Genomic Signal Processing Edward R Dougherty Department of

![Classifier Design Error E[ n] E[ n, C] opt, C opt N 1 N Classifier Design Error E[ n] E[ n, C] opt, C opt N 1 N](https://slidetodoc.com/presentation_image_h2/58fab06d4268831a3cf7c73fff1d30cf/image-15.jpg)
![How Many Features? Misclassification error • Peeking phenomenon E[εd, n] εd, n εd number How Many Features? Misclassification error • Peeking phenomenon E[εd, n] εd, n εd number](https://slidetodoc.com/presentation_image_h2/58fab06d4268831a3cf7c73fff1d30cf/image-19.jpg)
![Is Cross-validation Reliable? • The preceding unbiasedness tells us that Expectation[CV estimate error] 0 Is Cross-validation Reliable? • The preceding unbiasedness tells us that Expectation[CV estimate error] 0](https://slidetodoc.com/presentation_image_h2/58fab06d4268831a3cf7c73fff1d30cf/image-23.jpg)



- Slides: 57

Genomic Signal Processing Edward R. Dougherty Department of Electrical Engineering, Texas A&M University Division of Computational Biology, Translational Genomics Research Institute Department of Pathology, University of Texas, M. D. Anderson Cancer Center 6/6/2021 http: //gsp. tamu. edu 1

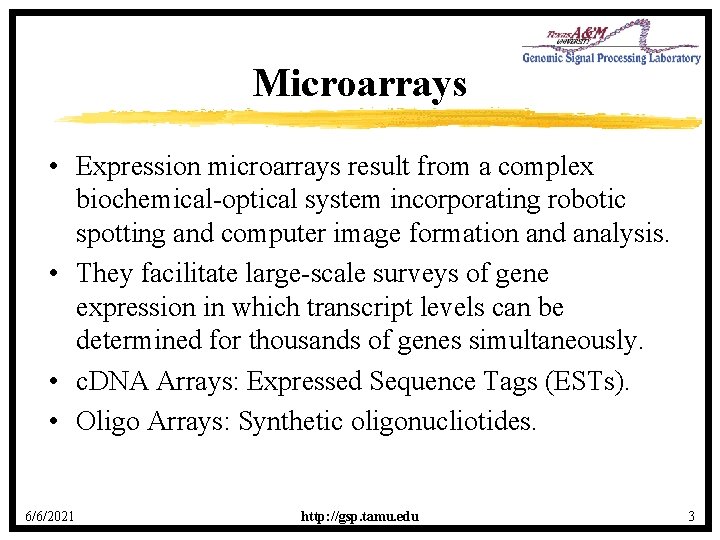
Genome-wide Data Analysis • One way of gaining insight into a gene’s role in cellular activity is to study its expression pattern in a variety of circumstances and contexts, as it responds to its environment and to the action of other genes. • As the link in the DNA → RNA → Protein chain of the central dogma, m. RNA carries a great deal of information regarding cellular function. 6/6/2021 http: //gsp. tamu. edu 2

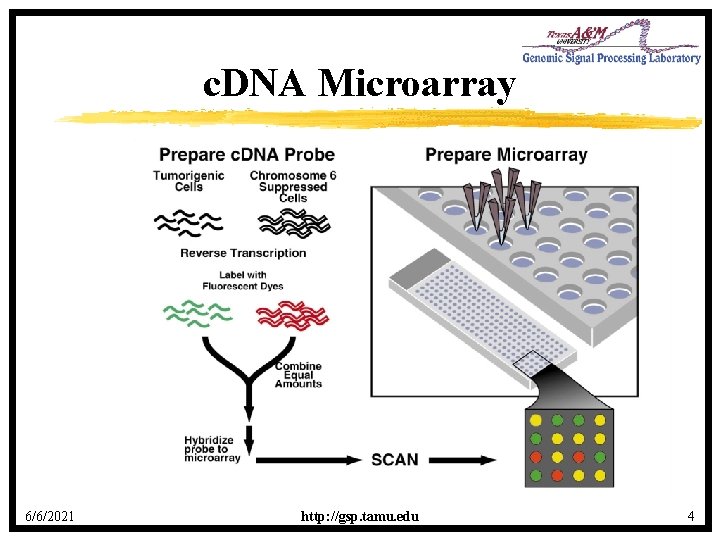
Microarrays • Expression microarrays result from a complex biochemical-optical system incorporating robotic spotting and computer image formation and analysis. • They facilitate large-scale surveys of gene expression in which transcript levels can be determined for thousands of genes simultaneously. • c. DNA Arrays: Expressed Sequence Tags (ESTs). • Oligo Arrays: Synthetic oligonucliotides. 6/6/2021 http: //gsp. tamu. edu 3

c. DNA Microarray 6/6/2021 http: //gsp. tamu. edu 4

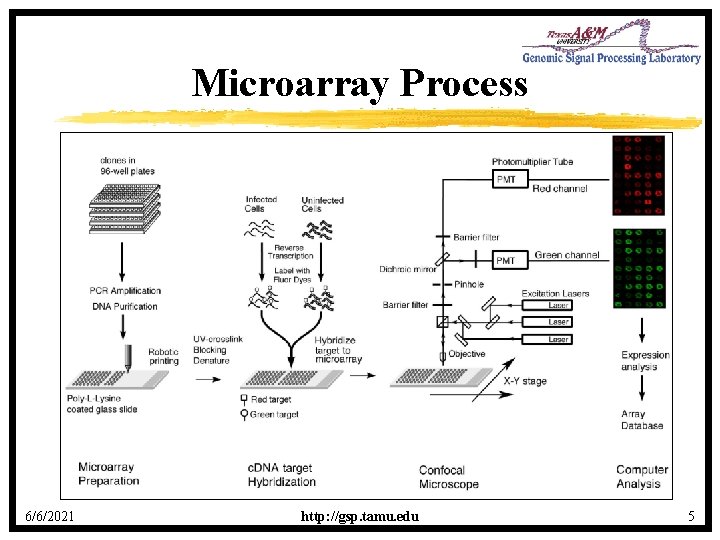
Microarray Process 6/6/2021 http: //gsp. tamu. edu 5

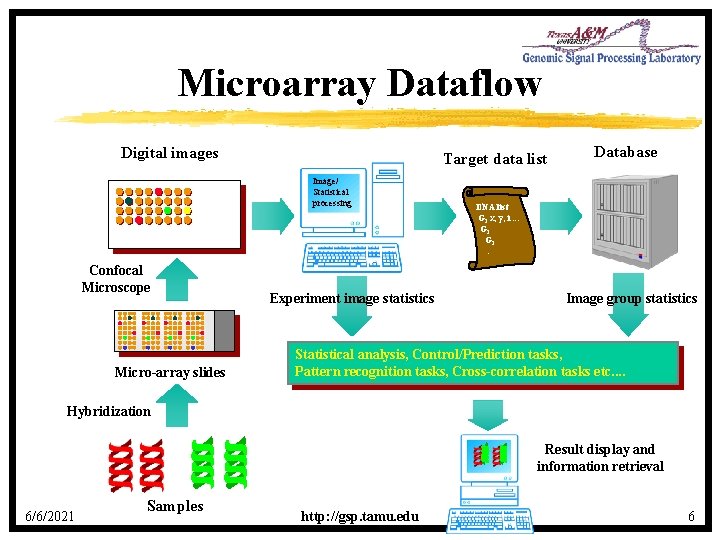
Microarray Dataflow Digital images Target data list Image/ Statistical processing Confocal Microscope Micro-array slides Experiment image statistics Database DNA list G 1 x, y, i. . . G 2 G 3 : Image group statistics Statistical analysis, Control/Prediction tasks, Pattern recognition tasks, Cross-correlation tasks etc. . Hybridization Result display and information retrieval 6/6/2021 Samples http: //gsp. tamu. edu 6

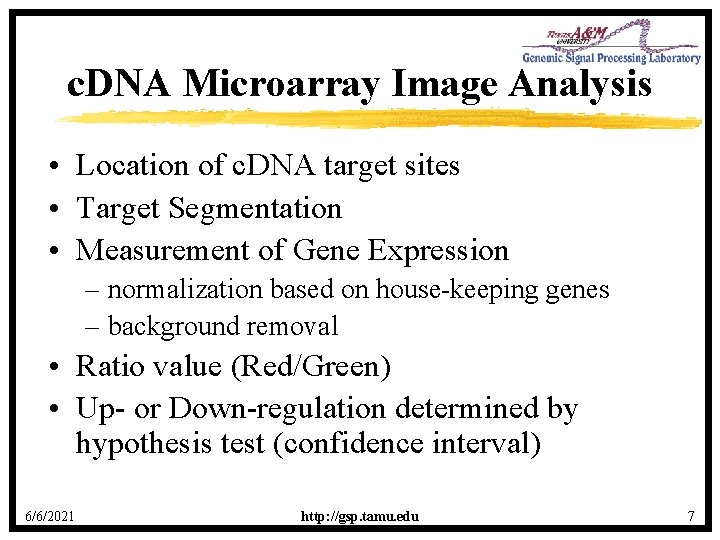
c. DNA Microarray Image Analysis • Location of c. DNA target sites • Target Segmentation • Measurement of Gene Expression – normalization based on house-keeping genes – background removal • Ratio value (Red/Green) • Up- or Down-regulation determined by hypothesis test (confidence interval) 6/6/2021 http: //gsp. tamu. edu 7

