p 53 guardian or the genome guardian of









- Slides: 57

p 53 - guardian or the genome + guardian of the tissuse

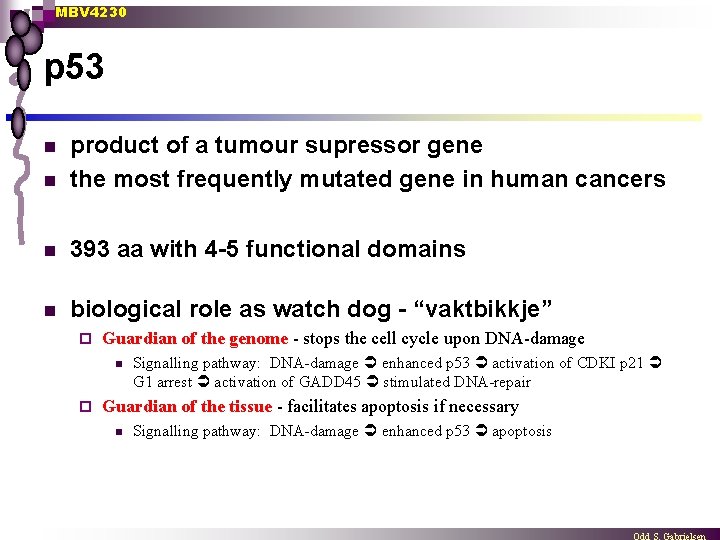
MBV 4230 p 53 n product of a tumour supressor gene the most frequently mutated gene in human cancers n 393 aa with 4 -5 functional domains n biological role as watch dog - “vaktbikkje” n ¨ Guardian of the genome - stops the cell cycle upon DNA-damage n ¨ Signalling pathway: DNA-damage enhanced p 53 activation of CDKI p 21 G 1 arrest activation of GADD 45 stimulated DNA-repair Guardian of the tissue - facilitates apoptosis if necessary n Signalling pathway: DNA-damage enhanced p 53 apoptosis

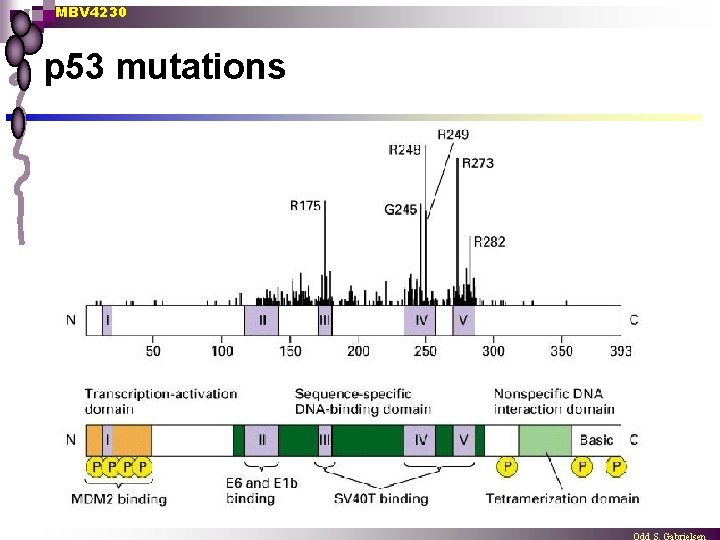
MBV 4230 p 53 mutations

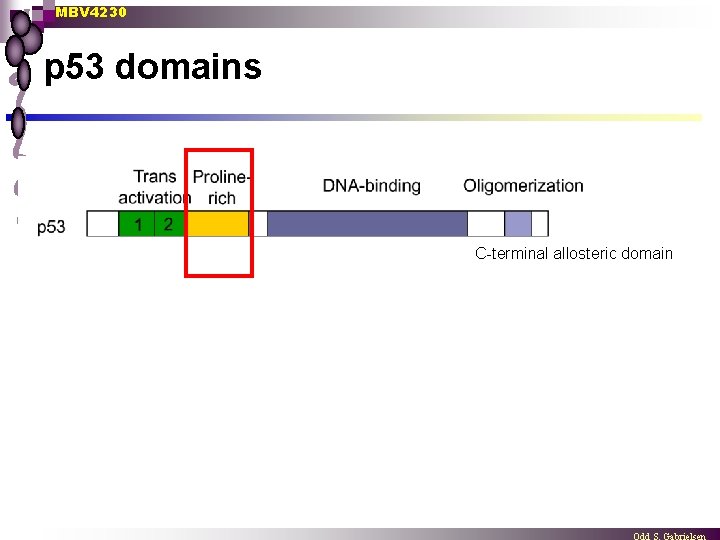
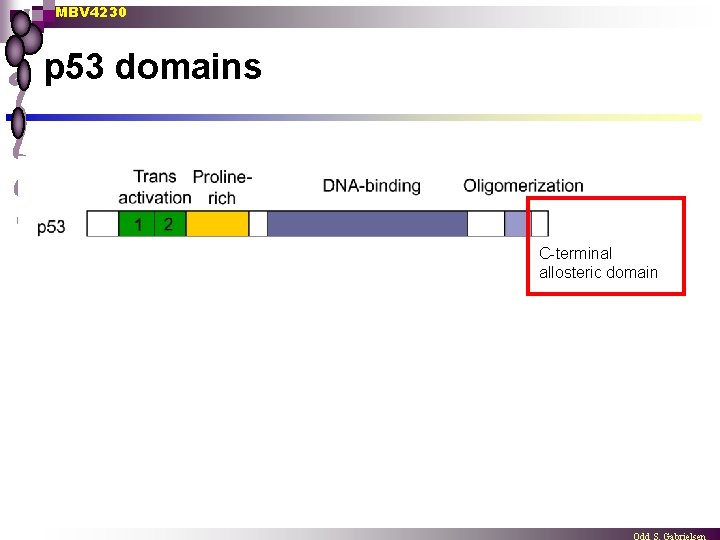
p 53 - protein domains

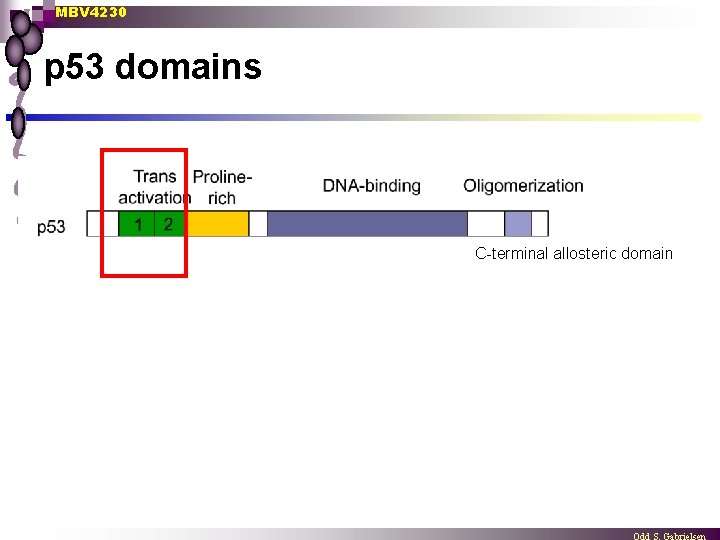
MBV 4230 p 53 domains C-terminal allosteric domain

