Genome Evolution Amos Tanay 2012 Genome evolution Lecture




- Slides: 34

Genome Evolution. Amos Tanay 2012 Genome evolution: Lecture 12: Evolution of regulatory sequences

Genome Evolution. Amos Tanay 2012 Beyond Protein Coding Sequences Non coding fraction of the genome: • E. coli : 12% • Yeast : 27% • Fly : 76% • Human : 97. 6% How biological functions of non-coding sequence can be defined?

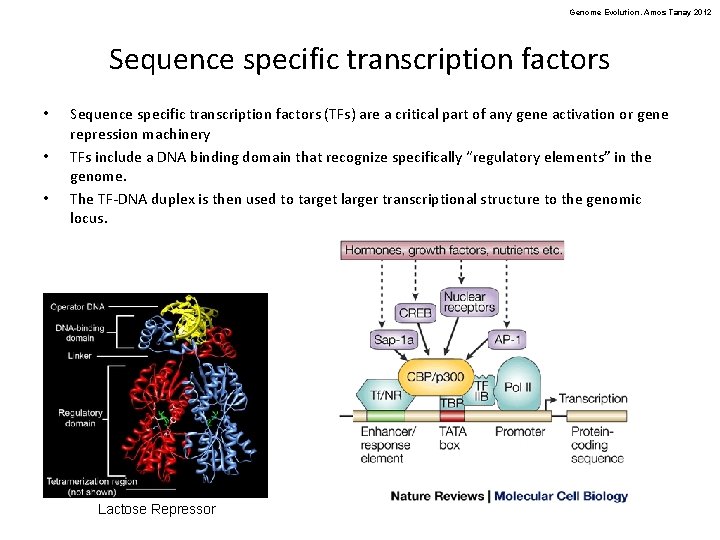
Genome Evolution. Amos Tanay 2012 Sequence specific transcription factors • • • Sequence specific transcription factors (TFs) are a critical part of any gene activation or gene repression machinery TFs include a DNA binding domain that recognize specifically “regulatory elements” in the genome. The TF-DNA duplex is then used to target larger transcriptional structure to the genomic locus. Lactose Repressor

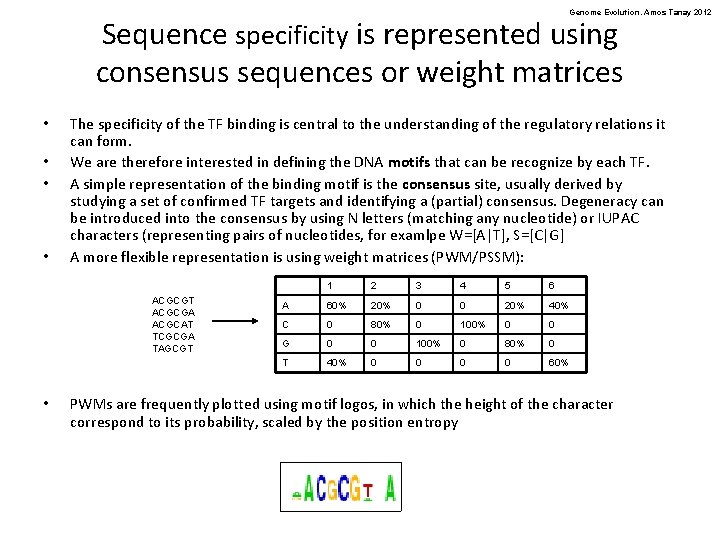
Genome Evolution. Amos Tanay 2012 Sequence specificity is represented using consensus sequences or weight matrices • • The specificity of the TF binding is central to the understanding of the regulatory relations it can form. We are therefore interested in defining the DNA motifs that can be recognize by each TF. A simple representation of the binding motif is the consensus site, usually derived by studying a set of confirmed TF targets and identifying a (partial) consensus. Degeneracy can be introduced into the consensus by using N letters (matching any nucleotide) or IUPAC characters (representing pairs of nucleotides, for examlpe W=[A|T], S=[C|G] A more flexible representation is using weight matrices (PWM/PSSM): ACGCGT ACGCGA ACGCAT TCGCGA TAGCGT • 1 2 3 4 5 6 A 60% 20% 0 0 20% 40% C 0 80% 0 100% 0 0 G 0 0 100% 0 80% 0 T 40% 0 0 60% PWMs are frequently plotted using motif logos, in which the height of the character correspond to its probability, scaled by the position entropy

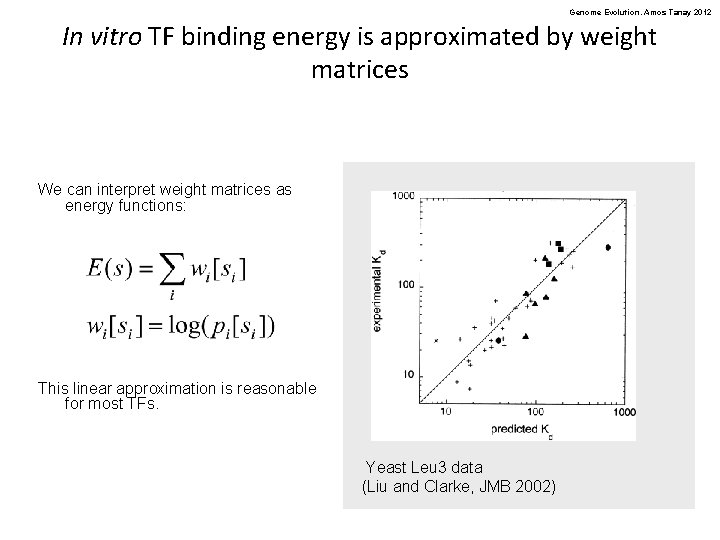
Genome Evolution. Amos Tanay 2012 In vitro TF binding energy is approximated by weight matrices We can interpret weight matrices as energy functions: This linear approximation is reasonable for most TFs. Yeast Leu 3 data (Liu and Clarke, JMB 2002)

