High Performance Computing for genomic applications Using genomic







- Slides: 17

High Performance Computing for genomic applications Using genomic software on Euler Scientific IT Services Michal Okoniewski, Samuel Fux, Manuel Kohler ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 1

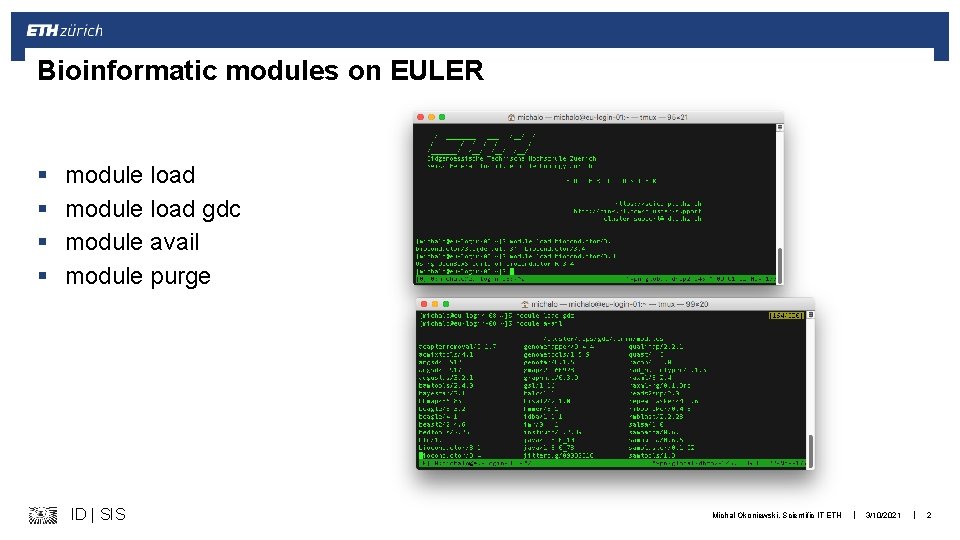
Bioinformatic modules on EULER § § module load gdc module avail module purge ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 2

Bioinformatic software jungle - categories § Data processing and convertion: samtools, picard, … § Aligners: bwa, bowtie, SHRi. MP, … § RNA aligners: STAR, tophat, subjunc § Transcriptome aligners: kallistio, sailfish, RSEM, … § Old-style aligners: Blast, Blat, VMATCH § De-novo assemblers: trinity, velvet, spades, … § Feature extraction, counting: HTSeq, feature. Count § Transcript discovery: cufflinks § Specialized tools: MISO, blast 2 go… ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 3

