Plan 23 2 2004 Eukaryot DNA replikasjon 1



























- Slides: 64

Plan 23. 2. 2004 Eukaryot DNA replikasjon 1. Replikasjonsorigins 2. Enzymologi 3. Initiering og Regulering av DNA replikasjon


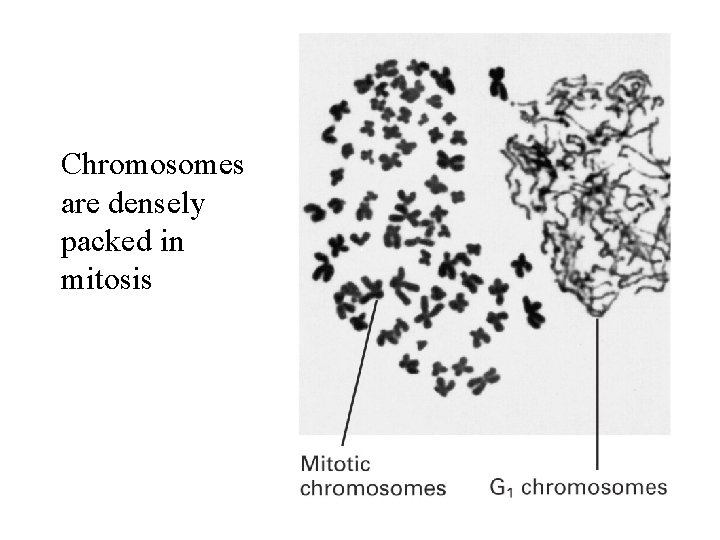
Chromosomes are densely packed in mitosis

The accuracy of DNA replication is seen in the quality of the product Fertilised Egg Product

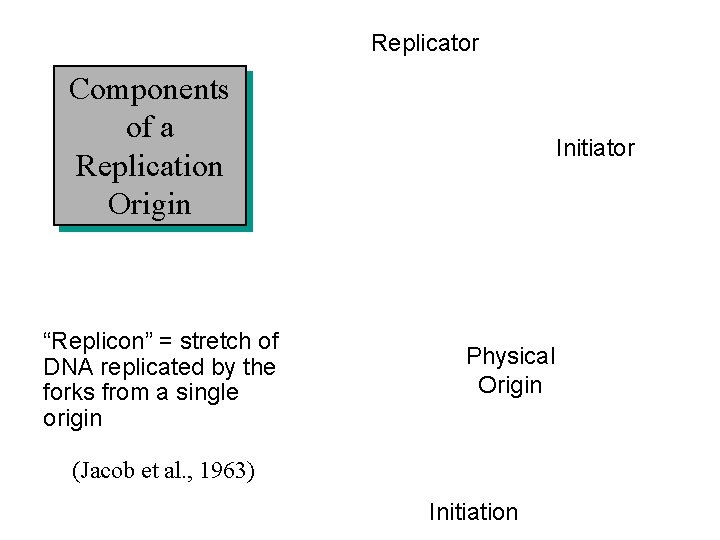
Replicator Components of a Replication Origin “Replicon” = stretch of DNA replicated by the forks from a single origin Initiator Physical Origin (Jacob et al. , 1963) Initiation

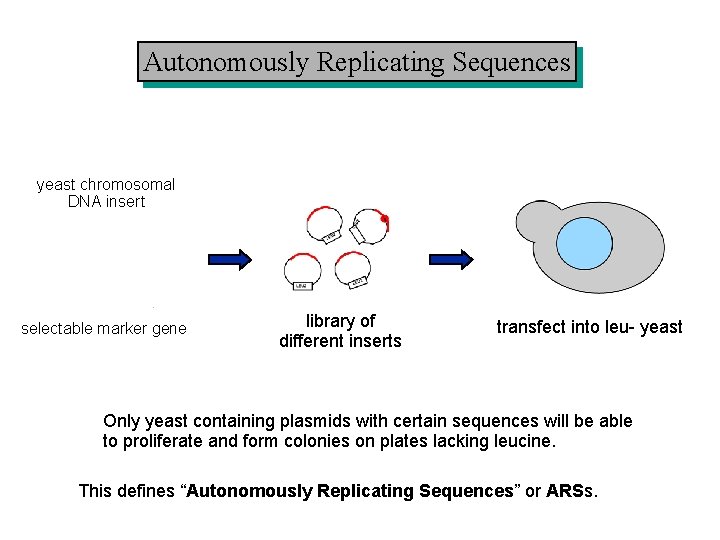
Autonomously Replicating Sequences yeast chromosomal DNA insert selectable marker gene library of different inserts transfect into leu- yeast Only yeast containing plasmids with certain sequences will be able to proliferate and form colonies on plates lacking leucine. This defines “Autonomously Replicating Sequences” or ARSs.

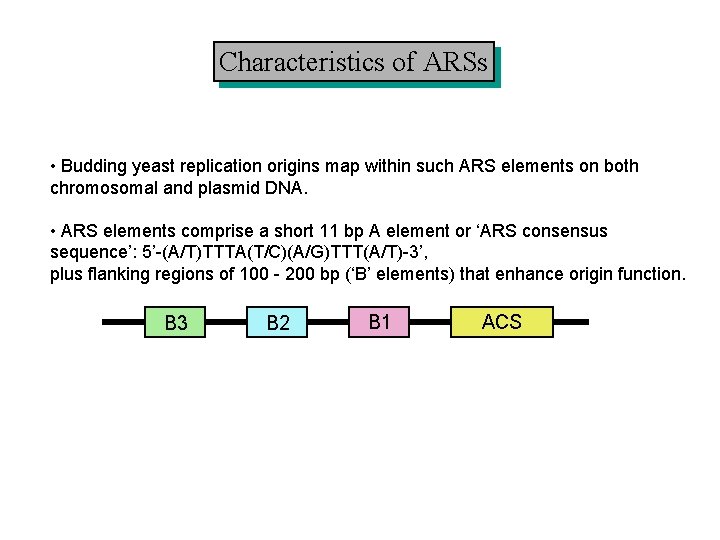
Characteristics of ARSs • Budding yeast replication origins map within such ARS elements on both chromosomal and plasmid DNA. • ARS elements comprise a short 11 bp A element or ‘ARS consensus sequence’: 5’-(A/T)TTTA(T/C)(A/G)TTT(A/T)-3’, plus flanking regions of 100 - 200 bp (‘B’ elements) that enhance origin function. B 3 B 2 B 1 ACS

Which proteins bind to and define eukaryotic replication origins?


ORC ORIGIN RECOGNITION COMPLEX - ORC ble identifisert som et proteinkompleks som bandt seg til ARS konsensus sekvens. - ORC består av seks forskjellige proteiner. - ORC er nødvendig for initiering av replikasjon og er bundet til ARS gjennom hele cellesyklus. - ORC homologer finnes i alle eukaryoter, til og med i archae

Replication origins in metazoans (somatic cells) The structure of replication origins in higher eukaryotes is unclear. • • Small extrachromosomal DNA sequences replicate poorly, even when carrying >10 kb genomic DNA known to act as origins when in the chromosome. • Replication initiates at specific regions at a characteristic time in S phase. Both place and timing may change with cell type. • Replication forks can potentially initiate at a number of different sites throughout an “initiation zone” that may extend over >10 kb.