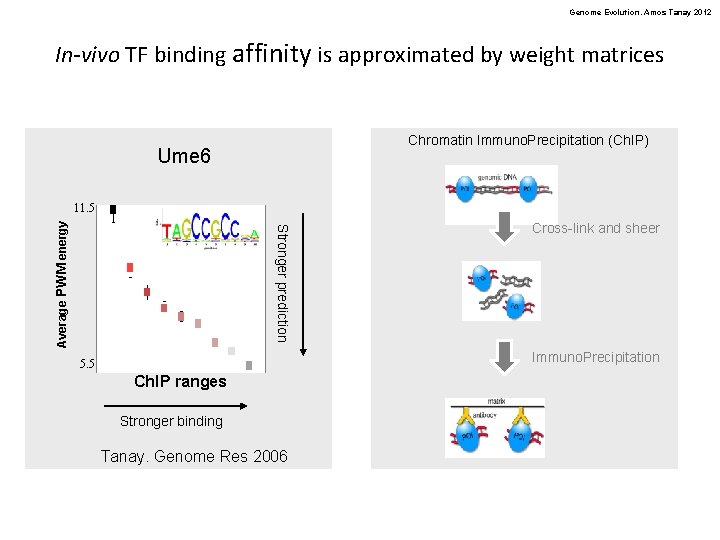
Genome Evolution. Amos Tanay 2012 In-vivo TF binding affinity is approximated by weight matrices Chromatin Immuno. Precipitation (Ch. IP) Ume 6 • s Stronger prediction Average PWM energy 11. 5 Cross-link and sheer • s Immuno. Precipitation 5. 5 Ch. IP ranges Stronger binding Tanay. Genome Res 2006

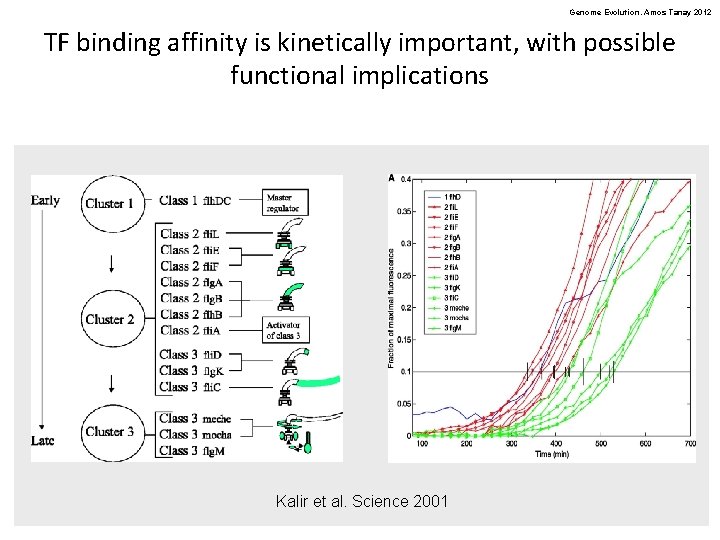
Genome Evolution. Amos Tanay 2012 TF binding affinity is kinetically important, with possible functional implications Kalir et al. Science 2001

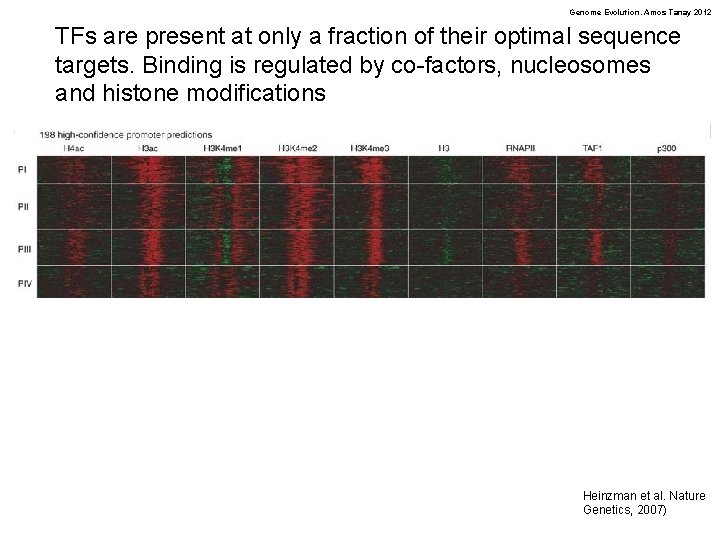
Genome Evolution. Amos Tanay 2012 TFs are present at only a fraction of their optimal sequence targets. Binding is regulated by co-factors, nucleosomes and histone modifications Heinzman et al. Nature Genetics, 2007)

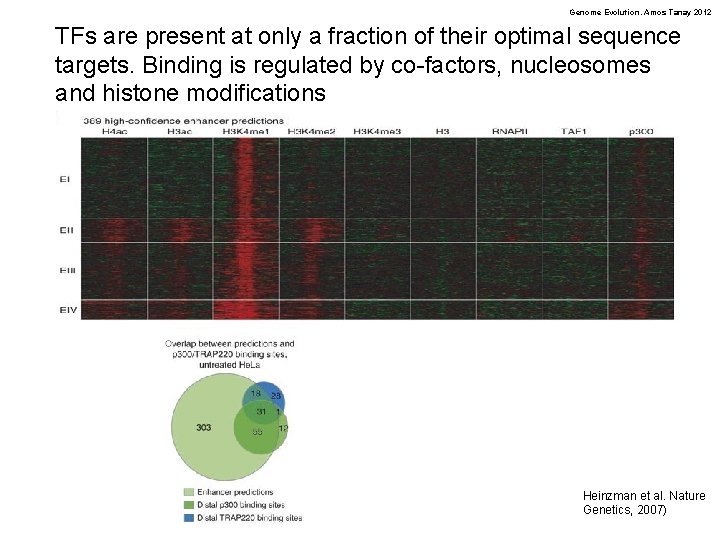
Genome Evolution. Amos Tanay 2012 TFs are present at only a fraction of their optimal sequence targets. Binding is regulated by co-factors, nucleosomes and histone modifications Heinzman et al. Nature Genetics, 2007)

