EE 150 a Genomic Signal and Information Processing



![Markov Chain Model Cont’d Let i=[ i, 1 i, 2 … i, k+2]T be Markov Chain Model Cont’d Let i=[ i, 1 i, 2 … i, k+2]T be](https://slidetodoc.com/presentation_image_h2/cfe2e81a85c82c4596dc9543ab723ac7/image-18.jpg)

- Slides: 27

EE 150 a – Genomic Signal and Information Processing On DNA Microarrays Technology October 12, 2004

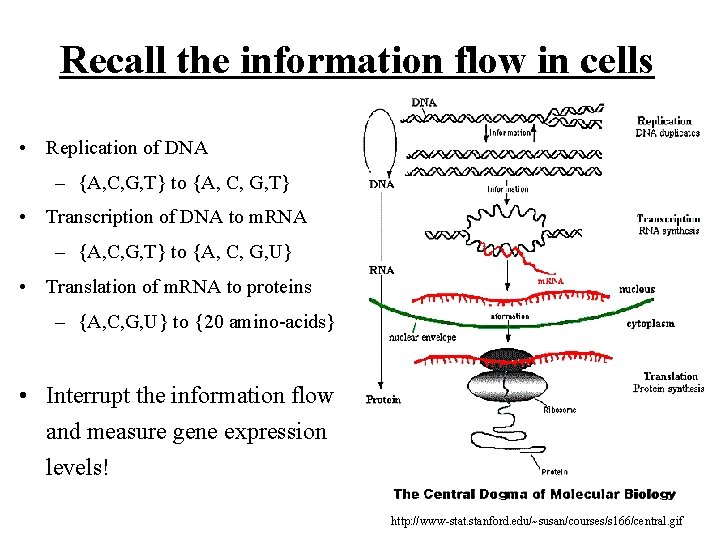
Recall the information flow in cells • Replication of DNA – {A, C, G, T} to {A, C, G, T} • Transcription of DNA to m. RNA – {A, C, G, T} to {A, C, G, U} • Translation of m. RNA to proteins – {A, C, G, U} to {20 amino-acids} • Interrupt the information flow and measure gene expression levels! http: //www-stat. stanford. edu/~susan/courses/s 166/central. gif

Gene Microarrays • A medium for matching known and unknown sequences of nucleotides based on hybridization (base-pairing: A-T, C-G) • Applications – identification of a sequence (gene or gene mutation) – determination of expression level (abundance) of genes – verification of computationally determined genes • Enables massively parallel gene expression studies • Two types of molecules take part in the experiments: – probes, orderly arranged on an array – targets, the unknown samples to be detected