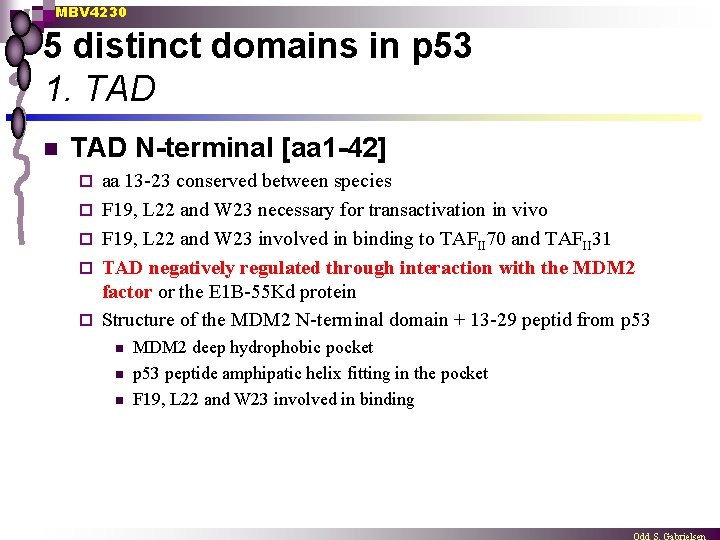
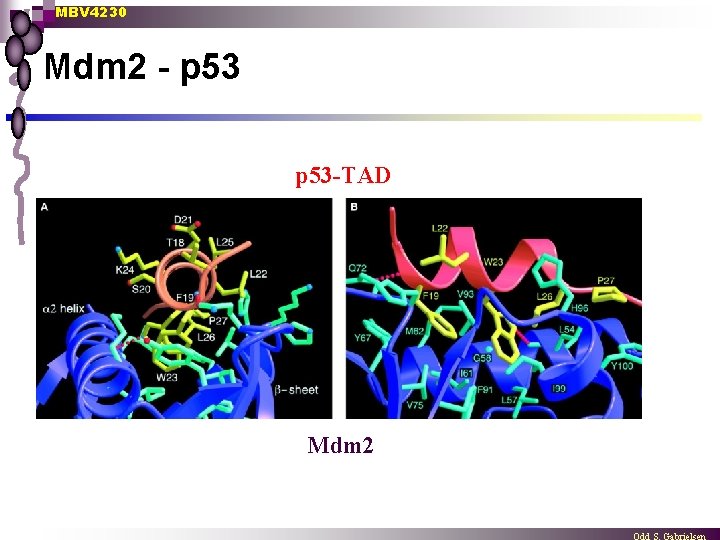
MBV 4230 5 distinct domains in p 53 1. TAD n TAD N-terminal [aa 1 -42] ¨ ¨ ¨ aa 13 -23 conserved between species F 19, L 22 and W 23 necessary for transactivation in vivo F 19, L 22 and W 23 involved in binding to TAFII 70 and TAFII 31 TAD negatively regulated through interaction with the MDM 2 factor or the E 1 B-55 Kd protein Structure of the MDM 2 N-terminal domain + 13 -29 peptid from p 53 n n n MDM 2 deep hydrophobic pocket p 53 peptide amphipatic helix fitting in the pocket F 19, L 22 and W 23 involved in binding

MBV 4230 Mdm 2 - p 53 -TAD Mdm 2

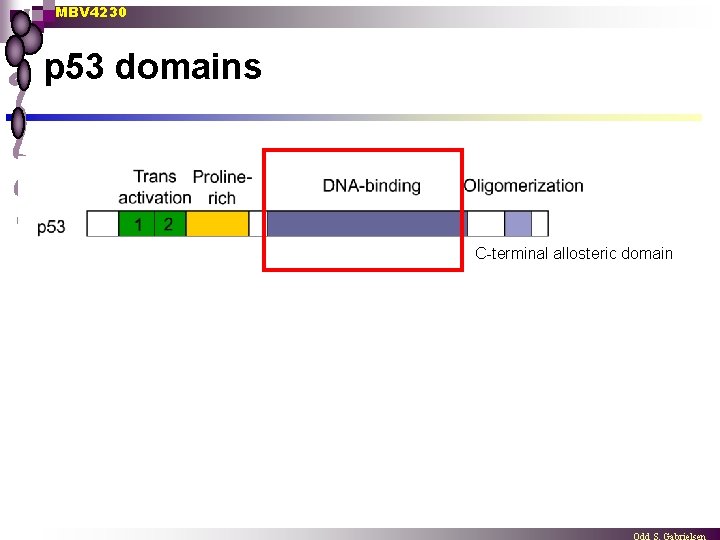
MBV 4230 p 53 domains C-terminal allosteric domain

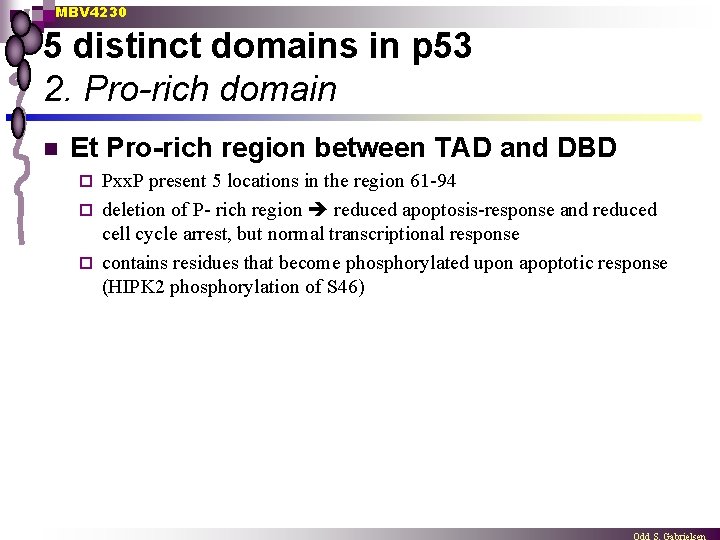
MBV 4230 5 distinct domains in p 53 2. Pro-rich domain n Et Pro-rich region between TAD and DBD Pxx. P present 5 locations in the region 61 -94 ¨ deletion of P- rich region reduced apoptosis-response and reduced cell cycle arrest, but normal transcriptional response ¨ contains residues that become phosphorylated upon apoptotic response (HIPK 2 phosphorylation of S 46) ¨

MBV 4230 p 53 domains C-terminal allosteric domain

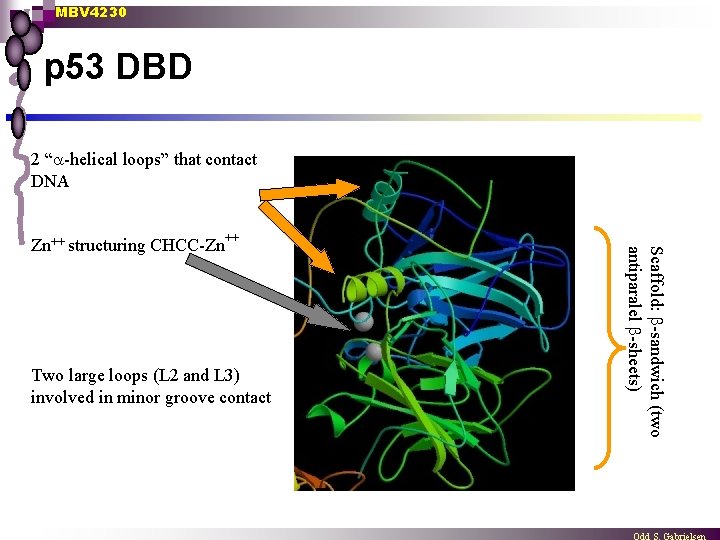
MBV 4230 p 53 DBD 2 “ -helical loops” that contact DNA Two large loops (L 2 and L 3) involved in minor groove contact Scaffold: -sandwich (two antiparalel -sheets) Zn++ structuring CHCC-Zn++

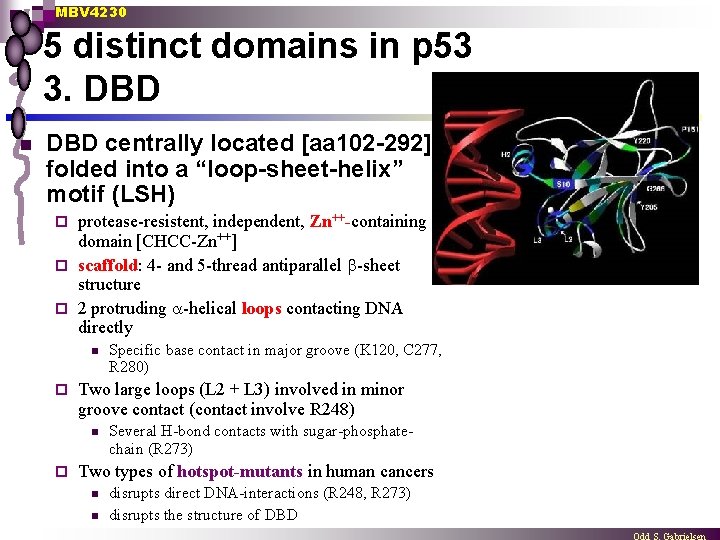
MBV 4230 5 distinct domains in p 53 3. DBD n DBD centrally located [aa 102 -292] folded into a “loop-sheet-helix” motif (LSH) protease-resistent, independent, Zn++-containing domain [CHCC-Zn++] ¨ scaffold: 4 - and 5 -thread antiparallel -sheet structure ¨ 2 protruding -helical loops contacting DNA directly ¨ n ¨ Two large loops (L 2 + L 3) involved in minor groove contact (contact involve R 248) n ¨ Specific base contact in major groove (K 120, C 277, R 280) Several H-bond contacts with sugar-phosphatechain (R 273) Two types of hotspot-mutants in human cancers n n disrupts direct DNA-interactions (R 248, R 273) disrupts the structure of DBD

