Gene Finding and Sequence Annotation Lecture 3 Gene










- Slides: 51

Gene Finding and Sequence Annotation Lecture 3. Gene Finding and Sequence Annotation

Objectives of this lecture • Introduce you to basic concepts and approaches of gene finding • Show you differences between gene prediction for prokaryotic and eukaryotic genomes • Show you which sequence features can be used to identify genes • Introduce you gene finding methods • Briefly discuss the evaluation of gene finding methods This lecture will get you familiar with several important concepts of gene prediction, which will help you to recognize some important pitfalls and to make an informed choice for specific software applications. Lecture 3. Gene Finding and Sequence Annotation

Gene Prediction: Computational Challenge >Genomics DNA……. . atgcatgcggctatgctaagctgggatccgatgacaat gcatgcggctatgctaatgcggctatgcaagctgggatccgatgactatgct aagctgggatccgatgacaatgcggctatgctaatggtcttgggattt accttggaatgctaagctgggatccgatgacaatgcggctatgctaatgaat ggtcttgggatttaccttggaatatgctaatgcggctatgctaagctgggat ccgatgacaatgcggctatgctaatgcggctatgcaagctgggatccg atgactatgctaagctgcggctatgctaatgcggctatgctaagctgggatc cgatgacaatgcggctatgctaatgcggctatgcaagctgggatcctg cggctatgctaatggtcttgggatttaccttggaatgctaagctgggatccg atgacaatgcggctatgctaatggtcttgggatttaccttggaatatg ctaatgcatgcggctatgctaagctgggat ccgatgacaatgcggctatgctaatgcggctatgcaagctgggatccg atgactatgctaagctgcggctatgctaatgcggctatgctaagctcatgcg gctatgctaagctgggaatgcggctatgctaagctgggatccgatgacaatg catgcggctatgctaatgcggctatgcaagctgggatccgatgactatgcta agctgcggctatgctaatgcggctatgctaagctcggctatgctaatg gtcttgggatttaccttggaatgctaagctgggatccgatgacaatgcggct atgctaatggtcttgggatttaccttggaatatgctaatgcggctatg ctaagctgggaatgcggctatgctaagctgggatccgatgacaatgcg gctatgctaatgcggctatgcaagctgggatccgatgactatgctaagctgc ggctatgctaatgcggctatgctaagctcatgcgg Where is gene?

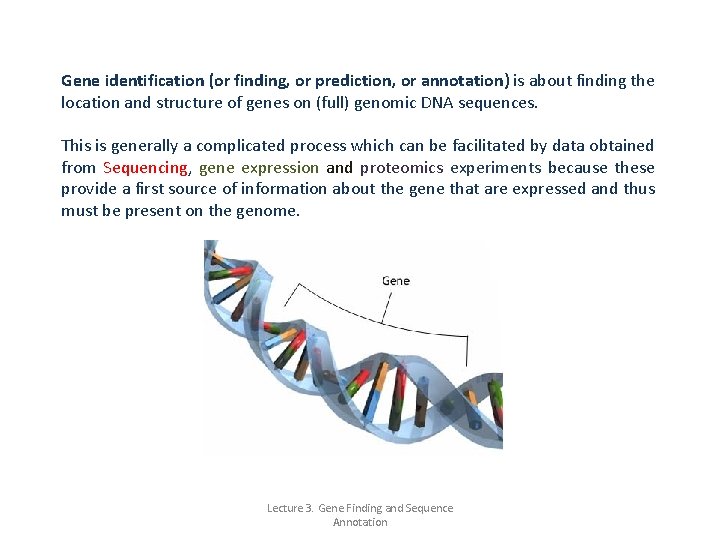
Gene identification (or finding, or prediction, or annotation) is about finding the location and structure of genes on (full) genomic DNA sequences. This is generally a complicated process which can be facilitated by data obtained from Sequencing, gene expression and proteomics experiments because these provide a first source of information about the gene that are expressed and thus must be present on the genome. Lecture 3. Gene Finding and Sequence Annotation