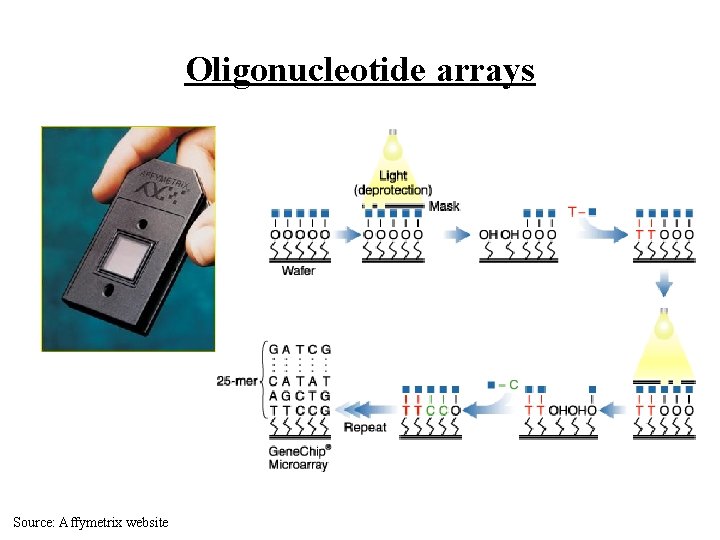
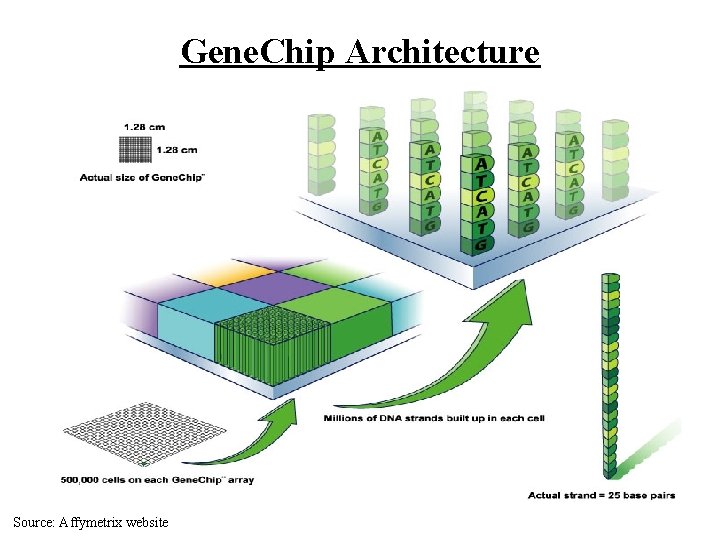
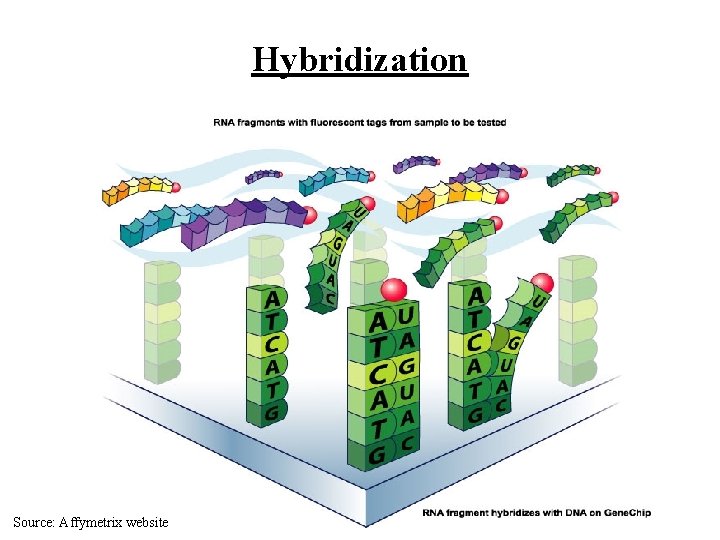
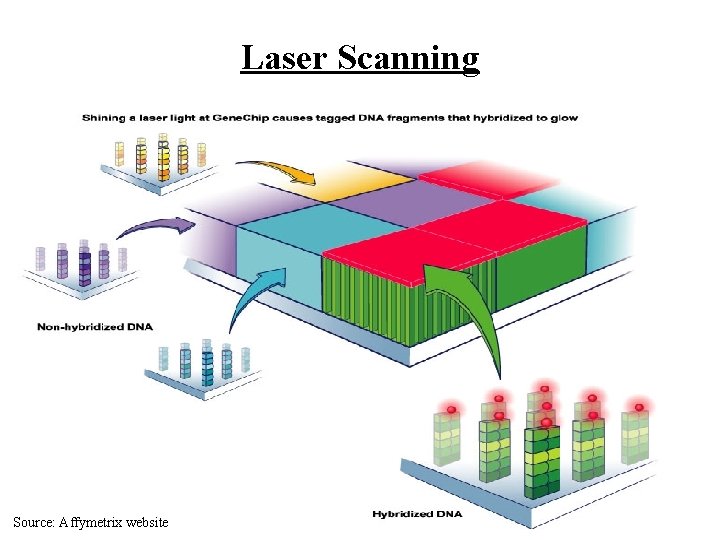
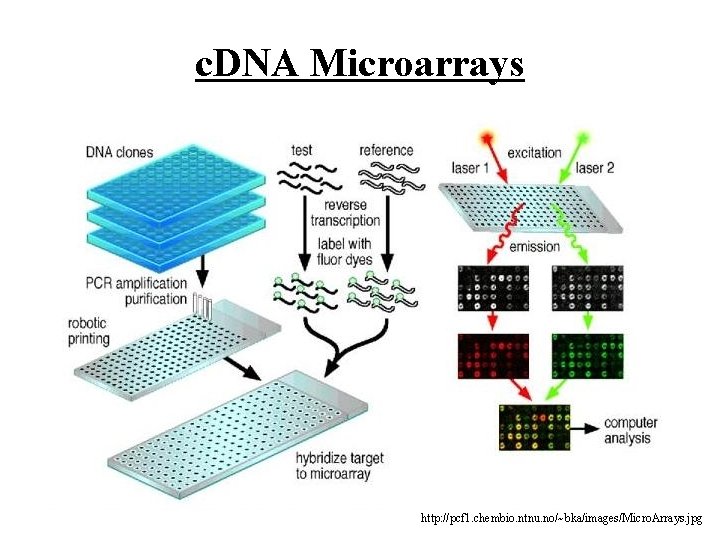
Microarray Technologies • Oligonucleotide arrays (Affymetrix Gene. Chips) – probes are photo-etched on a chip (20 -80 nucleotides) – dye-labeled m. RNA is hybridized to the chip – laser scanning is used to detect gene expression levels (i. e. , amount of m. RNA) • c. DNA arrays – complementary DNA (c. DNA) sequences “spotted” on arrays (500 -1000 nucleotides) – dye-labeled m. RNA is hybridized to the chip (2 types!) – laser scanning is used to detect gene expression levels • There are various hybrids of the two technologies above