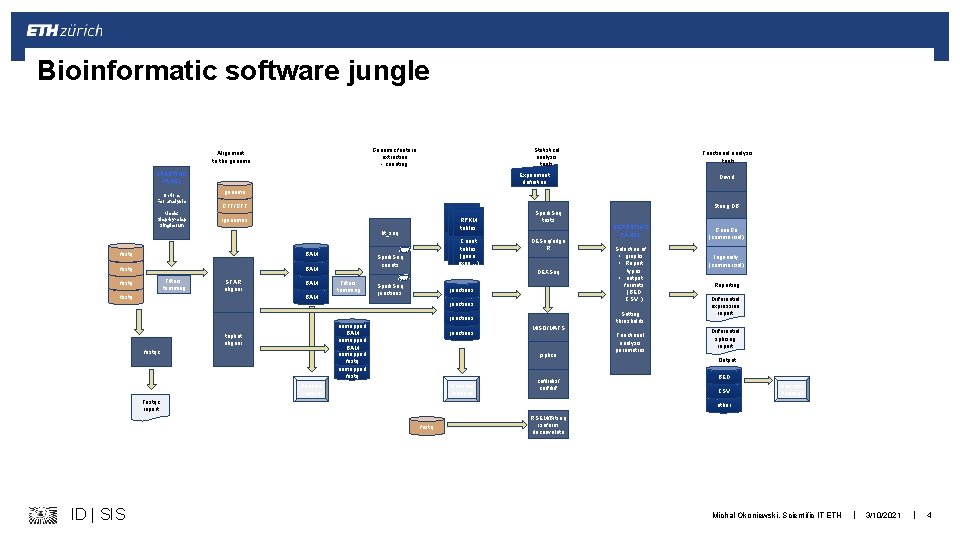
Bioinformatic software jungle Genomic feature extraction - counting Alignment to the genome Statistical analysis tools STARTING PANEL Options For analysis Mode: Step-by-step Single-run Experiment definition genome String DB RPKM tables igenomes fastq BAM Filters, trimming STAR aligner fastq BAM Count tables table (gene, exon…) Spark. Seq counts Spark. Seq tests DESeq/edge R DEXSeq Filters, trimming BAM Spark. Seq junctions unmapped BAM unmapped fastq tophat aligner fastqc junctions Genome browser Selection of • graphs • Report types • output formats (BED CSV. . ) MISO/MATS jsplice Genome browser REPORTING PANEL Setting thresholds junctions cufflinks/ cuffdiff Fastqc report Functional analysis parametres Gene. Go (commercial) Ingenuity (commercial) Reporting Differential expression report Differential splicing report Output BED CSV Genome browser other fastq ID | SIS David GTF/GFF ht_seq fastq Functional analysis tools RSEM/Bitseq isoform deconvolute Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 4

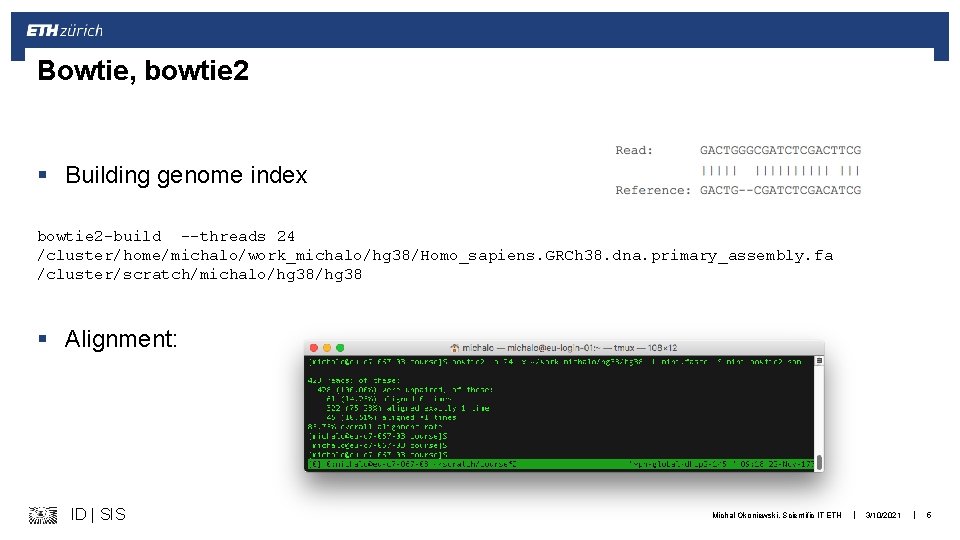
Bowtie, bowtie 2 § Building genome index bowtie 2 -build --threads 24 /cluster/home/michalo/work_michalo/hg 38/Homo_sapiens. GRCh 38. dna. primary_assembly. fa /cluster/scratch/michalo/hg 38 § Alignment: ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 5

Tophat § Classic splice-aware aligner § Uses bowtie 2 as engine, so also bowtie 2 index tophat -p 24 -o tophat_out --library-type fr-firststrand ~/work_michalo/hg 38 mini. fastq. gz § Manual: http: //ccb. jhu. edu/software/tophat/index. shtml ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 6