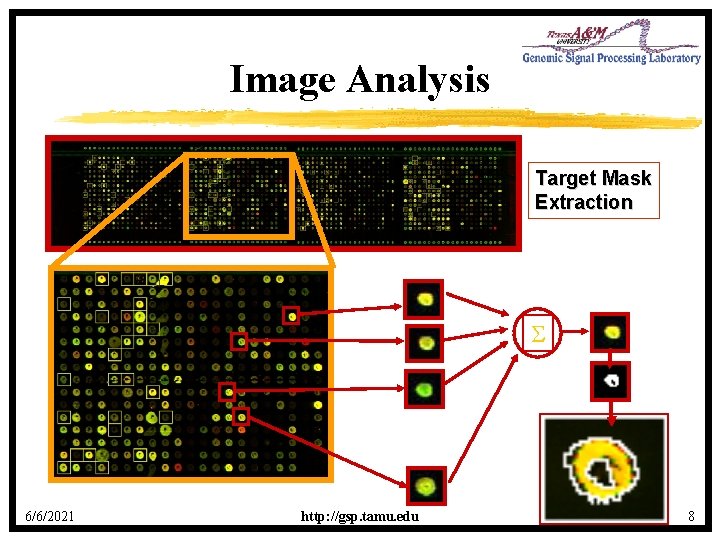
Image Analysis Target Mask Extraction 6/6/2021 http: //gsp. tamu. edu 8

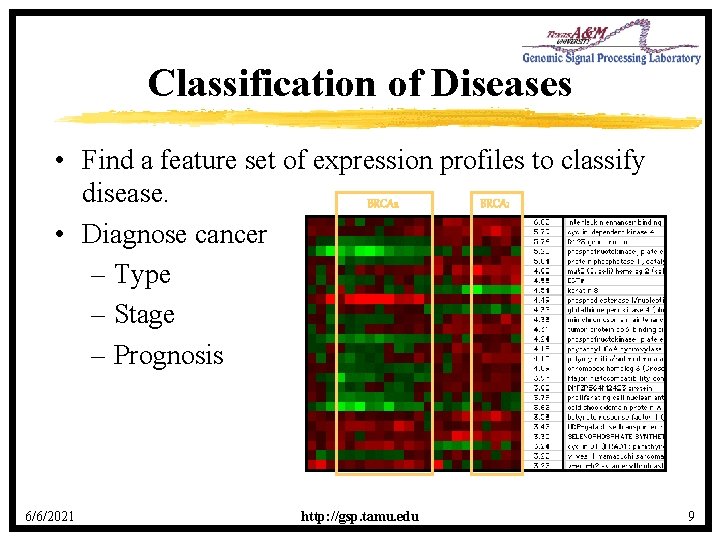
Classification of Diseases • Find a feature set of expression profiles to classify disease. BRCA 2 BRCA 1 • Diagnose cancer – Type – Stage – Prognosis 6/6/2021 http: //gsp. tamu. edu 9

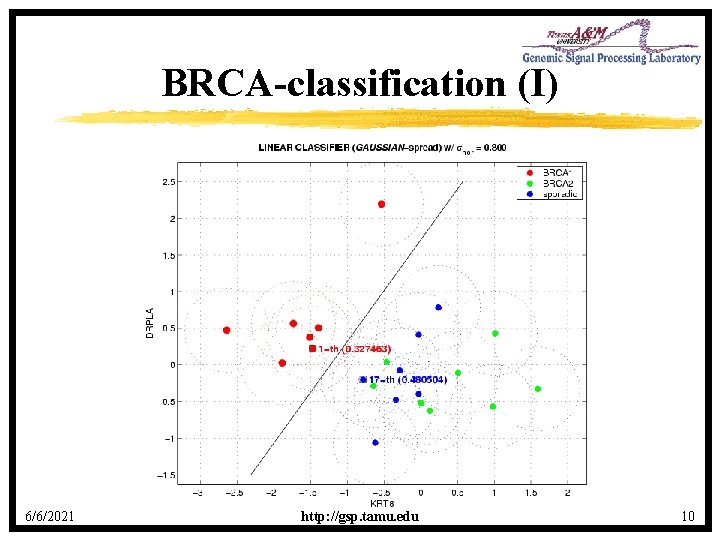
BRCA-classification (I) 6/6/2021 http: //gsp. tamu. edu 10

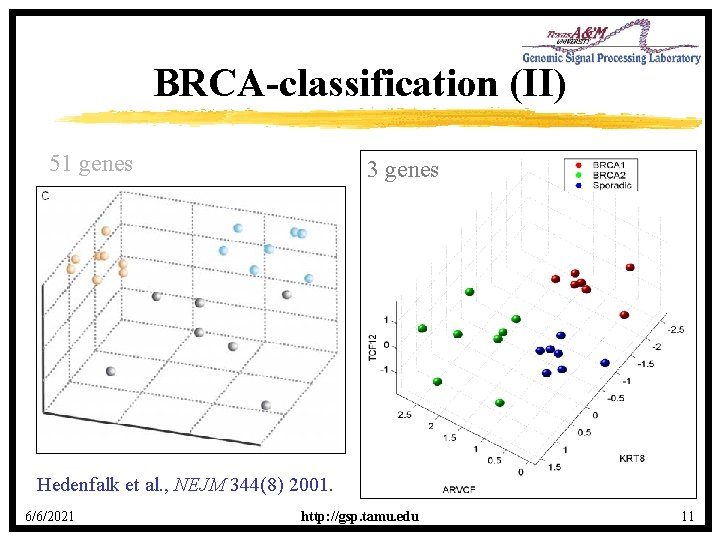
BRCA-classification (II) 51 genes 3 genes Hedenfalk et al. , NEJM 344(8) 2001. 6/6/2021 http: //gsp. tamu. edu 11

Small-Sample Issues • Imprecise classifier design: designed classifier can be a poor estimate of the optimal classifier. • Poor error estimation owing to no test data. • Poor feature selection. ü Dougherty, E. R. , "Small Sample Issues for Microarray-Based Classification, " Comparative and Functional Genomics, Vol. 2, 28 -34, 2001. 6/6/2021 http: //gsp. tamu. edu 12

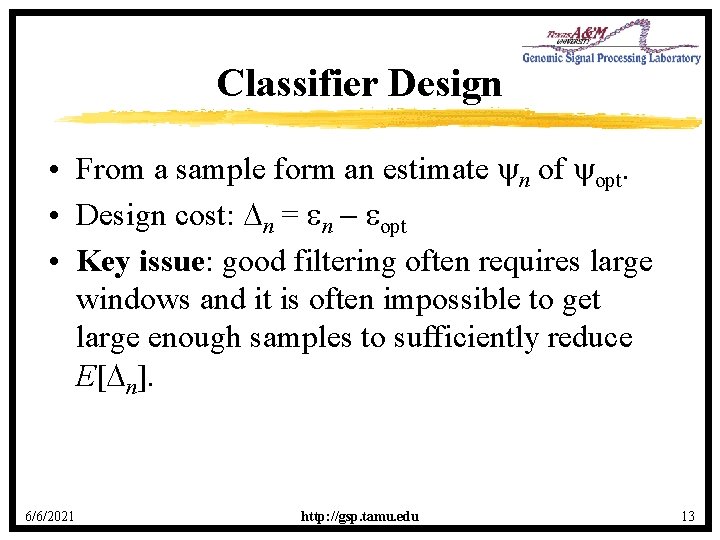
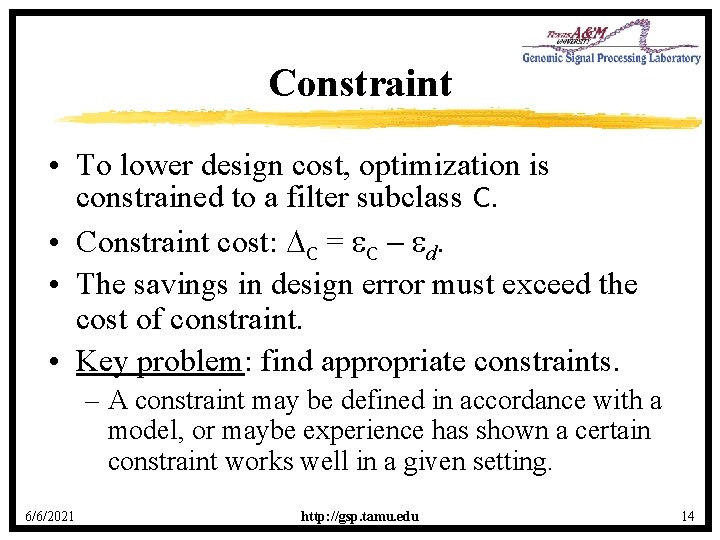
Classifier Design • From a sample form an estimate n of opt. • Design cost: n = n opt • Key issue: good filtering often requires large windows and it is often impossible to get large enough samples to sufficiently reduce E[ n]. 6/6/2021 http: //gsp. tamu. edu 13