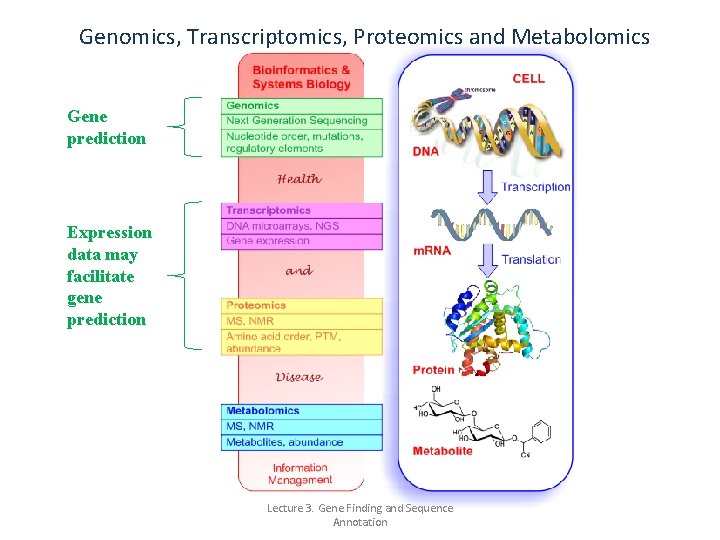
Genomics, Transcriptomics, Proteomics and Metabolomics Gene prediction Expression data may facilitate gene prediction Lecture 3. Gene Finding and Sequence Annotation

Why Gene Prediction/finding/searching? With the advent of next generation sequencing it has become fairly easy to generate full genome sequences. The real challenge is the annotation of these sequences (see next slide), i. e. , providing a full description of the genome that lists all genes and other structures on the genome. Lecture 3. Gene Finding and Sequence Annotation

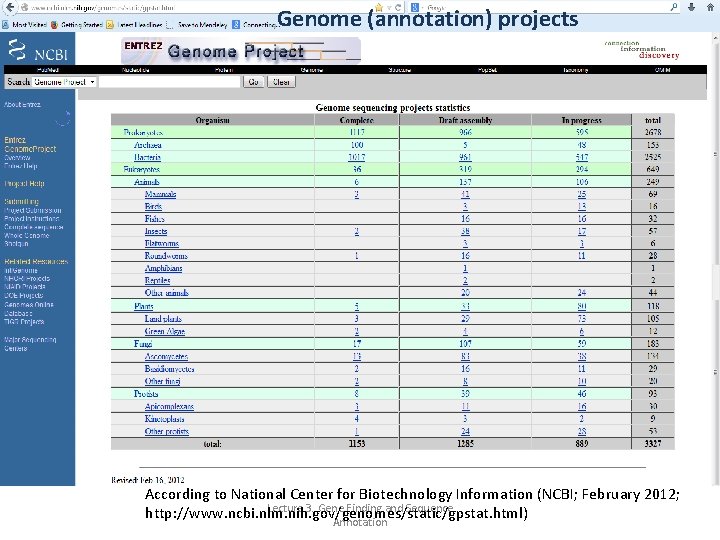
Genome (annotation) projects According to National Center for Biotechnology Information (NCBI; February 2012; Lecture 3. Gene Finding and Sequence http: //www. ncbi. nlm. nih. gov/genomes/static/gpstat. html) Annotation

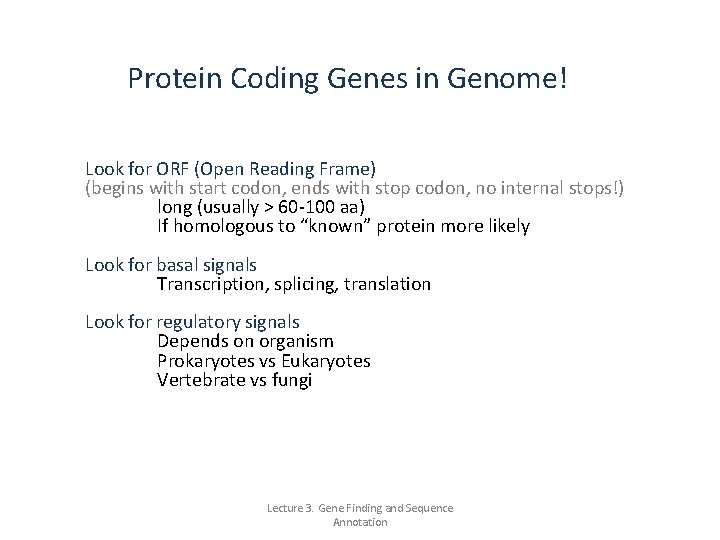
Protein Coding Genes in Genome! Look for ORF (Open Reading Frame) (begins with start codon, ends with stop codon, no internal stops!) long (usually > 60 -100 aa) If homologous to “known” protein more likely Look for basal signals Transcription, splicing, translation Look for regulatory signals Depends on organism Prokaryotes vs Eukaryotes Vertebrate vs fungi Lecture 3. Gene Finding and Sequence Annotation