Oligonucleotide arrays Source: Affymetrix website

Gene. Chip Architecture Source: Affymetrix website

Hybridization Source: Affymetrix website

Laser Scanning Source: Affymetrix website

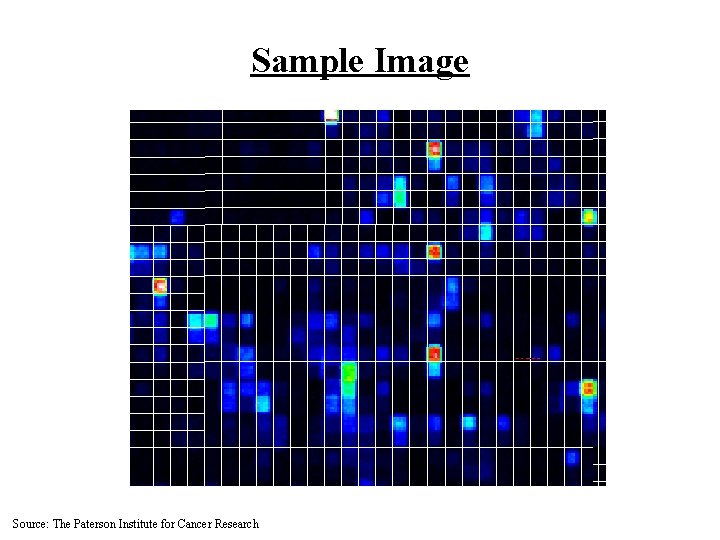
Sample Image Source: The Paterson Institute for Cancer Research

Competing Microarray Technologies • So far considered oligonucleotide arrays: – automated, on-chip design – light dispersion may cause problems – short probes, 20 -80 • c. DNA microarrays are another technology: – longer probes obtained via PCR, polymerase chain reaction – [sidenote: what is optimal length? ] – probes grown in a lab, robot printing – two types of targets – control and test

c. DNA Microarrays http: //pcf 1. chembio. ntnu. no/~bka/images/Micro. Arrays. jpg

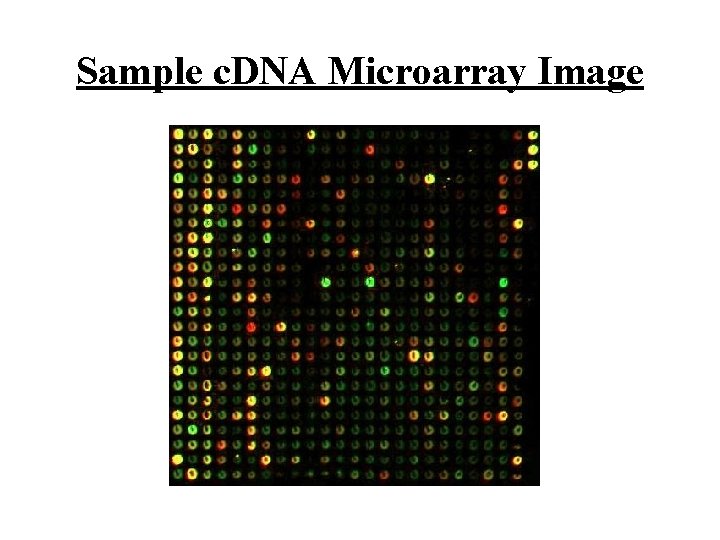
Sample c. DNA Microarray Image

Some Design Issues • Photo-etching based design: unwanted light exposure – border minimization – the probes are 20 -80 long • Hybridization: binding of a target to its perfect complement • However, when a probe differs from a target by a small number of bases, it still may bind • This non-specific binding (cross-hybridization) is a source of measurement noise • In special cases (e. g. , arrays for gene detection), designer has a lot of control over the landscape of the probes on the array