The ‘Origin Number’ Paradox E. coli: Genome, 4 Mb = 4 x 106 bp Fork rate approx. 800 bp / sec Replication time approx. 40 minutes = 2, 400 secs Amount replicated by 2 forks in 40 mins = 2 x 2400 x 800 = 3, 840, 000 bp (~4 Mb) Eukaryotes Genome 20 Mb (yeast) up to 6, 000 Mb (human) Fork rate 10 bp / sec (frog) - 50 bp / sec (mammal) Amount replicated by 2 forks in 8 hr (human cells) = 2 x 50 x 28, 800 = 2, 880, 000 (~ 3 Mb, a 2, 000 -fold deficit) 46 chromosomes (human cells) - with one origin per chromosome, at least 92 replication forks gives approx. 140 Mb replicated in 8 hours (still a 40 -fold deficit)

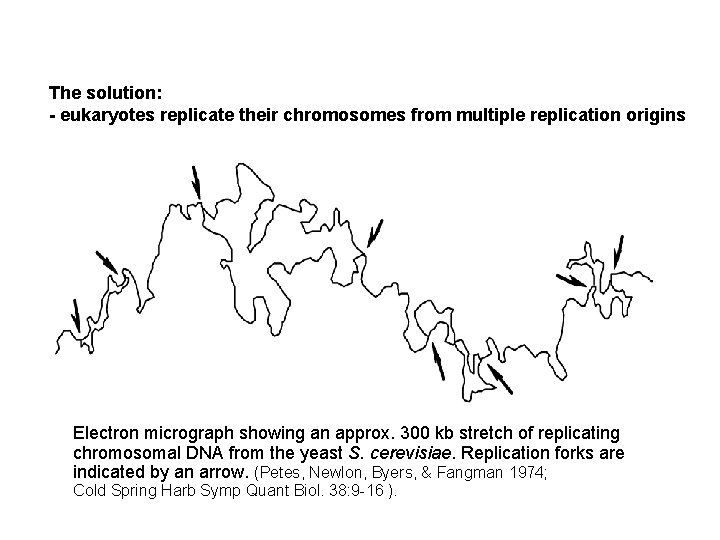
The solution: - eukaryotes replicate their chromosomes from multiple replication origins Electron micrograph showing an approx. 300 kb stretch of replicating chromosomal DNA from the yeast S. cerevisiae. Replication forks are indicated by an arrow. (Petes, Newlon, Byers, & Fangman 1974; Cold Spring Harb Symp Quant Biol. 38: 9 -16 ).

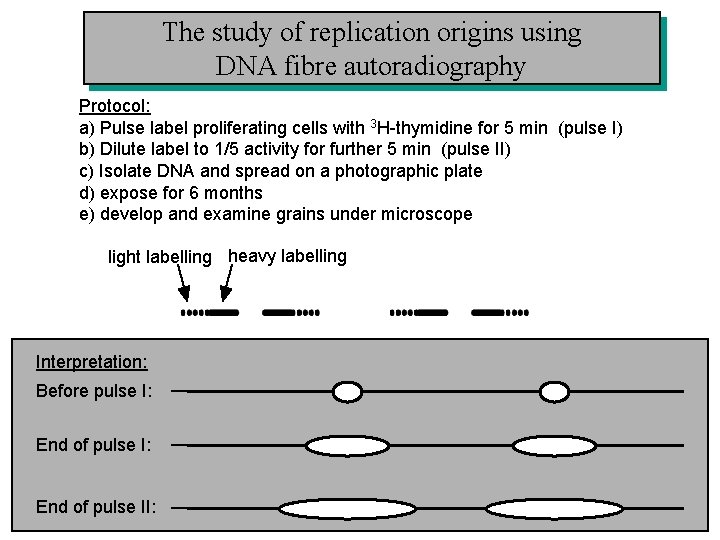
The study of replication origins using DNA fibre autoradiography Protocol: a) Pulse label proliferating cells with 3 H-thymidine for 5 min (pulse I) b) Dilute label to 1/5 activity for further 5 min (pulse II) c) Isolate DNA and spread on a photographic plate d) expose for 6 months e) develop and examine grains under microscope light labelling heavy labelling Interpretation: Before pulse I: End of pulse II:

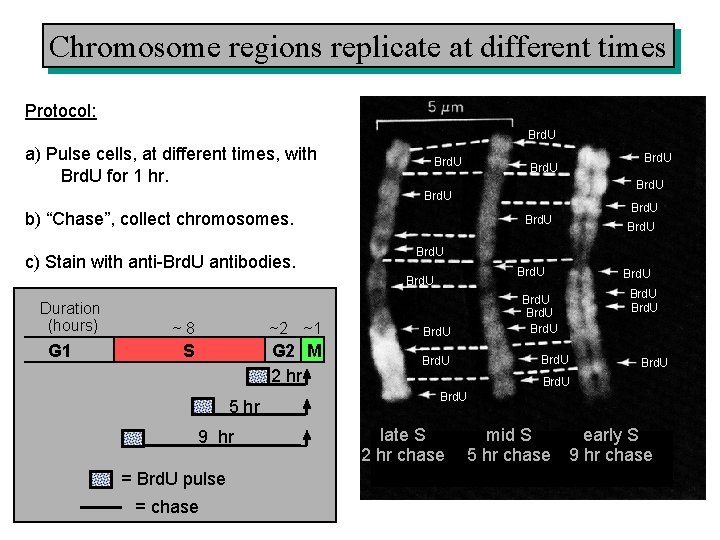
Chromosome regions replicate at different times Protocol: Brd. U a) Pulse cells, at different times, with Brd. U for 1 hr. Brd. U b) “Chase”, collect chromosomes. c) Stain with anti-Brd. U antibodies. Brd. U G 1 Brd. U ~8 ~2 ~1 Brd. U S G 2 M 2 hr Brd. U 5 hr 9 hr = Brd. U pulse = chase Brd. U Duration (hours) Brd. U Brd. U late S 2 hr chase mid S early S 5 hr chase 9 hr chase

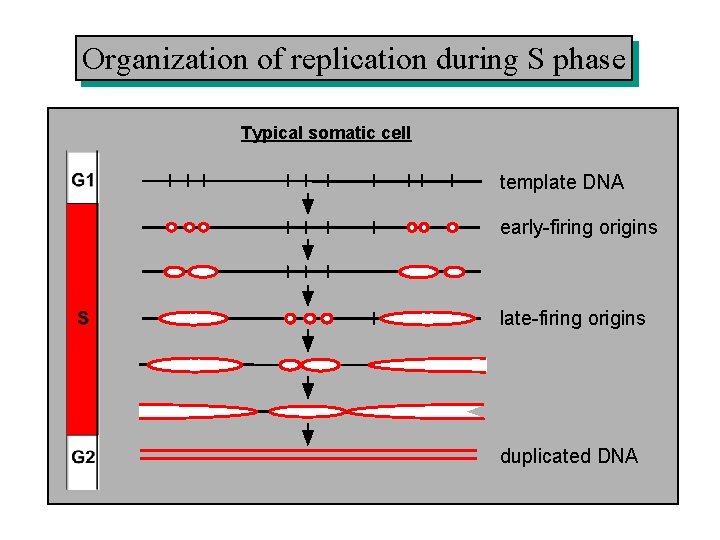
Organization of replication during S phase Typical somatic cell template DNA early-firing origins late-firing origins duplicated DNA