Why and How Annotation? • This Increase in number of whole-genome sequences make it necessary • These are analyzed to identify protein-coding genes AND other genetic elements • Often some experimental data available to assist in this task – E. g. , previously characterized genes, gene products, ESTs – Sequences of genes and products (from other organisms) can be aligned to identify translated regions • Set of genes from alignment only will be incomplete – Features such as repeat and control sequences will be missing • Therefore, computational methods have been developed to Lectureother 3. Gene Finding and Sequence characterize genes and features: ANNOTATION Annotation

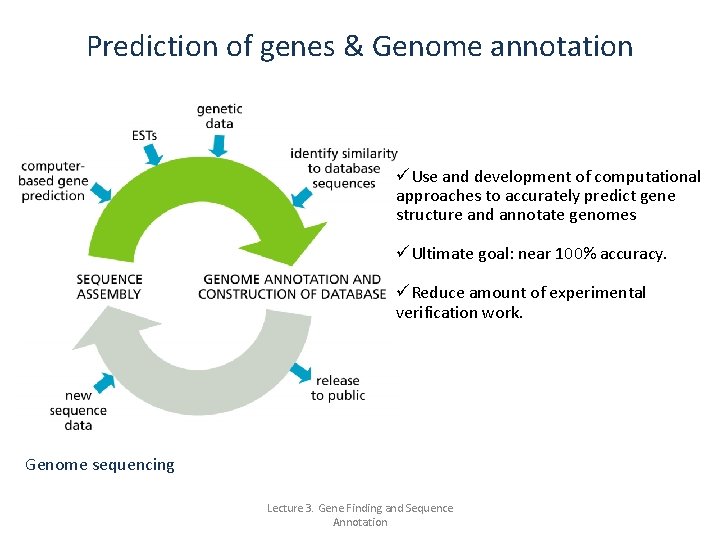
Prediction of genes & Genome annotation üUse and development of computational approaches to accurately predict gene structure and annotate genomes üUltimate goal: near 100% accuracy. üReduce amount of experimental verification work. Genome sequencing Lecture 3. Gene Finding and Sequence Annotation

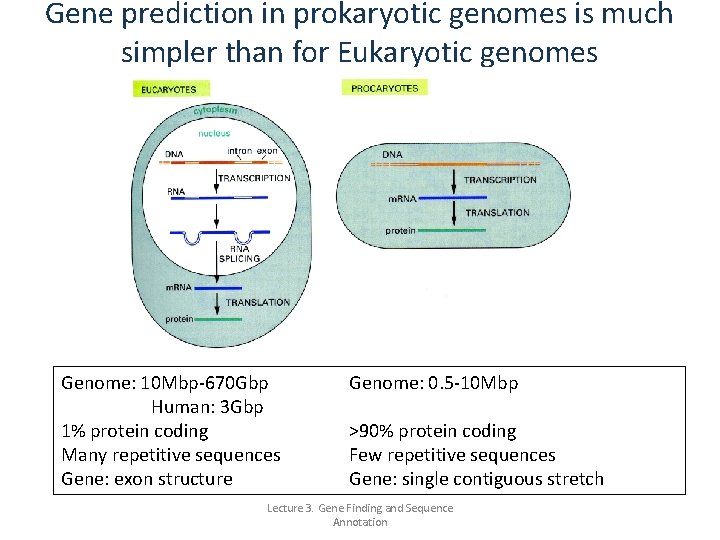
Gene prediction in prokaryotic genomes is much simpler than for Eukaryotic genomes Genome: 10 Mbp-670 Gbp Human: 3 Gbp 1% protein coding Many repetitive sequences Gene: exon structure Genome: 0. 5 -10 Mbp >90% protein coding Few repetitive sequences Gene: single contiguous stretch Lecture 3. Gene Finding and Sequence Annotation