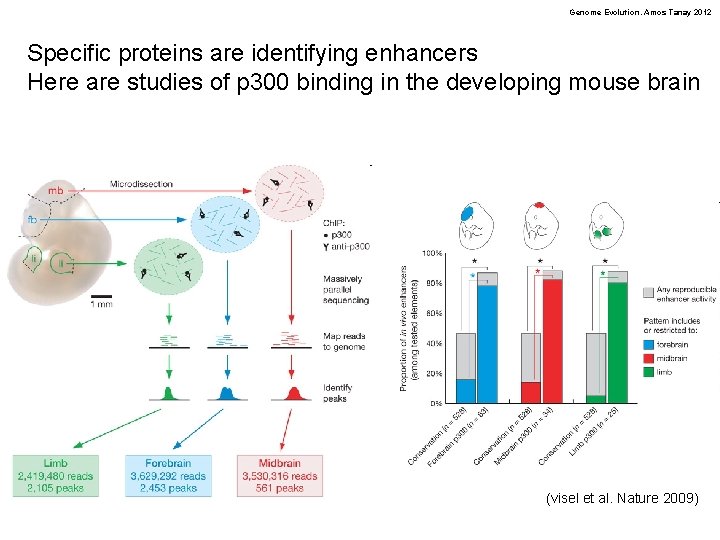
Genome Evolution. Amos Tanay 2012 Specific proteins are identifying enhancers Here are studies of p 300 binding in the developing mouse brain (visel et al. Nature 2009)

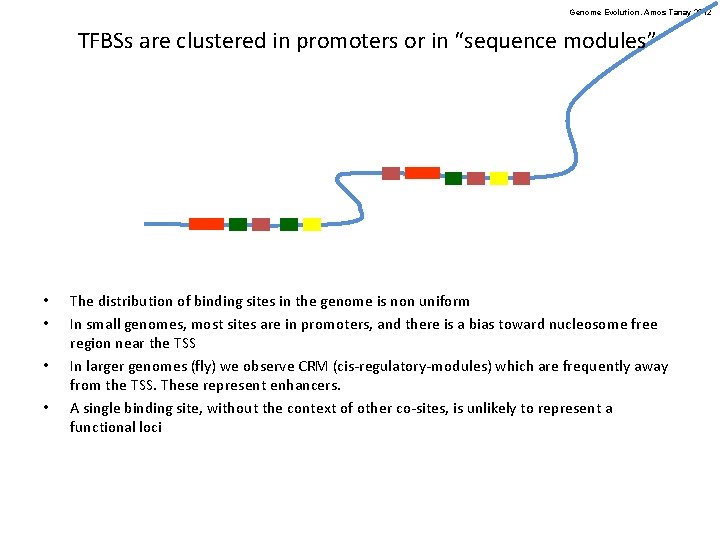
Genome Evolution. Amos Tanay 2012 TFBSs are clustered in promoters or in “sequence modules” • • The distribution of binding sites in the genome is non uniform In small genomes, most sites are in promoters, and there is a bias toward nucleosome free region near the TSS In larger genomes (fly) we observe CRM (cis-regulatory-modules) which are frequently away from the TSS. These represent enhancers. A single binding site, without the context of other co-sites, is unlikely to represent a functional loci

Genome Evolution. Amos Tanay 2012 Discriminative scores for motifs • • • So far we used a generative probabilistic model to learn PWMs The model was designed to generate the data from parameters We assumed that TFBSs are distributed differently than some fixed background model • If our background model is wrong, we will get the wrong motifs. . • • A different scoring approach try to maximize the discriminative power of the motif model. We will not go here into the details of discriminative vs. generative models, but we shall exemplify the discriminative approach for PWMs. Lousy discriminator High specificity discriminator High sensitivity discriminator

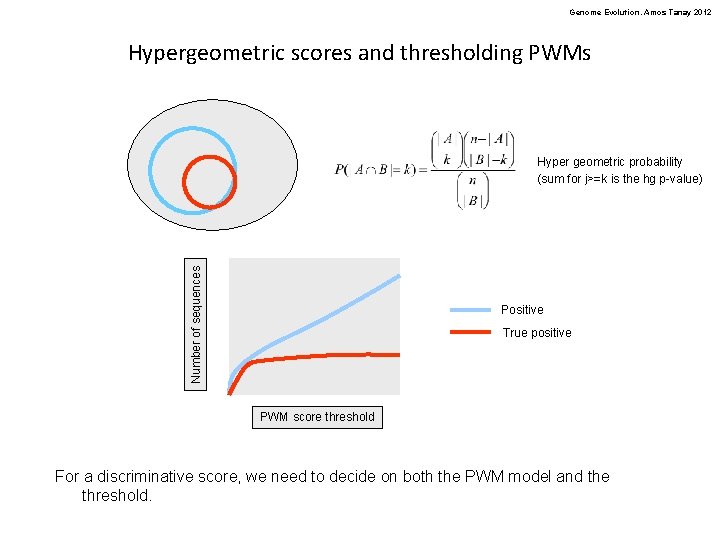
Genome Evolution. Amos Tanay 2012 Hypergeometric scores and thresholding PWMs Number of sequences Hyper geometric probability (sum for j>=k is the hg p-value) Positive True positive PWM score threshold For a discriminative score, we need to decide on both the PWM model and the threshold.