Constraint • To lower design cost, optimization is constrained to a filter subclass C. • Constraint cost: C = C d. • The savings in design error must exceed the cost of constraint. • Key problem: find appropriate constraints. – A constraint may be defined in accordance with a model, or maybe experience has shown a certain constraint works well in a given setting. 6/6/2021 http: //gsp. tamu. edu 14
![Classifier Design Error E n E n C opt C opt N 1 N Classifier Design Error E[ n] E[ n, C] opt, C opt N 1 N](https://slidetodoc.com/presentation_image_h2/58fab06d4268831a3cf7c73fff1d30cf/image-15.jpg)
Classifier Design Error E[ n] E[ n, C] opt, C opt N 1 N 0 N 2 Sample size, N 6/6/2021 http: //gsp. tamu. edu 15

Regularization • Regularized Discriminant Analysis for QDA – weight covariance estimates towards pooled covariance for LDA (Titterington, Friedman) • Error Regularization: error + complexity penalty – VC dimension, MDL (Tabus & Astola) • Noise Injection – random perturbation (Sietsma, et al. ); Monte Carlo injection (Skurichina, et al. ); analytic injection (Kim, et al. ) 6/6/2021 http: //gsp. tamu. edu 16

LDA with Different Noise Injections 6/6/2021 http: //gsp. tamu. edu 17

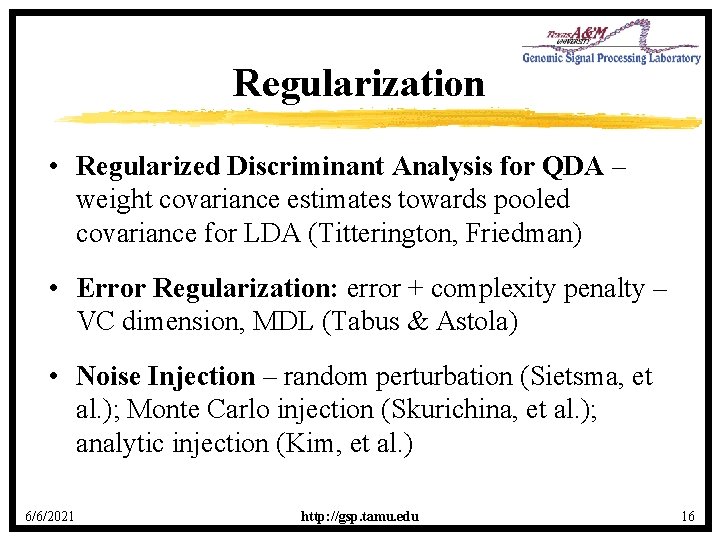
Feature-Selection Problem · Select a subset of k features from a set of n features with minimum error among all subsets of size k. · Cover and van Campenhout Theorem: All k-element subsets must be checked. · Heuristic suboptimal algorithms have been proposed to circumvent the full combinatorial search. · Issues · Mathematical analysis of algorithms · Impact of error estimation · Validation – Does algorithm outperform SFFS? 6/6/2021 http: //gsp. tamu. edu 18
![How Many Features Misclassification error Peeking phenomenon Eεd n εd n εd number How Many Features? Misclassification error • Peeking phenomenon E[εd, n] εd, n εd number](https://slidetodoc.com/presentation_image_h2/58fab06d4268831a3cf7c73fff1d30cf/image-19.jpg)
How Many Features? Misclassification error • Peeking phenomenon E[εd, n] εd, n εd number of variables, d 6/6/2021 http: //gsp. tamu. edu 19

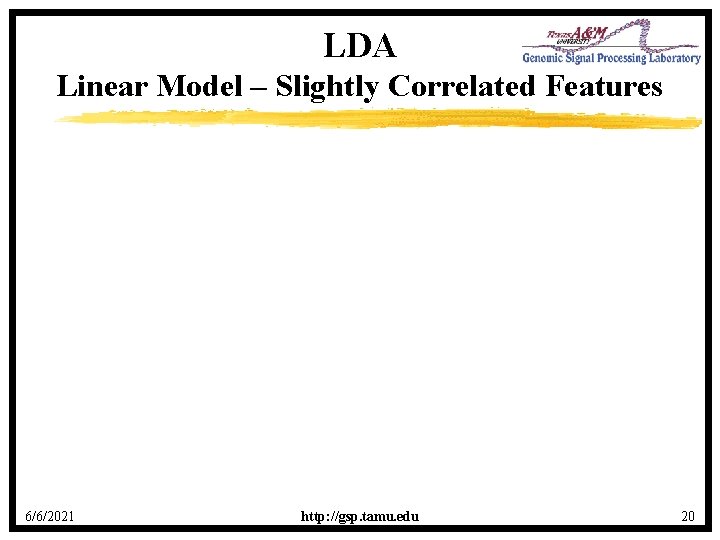
LDA Linear Model – Slightly Correlated Features 6/6/2021 http: //gsp. tamu. edu 20

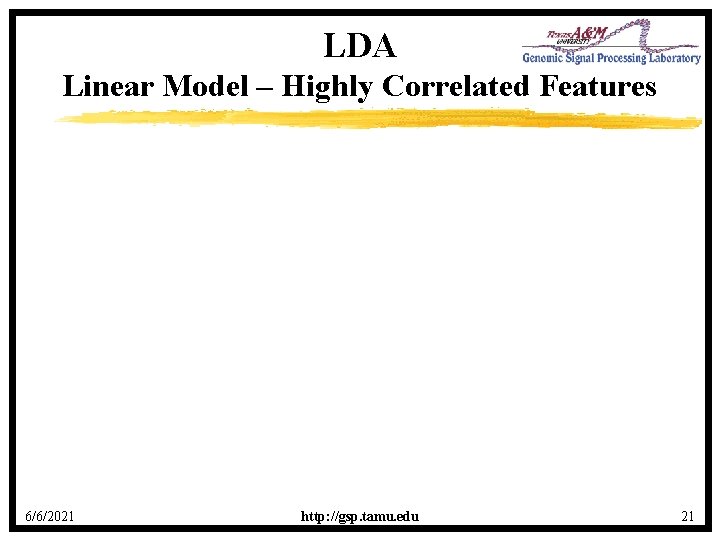
LDA Linear Model – Highly Correlated Features 6/6/2021 http: //gsp. tamu. edu 21

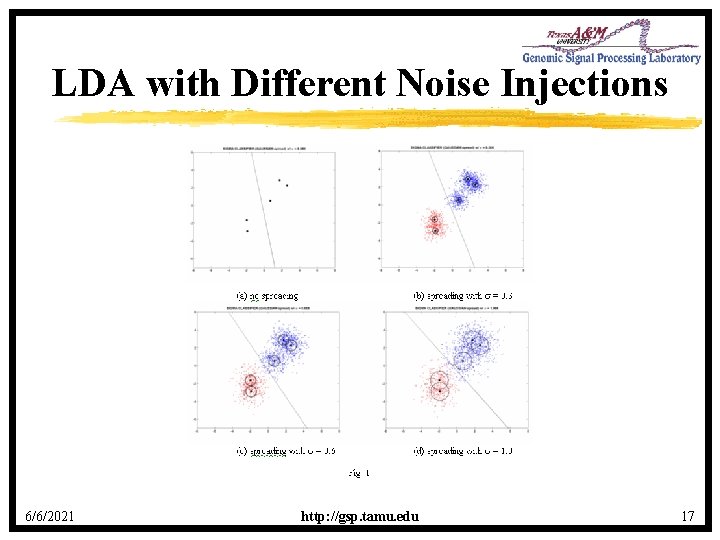
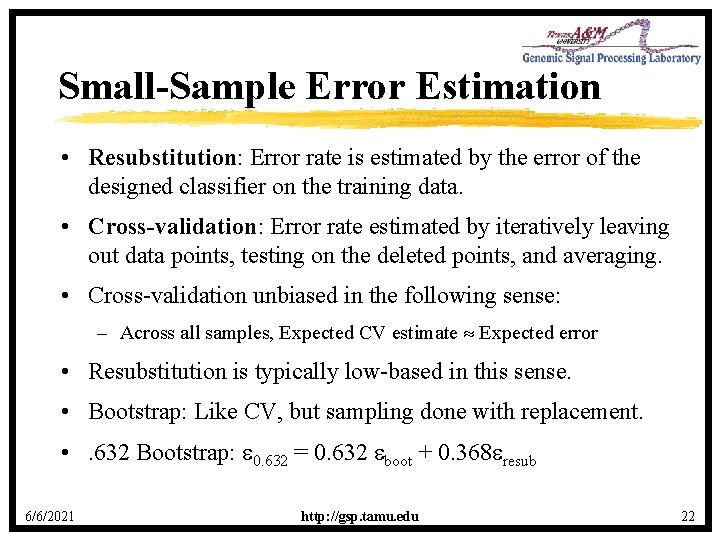
Small-Sample Error Estimation • Resubstitution: Error rate is estimated by the error of the designed classifier on the training data. • Cross-validation: Error rate estimated by iteratively leaving out data points, testing on the deleted points, and averaging. • Cross-validation unbiased in the following sense: – Across all samples, Expected CV estimate Expected error • Resubstitution is typically low-based in this sense. • Bootstrap: Like CV, but sampling done with replacement. • . 632 Bootstrap: 0. 632 = 0. 632 boot + 0. 368 resub 6/6/2021 http: //gsp. tamu. edu 22
![Is Crossvalidation Reliable The preceding unbiasedness tells us that ExpectationCV estimate error 0 Is Cross-validation Reliable? • The preceding unbiasedness tells us that Expectation[CV estimate error] 0](https://slidetodoc.com/presentation_image_h2/58fab06d4268831a3cf7c73fff1d30cf/image-23.jpg)
Is Cross-validation Reliable? • The preceding unbiasedness tells us that Expectation[CV estimate error] 0 • But this may say little about the number we are interested in, Expectation[|CV estimate error|] unless CV variance is small – not for small samples. • Deviation: |estimate error| 6/6/2021 http: //gsp. tamu. edu 23