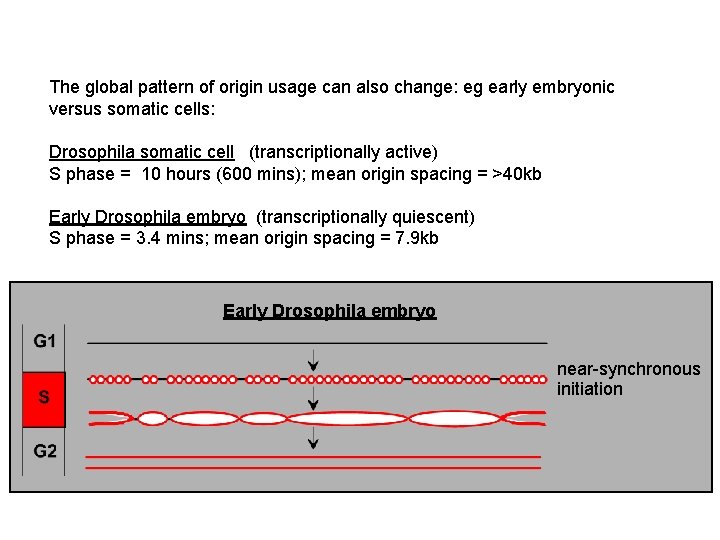
The global pattern of origin usage can also change: eg early embryonic versus somatic cells: Drosophila somatic cell (transcriptionally active) S phase = 10 hours (600 mins); mean origin spacing = >40 kb Early Drosophila embryo (transcriptionally quiescent) S phase = 3. 4 mins; mean origin spacing = 7. 9 kb Early Drosophila embryo near-synchronous initiation

What determines origin usage? The Jesuit principle: ”Many are called – few are chosen”

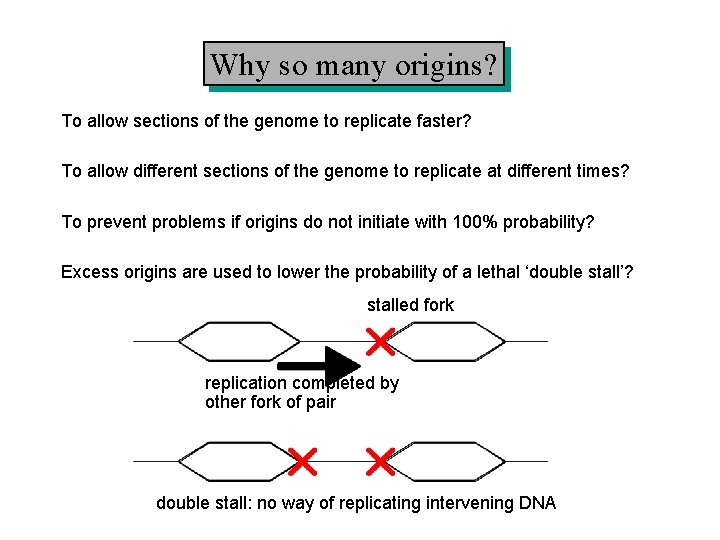
Why so many origins? To allow sections of the genome to replicate faster? To allow different sections of the genome to replicate at different times? To prevent problems if origins do not initiate with 100% probability? Excess origins are used to lower the probability of a lethal ‘double stall’? stalled fork replication completed by other fork of pair double stall: no way of replicating intervening DNA

Facts I • Rate of progression of replication forks is fairly constant for a given organism • Forks generally stop only when they encounter an oppositely moving fork • Chromosome replication is regulated mainly through control of the initiation of new replication forks For example: -by regulating the number and spacing of origins that fire eg. during development -by regulating the time during S phase at which different origins are activated

Facts II • In somatic mammalian cells, most inter-origin distances (replicon sizes) are between 30 - 300 kb (ie would take 5 - 50 min to replicate completely). • Some adjacent origins (“origin clusters”, typically 2 - 5 origins) initiate synchronously • Different origins / origin clusters initiate at different times during S phase Typical mammalian cell replicates 6, 000 Mb in 8 hr = 6 x 109 ÷ 28, 800 bp/sec ie. ~200, 000 bp/sec For fork rate of 50 bp / sec = 200, 000 ÷ 50 ~ 4, 000 forks active at any given time in S phase


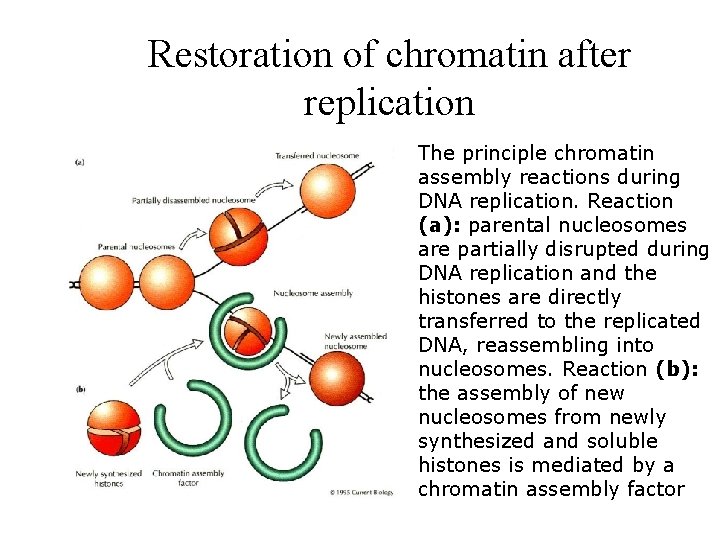
Restoration of chromatin after replication The principle chromatin assembly reactions during DNA replication. Reaction (a): parental nucleosomes are partially disrupted during DNA replication and the histones are directly transferred to the replicated DNA, reassembling into nucleosomes. Reaction (b): the assembly of new nucleosomes from newly synthesized and soluble histones is mediated by a chromatin assembly factor


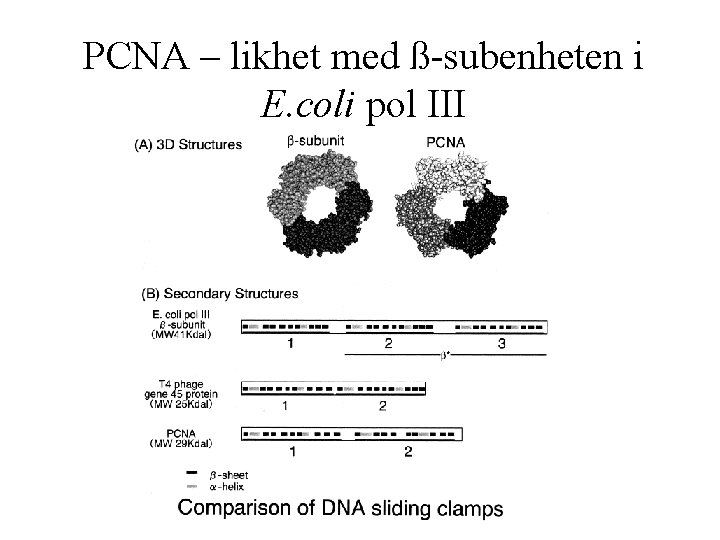
PCNA – likhet med ß-subenheten i E. coli pol III