Genome Evolution. Amos Tanay 2012 Constructing a weight matrix from aligned TFBSs is trivial • This is done by counting (or “voting”) • Several databases (e. g. , TRANSFAC, JASPAR) contain matrices that were constructed from a set of curated and validated binding site • Validated site: usually using “promoter bashing” – testing reported constructs with and without the putative site Transfac 7. 0/11. 3 have 400/830 different PWMs, based on more than 11, 000 papers However, there are no real different 830 matrices out there – the real binding repertoire in nature is still somewhat unclear

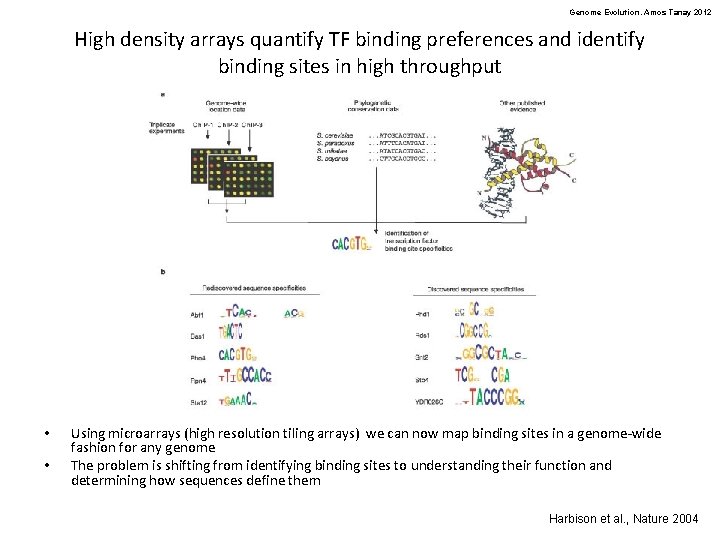
Genome Evolution. Amos Tanay 2012 High density arrays quantify TF binding preferences and identify binding sites in high throughput • • Using microarrays (high resolution tiling arrays) we can now map binding sites in a genome-wide fashion for any genome The problem is shifting from identifying binding sites to understanding their function and determining how sequences define them Harbison et al. , Nature 2004

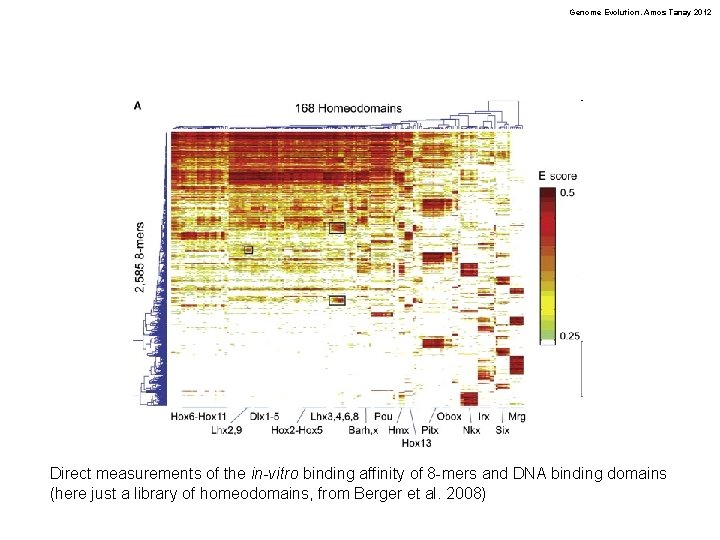
Genome Evolution. Amos Tanay 2012 Direct measurements of the in-vitro binding affinity of 8 -mers and DNA binding domains (here just a library of homeodomains, from Berger et al. 2008)

Genome Evolution. Amos Tanay 2012 Profiling binding affinity to the entire k-mer spectrum provide direct quantification of in-vitro affinity (Badis et al. , 2009) 104 TFs 8 -mers Heatmap of 2 D hierarchical agglomerative clustering analysis of 4740 ungapped 8 mers over 104 nonredundant TFs, with both 8 - mers and proteins clustered using averaged E-score from the two different array designs.

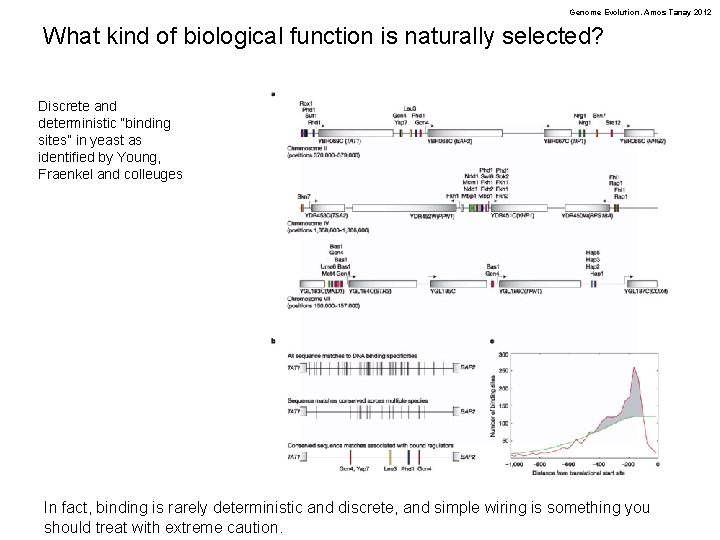
Genome Evolution. Amos Tanay 2012 What kind of biological function is naturally selected? Discrete and deterministic “binding sites” in yeast as identified by Young, Fraenkel and colleuges In fact, binding is rarely deterministic and discrete, and simple wiring is something you should treat with extreme caution.