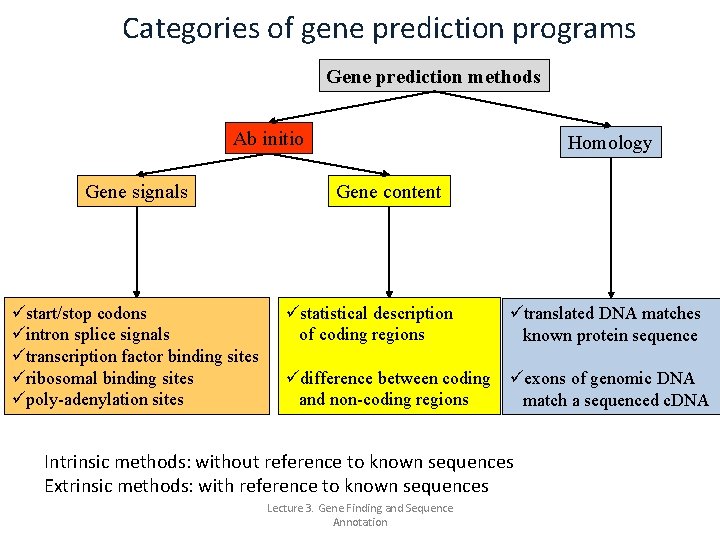
Gene prediction methods There exist several classes of gene prediction methods: >methods are based on homology. Homology between protein or DNA sequences is defined in terms of shared ancestry. Two segments of DNA can have shared ancestry because of either a speciation event (orthologs) or a duplication event (paralogs). In gene identification you can compare known DNA/m. RNA sequences to a newly obtained genome sequence to obtain information about the location of a gene (and its structure) on the genome. >Other methods are ‘ab initio’. These methods don’t use existing experimental data (e. g. , sequence data as in homology searching) but apply algorithms to identify gene signals in the DNA which may indicate the presence of a gene, or they determine the composition (gene content) of a piece of DNA, which may also give clues about the existence of a gene in a particular region of DNA. Lecture 3. Gene Finding and Sequence Annotation

Categories of gene prediction programs Gene prediction methods Ab initio Gene signals üstart/stop codons üintron splice signals ütranscription factor binding sites üribosomal binding sites üpoly-adenylation sites Homology Gene content üstatistical description of coding regions ütranslated DNA matches known protein sequence üdifference between coding üexons of genomic DNA and non-coding regions match a sequenced c. DNA Intrinsic methods: without reference to known sequences Extrinsic methods: with reference to known sequences Lecture 3. Gene Finding and Sequence Annotation

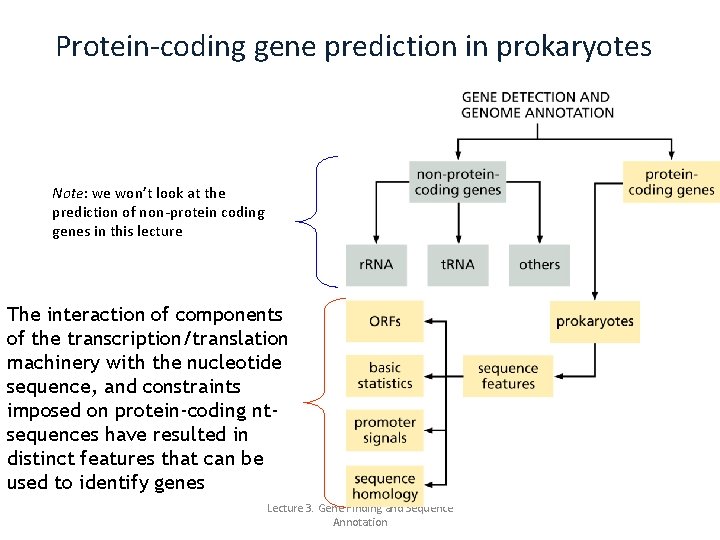
Protein-coding gene prediction in prokaryotes Note: we won’t look at the prediction of non-protein coding genes in this lecture The interaction of components of the transcription/translation machinery with the nucleotide sequence, and constraints imposed on protein-coding ntsequences have resulted in distinct features that can be used to identify genes Lecture 3. Gene Finding and Sequence Annotation