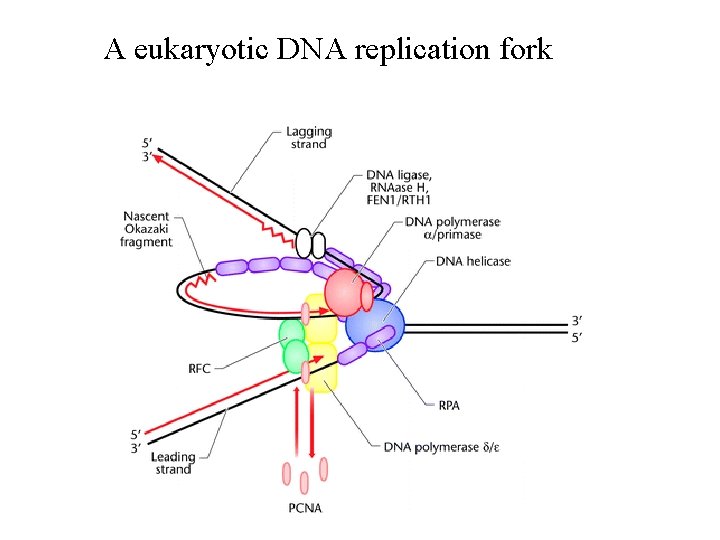
A eukaryotic DNA replication fork

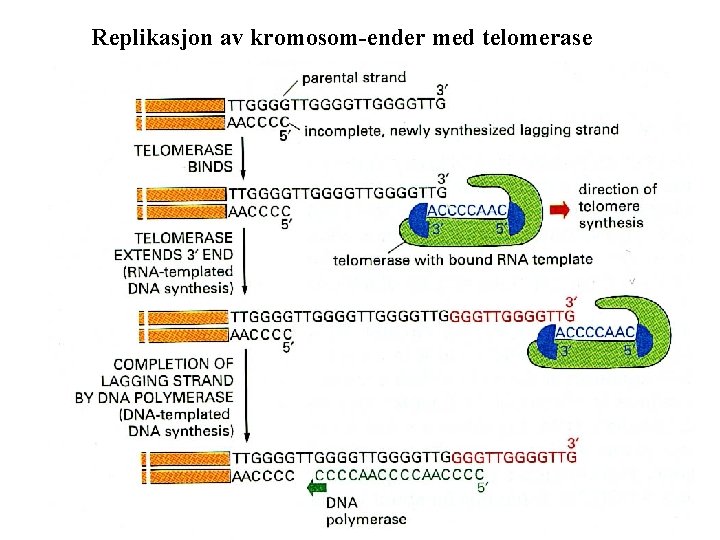
Replikasjon av kromosom-ender med telomerase

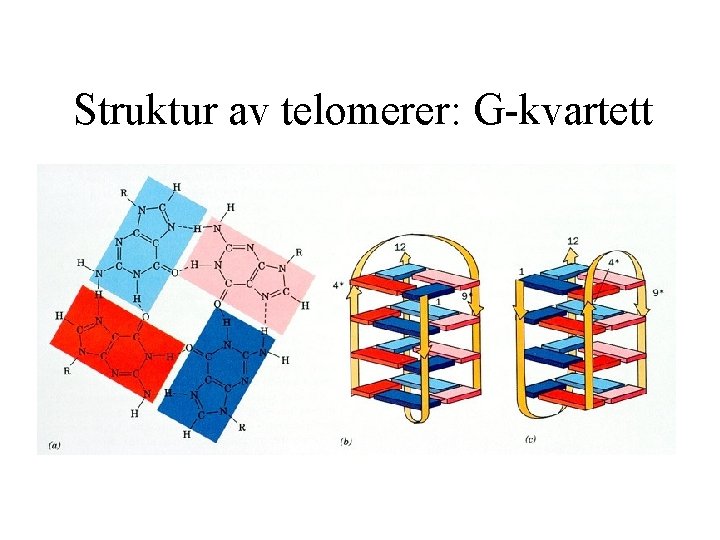
Struktur av telomerer: G-kvartett

Initiering av DNA replikasjon Regulering av DNA replikasjon


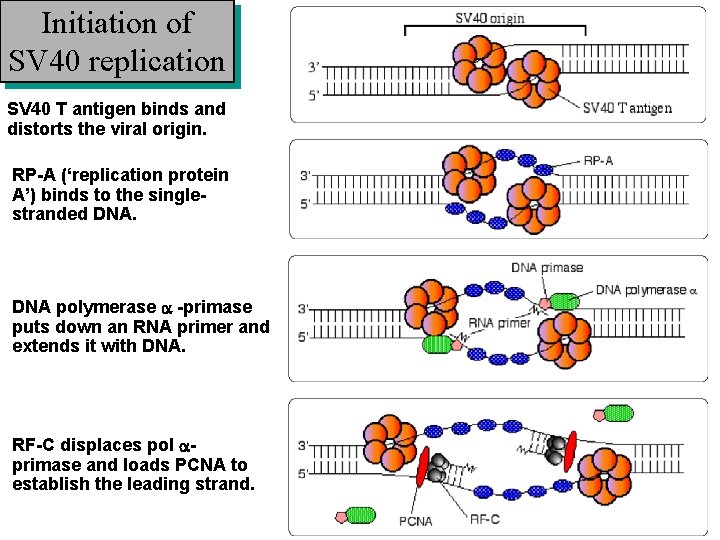
Initiation of SV 40 replication SV 40 T antigen binds and distorts the viral origin. RP-A (‘replication protein A’) binds to the singlestranded DNA polymerase a -primase puts down an RNA primer and extends it with DNA. RF-C displaces pol aprimase and loads PCNA to establish the leading strand.

