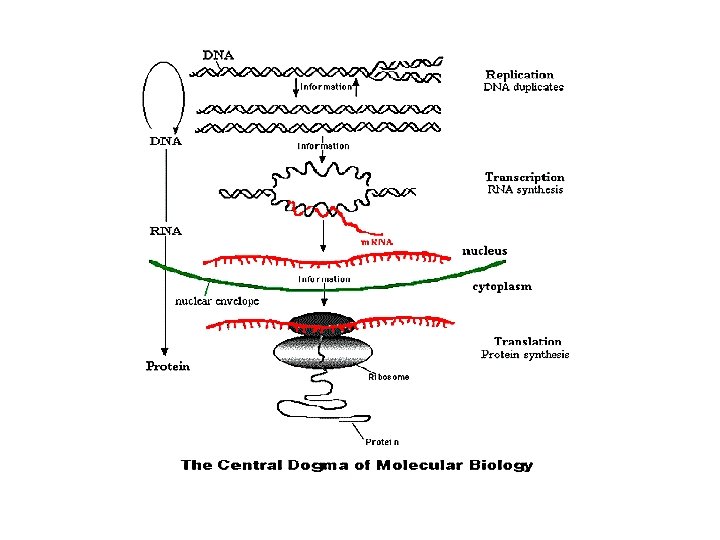
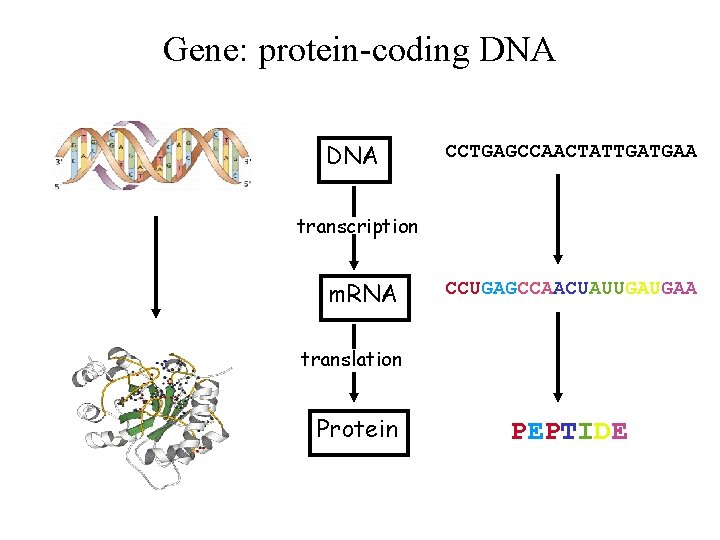
Gene: protein-coding DNA CCTGAGCCAACTATTGATGAA transcription m. RNA CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE

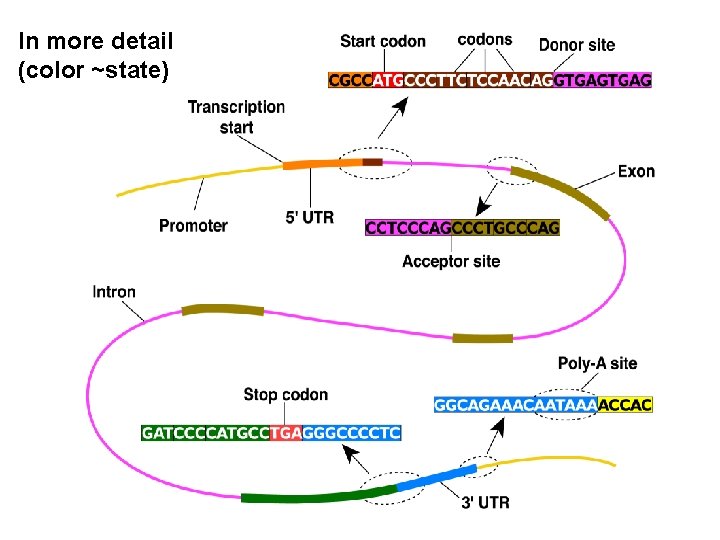
In more detail (color ~state)

Signal Processing for Genomic Sequences

The Data Set

The Problem • Genomic information is digital letters A, T, C and G • Signal processing deals with numerical sequences, character strings have to be mapped into one or more numerical sequences • Identification of protein coding regions • Prediction of whether or not a given DNA segment is a part of a protein coding region • Prediction of the proper reading frame • Comparing to traditional methods, signal processing methods are much quicker, and can be even more accurate in some cases.