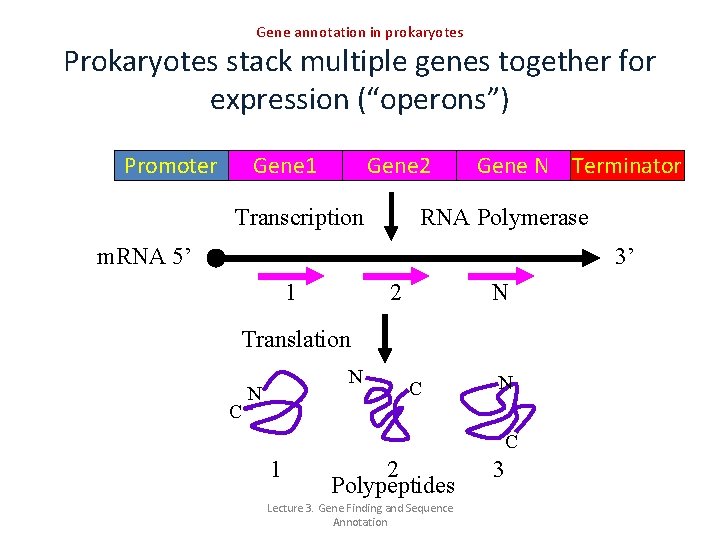
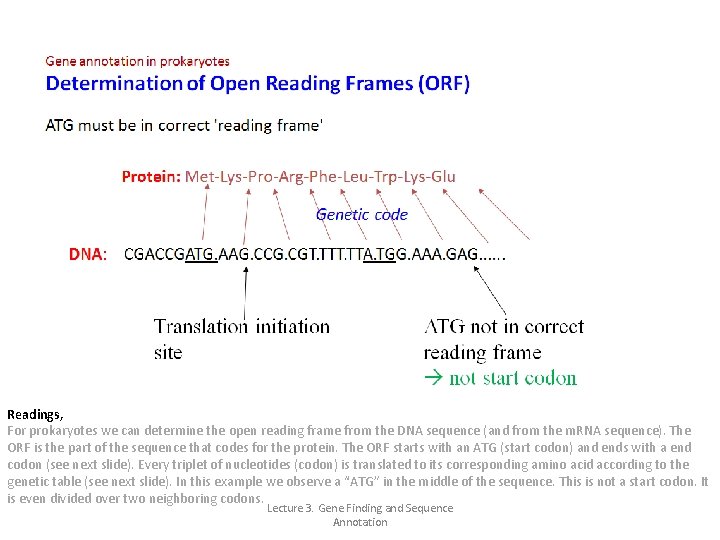
Gene annotation in prokaryotes Prokaryotes stack multiple genes together for expression (“operons”) Promoter Gene 1 Gene 2 Transcription Gene N Terminator RNA Polymerase m. RNA 5’ 3’ 1 2 N Translation C N N C 1 2 Polypeptides Lecture 3. Gene Finding and Sequence Annotation 3

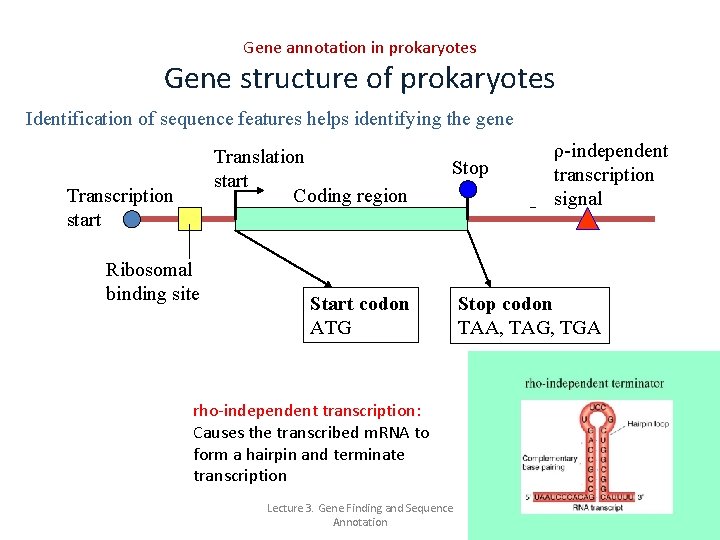
Gene annotation in prokaryotes Gene structure of prokaryotes Identification of sequence features helps identifying the gene Translation start Coding region Transcription start Ribosomal binding site Stop Start codon ATG rho-independent transcription: Causes the transcribed m. RNA to form a hairpin and terminate transcription Lecture 3. Gene Finding and Sequence Annotation ρ-independent transcription signal Stop codon TAA, TAG, TGA