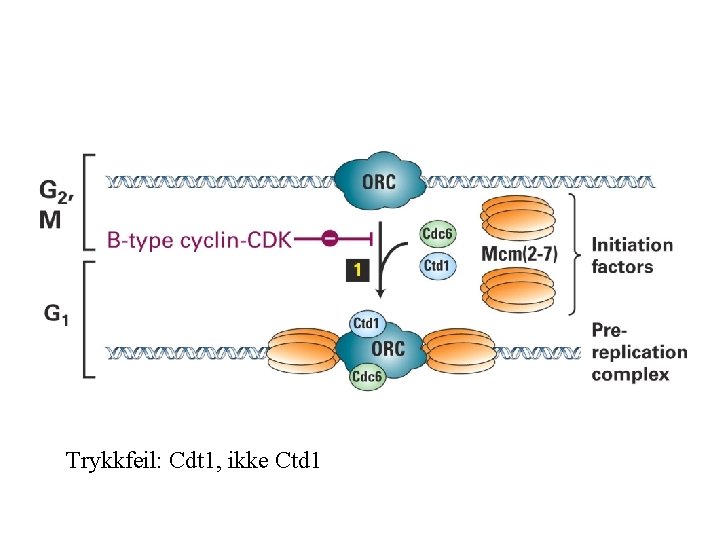
Trykkfeil: Cdt 1, ikke Ctd 1






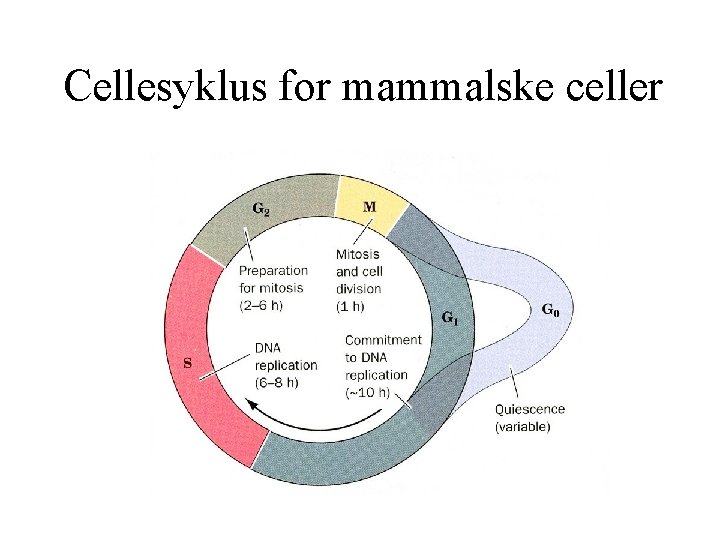
Cellesyklus for mammalske celler

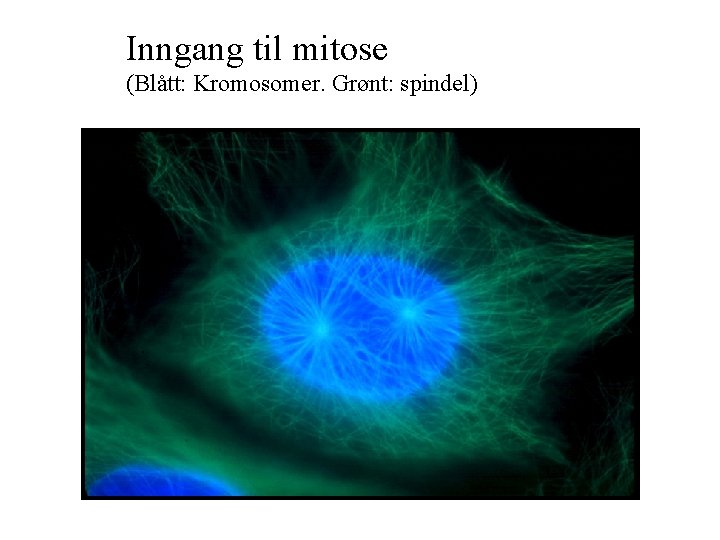
Inngang til mitose (Blått: Kromosomer. Grønt: spindel)

Kromosomene kondenseres

Spindeltrådene (mikrotubuli) fester seg på kromsomene (sentromerer)

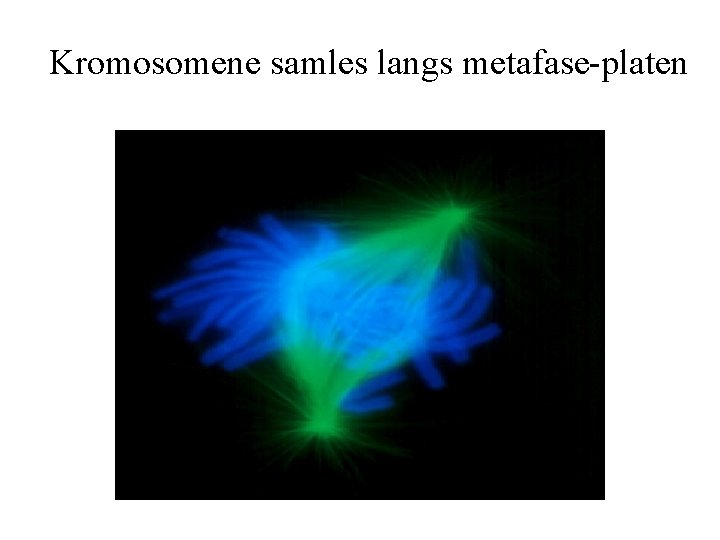
Kromosomene samles langs metafase-platen

Mikrotubuli separerer kromosomene


LICENSING OF DNA REPLICATION

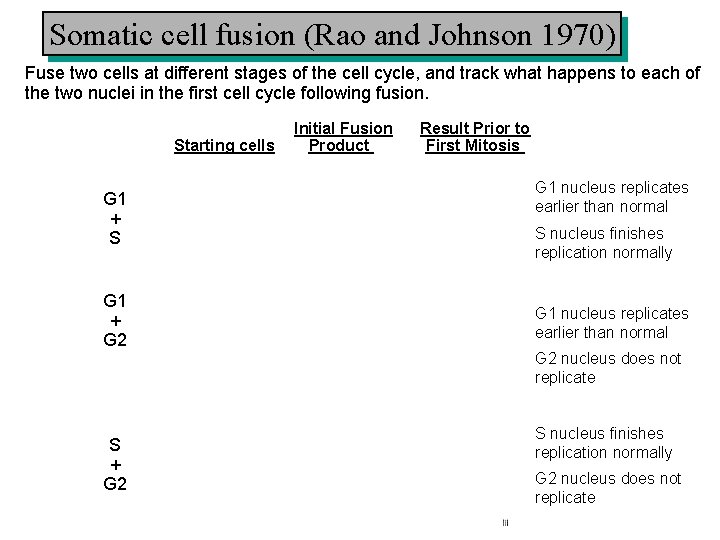
Somatic cell fusion (Rao and Johnson 1970) Fuse two cells at different stages of the cell cycle, and track what happens to each of the two nuclei in the first cell cycle following fusion. Starting cells G 1 + S G 1 + G 2 S + G 2 Initial Fusion Product Result Prior to First Mitosis G 1 nucleus replicates earlier than normal S nucleus finishes replication normally G 1 nucleus replicates earlier than normal G 2 nucleus does not replicate S nucleus finishes replication normally G 2 nucleus does not replicate