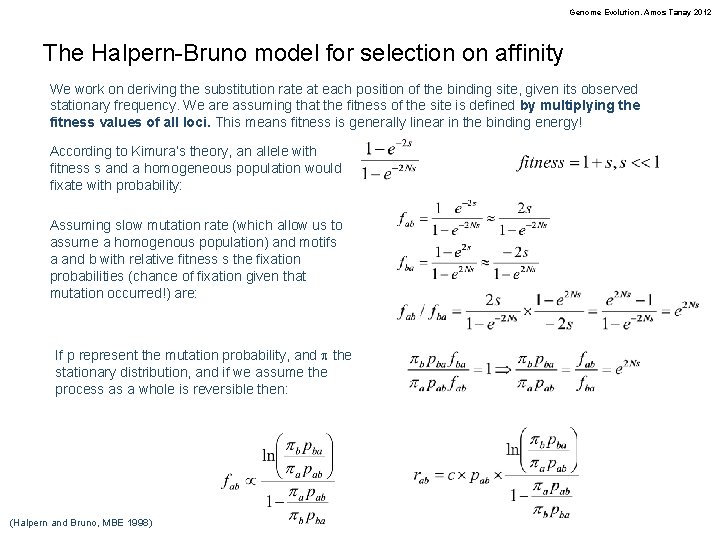
Genome Evolution. Amos Tanay 2012 The Halpern-Bruno model for selection on affinity We work on deriving the substitution rate at each position of the binding site, given its observed stationary frequency. We are assuming that the fitness of the site is defined by multiplying the fitness values of all loci. This means fitness is generally linear in the binding energy! According to Kimura’s theory, an allele with fitness s and a homogeneous population would fixate with probability: Assuming slow mutation rate (which allow us to assume a homogenous population) and motifs a and b with relative fitness s the fixation probabilities (chance of fixation given that mutation occurred!) are: If p represent the mutation probability, and p the stationary distribution, and if we assume the process as a whole is reversible then: (Halpern and Bruno, MBE 1998)

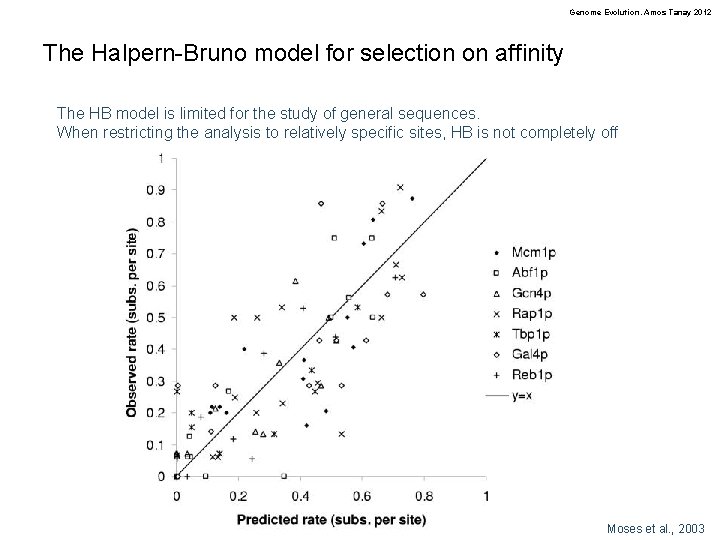
Genome Evolution. Amos Tanay 2012 The Halpern-Bruno model for selection on affinity The HB model is limited for the study of general sequences. When restricting the analysis to relatively specific sites, HB is not completely off Moses et al. , 2003

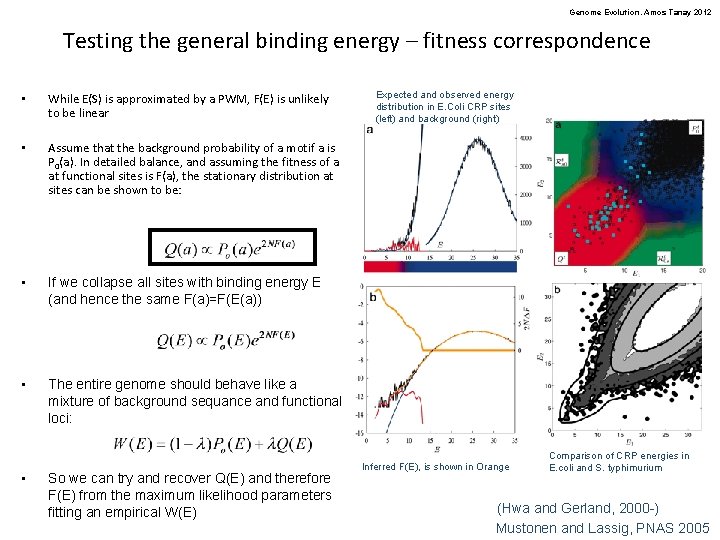
Genome Evolution. Amos Tanay 2012 Testing the general binding energy – fitness correspondence • While E(S) is approximated by a PWM, F(E) is unlikely to be linear • Assume that the background probability of a motif a is P 0(a). In detailed balance, and assuming the fitness of a at functional sites is F(a), the stationary distribution at sites can be shown to be: • If we collapse all sites with binding energy E (and hence the same F(a)=F(E(a)) • The entire genome should behave like a mixture of background sequance and functional loci: • So we can try and recover Q(E) and therefore F(E) from the maximum likelihood parameters fitting an empirical W(E) Expected and observed energy distribution in E. Coli CRP sites (left) and background (right) Inferred F(E), is shown in Orange Comparison of CRP energies in E. coli and S. typhimurium (Hwa and Gerland, 2000 -) Mustonen and Lassig, PNAS 2005

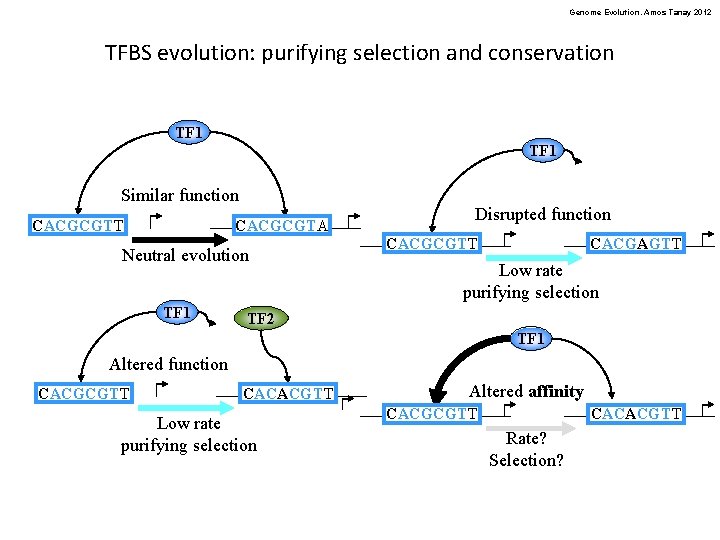
Genome Evolution. Amos Tanay 2012 TFBS evolution: purifying selection and conservation TF 1 Similar function CACGCGTT CACGCGTA Neutral evolution TF 1 Disrupted function CACGCGTT CACGAGTT Low rate purifying selection TF 2 TF 1 Altered function CACGCGTT CACACGTT Low rate purifying selection Altered affinity CACGCGTT CACACGTT Rate? Selection?