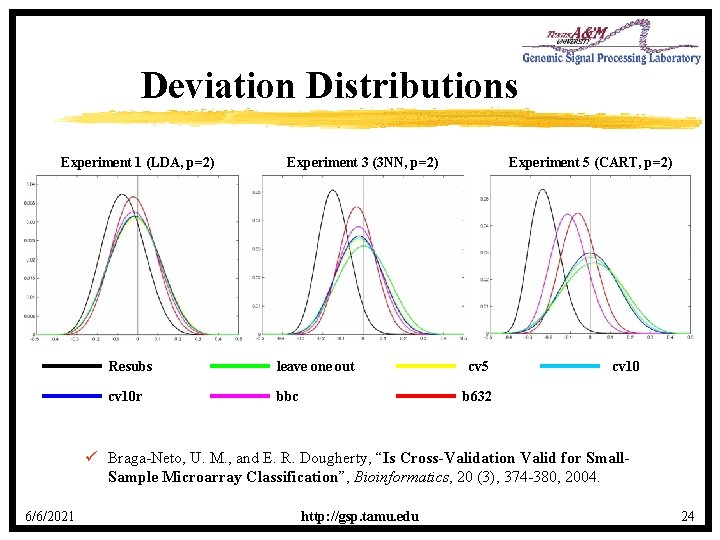
Deviation Distributions Experiment 1 (LDA, p=2) Experiment 3 (3 NN, p=2) Resubs leave one out cv 10 r bbc Experiment 5 (CART, p=2) cv 5 cv 10 b 632 ü Braga-Neto, U. M. , and E. R. Dougherty, “Is Cross-Validation Valid for Small. Sample Microarray Classification”, Bioinformatics, 20 (3), 374 -380, 2004. 6/6/2021 http: //gsp. tamu. edu 24

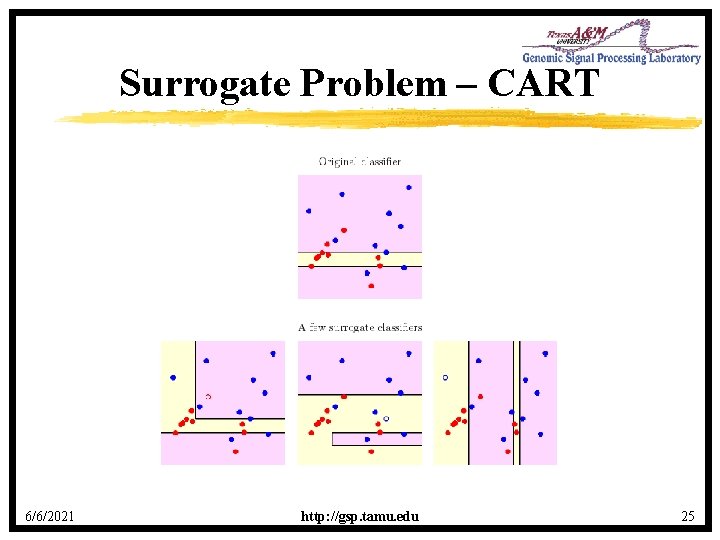
Surrogate Problem – CART 6/6/2021 http: //gsp. tamu. edu 25

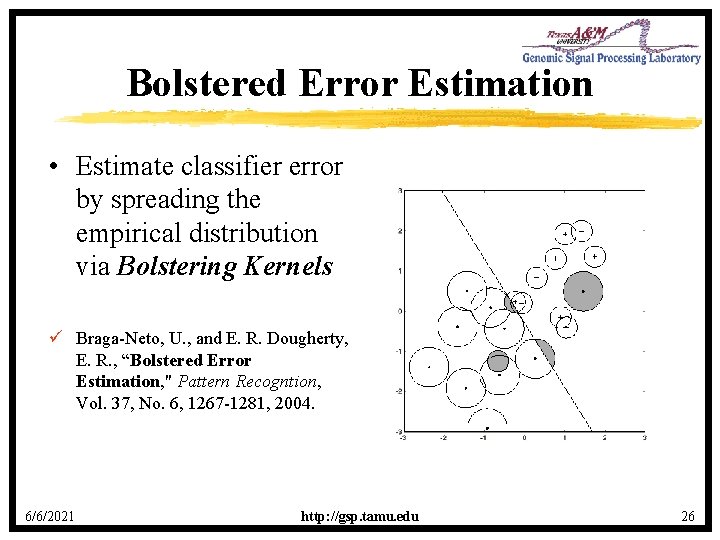
Bolstered Error Estimation • Estimate classifier error by spreading the empirical distribution via Bolstering Kernels ü Braga-Neto, U. , and E. R. Dougherty, E. R. , “Bolstered Error Estimation, " Pattern Recogntion, Vol. 37, No. 6, 1267 -1281, 2004. 6/6/2021 http: //gsp. tamu. edu 26

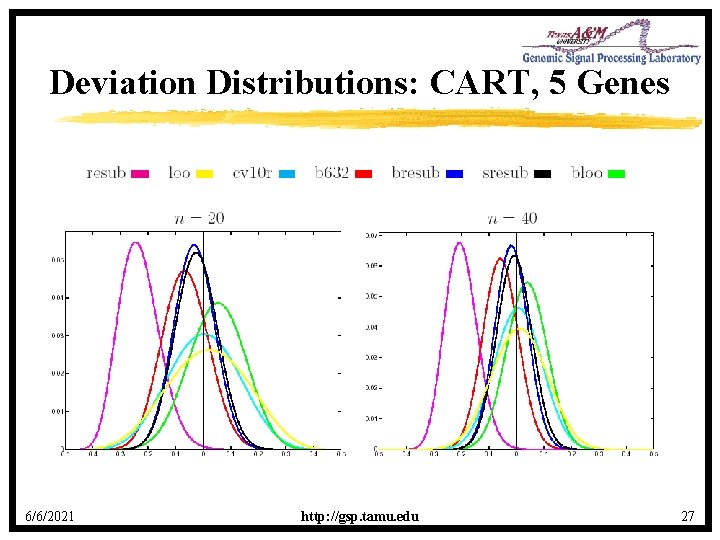
Deviation Distributions: CART, 5 Genes 6/6/2021 http: //gsp. tamu. edu 27

Salient Points for Small Samples • Beware of complex classifiers. • Keep feature sets small. – Yields smaller design errors – Fosters biological interpretation • Avoid cross-validation – where possible. • Recognize the heavy influence of the feature-label distribution and classification rule. • Report a list of classifiers and feature sets for analysis. • Issues: Analysis of classifier and feature-selection performance – – Better error estimation Mathematical analysis of error estimators – Braga-Neto, U. , and E. Dougherty, “Exact Performance Measures and Distributions of Error Estimators for Discrete Classifiers, ” Pattern Recognition, in press. 6/6/2021 http: //gsp. tamu. edu 28

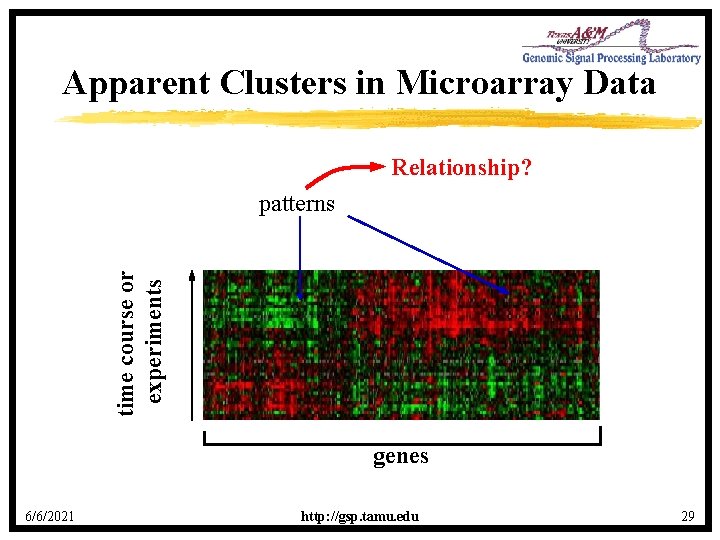
Apparent Clusters in Microarray Data Relationship? time course or experiments patterns genes 6/6/2021 http: //gsp. tamu. edu 29