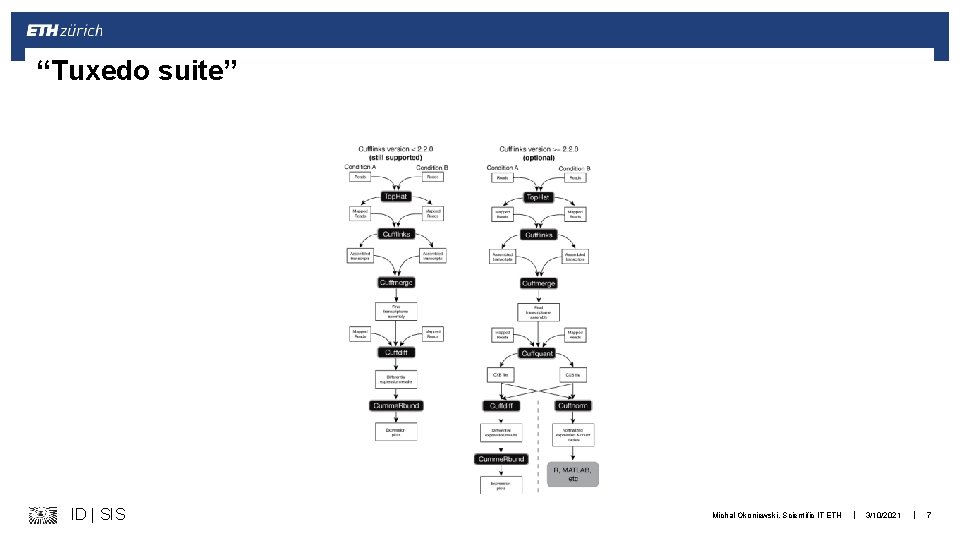
“Tuxedo suite” ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 7

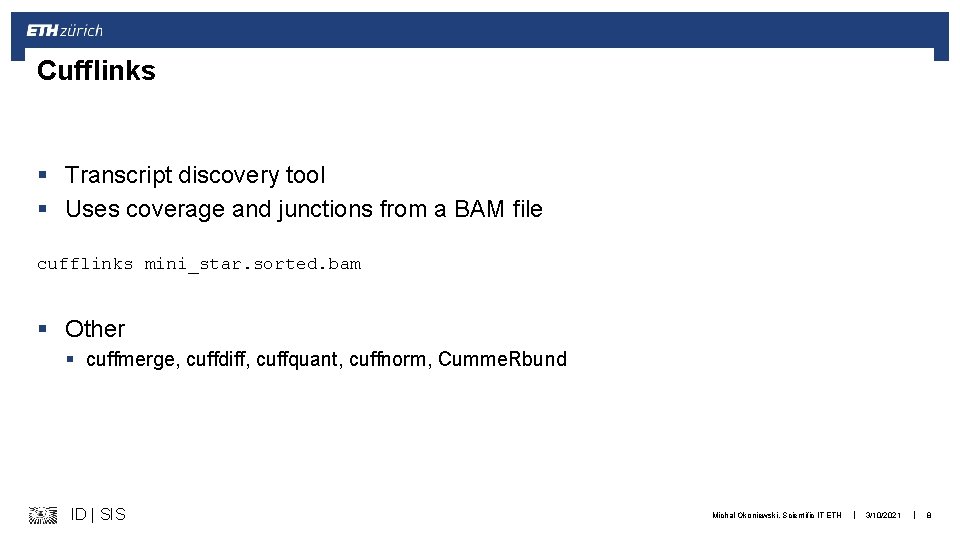
Cufflinks § Transcript discovery tool § Uses coverage and junctions from a BAM file cufflinks mini_star. sorted. bam § Other § cuffmerge, cuffdiff, cuffquant, cuffnorm, Cumme. Rbund ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 8