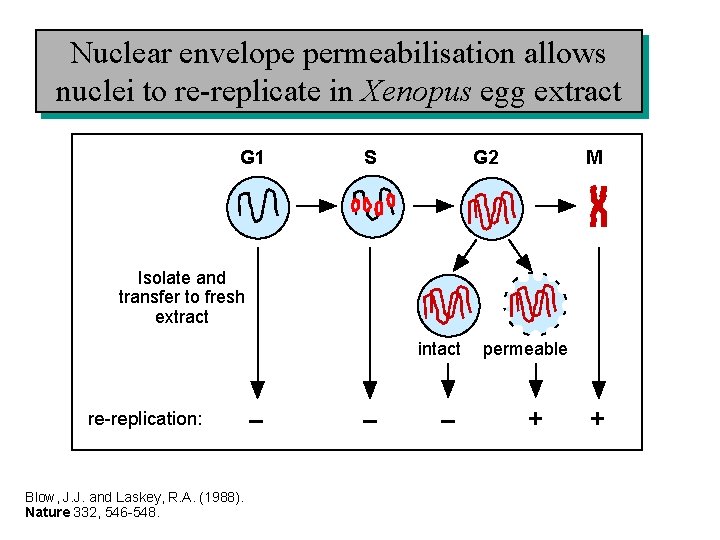
Nuclear envelope permeabilisation allows nuclei to re-replicate in Xenopus egg extract G 1 S G 2 M Isolate and transfer to fresh extract intact re-replication: Blow, J. J. and Laskey, R. A. (1988). Nature 332, 546 -548. – – – permeable + +

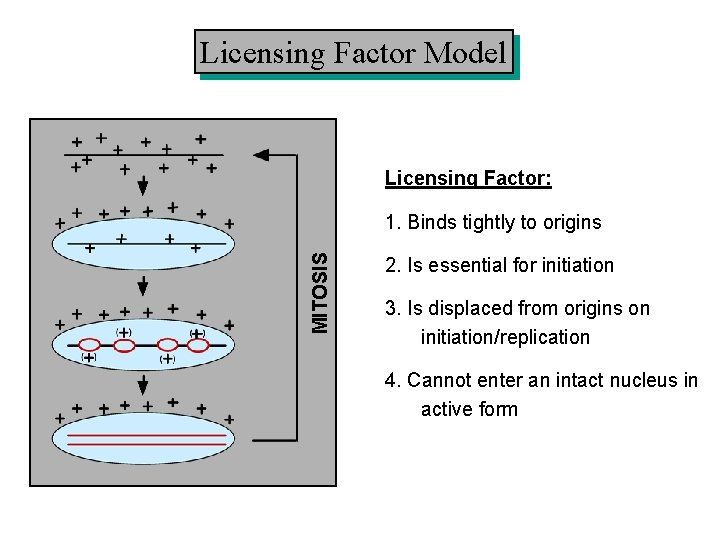
Licensing Factor Model Licensing Factor: MITOSIS 1. Binds tightly to origins 2. Is essential for initiation 3. Is displaced from origins on initiation/replication 4. Cannot enter an intact nucleus in active form

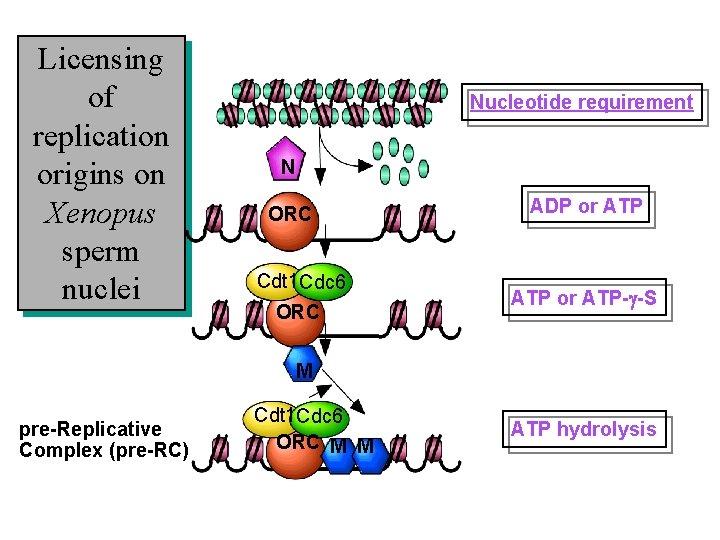
Licensing of replication origins on Xenopus sperm nuclei Nucleotide requirement N ORC Cdt 1 Cdc 6 ORC ADP or ATP-g-S M pre-Replicative Complex (pre-RC) Cdt 1 Cdc 6 ORC M M ATP hydrolysis

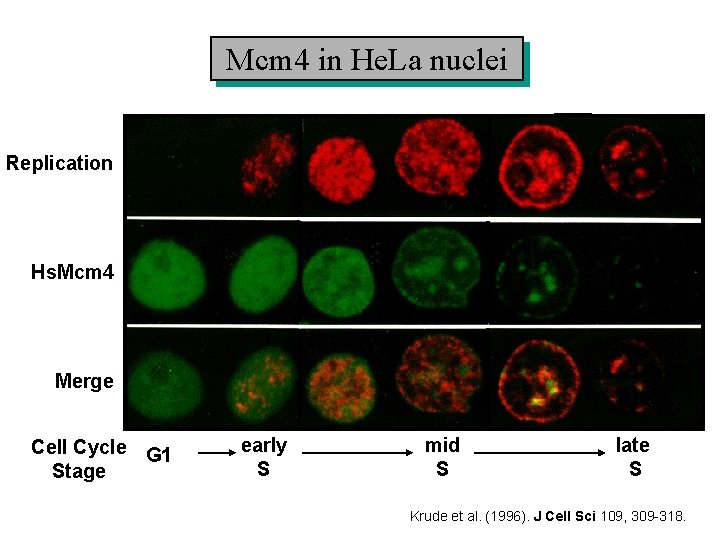
Mcm 4 in He. La nuclei Replication Hs. Mcm 4 Merge Cell Cycle G 1 Stage early S mid S late S Krude et al. (1996). J Cell Sci 109, 309 -318.

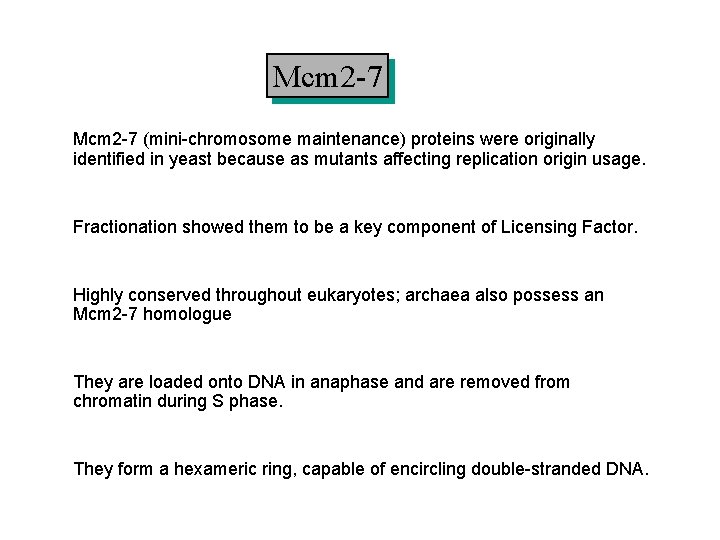
Mcm 2 -7 (mini-chromosome maintenance) proteins were originally identified in yeast because as mutants affecting replication origin usage. Fractionation showed them to be a key component of Licensing Factor. Highly conserved throughout eukaryotes; archaea also possess an Mcm 2 -7 homologue They are loaded onto DNA in anaphase and are removed from chromatin during S phase. They form a hexameric ring, capable of encircling double-stranded DNA.