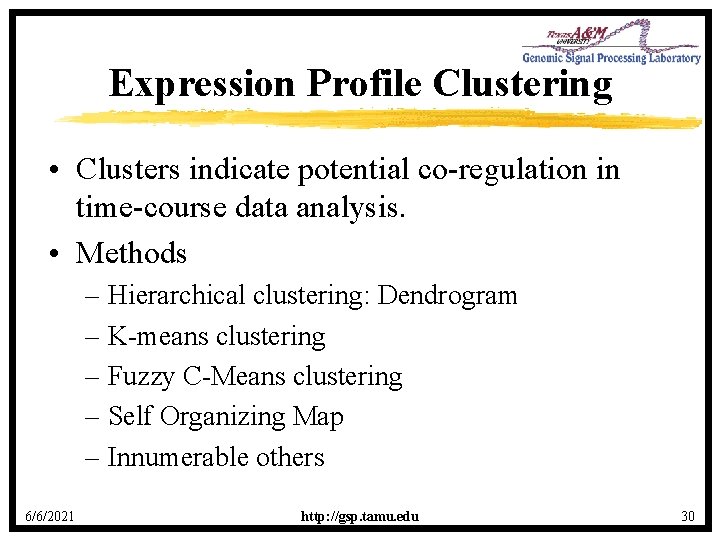
Expression Profile Clustering • Clusters indicate potential co-regulation in time-course data analysis. • Methods – Hierarchical clustering: Dendrogram – K-means clustering – Fuzzy C-Means clustering – Self Organizing Map – Innumerable others 6/6/2021 http: //gsp. tamu. edu 30

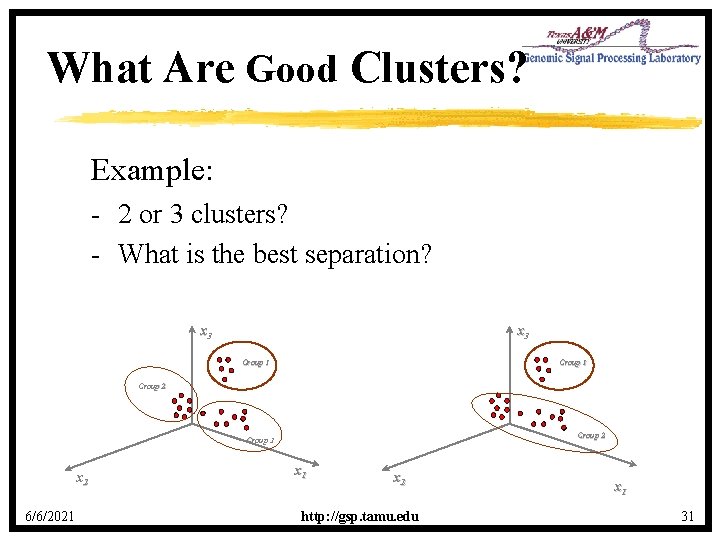
What Are Good Clusters? Example: - 2 or 3 clusters? - What is the best separation? x 3 Group 1 Group 2 Group 3 x 2 6/6/2021 x 2 http: //gsp. tamu. edu x 1 31

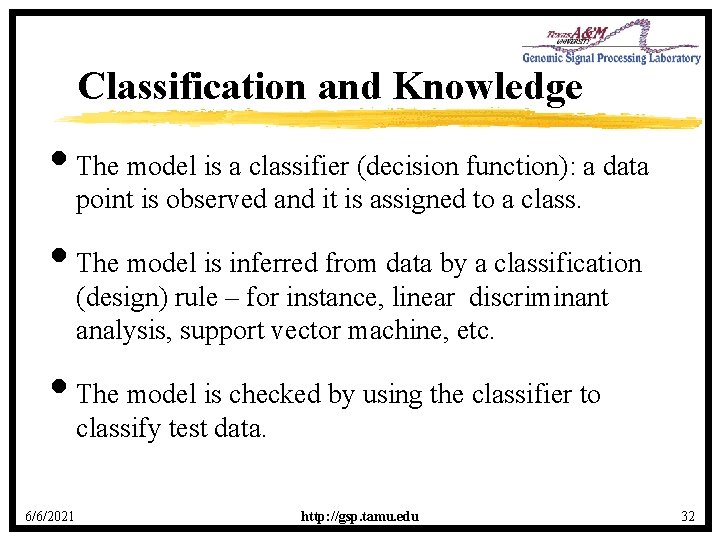
Classification and Knowledge • The model is a classifier (decision function): a data point is observed and it is assigned to a class. • The model is inferred from data by a classification (design) rule – for instance, linear discriminant analysis, support vector machine, etc. • The model is checked by using the classifier to classify test data. 6/6/2021 http: //gsp. tamu. edu 32

The Clustering Problem Jain et al. : “Clustering is subjective and validation measures are not general, they depend upon assumptions one is willing to accept. ” 6/6/2021 http: //gsp. tamu. edu 33

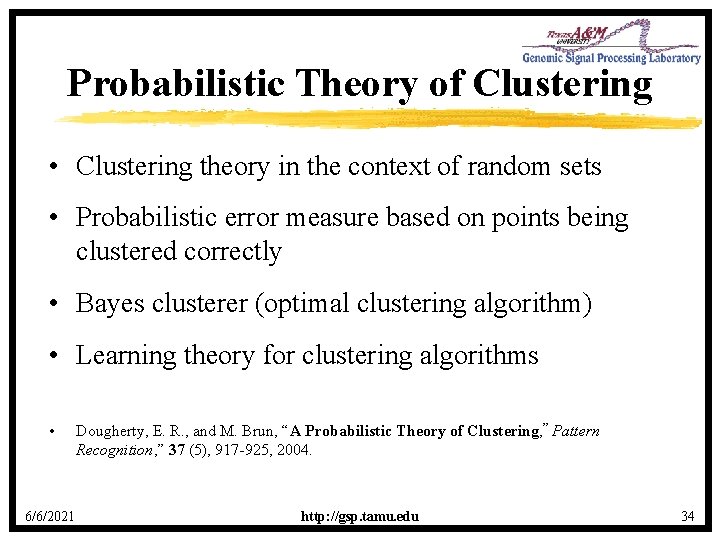
Probabilistic Theory of Clustering • Clustering theory in the context of random sets • Probabilistic error measure based on points being clustered correctly • Bayes clusterer (optimal clustering algorithm) • Learning theory for clustering algorithms • 6/6/2021 Dougherty, E. R. , and M. Brun, “A Probabilistic Theory of Clustering, ” Pattern Recognition, ” 37 (5), 917 -925, 2004. http: //gsp. tamu. edu 34

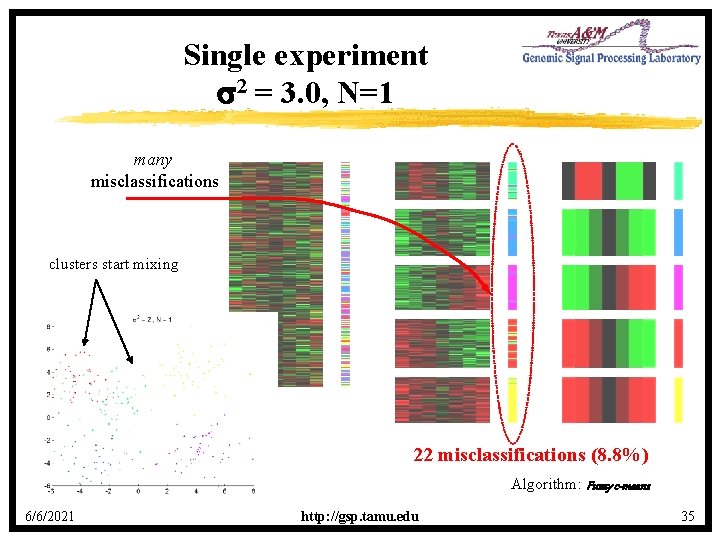
Single experiment s 2 = 3. 0, N=1 many misclassifications clusters start mixing 22 misclassifications (8. 8%) Algorithm: Fuzzy c-means 6/6/2021 http: //gsp. tamu. edu 35