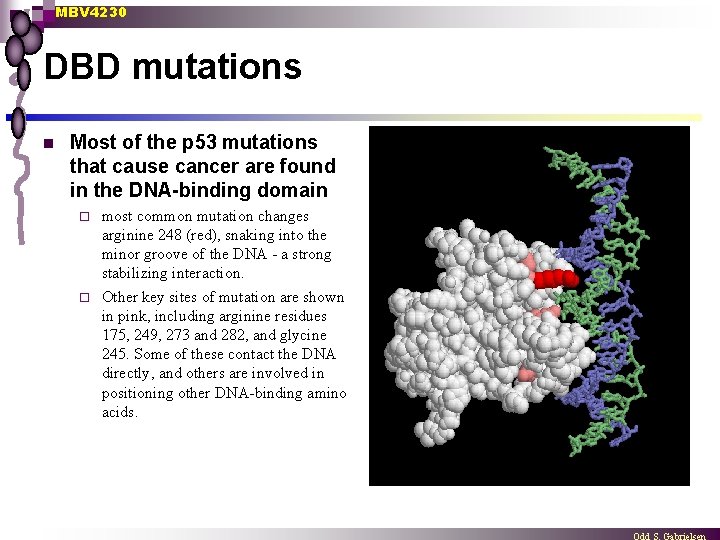
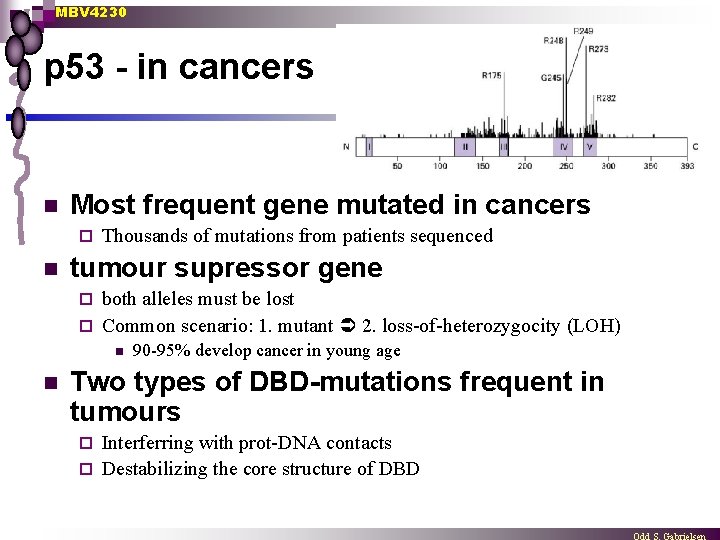
MBV 4230 DBD mutations n Most of the p 53 mutations that cause cancer are found in the DNA-binding domain most common mutation changes arginine 248 (red), snaking into the minor groove of the DNA - a strong stabilizing interaction. ¨ Other key sites of mutation are shown in pink, including arginine residues 175, 249, 273 and 282, and glycine 245. Some of these contact the DNA directly, and others are involved in positioning other DNA-binding amino acids. ¨

MBV 4230 5 distinct domains in p 53 3. DBD n binds DNA as tetramer (dimer of dimer) ¨ DNA recognition sequence reflects this: 4 x RRRCW arranged like this:

MBV 4230 p 53 domains C-terminal allosteric domain

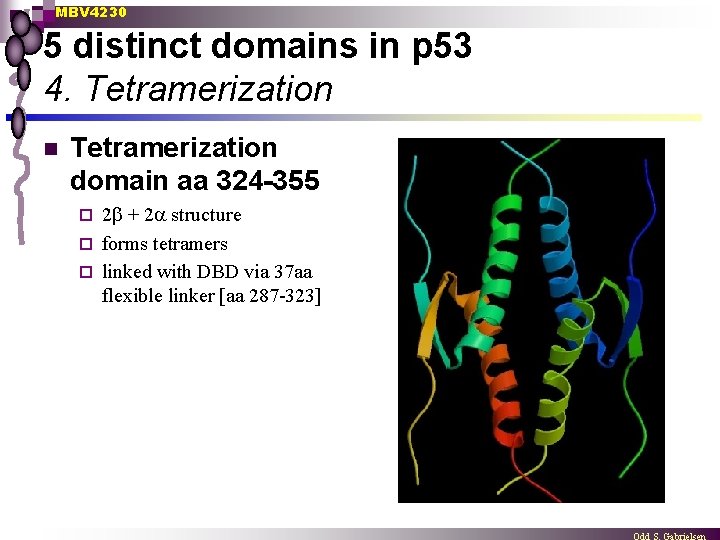
MBV 4230 5 distinct domains in p 53 4. Tetramerization n Tetramerization domain aa 324 -355 2 + 2 structure ¨ forms tetramers ¨ linked with DBD via 37 aa flexible linker [aa 287 -323] ¨

MBV 4230 p 53 domains C-terminal allosteric domain

MBV 4230 5 distinct domains in p 53 5. C-terminal allosteric domain n DNA/RNA-binding C-terminal (last 26 aa) open protease-sensitive domain ¨ Basic region ¨ binds DNA and RNA non-specifically and can stimulate annealing n binds DNA ends, internal loops or other loose ends from damaged duplexes ¨ possible function: (allo)steric regulator of specific DNA-binding n p 53 appears to be present in a latent form inactive in seq. spec. DNAbinding n Several events in the C-terminal can reactivate p 53 s central DBD ¨ deletion of basic C-terminal ¨ phosphorylation of S 378 with PKC ¨ phosphorylation of S 392 with CK 2 ¨ binding of C-terminal antibody PAb 421 ¨ small singlestranded DNA oligos ¨

Activation of p 53 - upstream inputs

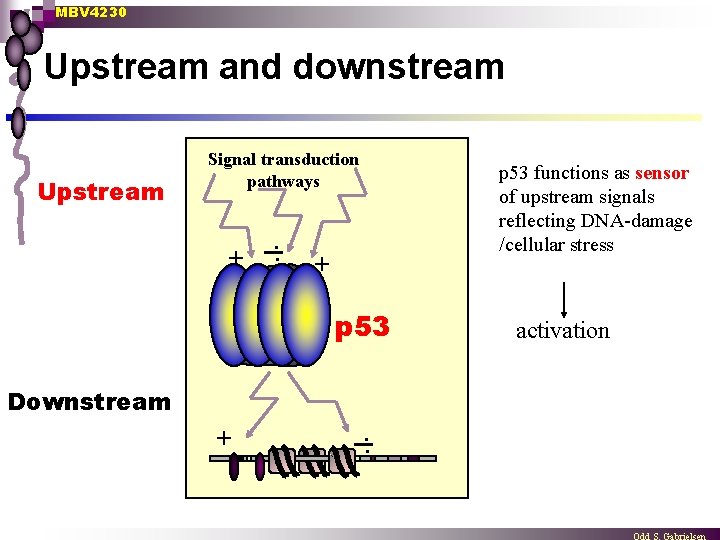
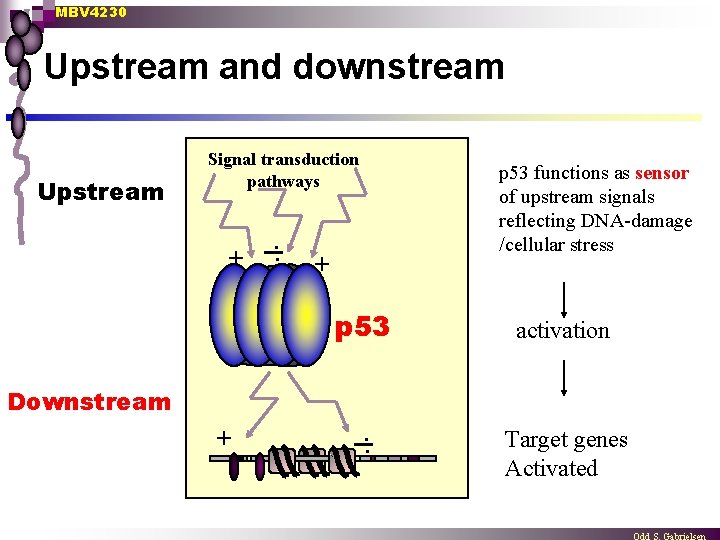
MBV 4230 Upstream and downstream Upstream Signal transduction pathways + . . p 53 functions as sensor of upstream signals reflecting DNA-damage /cellular stress + p 53 Downstream + . . activation

MBV 4230 Activation of p 53 - what happens? n DNA-damage/stress ¨ 1. n n n activation of latent p 53 [latent form active form] enhanced DNA-binding activity probably also enhanced transactivation activity post-translational modifications ¨ 2. stabillization and a rapid increase in protein level activation of response n ¨ activation level increases 10 -100 x Since enhanced levels of p 53 may lead to cell cycle-arrest and apoptosis, it is of critical importance that normal cells keep their p 53 levels low