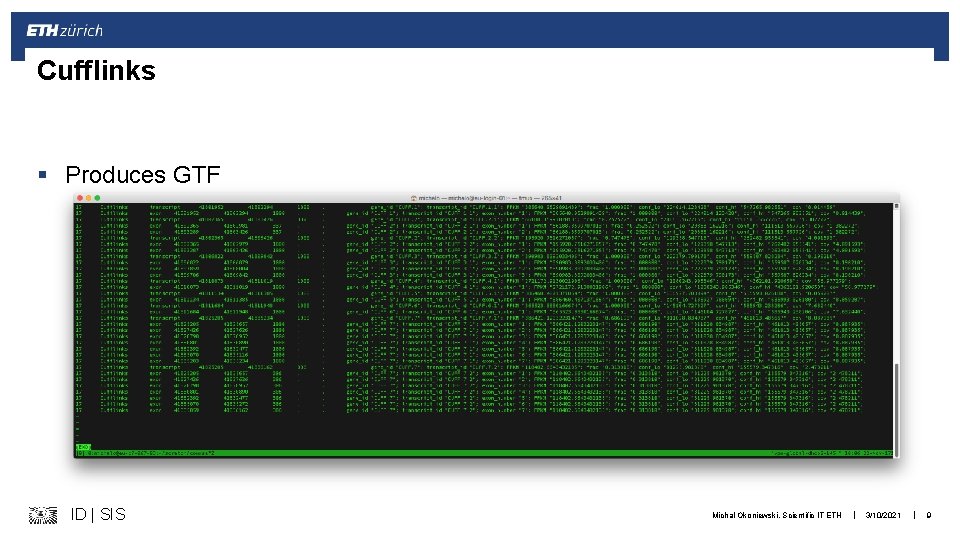
Cufflinks § Produces GTF ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 9

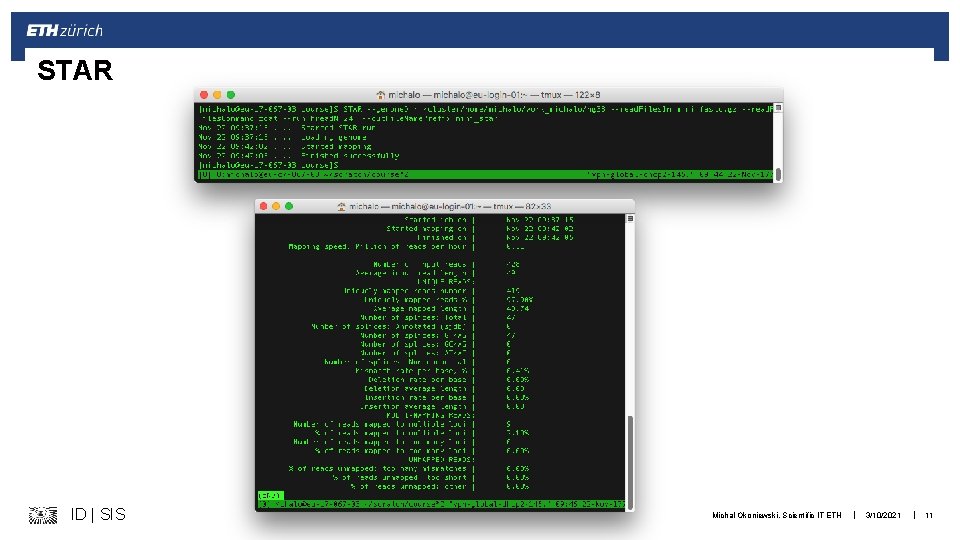
STAR § § Splice aware aligner, loading index into memory Results similar to tophat, but faster --genome. Load. And. Keep With specific options, can produce BAM and do the counting too § https: //github. com/alexdobin/STAR/blob/master/doc/STARmanual. pdf ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 10