Dealing with Measurement Noise • Recent models of microarray noise – measurements reveal signal-dependent noise (i. e. , shot-noise) as the major component – additional Gaussian-like noise due to sample preparation, image scanning, etc. • Image processing assumes image background noise – attempts to subtract it – sets up thresholds • Lack of models of processes on microarrays

Probabilistic DNA Microarray Model • Consider an m£m DNA microarray, with m 2 unique types of nucleotide probes • A total of N molecules of n different types of c. DNA targets with concentrations c 1, …, cn, is applied to the microarray • Measurement is taken after the system reached chemical equilibrium • Our goal: from the scanned image, estimate the concentrations

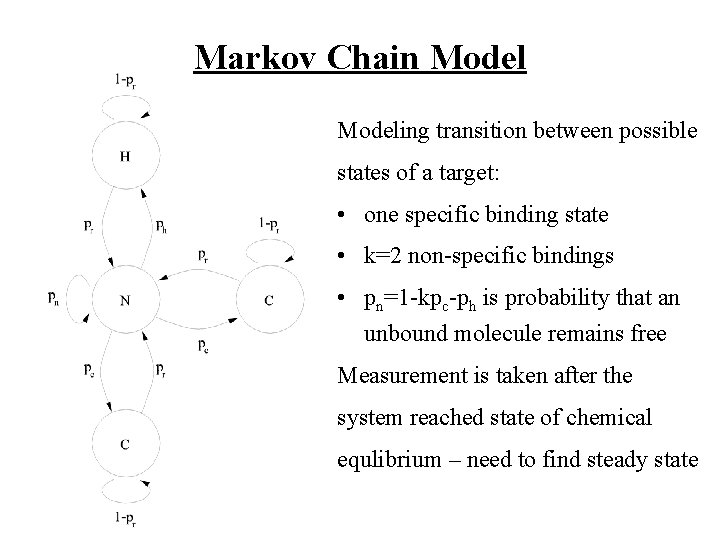
DNA Microarray Model Cont’d • Each target may hybridize to only one type of probe • There are k non-specific bindings • Model diffusion of unbound molecules by random walk; distribution of unbound molecules uniform on the array – justified by reported experimental results • Assume known probabilities of hybridization and crosshybridization – Theoretically: from melting temperature – Experimentally: measurements (e. g. , from control target samples)

Markov Chain Modeling transition between possible states of a target: • one specific binding state • k=2 non-specific bindings • pn=1 -kpc-ph is probability that an unbound molecule remains free Measurement is taken after the system reached state of chemical equlibrium – need to find steady state
![Markov Chain Model Contd Let i i 1 i 2 i k2T be Markov Chain Model Cont’d Let i=[ i, 1 i, 2 … i, k+2]T be](https://slidetodoc.com/presentation_image_h2/cfe2e81a85c82c4596dc9543ab723ac7/image-18.jpg)
Markov Chain Model Cont’d Let i=[ i, 1 i, 2 … i, k+2]T be a vector whose components are numbers of the type i targets that are in one of the k+2 states of the Markov chain • i, 1 is the # of hybridized molecules • i, j, 2 < j · k+2 is # of cross-hybrid. Note that k=1 k+2 i, k=ci for every i.

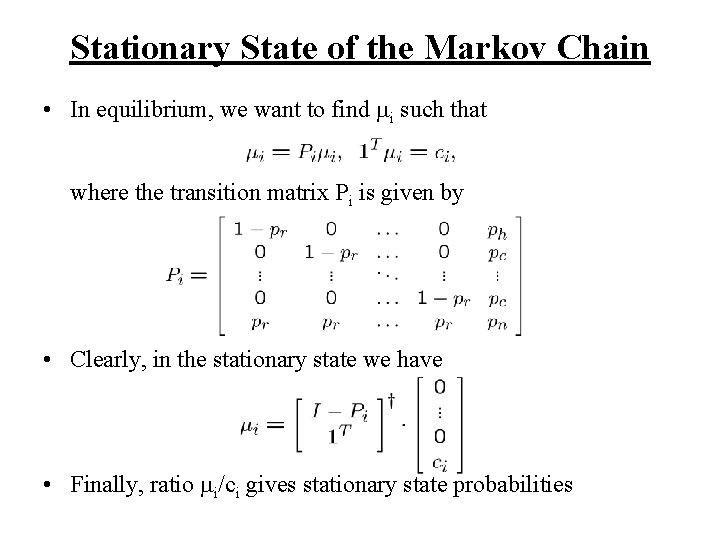
Stationary State of the Markov Chain • In equilibrium, we want to find i such that where the transition matrix Pi is given by • Clearly, in the stationary state we have • Finally, ratio i/ci gives stationary state probabilities