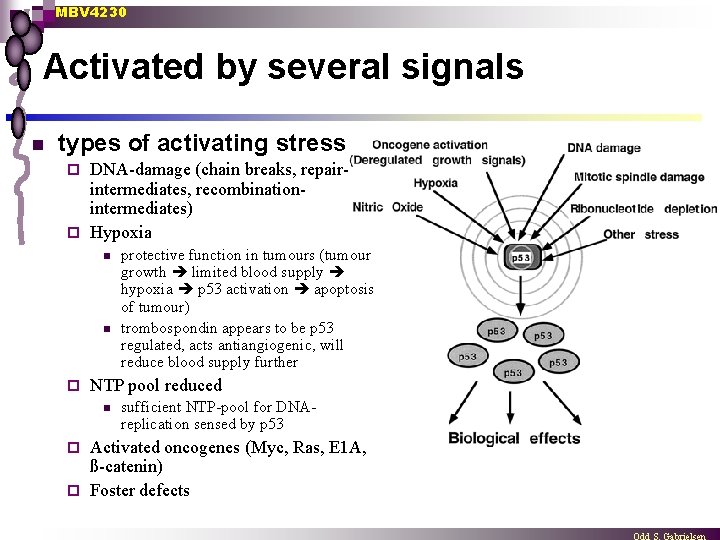
MBV 4230 Activated by several signals n types of activating stress DNA-damage (chain breaks, repairintermediates, recombinationintermediates) ¨ Hypoxia ¨ n n ¨ protective function in tumours (tumour growth limited blood supply hypoxia p 53 activation apoptosis of tumour) trombospondin appears to be p 53 regulated, acts antiangiogenic, will reduce blood supply further NTP pool reduced n sufficient NTP-pool for DNAreplication sensed by p 53 Activated oncogenes (Myc, Ras, E 1 A, ß-catenin) ¨ Foster defects ¨

MBV 4230 The key to stabillization: the MDM 2 -p 53 coupling n MDM 2 associates with p 53 s TAD (aa 17 -27) n MDM 2 -binding leads to ¨ 1. Repression of transactivation ¨ 2. Destabillization of p 53 since MDM 2 -binding stimulates degradation of p 53 n mdm 2 knock-out = lethal, rescued by simultaneous deletion of p 53

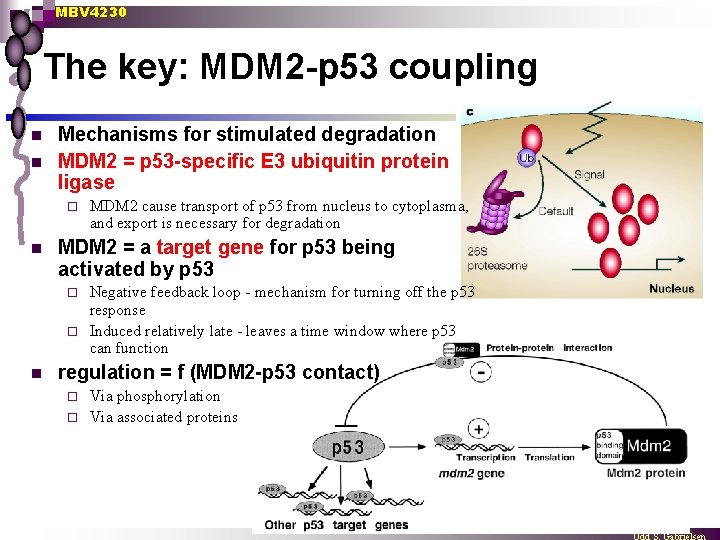
MBV 4230 The key: MDM 2 -p 53 coupling n n Mechanisms for stimulated degradation MDM 2 = p 53 -specific E 3 ubiquitin protein ligase ¨ n MDM 2 cause transport of p 53 from nucleus to cytoplasma, and export is necessary for degradation MDM 2 = a target gene for p 53 being activated by p 53 Negative feedback loop - mechanism for turning off the p 53 response ¨ Induced relatively late - leaves a time window where p 53 can function ¨ n regulation = f (MDM 2 -p 53 contact) Via phosphorylation ¨ Via associated proteins ¨

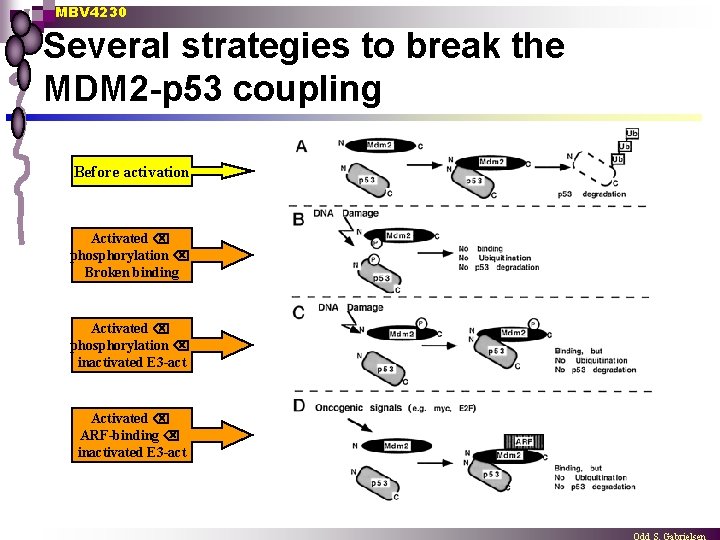
MBV 4230 Several strategies to break the MDM 2 -p 53 coupling Before activation Activated phosphorylation Broken binding Activated phosphorylation inactivated E 3 -act Activated ARF-binding inactivated E 3 -act

MBV 4230 Recent news n More E 3 enzymes suggesting ubiquitylation independent of Mdm 2

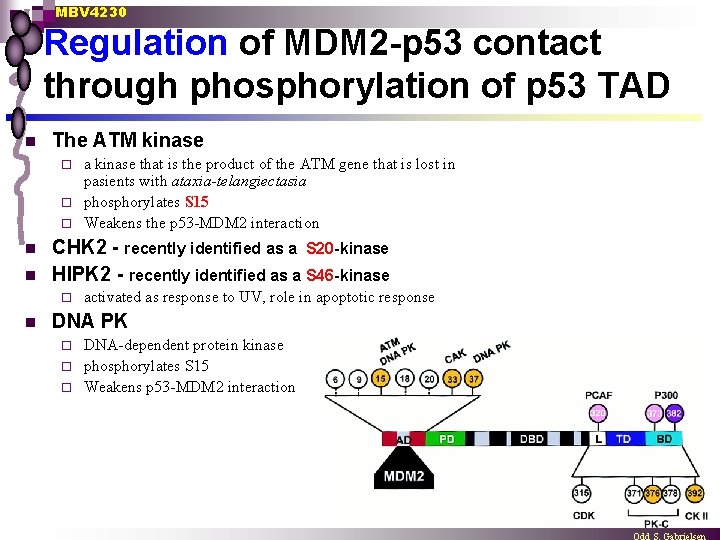
MBV 4230 Regulation of MDM 2 -p 53 contact through phosphorylation of p 53 TAD n The ATM kinase a kinase that is the product of the ATM gene that is lost in pasients with ataxia-telangiectasia ¨ phosphorylates S 15 ¨ Weakens the p 53 -MDM 2 interaction ¨ n n CHK 2 - recently identified as a S 20 -kinase HIPK 2 - recently identified as a S 46 -kinase ¨ n activated as response to UV, role in apoptotic response DNA PK DNA-dependent protein kinase ¨ phosphorylates S 15 ¨ Weakens p 53 -MDM 2 interaction ¨

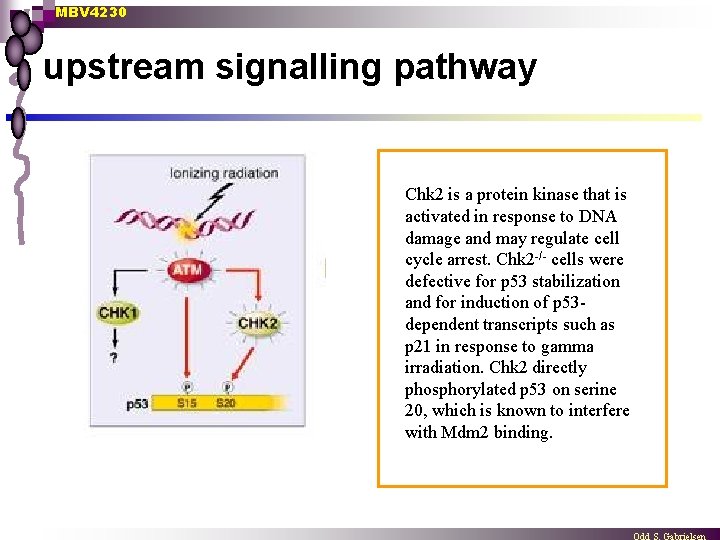
MBV 4230 upstream signalling pathway Chk 2 is a protein kinase that is activated in response to DNA damage and may regulate cell cycle arrest. Chk 2 -/- cells were defective for p 53 stabilization and for induction of p 53 dependent transcripts such as p 21 in response to gamma irradiation. Chk 2 directly phosphorylated p 53 on serine 20, which is known to interfere with Mdm 2 binding.