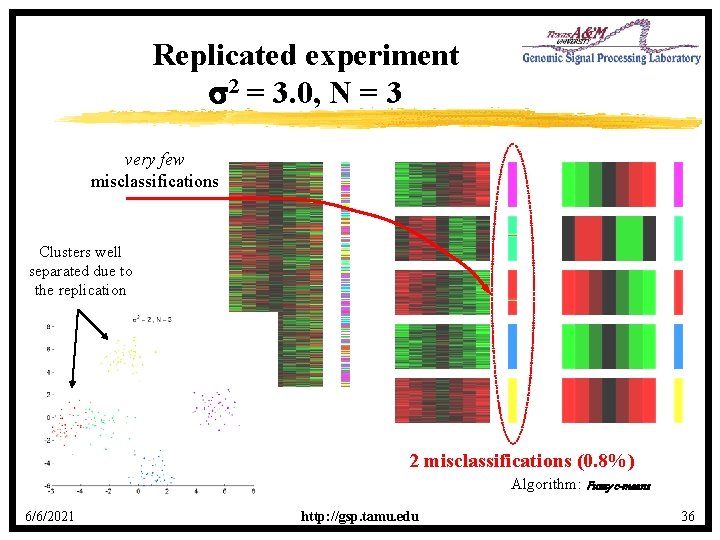
Replicated experiment s 2 = 3. 0, N = 3 very few misclassifications Clusters well separated due to the replication 2 misclassifications (0. 8%) Algorithm: Fuzzy c-means 6/6/2021 http: //gsp. tamu. edu 36

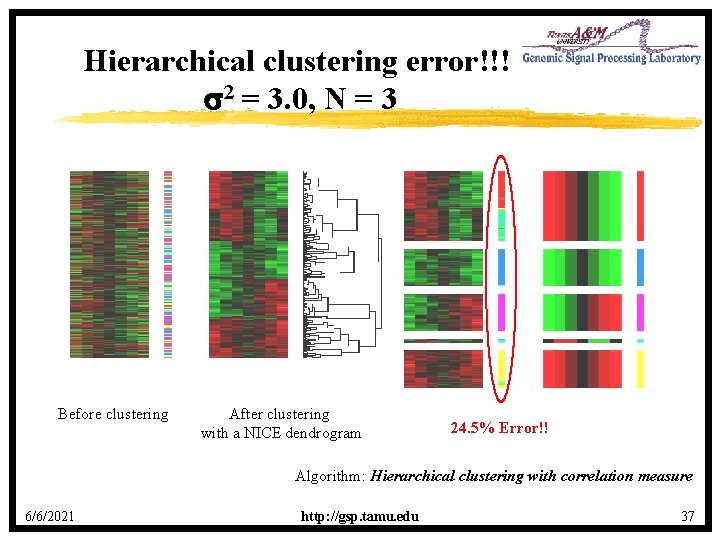
Hierarchical clustering error!!! s 2 = 3. 0, N = 3 Before clustering After clustering with a NICE dendrogram 24. 5% Error!! Algorithm: Hierarchical clustering with correlation measure 6/6/2021 http: //gsp. tamu. edu 37

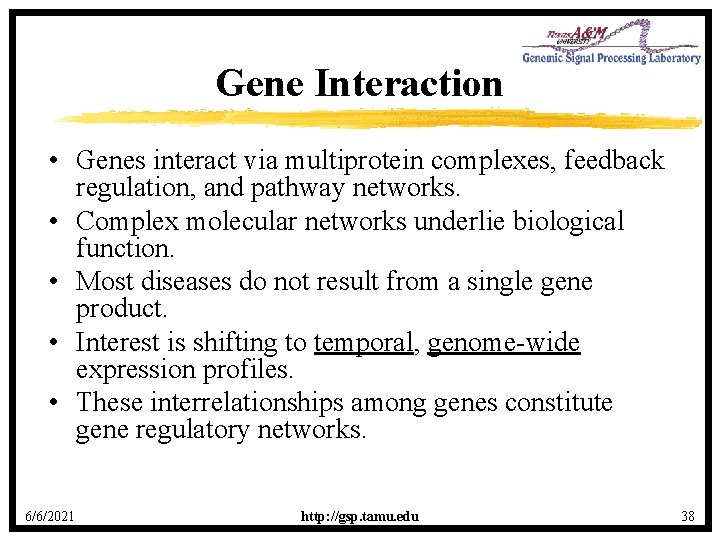
Gene Interaction • Genes interact via multiprotein complexes, feedback regulation, and pathway networks. • Complex molecular networks underlie biological function. • Most diseases do not result from a single gene product. • Interest is shifting to temporal, genome-wide expression profiles. • These interrelationships among genes constitute gene regulatory networks. 6/6/2021 http: //gsp. tamu. edu 38

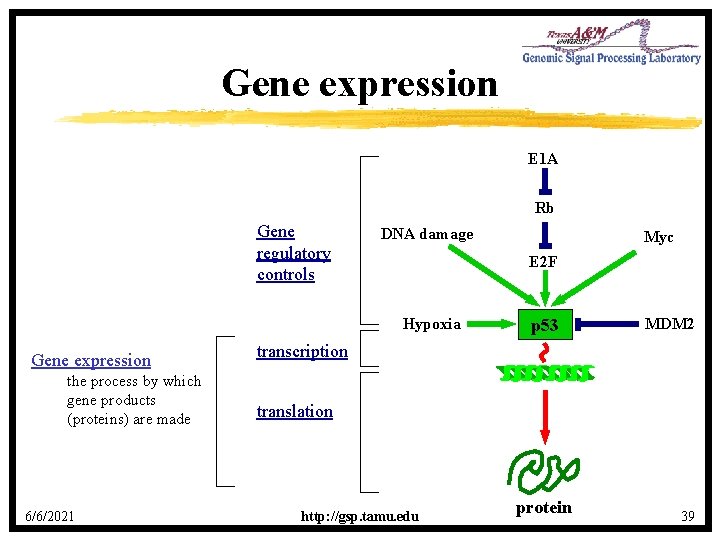
Gene expression E 1 A Rb Gene regulatory controls DNA damage E 2 F Hypoxia Gene expression the process by which gene products (proteins) are made 6/6/2021 Myc p 53 MDM 2 transcription translation http: //gsp. tamu. edu protein 39

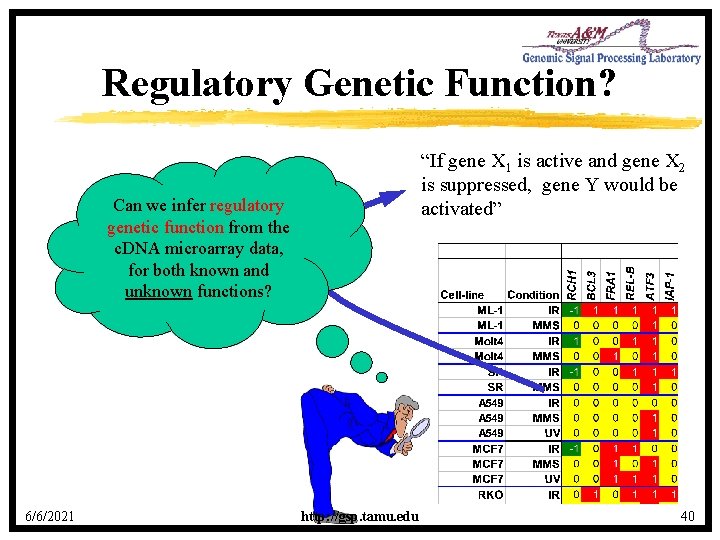
Regulatory Genetic Function? “If gene X 1 is active and gene X 2 is suppressed, gene Y would be activated” Can we infer regulatory genetic function from the c. DNA microarray data, for both known and unknown functions? 6/6/2021 http: //gsp. tamu. edu 40

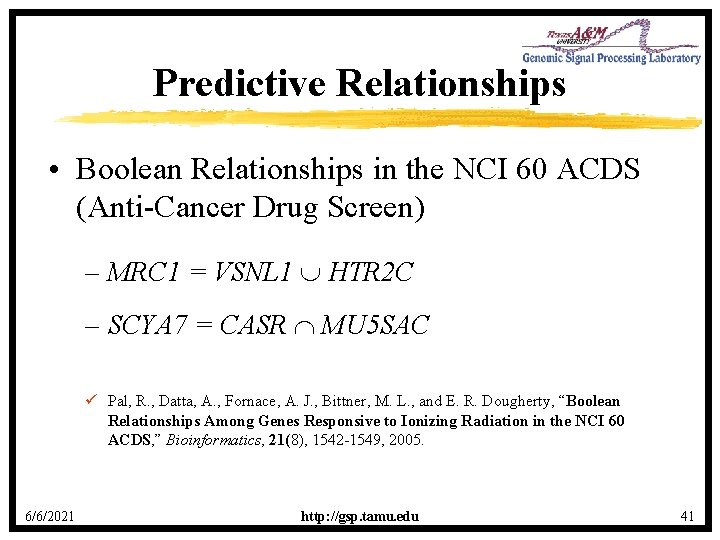
Predictive Relationships • Boolean Relationships in the NCI 60 ACDS (Anti-Cancer Drug Screen) – MRC 1 = VSNL 1 HTR 2 C – SCYA 7 = CASR MU 5 SAC ü Pal, R. , Datta, A. , Fornace, A. J. , Bittner, M. L. , and E. R. Dougherty, “Boolean Relationships Among Genes Responsive to Ionizing Radiation in the NCI 60 ACDS, ” Bioinformatics, 21(8), 1542 -1549, 2005. 6/6/2021 http: //gsp. tamu. edu 41