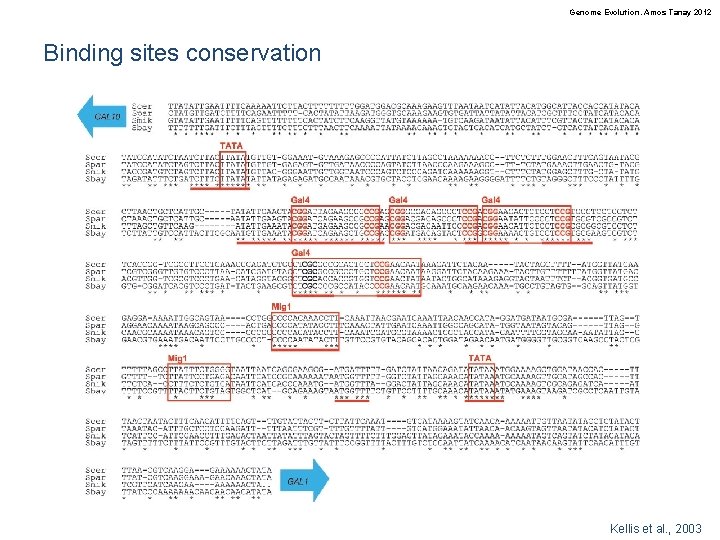
Genome Evolution. Amos Tanay 2012 Binding sites conservation Kellis et al. , 2003

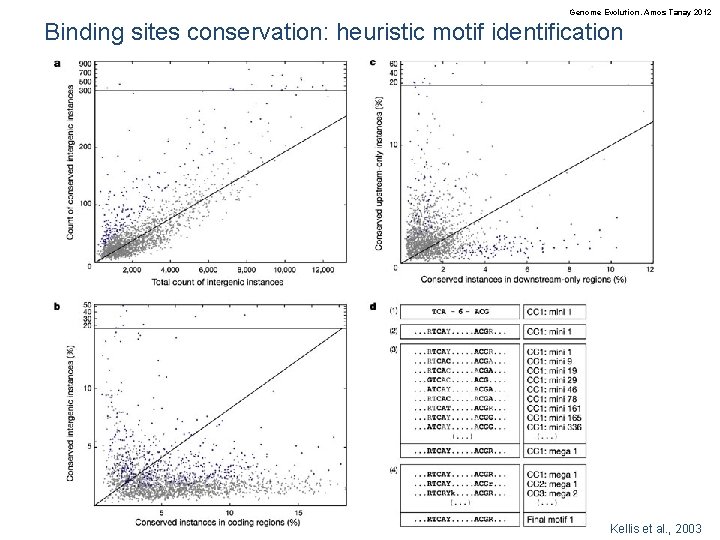
Genome Evolution. Amos Tanay 2012 Binding sites conservation: heuristic motif identification Kellis et al. , 2003

Genome Evolution. Amos Tanay 2012 Analyzing k-mer evolutionary dynamics • Instead of trying to identify conserved motifs try to infer the evolutionary rate of substitution between pairs of k-mers • Start from a multiple alignment and reconstruct ancestral sequences (assuming site independence, or even max parsimony) • Now estimate the number of substitution between pairs of 8 -mers, compare this number to the number expected by the background model • Do it for a lot of sequence, so that statistics on the difference between observed and expected substitutions can be derived

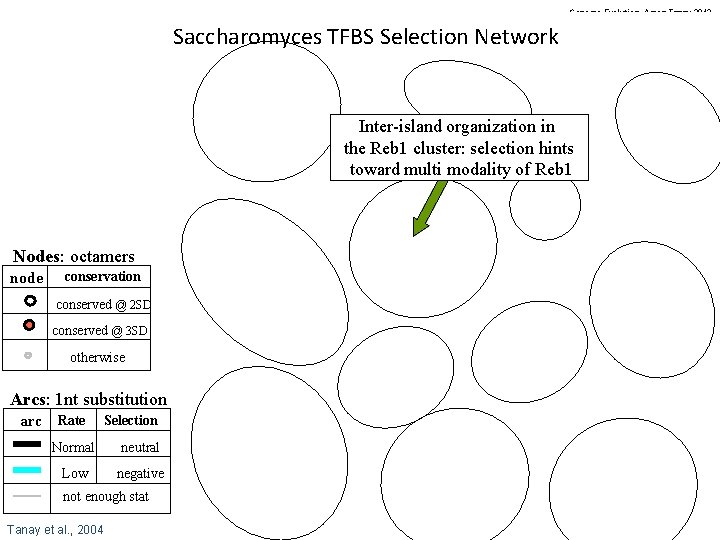
Genome Evolution. Amos Tanay 2012 Saccharomyces TFBS Selection Network Inter-island organization in the Reb 1 cluster: selection hints toward multi modality of Reb 1 Nodes: octamers node conservation conserved @ 2 SD conserved @ 3 SD otherwise Arcs: 1 nt substitution arc Rate Selection Normal neutral Low negative not enough stat Tanay et al. , 2004