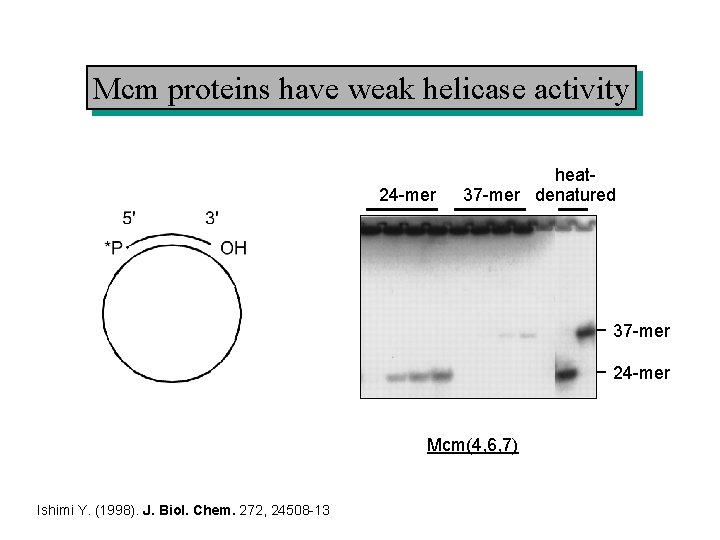
Mcm proteins have weak helicase activity 24 -mer heat 37 -mer denatured 37 -mer 24 -mer Mcm(4, 6, 7) Ishimi Y. (1998). J. Biol. Chem. 272, 24508 -13

Do Mcm 2 -7 provide the fork helicase? 1. Mcm proteins have weak helicase activity (can unwind double-stranded DNA. 2. DNA synthesis stops rapidly if Mcm 2 -7 proteins are degraded. 3. Chromatin immunoprecipitation shows Mcm 2 -7 proteins at the fork. But. . 4. There is ~20 -fold excess of Mcm 2 -7 over origins. 5. Immunofluorescence shows no major co-localisation of Mcm 2 -7 and sites of DNA synthesis.

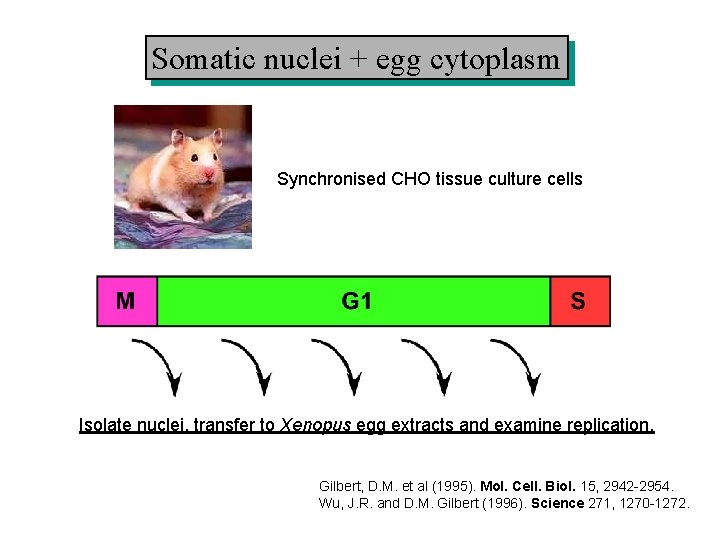
Somatic nuclei + egg cytoplasm Synchronised CHO tissue culture cells Isolate nuclei, transfer to Xenopus egg extracts and examine replication. Gilbert, D. M. et al (1995). Mol. Cell. Biol. 15, 2942 -2954. Wu, J. R. and D. M. Gilbert (1996). Science 271, 1270 -1272.

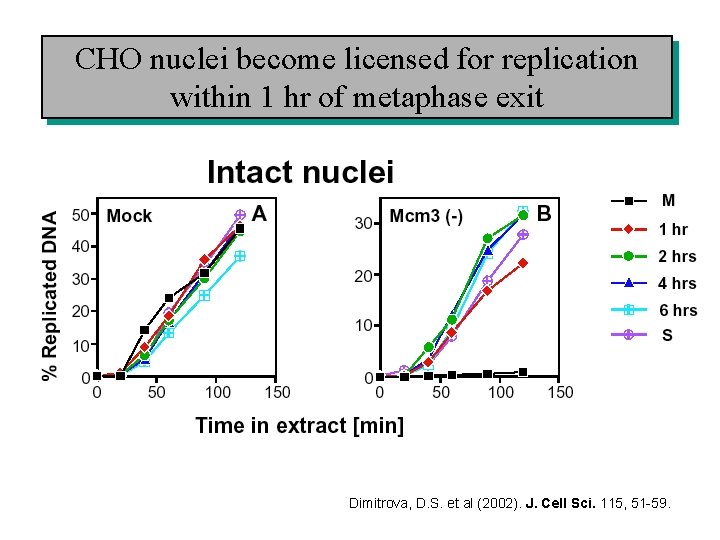
CHO nuclei become licensed for replication within 1 hr of metaphase exit Dimitrova, D. S. et al (2002). J. Cell Sci. 115, 51 -59.