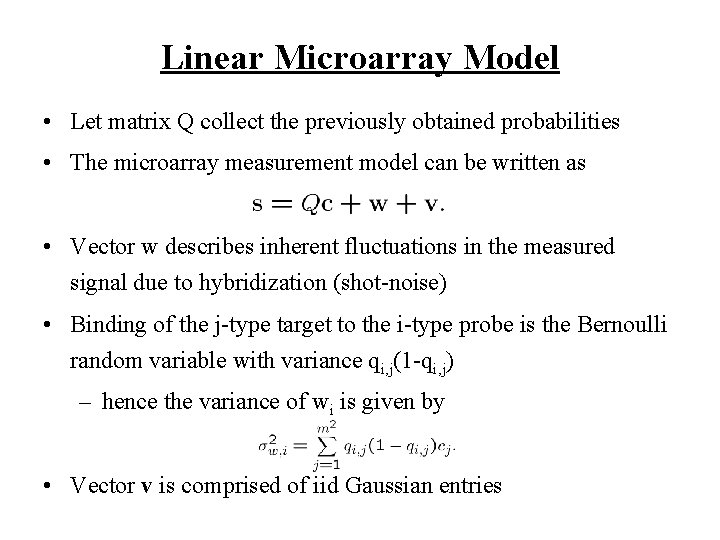
Linear Microarray Model • Let matrix Q collect the previously obtained probabilities • The microarray measurement model can be written as • Vector w describes inherent fluctuations in the measured signal due to hybridization (shot-noise) • Binding of the j-type target to the i-type probe is the Bernoulli random variable with variance qi, j(1 -qi, j) – hence the variance of wi is given by • Vector v is comprised of iid Gaussian entries

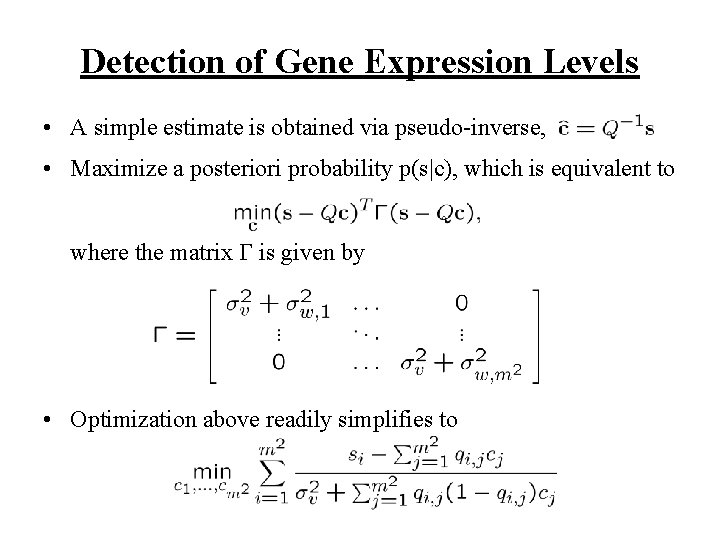
Detection of Gene Expression Levels • A simple estimate is obtained via pseudo-inverse, • Maximize a posteriori probability p(s|c), which is equivalent to where the matrix is given by • Optimization above readily simplifies to