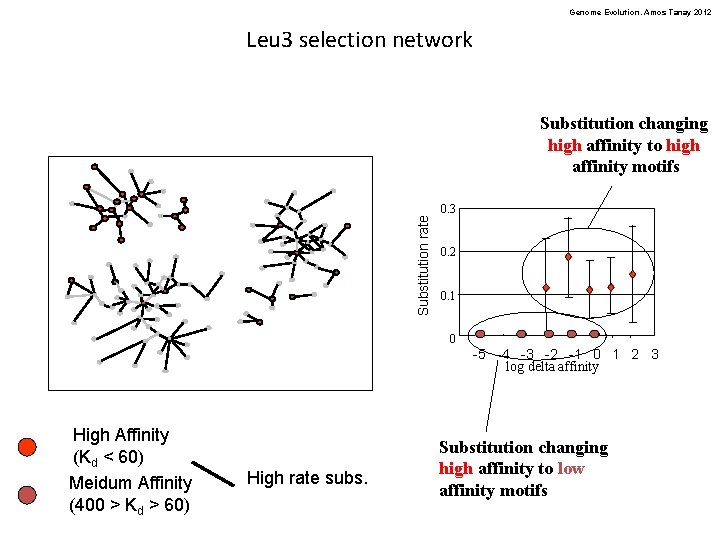
Genome Evolution. Amos Tanay 2012 Leu 3 selection network Substitution rate Substitution changing high affinity to high affinity motifs 0. 3 0. 2 0. 1 0 -5 -4 -3 -2 -1 0 1 2 3 log delta affinity High Affinity (Kd < 60) Meidum Affinity (400 > Kd > 60) High rate subs. Substitution changing high affinity to low affinity motifs

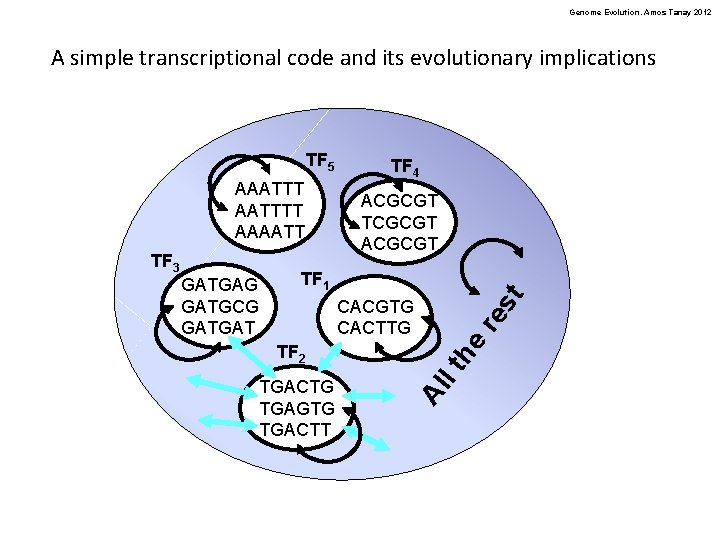
Genome Evolution. Amos Tanay 2012 A simple transcriptional code and its evolutionary implications TF 5 AAATTTT AAAATT st TF 1 TGACTG TGAGTG TGACTT he TF 2 re CACGTG CACTTG lt GATGAG GATGCG GATGAT ACGCGT TCGCGT Al TF 3 TF 4

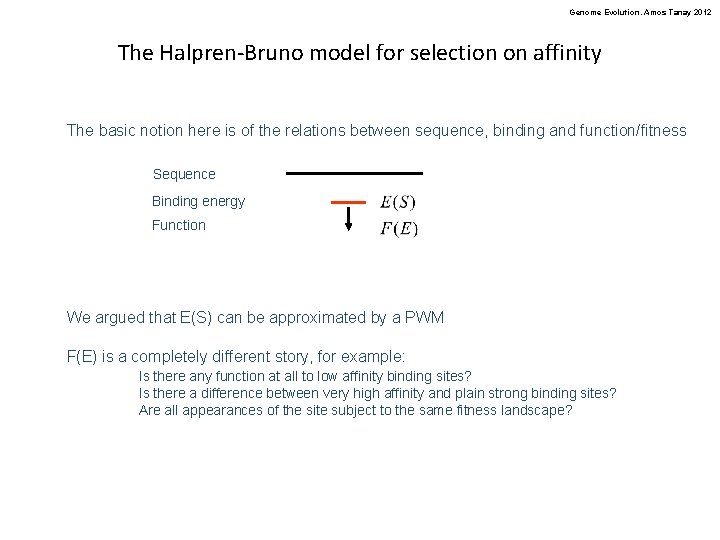
Genome Evolution. Amos Tanay 2012 The Halpren-Bruno model for selection on affinity The basic notion here is of the relations between sequence, binding and function/fitness Sequence Binding energy Function We argued that E(S) can be approximated by a PWM F(E) is a completely different story, for example: Is there any function at all to low affinity binding sites? Is there a difference between very high affinity and plain strong binding sites? Are all appearances of the site subject to the same fitness landscape?

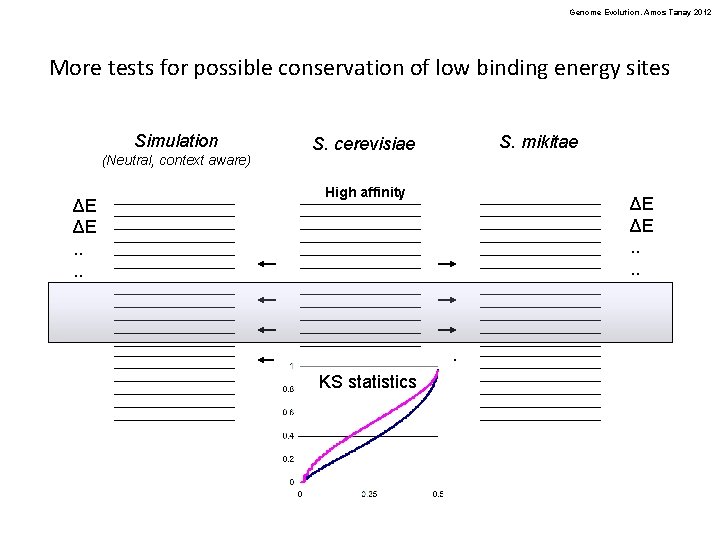
Genome Evolution. Amos Tanay 2012 More tests for possible conservation of low binding energy sites Simulation (Neutral, context aware) ΔE ΔE. . S. cerevisiae High affinity KS statistics Low affinity S. mikitae ΔE ΔE. .