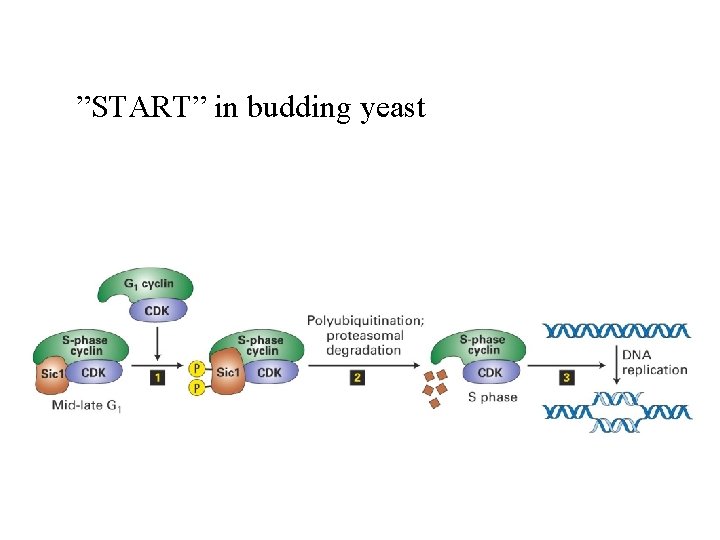
”START” in budding yeast

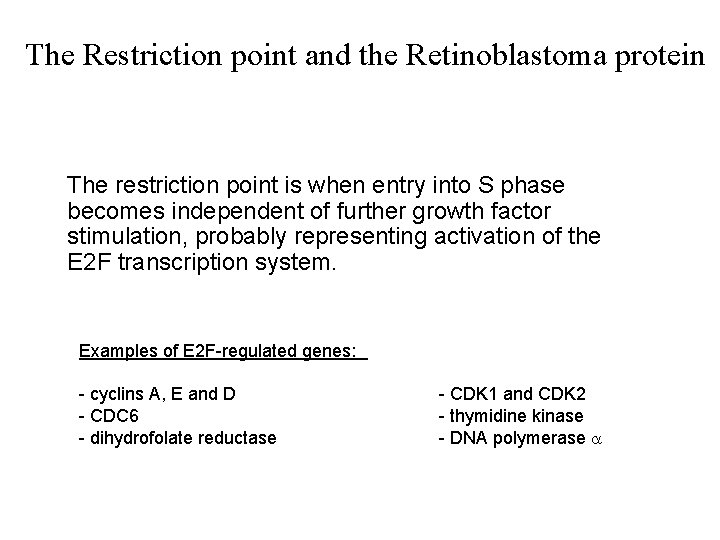
The Restriction point and the Retinoblastoma protein The restriction point is when entry into S phase becomes independent of further growth factor stimulation, probably representing activation of the E 2 F transcription system. Examples of E 2 F-regulated genes: - cyclins A, E and D - CDC 6 - dihydrofolate reductase - CDK 1 and CDK 2 - thymidine kinase - DNA polymerase a

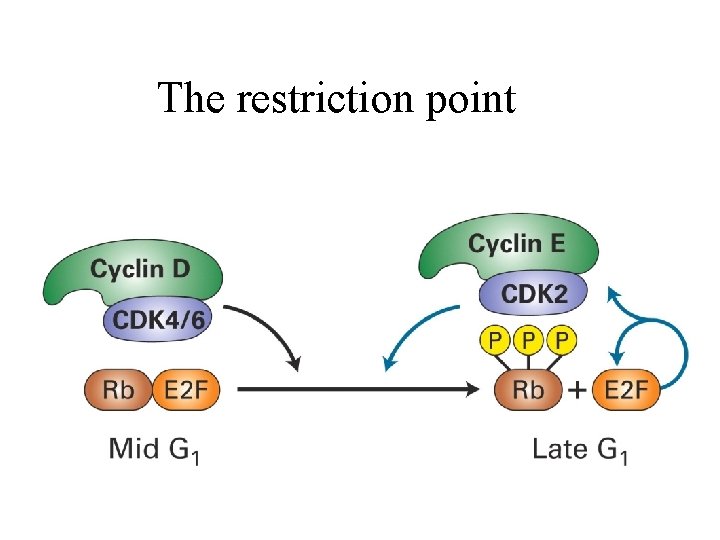
The restriction point

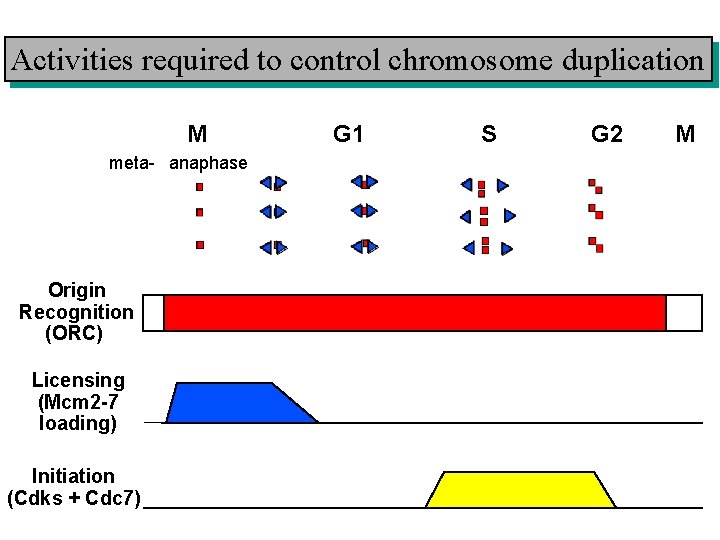
Activities required to control chromosome duplication M meta- anaphase Origin Recognition (ORC) Licensing (Mcm 2 -7 loading) Initiation (Cdks + Cdc 7) G 1 S G 2 M

In somatic cells, transcriptionally active euchromatin replicates early, transcriptionally inactive whilst heterochromatin replicates late. No heterochromatic regions are typically seen in the nuclei of the early Xenopus embryo.

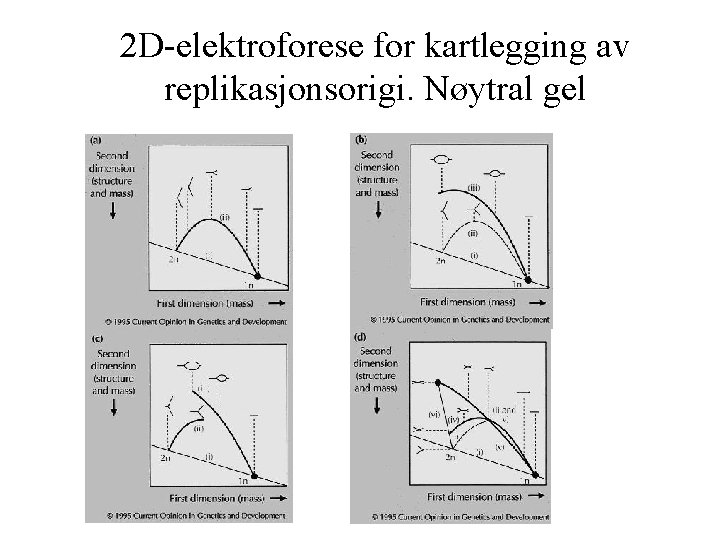
2 D-elektroforese for kartlegging av replikasjonsorigi. Nøytral gel

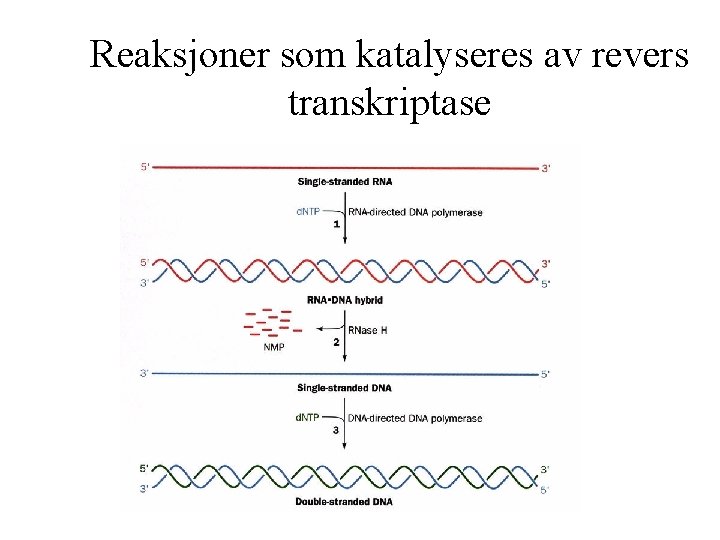
Reaksjoner som katalyseres av revers transkriptase