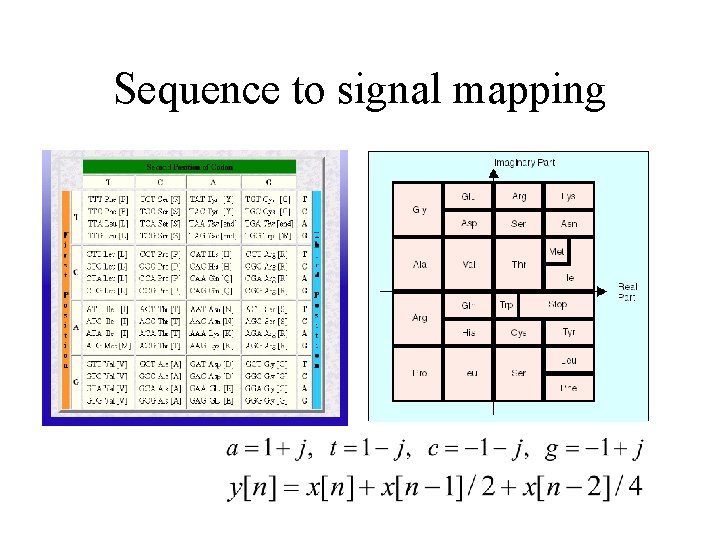
Sequence to signal mapping

Signal Analysis • Spectral analysis (Fourier transform, periodogram) • Spectrogram • Wavelet analysis • HMT: wavelet-based Hidden Markov Tree • Spectral envelope (using optimal string to numerical value mapping)

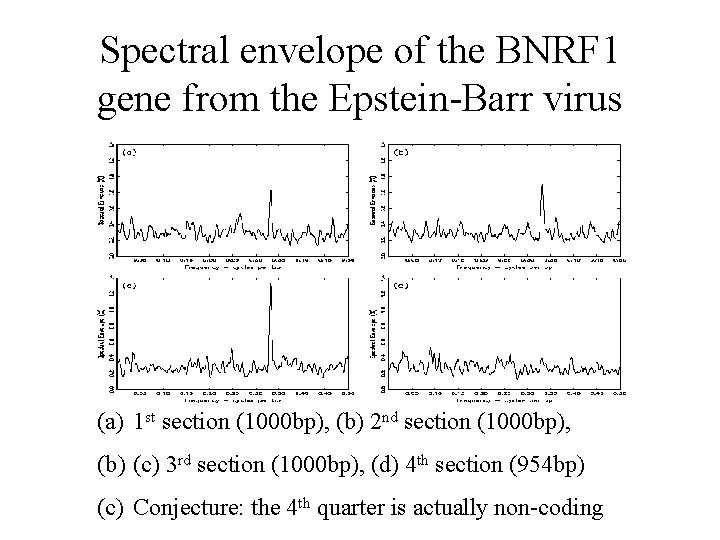
Spectral envelope of the BNRF 1 gene from the Epstein-Barr virus (a) 1 st section (1000 bp), (b) 2 nd section (1000 bp), (b) (c) 3 rd section (1000 bp), (d) 4 th section (954 bp) (c) Conjecture: the 4 th quarter is actually non-coding

Signal Processing for Gene Expression

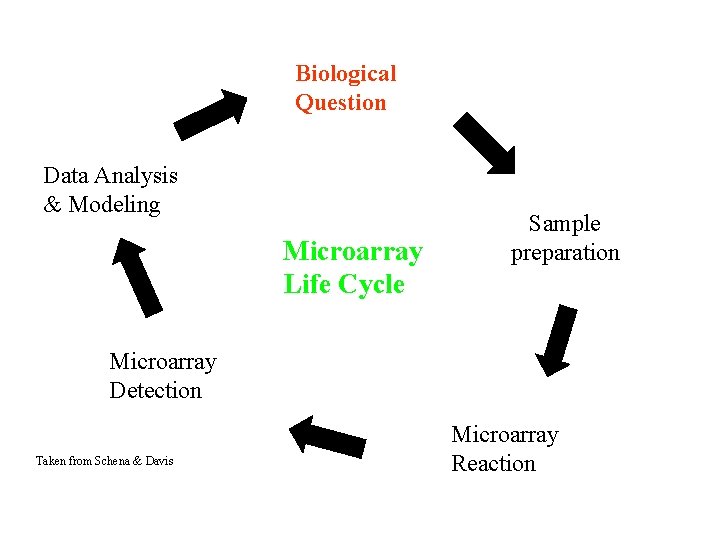
Biological Question Data Analysis & Modeling Microarray Life Cycle Sample preparation Microarray Detection Taken from Schena & Davis Microarray Reaction