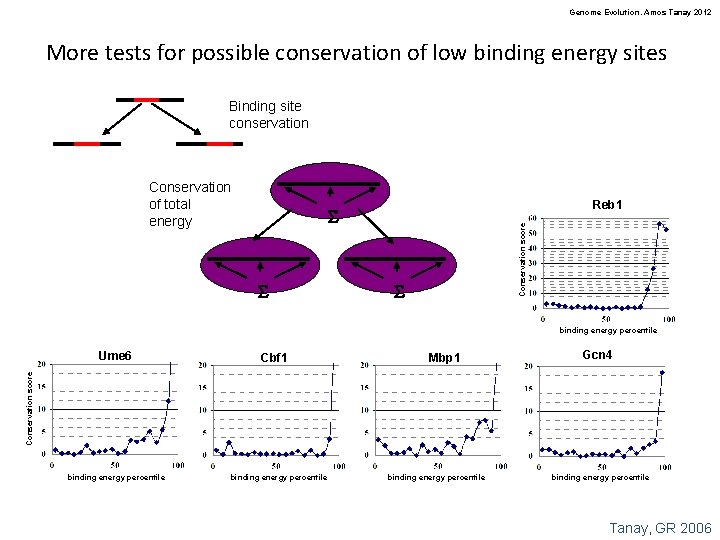
Genome Evolution. Amos Tanay 2012 More tests for possible conservation of low binding energy sites Binding site conservation Conservation of total energy Reb 1 S Conservation score S S binding energy percentile Cbf 1 Mbp 1 Gcn 4 Conservation score Ume 6 binding energy percentile Tanay, GR 2006

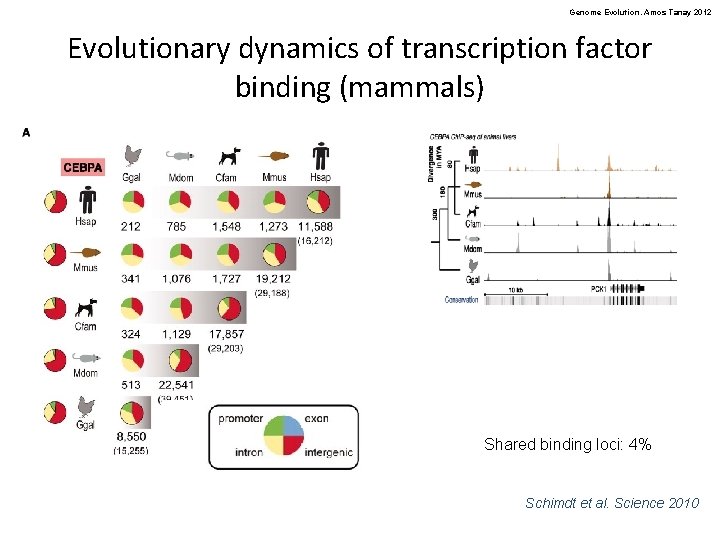
Genome Evolution. Amos Tanay 2012 Evolutionary dynamics of transcription factor binding (mammals) Shared binding loci: 4% Schimdt et al. Science 2010

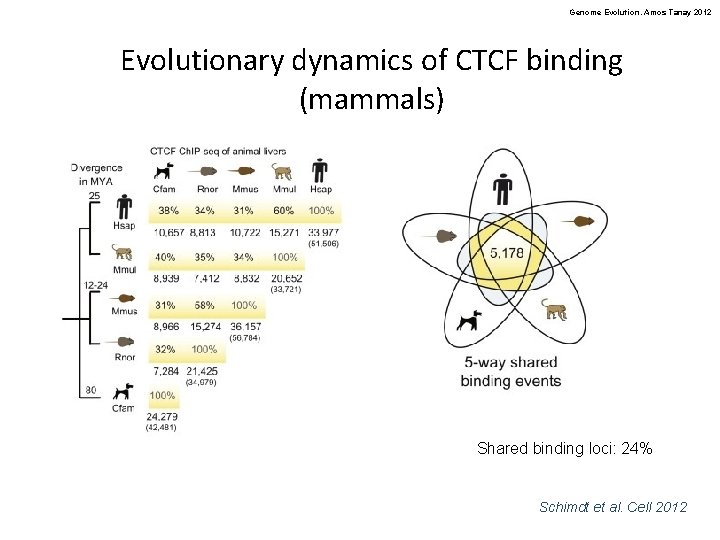
Genome Evolution. Amos Tanay 2012 Evolutionary dynamics of CTCF binding (mammals) Shared binding loci: 24% Schimdt et al. Cell 2012

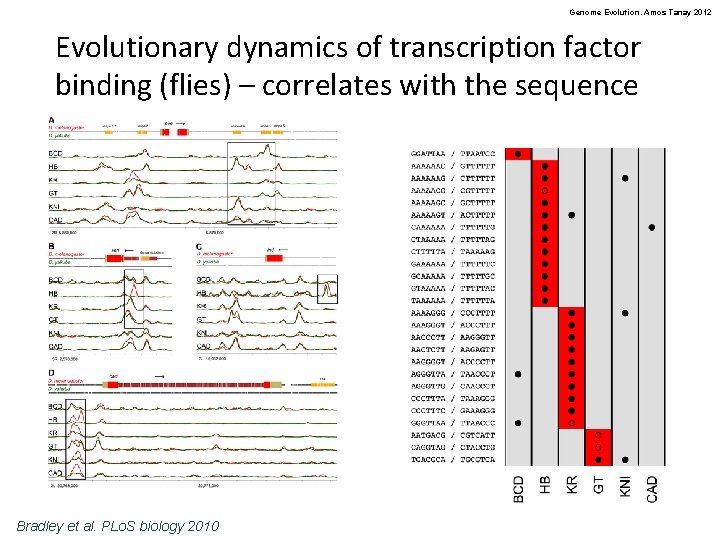
Genome Evolution. Amos Tanay 2012 Evolutionary dynamics of transcription factor binding (flies) – correlates with the sequence Bradley et al. PLo. S biology 2010