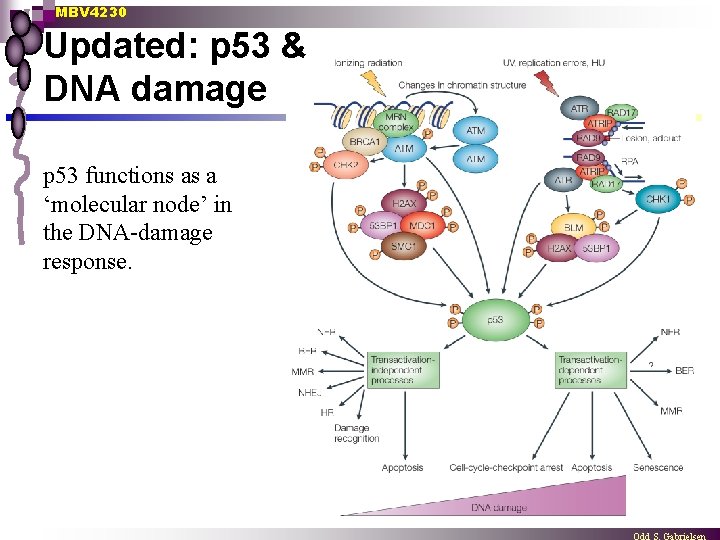
MBV 4230 Updated: p 53 & DNA damage p 53 functions as a ‘molecular node’ in the DNA-damage response.

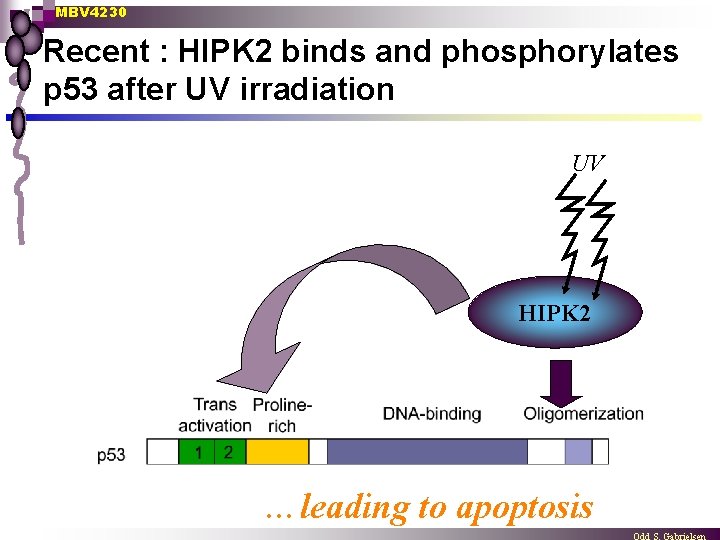
MBV 4230 Recent : HIPK 2 binds and phosphorylates p 53 after UV irradiation UV HIPK 2 …leading to apoptosis

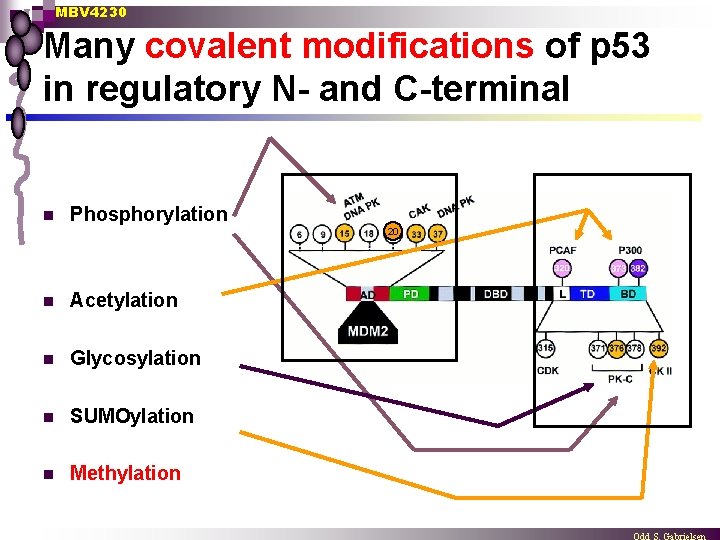
MBV 4230 Many covalent modifications of p 53 in regulatory N- and C-terminal n Phosphorylation 20 n Acetylation n Glycosylation n SUMOylation n Methylation

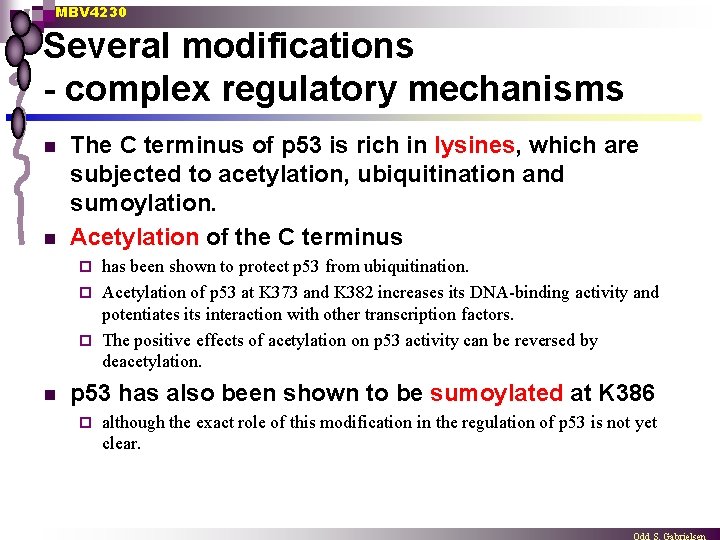
MBV 4230 Several modifications - complex regulatory mechanisms n n The C terminus of p 53 is rich in lysines, which are subjected to acetylation, ubiquitination and sumoylation. Acetylation of the C terminus has been shown to protect p 53 from ubiquitination. ¨ Acetylation of p 53 at K 373 and K 382 increases its DNA-binding activity and potentiates its interaction with other transcription factors. ¨ The positive effects of acetylation on p 53 activity can be reversed by deacetylation. ¨ n p 53 has also been shown to be sumoylated at K 386 ¨ although the exact role of this modification in the regulation of p 53 is not yet clear.

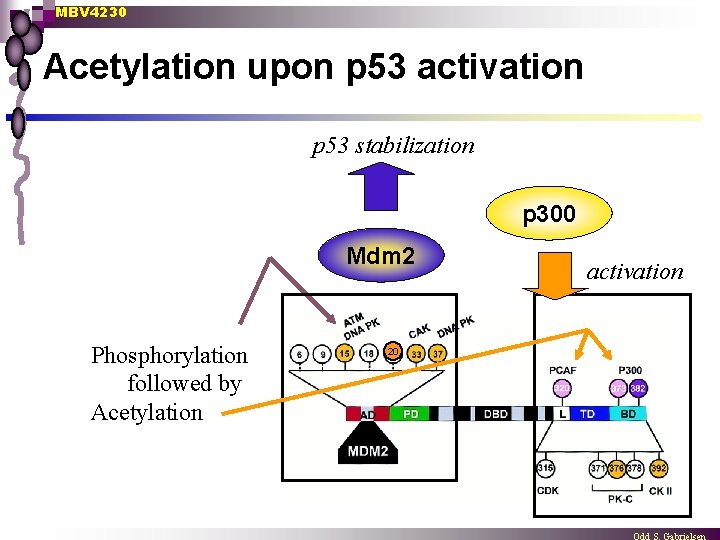
MBV 4230 Acetylation upon p 53 activation p 53 stabilization p 300 Mdm 2 Phosphorylation followed by Acetylation 20 activation

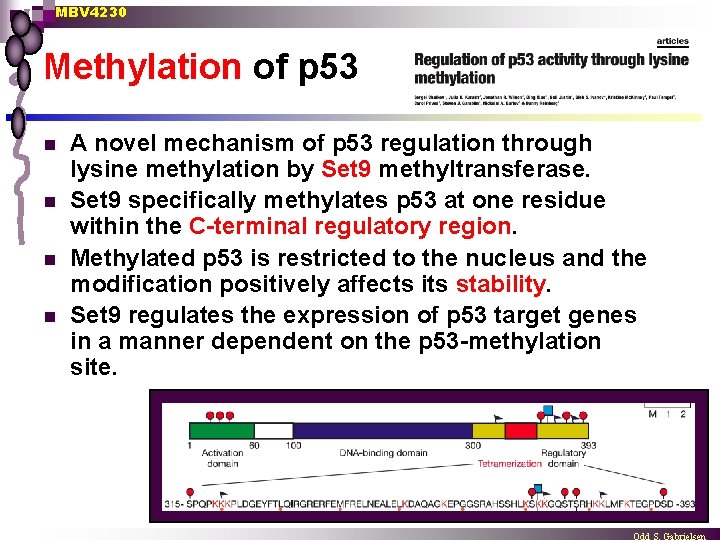
MBV 4230 Methylation of p 53 n n A novel mechanism of p 53 regulation through lysine methylation by Set 9 methyltransferase. Set 9 specifically methylates p 53 at one residue within the C-terminal regulatory region. Methylated p 53 is restricted to the nucleus and the modification positively affects its stability. Set 9 regulates the expression of p 53 target genes in a manner dependent on the p 53 -methylation site.