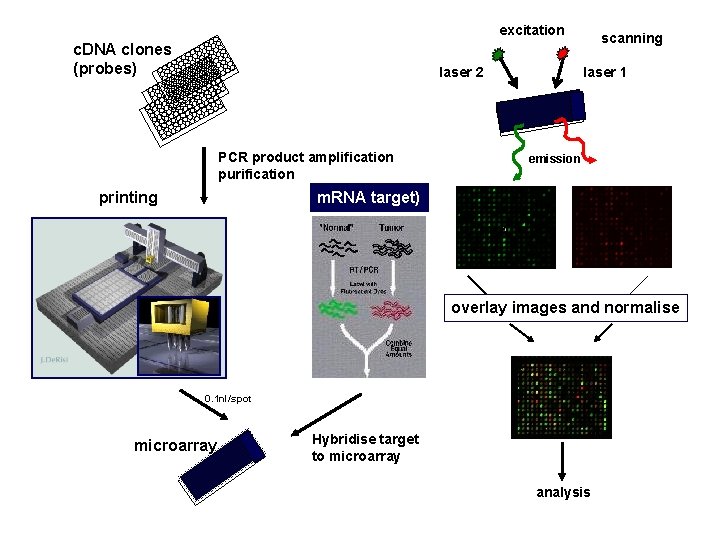
excitation c. DNA clones (probes) laser 2 PCR product amplification purification printing scanning laser 1 emission m. RNA target) overlay images and normalise 0. 1 nl/spot microarray Hybridise target to microarray analysis


Image Segmentation • Simple way: fixed circle method • Advanced: fast marching level set segmentation Advanced Fixed circle

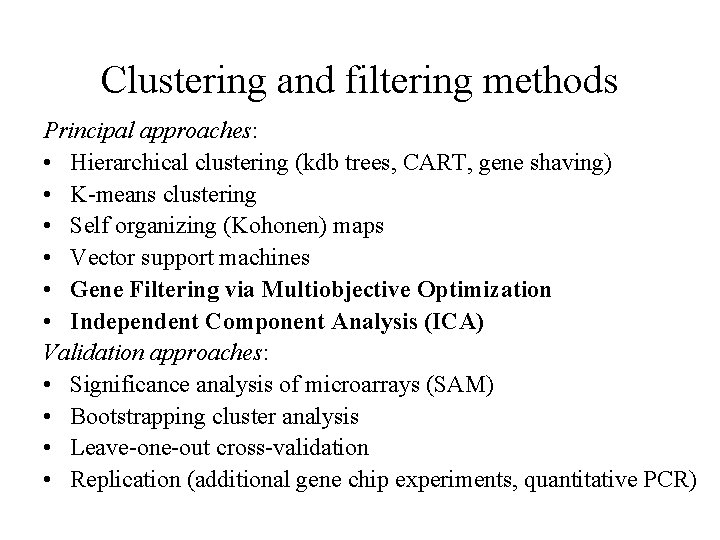
Clustering and filtering methods Principal approaches: • Hierarchical clustering (kdb trees, CART, gene shaving) • K-means clustering • Self organizing (Kohonen) maps • Vector support machines • Gene Filtering via Multiobjective Optimization • Independent Component Analysis (ICA) Validation approaches: • Significance analysis of microarrays (SAM) • Bootstrapping cluster analysis • Leave-one-out cross-validation • Replication (additional gene chip experiments, quantitative PCR)

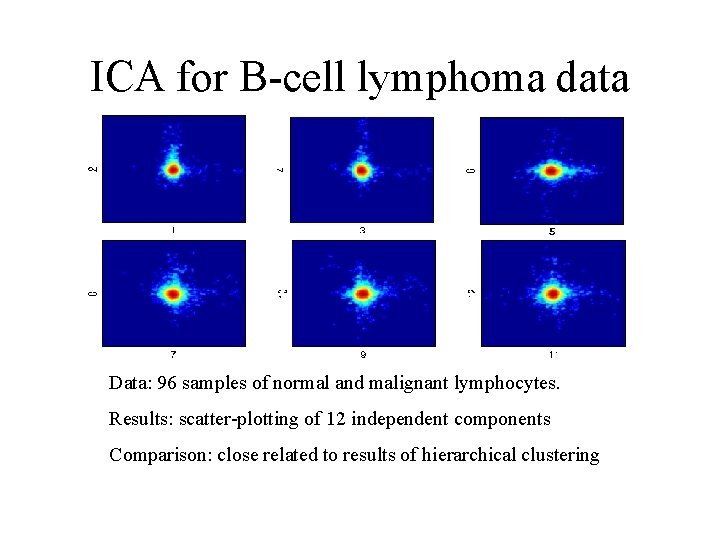
ICA for B-cell lymphoma data Data: 96 samples of normal and malignant lymphocytes. Results: scatter-plotting of 12 independent components Comparison: close related to results of hierarchical clustering

Resources and Co-operations Resources: databases on the internet such as • Gene. Bank • Protein. Bank • Some small databases of microarray data Co-operations in need: • First hand microarray data • Biological experiment for validation

Challenges and Future Work • Genomic signal processing opens a new signal processing frontier • Sequence analysis: symbolic or categorical signal, classical signal processing methods are not directly applicable • Increasingly high dimensionality of genetic data sets and the complexity involved call for fast and high throughput implementations of genomic signal processing algorithms • Future work: spectral analysis of DNA sequence and data clustering of microarray data. Modify classical signal processing methods, and develop new ones.