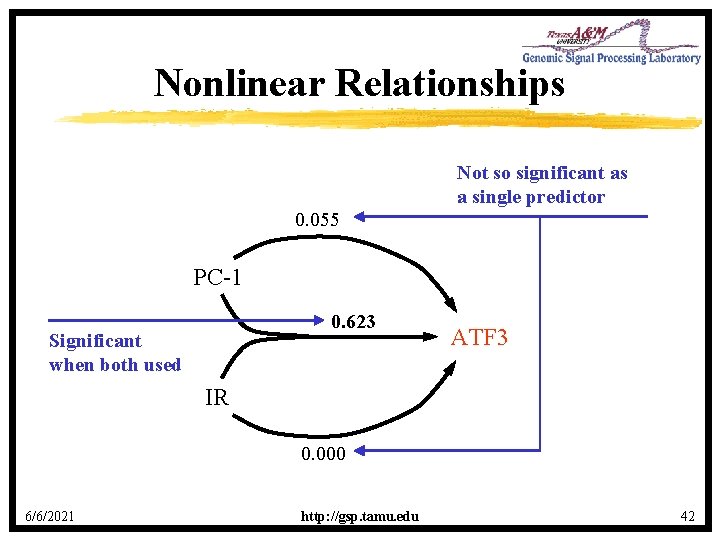
Nonlinear Relationships Not so significant as a single predictor 0. 055 PC-1 0. 623 Significant when both used ATF 3 IR 0. 000 6/6/2021 http: //gsp. tamu. edu 42

Goals of Dynamical Modeling • • Prediction of new targets based on pathway context. Stress and toxic response mechanisms. Off-target effects of therapeutic compounds. Characterization of disease states by dynamic behavior. • Gene- and protein-expression signatures for diagnostics. • Regulatory analysis for therapeutic intervention. 6/6/2021 http: //gsp. tamu. edu 43

Regulatory Modeling • Find analytical tools for expression data that can detect multivariate influences on decision-making produced by complex genetic networks. • Genomic signals must be processed to characterize their regulatory effects and their relationship to changes at both the genotypic and phenotypic levels. • Given a model, discover ways to intervene in its dynamics to obtain desired behavior. 6/6/2021 http: //gsp. tamu. edu 44

Model Properties • Incorporate rule-based dependencies between genes. – Rule-based dependencies may constitute important biological information. • Allow systematic study of global network dynamics. – In particular, individual gene effects on long-run network behavior. • Cope with uncertainty. – Small sample size, noisy measurements, robustness – System must be open to external latent variables 6/6/2021 http: //gsp. tamu. edu 45

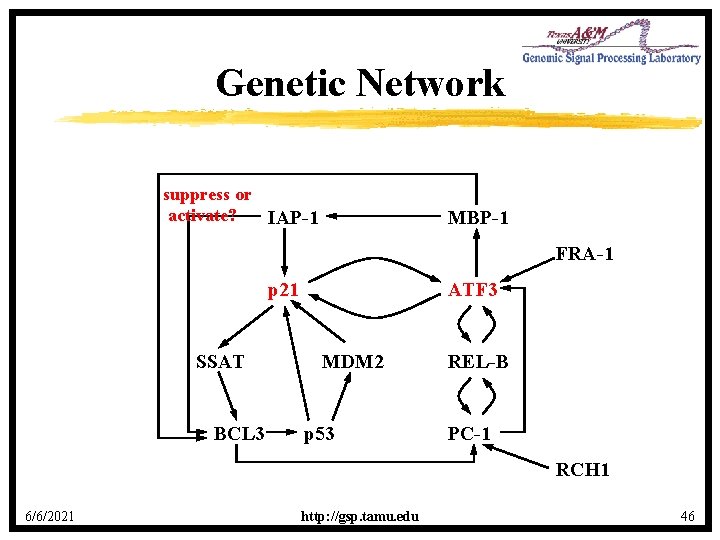
Genetic Network suppress or activate? IAP-1 MBP-1 FRA-1 p 21 SSAT BCL 3 ATF 3 MDM 2 p 53 REL-B PC-1 RCH 1 6/6/2021 http: //gsp. tamu. edu 46

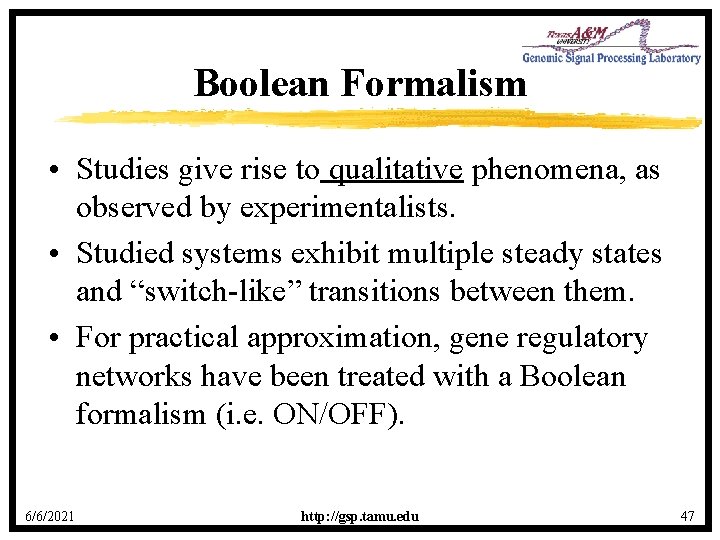
Boolean Formalism • Studies give rise to qualitative phenomena, as observed by experimentalists. • Studied systems exhibit multiple steady states and “switch-like” transitions between them. • For practical approximation, gene regulatory networks have been treated with a Boolean formalism (i. e. ON/OFF). 6/6/2021 http: //gsp. tamu. edu 47

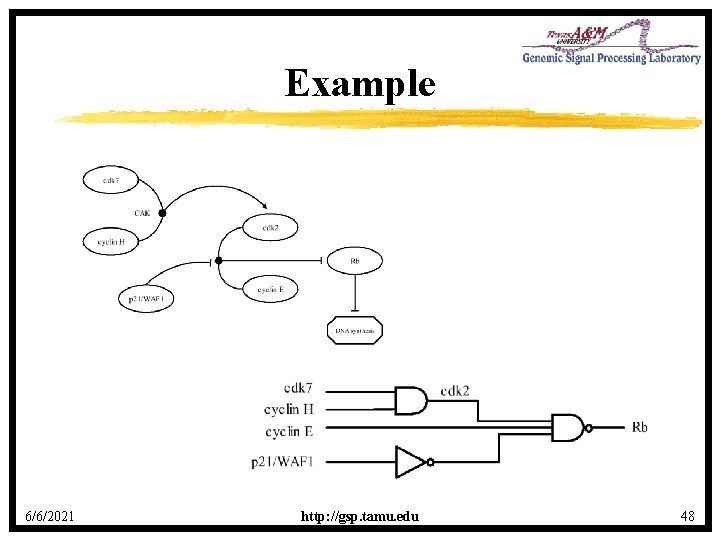
Example 6/6/2021 http: //gsp. tamu. edu 48

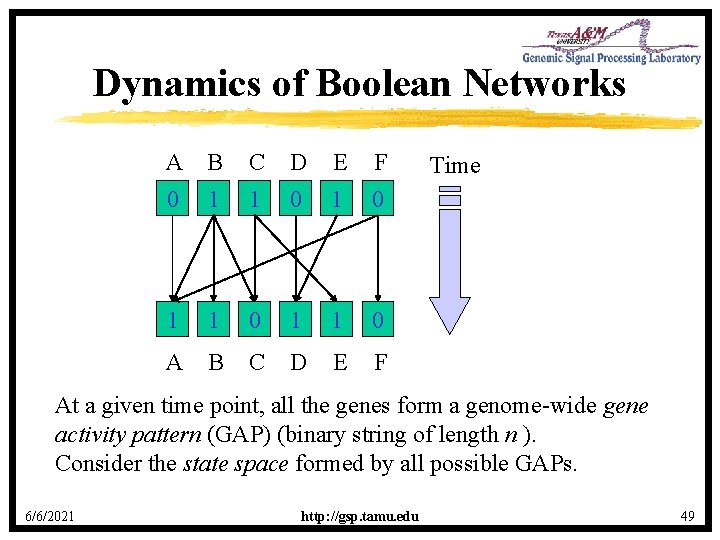
Dynamics of Boolean Networks A B C D E F 0 1 1 0 A B C D E F Time At a given time point, all the genes form a genome-wide gene activity pattern (GAP) (binary string of length n ). Consider the state space formed by all possible GAPs. 6/6/2021 http: //gsp. tamu. edu 49