STAR ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 11

subread § Includes subjunc similar to STAR and feature. Counts § Building index subread-buildindex -o /cluster/home/michalo/work_michalo/hg 38/subread_index/hg 38 /cluster/home/michalo/work_michalo/hg 38/Homo_sapiens. GRCh 38. dna. primary_assembly. fa § Alignment subread -T 24 -i /cluster/home/michalo/work_michalo/hg 38/subread_index/hg 38 -r mini. fastq -o mapped_reads_subjunc/mini. bam subjunc -T 24 -i /cluster/home/michalo/work_michalo/hg 38/subread_index/hg 38 -r mini. fastq -o mapped_reads_subjunc/mini. bam § http: //bioinf. wehi. edu. au/subread-package/Subread. Users. Guide. pdf ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 12

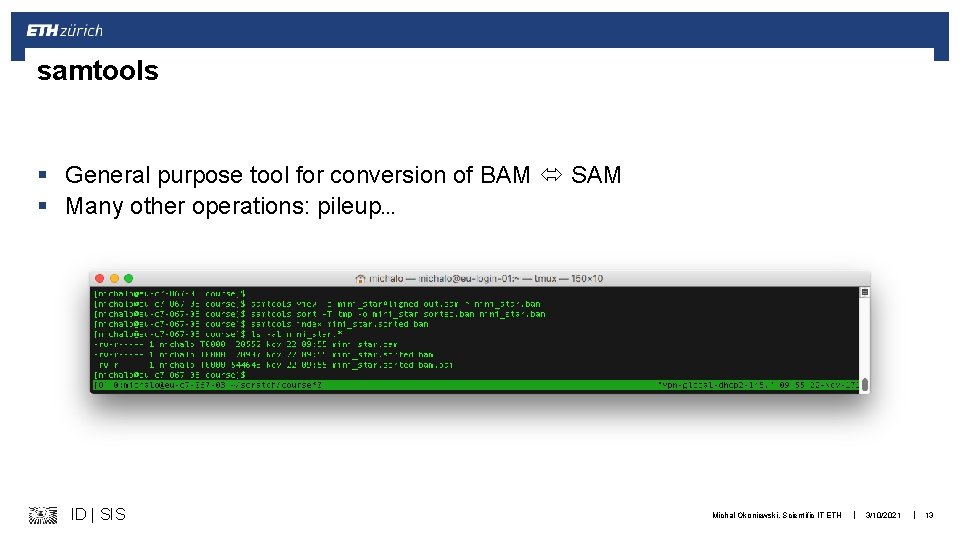
samtools § General purpose tool for conversion of BAM SAM § Many other operations: pileup… ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 13

feature. Counts § Fast and flexible counting in genomic features feature. Counts -M -s 2 -T 24 -t gene -g gene_id -a /cluster/home/michalo/work_michalo/hg 38/Homo_sapiens. GRCh 38. 86. chr. gtf -o mini. cnt mini_star. sorted. bam § Important options: ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 14

feature. Counts ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 15

GATK § GATK is a genomic toolbox for various operations related mainly to genomic variants calling § Operations include producing a variant file *. vcf from an alignment file *. bam module load gcc/4. 8. 2 gdc java/1. 8. 0_73 gatk/3. 5 java -jar Genome. Analysis. TK. jar -T Unified. Genotyper -R ref/human_g 1 k_b 37_20. fasta -I bams/exp_design/NA 12878_wgs_20. bam -o sandbox/NA 12878_wgs_20_UG_calls. vcf -glm BOTH -L 20: 10, 000 -10, 200, 000 https: //software. broadinstitute. org/gatk/documentation/tooldocs/current/ https: //software. broadinstitute. org/gatk/documentation/topic? name=tutorials http: //gatkforums. broadinstitute. org/gatk/discussion/7869/howto-discover-variantswith-gatk-a-gatk-workshop-tutorial ID | SIS Michal Okoniewski, Scientific IT ETH | 3/10/2021 | 16

Thank you! Using genomic software on Euler