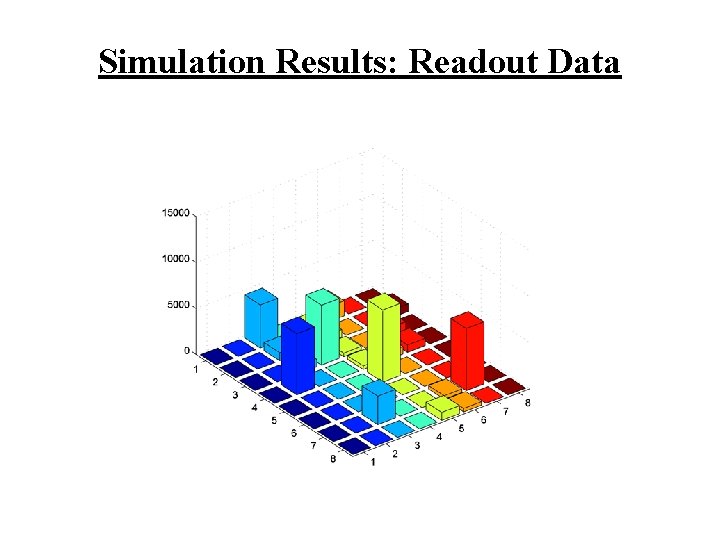
Simulation Results • Consider an 8£ 8 array (m=8) • Apply n=6 types of targets • Concentrations: [1 e 5 2 e 5 1 e 5 2 e 5] (N=1 e 6) • Assume the following probabilities: – hybridization – 0. 8 – cross-hybridization – 0. 1 – release – 0. 02 • Let k=3 (number of non-specific bindings) • Free molecules perform random walk on the array

Simulation Results: Readout Data

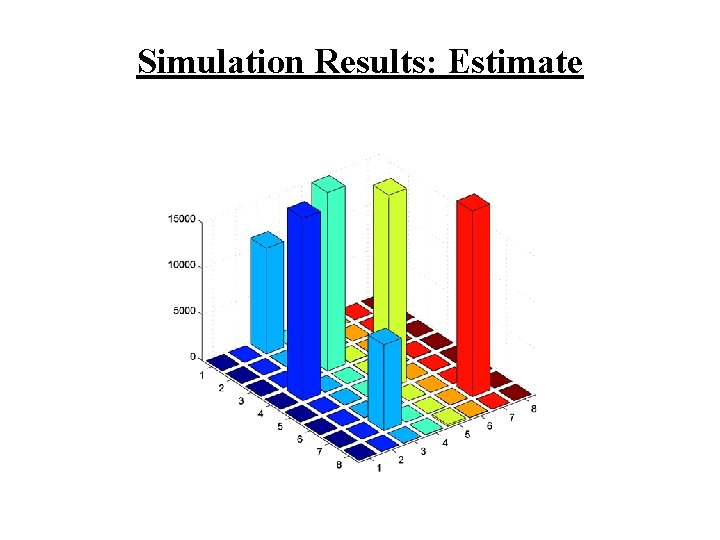
Simulation Results: Estimate

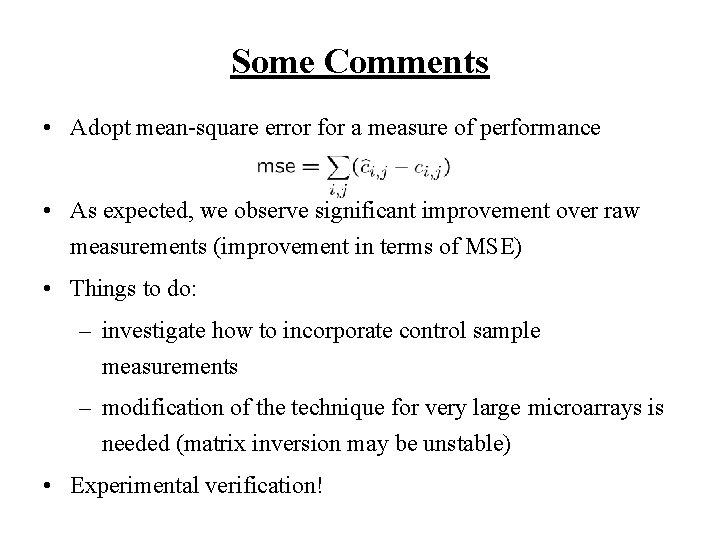
Some Comments • Adopt mean-square error for a measure of performance • As expected, we observe significant improvement over raw measurements (improvement in terms of MSE) • Things to do: – investigate how to incorporate control sample measurements – modification of the technique for very large microarrays is needed (matrix inversion may be unstable) • Experimental verification!

Why is this Estimation Problem Important? • Microarrays measure expression levels of thousands of gene simultaneously • Assume that we are taking samples at different times during a biological process • Cluster data in the expression level space – relatedness in biological function often implies similarity in expression behavior (and vice versa) – similar expression behavior indicates co-expression • Clustering of expression level data heavily depends on the measurements – better estimation may lead to different functionality conclusions

Summary • Microarray technologies are becoming of great importance for medicine and biology – understanding how the cell functions, effects on organism – towards diagnostics, personalized medicine • Plenty of interesting problems – combinatorial design techniques – statistical analysis of the data – signal processing / estimation