MBV 4230 Turning p 53 OFF - the h. Sir 2 link n Sir 2 - ”silent information regulator” conserved family identified in silencing in yeast ¨ function as NAD-dep deacetylase ¨

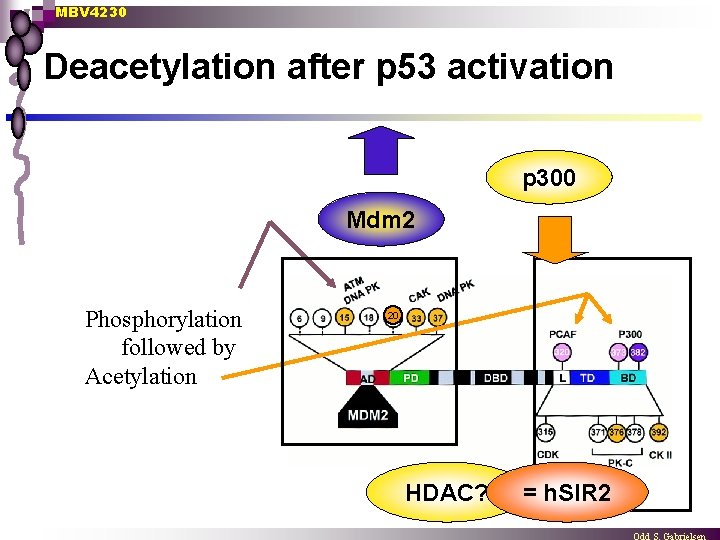
MBV 4230 Deacetylation after p 53 activation p 300 Mdm 2 Phosphorylation followed by Acetylation 20 HDAC? = h. SIR 2

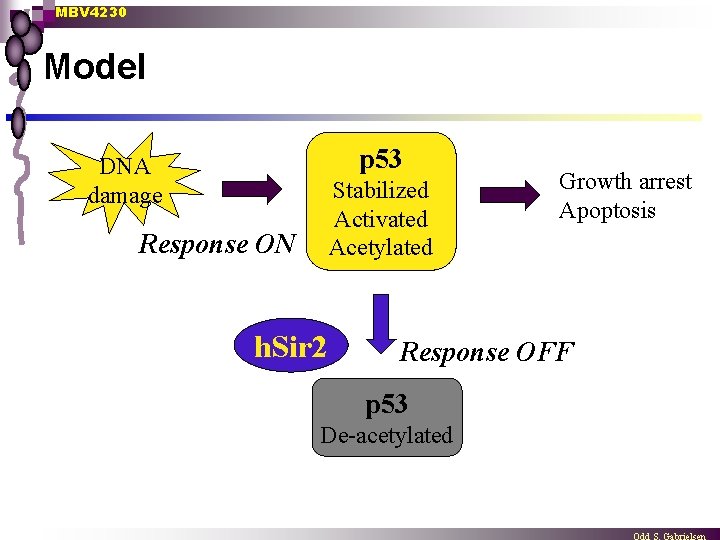
MBV 4230 Model p 53 DNA damage Stabilized Activated Acetylated Response ON h. Sir 2 Growth arrest Apoptosis Response OFF p 53 De-acetylated

Alternative activation - the ARF input

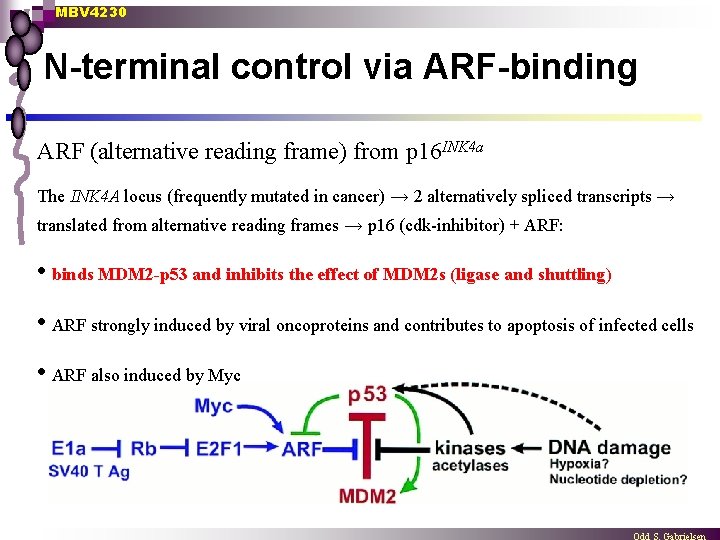
MBV 4230 N-terminal control via ARF-binding ARF (alternative reading frame) from p 16 INK 4 a The INK 4 A locus (frequently mutated in cancer) → 2 alternatively spliced transcripts → translated from alternative reading frames → p 16 (cdk-inhibitor) + ARF: • binds MDM 2 -p 53 and inhibits the effect of MDM 2 s (ligase and shuttling) • ARF strongly induced by viral oncoproteins and contributes to apoptosis of infected cells • ARF also induced by Myc

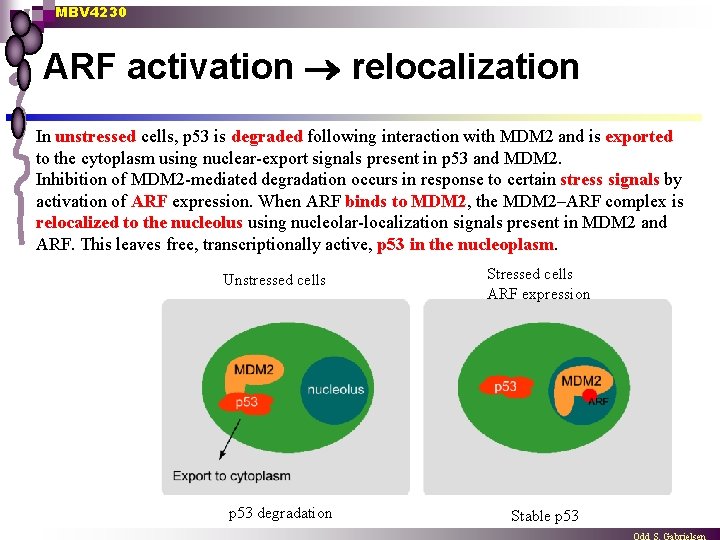
MBV 4230 ARF activation relocalization In unstressed cells, p 53 is degraded following interaction with MDM 2 and is exported to the cytoplasm using nuclear-export signals present in p 53 and MDM 2. Inhibition of MDM 2 -mediated degradation occurs in response to certain stress signals by activation of ARF expression. When ARF binds to MDM 2, the MDM 2–ARF complex is relocalized to the nucleolus using nucleolar-localization signals present in MDM 2 and ARF. This leaves free, transcriptionally active, p 53 in the nucleoplasm. Unstressed cells p 53 degradation Stressed cells ARF expression Stable p 53