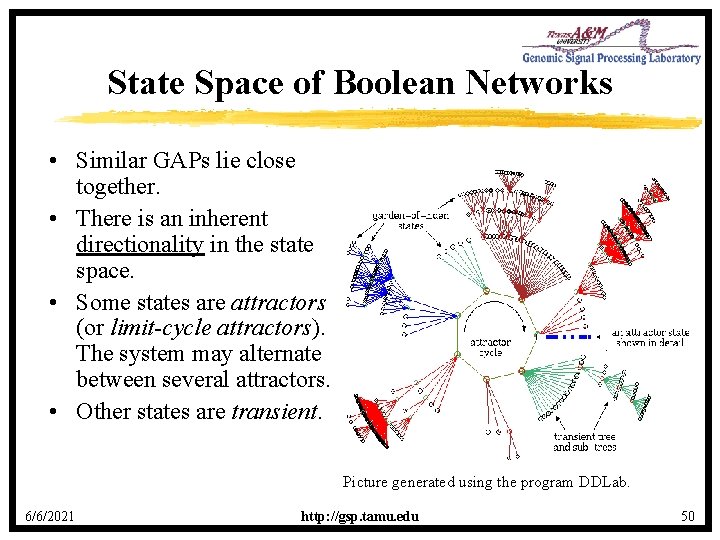
State Space of Boolean Networks • Similar GAPs lie close together. • There is an inherent directionality in the state space. • Some states are attractors (or limit-cycle attractors). The system may alternate between several attractors. • Other states are transient. Picture generated using the program DDLab. 6/6/2021 http: //gsp. tamu. edu 50

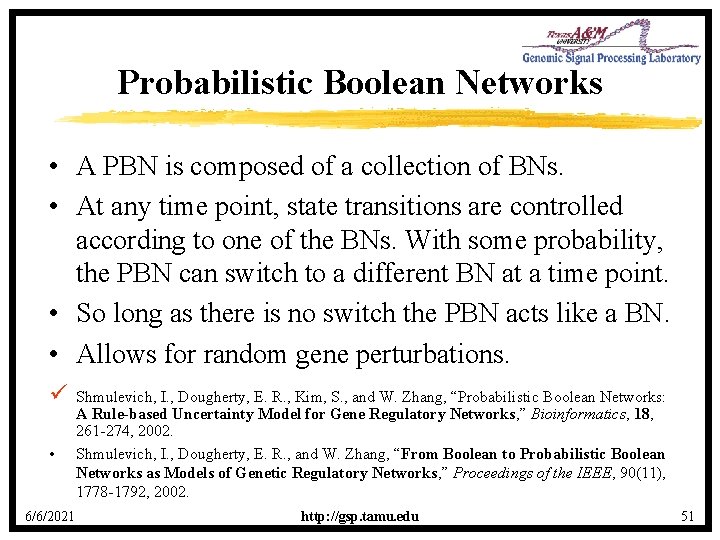
Probabilistic Boolean Networks • A PBN is composed of a collection of BNs. • At any time point, state transitions are controlled according to one of the BNs. With some probability, the PBN can switch to a different BN at a time point. • So long as there is no switch the PBN acts like a BN. • Allows for random gene perturbations. ü • 6/6/2021 Shmulevich, I. , Dougherty, E. R. , Kim, S. , and W. Zhang, “Probabilistic Boolean Networks: A Rule-based Uncertainty Model for Gene Regulatory Networks, ” Bioinformatics, 18, 261 -274, 2002. Shmulevich, I. , Dougherty, E. R. , and W. Zhang, “From Boolean to Probabilistic Boolean Networks as Models of Genetic Regulatory Networks, ” Proceedings of the IEEE, 90(11), 1778 -1792, 2002. http: //gsp. tamu. edu 51

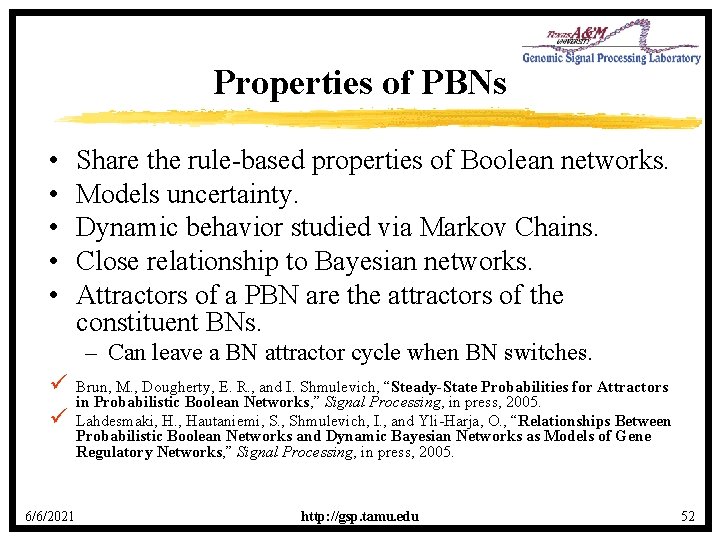
Properties of PBNs • • • Share the rule-based properties of Boolean networks. Models uncertainty. Dynamic behavior studied via Markov Chains. Close relationship to Bayesian networks. Attractors of a PBN are the attractors of the constituent BNs. – Can leave a BN attractor cycle when BN switches. ü ü 6/6/2021 Brun, M. , Dougherty, E. R. , and I. Shmulevich, “Steady-State Probabilities for Attractors in Probabilistic Boolean Networks, ” Signal Processing, in press, 2005. Lahdesmaki, H. , Hautaniemi, S. , Shmulevich, I. , and Yli-Harja, O. , “Relationships Between Probabilistic Boolean Networks and Dynamic Bayesian Networks as Models of Gene Regulatory Networks, ” Signal Processing, in press, 2005. http: //gsp. tamu. edu 52

Inference From Data • Key issues – Complex model – Limited data – Lack of appropriate time-course data for dynamics • Ill-posed inverse problem • Formalize inference by postulating criteria that constitute a solution space for the inverse problem. – Constraint criteria are composed of restrictions on the form of the network – biological, complexity. – Operational criteria are composed of relations that must be satisfied between the model and the data. 6/6/2021 http: //gsp. tamu. edu 53

Various Design Methods Proposed • Find genes with predictive capability for target gene (Co. D). • Use mutual-information clustering to find related genes. • Optimize connectivity in a Bayesian framework relative to the gene profiles in the data. • Find networks satisfying biologically related constraints such as limited attractor structure, transient time, and connectivity. • Assuming steady-state data, require data states to be attractors. • Assuming biological determinism within a given cellular context, design a PBN under the assumption that constituent BNs produce consistent data subsets in the sample data. 6/6/2021 http: //gsp. tamu. edu 54

Possible Intervention Goals • Minimize the mean first passage time to a desirable state. • Maximize the probability of reaching a desirable state before a certain fixed time. • Minimize the time needed to reach a desirable state with a given fixed probability. ü Shmulevich, I. , Dougherty, E. R. , and W. Zhang, “Gene Perturbation and Intervention in Probabilistic Boolean Networks, ” Bioinformatics, Vol. 18, 1319 -1331, 2002. ü Shmulevich, I. , Dougherty, E. R. , and W. Zhang, “Control of Stationary Behavior in Probabilistic Boolean Networks by Means of Structural Intervention, ” Biological Systems, Vol. 10, 431 -446, 2002. 6/6/2021 http: //gsp. tamu. edu 55

External Control in Gene Regulatory Networks • A PBN is a Markovian network: transition probabilities depend only on the previous state of the system. • Consider an external control variable and a cost function depending on the desirability of a state and cost of action. • Minimize the cost function by a sequence of control actions by using the classical method of dynamic programming. • Application: Design optimal treatment regime to drive the system away from states associated with cancer. ü Datta, A. , Choudhary, A. , Bittner, M. L. , and E. R. Dougherty, “External Control in Markovian Genetic Regulatory Networks, ” Machine Learning, 52 (1 -2), 169 -181, 2003. ü Pal, R. , Datta, A. , Bittner, M. L. , and E. R. Dougherty, “Intervention in Context-Sensitive Probabilistic Boolean Networks, ” Bioinformatics, 21(7), 1211 -1218, 2005. 6/6/2021 http: //gsp. tamu. edu 56

Collaborators q Texas A&M University – GSP Lab q q q q Aniruddha Datta Zixiang Xiong Erchin Serpedin Ivanov Jianping Hua Ulisses Braga-Neto Xiaobo Zhou q q q q Raymond Carroll Bani Mallick Tai Hsing Naisyin Wang q q q q 6/6/2021 Jeffrey Trent Michael Bittner Seungchan Kim Yoga Balagurunathan Marcel Brun Edward Suh Spyro Mousses Jaakko Astola Ioan Tabus q University of Sao Paulo q Translational Genomics Research Institute q Wei Zhang Ilya Shmulevich q Tampere University of Technology q q Yidong Chen Paul Meltzer q M. D. Anderson Cancer Center q Texas A&M University – Statistics q q NHGRI/NIH q Junior Barrera Ronaldo Hashimoto q Columbia University q Xiaodong Wang q Many Others and Students http: //gsp. tamu. edu 57