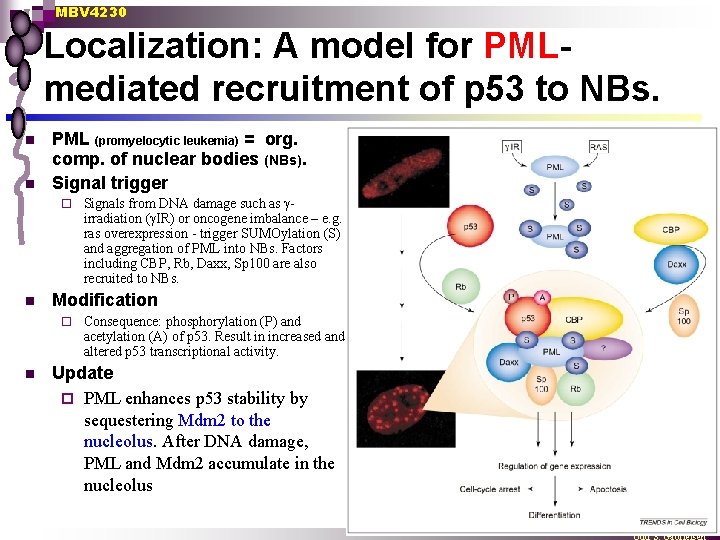
MBV 4230 Localization: A model for PMLmediated recruitment of p 53 to NBs. n n PML (promyelocytic leukemia) = org. comp. of nuclear bodies (NBs). Signal trigger ¨ n Modification ¨ n Signals from DNA damage such as girradiation (g. IR) or oncogene imbalance – e. g. ras overexpression - trigger SUMOylation (S) and aggregation of PML into NBs. Factors including CBP, Rb, Daxx, Sp 100 are also recruited to NBs. Consequence: phosphorylation (P) and acetylation (A) of p 53. Result in increased and altered p 53 transcriptional activity. Update ¨ PML enhances p 53 stability by sequestering Mdm 2 to the nucleolus. After DNA damage, PML and Mdm 2 accumulate in the nucleolus

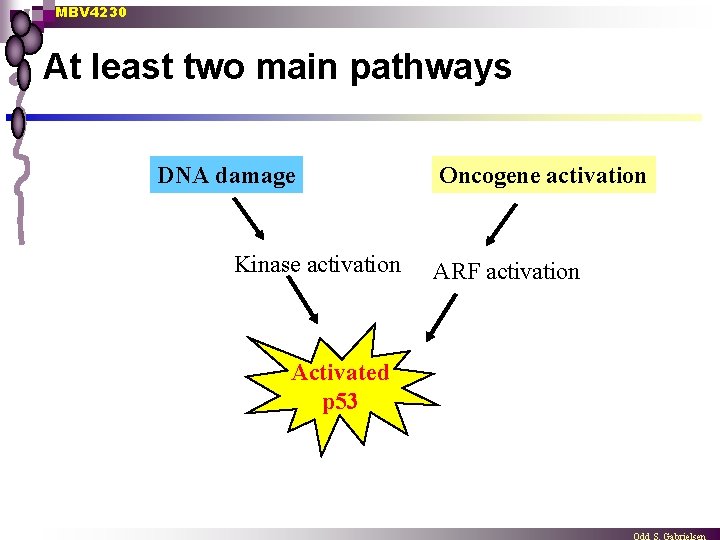
MBV 4230 At least two main pathways DNA damage Kinase activation Activated p 53 Oncogene activation ARF activation

MBV 4230 Multiple pathways - diverse responses n Multiple pathways exist to stabilize p 53 in response to different forms of stress ¨ n they may involve down-regulation of MDM 2 expression or regulation of the subcellular localization of p 53 or MDM 2. Target genes induced by gamma radiation, UV radiation, and the zinc-induced p 53 form distinct sets and subsets with a few genes in common to all these treatments.

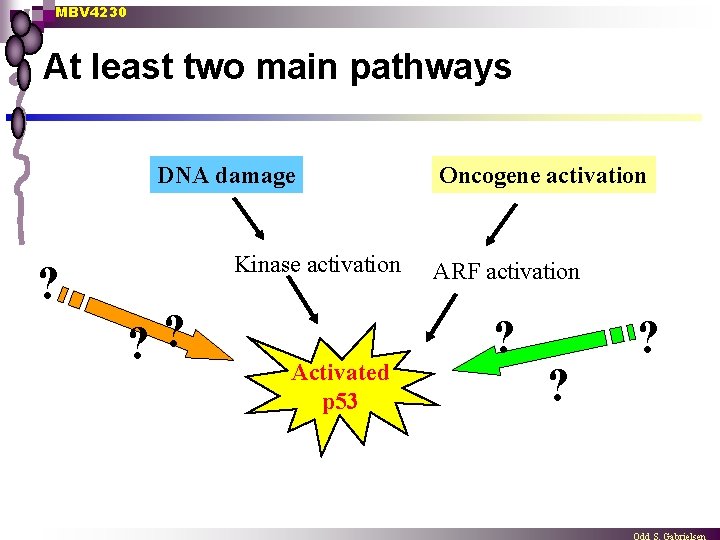
MBV 4230 At least two main pathways DNA damage ? Kinase activation ? ? Activated p 53 Oncogene activation ARF activation ? ? ?

The outcomes of activated p 53 - downstream effects

MBV 4230 Upstream and downstream Upstream Signal transduction pathways + . . p 53 functions as sensor of upstream signals reflecting DNA-damage /cellular stress + p 53 activation . . Target genes Activated Downstream +

MBV 4230 p 53 as signal transducer - downstream response n downstream consequences leading to repair of damage or apoptosis of damaged cell n Two main types of effects of activated p 53 n 1. Stop/regulation of the cell cycle 2. induction of apoptosis n

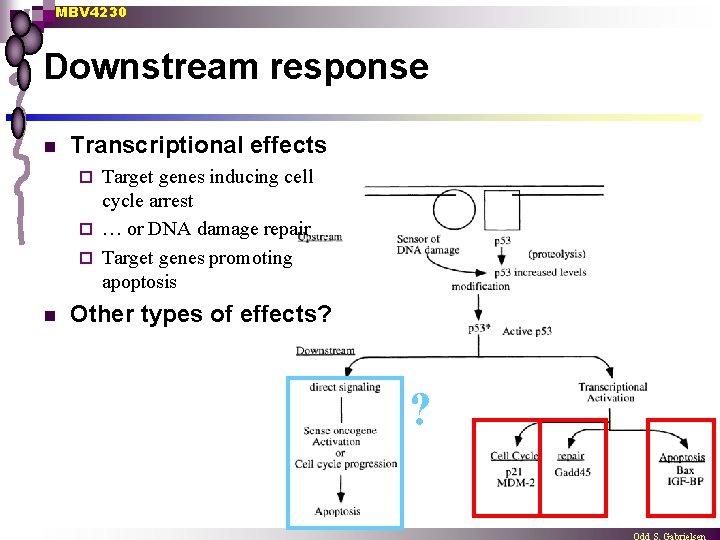
MBV 4230 Downstream response n Transcriptional effects Target genes inducing cell cycle arrest ¨ … or DNA damage repair ¨ Target genes promoting apoptosis ¨ n Other types of effects? ?

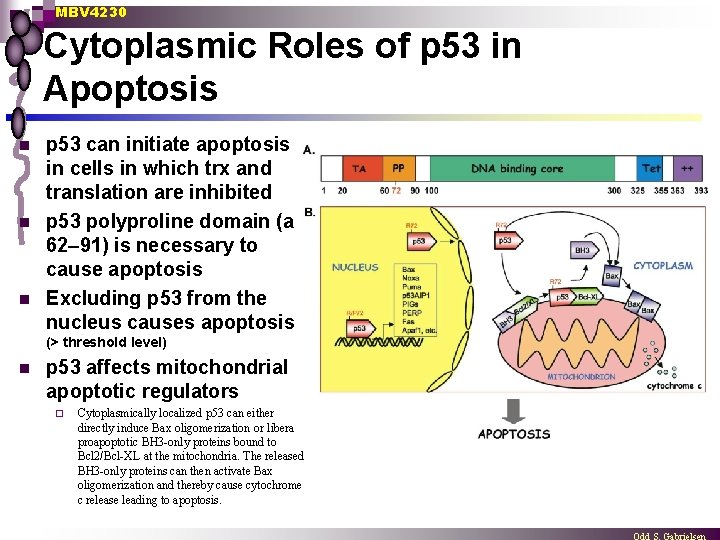
MBV 4230 Cytoplasmic Roles of p 53 in Apoptosis n n n p 53 can initiate apoptosis in cells in which trx and translation are inhibited p 53 polyproline domain (aa 62– 91) is necessary to cause apoptosis Excluding p 53 from the nucleus causes apoptosis (> threshold level) n p 53 affects mitochondrial apoptotic regulators ¨ Cytoplasmically localized p 53 can either directly induce Bax oligomerization or liberate proapoptotic BH 3 -only proteins bound to Bcl 2/Bcl-XL at the mitochondria. The released BH 3 -only proteins can then activate Bax oligomerization and thereby cause cytochrome c release leading to apoptosis.

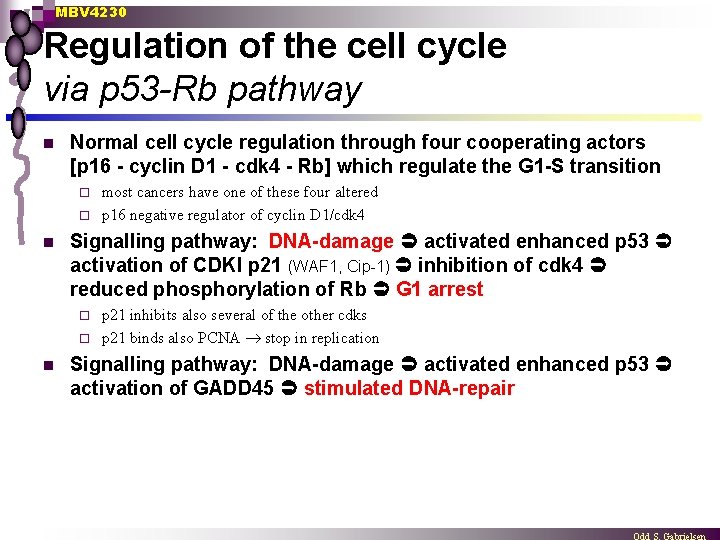
MBV 4230 Regulation of the cell cycle via p 53 -Rb pathway n Normal cell cycle regulation through four cooperating actors [p 16 - cyclin D 1 - cdk 4 - Rb] which regulate the G 1 -S transition most cancers have one of these four altered ¨ p 16 negative regulator of cyclin D 1/cdk 4 ¨ n Signalling pathway: DNA-damage activated enhanced p 53 activation of CDKI p 21 (WAF 1, Cip-1) inhibition of cdk 4 reduced phosphorylation of Rb G 1 arrest p 21 inhibits also several of the other cdks ¨ p 21 binds also PCNA stop in replication ¨ n Signalling pathway: DNA-damage activated enhanced p 53 activation of GADD 45 stimulated DNA-repair

MBV 4230 p 53 -Rb pathway DN Ad am ag e Apoptosis Cell cycle arrest

MBV 4230 p 53 as signal transducer - several downstream responses n n n p 53 also a role in preventing gene amplification p 53 also a role in G 2/M checkpoint Induces a separate ribonucleotide reductase (p 53 R 2) ¨ p 53 R 2 encodes a ribonucleotide reductase that is directly involved in the p 53 checkpoint for repair of damaged DNA.

MBV 4230 induction of apoptosis n n n DNA-damage in p 53+/+ cells apoptosis DNA-damage in p 53 -/- cells no apoptosis mechanisms far from fully understood Transcription activation necessary? only partially ¨ also TF-independent functions involved ¨ n n n Bax induced by p 53 - acts pro-apoptotic by counteracting Bcl 2 PERP is a novel effector of p 53 -dependent apoptosis p 53 AIP 1 (p 53 -regulated Apoptosis-Inducing Protein 1) ¨ upon severe DNA damage, Ser-46 on p 53 is phosphorylated and apoptosis is induced. In addition, substitution of Ser-46 inhibits the ability of p 53 to induce apoptosis and selectively blocks expression of p 53 AIP 1.

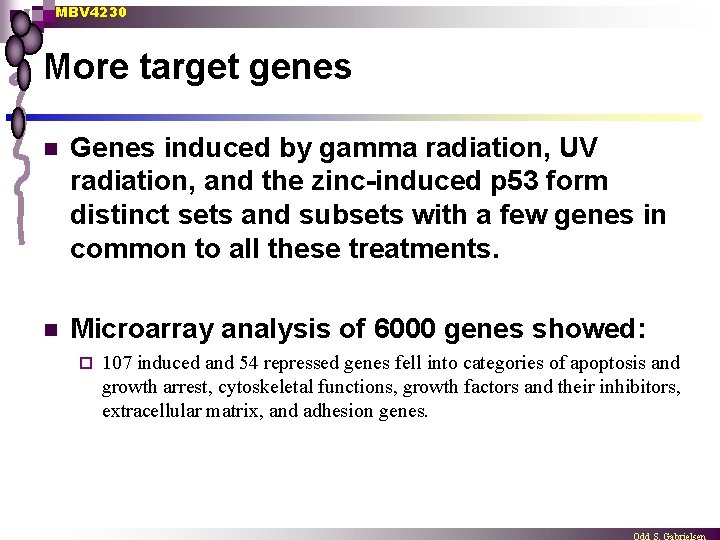
MBV 4230 More target genes n Genes induced by gamma radiation, UV radiation, and the zinc-induced p 53 form distinct sets and subsets with a few genes in common to all these treatments. n Microarray analysis of 6000 genes showed: ¨ 107 induced and 54 repressed genes fell into categories of apoptosis and growth arrest, cytoskeletal functions, growth factors and their inhibitors, extracellular matrix, and adhesion genes.

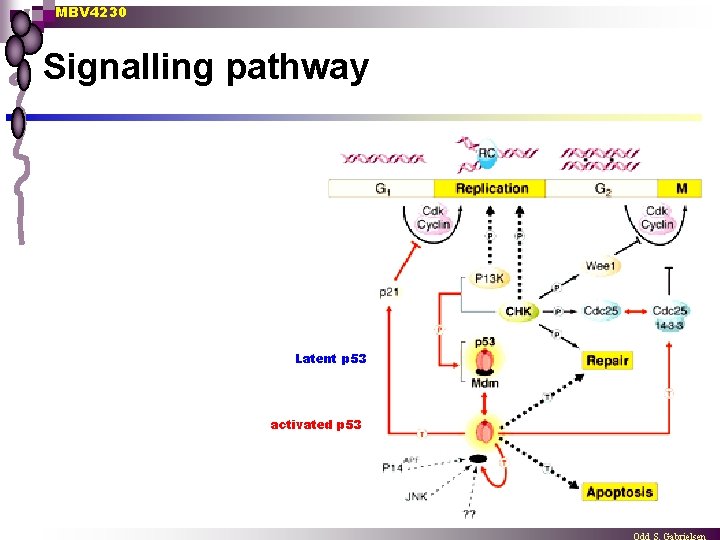
MBV 4230 Signalling pathway Latent p 53 activated p 53

p 53 and cancer

MBV 4230 p 53 - in cancers n Most frequent gene mutated in cancers ¨ n Thousands of mutations from patients sequenced tumour supressor gene both alleles must be lost ¨ Common scenario: 1. mutant 2. loss-of-heterozygocity (LOH) ¨ n n 90 -95% develop cancer in young age Two types of DBD-mutations frequent in tumours Interferring with prot-DNA contacts ¨ Destabilizing the core structure of DBD ¨
 Genome-to-genome distance calculator
Genome-to-genome distance calculator Maker annotation tutorial
Maker annotation tutorial Min-hash
Min-hash Img genome
Img genome 1000 genome project
1000 genome project Ucsc genome browser tutorial
Ucsc genome browser tutorial Translation
Translation National human genome research institute
National human genome research institute Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Tomato genome browser
Tomato genome browser Euphenics
Euphenics Innovation genome project
Innovation genome project Savant genome browser
Savant genome browser Chapter 15 the human genome answer key
Chapter 15 the human genome answer key Patric bioinformatics
Patric bioinformatics Savant genome browser
Savant genome browser Dna
Dna Section 14-3 human molecular genetics answer key
Section 14-3 human molecular genetics answer key Human genome size
Human genome size Gene finding
Gene finding Genome
Genome Genome assembly ppt
Genome assembly ppt Human genome project code
Human genome project code Genome adalah
Genome adalah Human genome structure
Human genome structure Artemis genome
Artemis genome Stanford
Stanford Genome mapping
Genome mapping Yale university poster
Yale university poster Chapter 14 the human genome
Chapter 14 the human genome National human genome research institute
National human genome research institute Human genome features
Human genome features Genome definition
Genome definition Genome is
Genome is Genome.gov
Genome.gov Genome identification
Genome identification Encode
Encode Genome klick
Genome klick Raspberry pi 3 model b specifications pdf
Raspberry pi 3 model b specifications pdf Shotgun sequencing
Shotgun sequencing Human genome project source code
Human genome project source code Genome project
Genome project Human genome size
Human genome size Igv genome browser
Igv genome browser Ap biol
Ap biol Human genome project
Human genome project Genome modification ustaz auni
Genome modification ustaz auni Vntrs vs strs
Vntrs vs strs Slidetodoc
Slidetodoc Plant genome research program
Plant genome research program Genome research limited
Genome research limited Genome sequencing
Genome sequencing History of sequencing
History of sequencing Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Shotgun sequencing
Shotgun sequencing Office of public guardian nebraska
Office of public guardian nebraska The guardian
The guardian