Readings, For prokaryotes we can determine the open reading frame from the DNA sequence (and from the m. RNA sequence). The ORF is the part of the sequence that codes for the protein. The ORF starts with an ATG (start codon) and ends with a end codon (see next slide). Every triplet of nucleotides (codon) is translated to its corresponding amino acid according to the genetic table (see next slide). In this example we observe a “ATG” in the middle of the sequence. This is not a start codon. It is even divided over two neighboring codons. Lecture 3. Gene Finding and Sequence Annotation

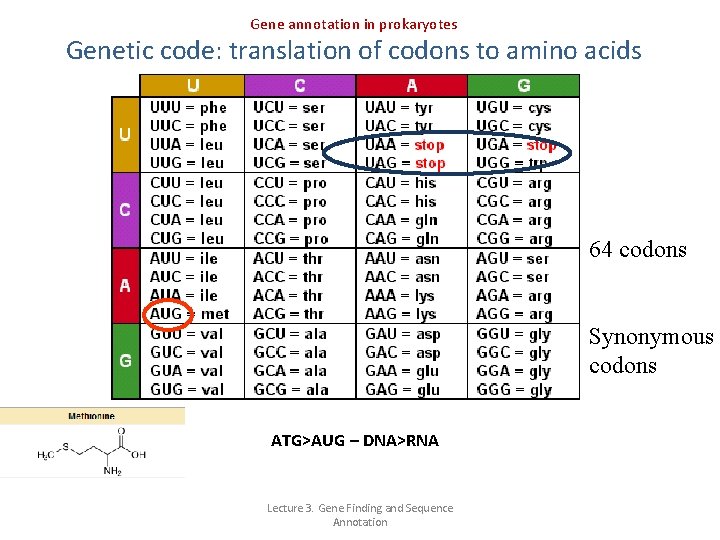
Gene annotation in prokaryotes Genetic code: translation of codons to amino acids 64 codons Synonymous codons ATG>AUG – DNA>RNA Lecture 3. Gene Finding and Sequence Annotation

Gene Prediction: Computational Challenge >Genomics DNA……. . atgcatgcggctatgctaagctgggatccgatgacaat gcatgcggctatgctaatgcggctatgcaagctgggatccgatgactatgct aagctgggatccgatgacaatgcggctatgctaatggtcttgggattt accttggaatgctaagctgggatccgatgacaatgcggctatgctaatgaat ggtcttgggatttaccttggaatatgctaatgcggctatgctaagctgggat ccgatgacaatgcggctatgctaatgcggctatgcaagctgggatccg atgactatgctaagctgcggctatgctaatgcggctatgctaagctgggatc cgatgacaatgcggctatgctaatgcggctatgcaagctgggatcctg cggctatgctaatggtcttgggatttaccttggaatgctaagctgggatccg atgacaatgcggctatgctaatggtcttgggatttaccttggaatatg ctaatgcatgcggctatgctaagctgggat ccgatgacaatgcggctatgctaatgcggctatgcaagctgggatccg atgactatgctaagctgcggctatgctaatgcggctatgctaagctcatgcg gctatgctaagctgggaatgcggctatgctaagctgggatccgatgacaatg catgcggctatgctaatgcggctatgcaagctgggatccgatgactatgcta agctgcggctatgctaatgcggctatgctaagctcggctatgctaatg gtcttgggatttaccttggaatgctaagctgggatccgatgacaatgcggct atgctaatggtcttgggatttaccttggaatatgctaatgcggctatg ctaagctgggaatgcggctatgctaagctgggatccgatgacaatgcg gctatgctaatgcggctatgcaagctgggatccgatgactatgctaagctgc ggctatgctaatgcggctatgctaagctcatgcgg Gene!

Microbial Gene Finding • Microbial genome tends to be gene rich (80%-90% of the sequence is coding) • The most reliable method – homology searches (e. g. using BLAST and/or FASTA) • Major problem – finding genes without known homologue.

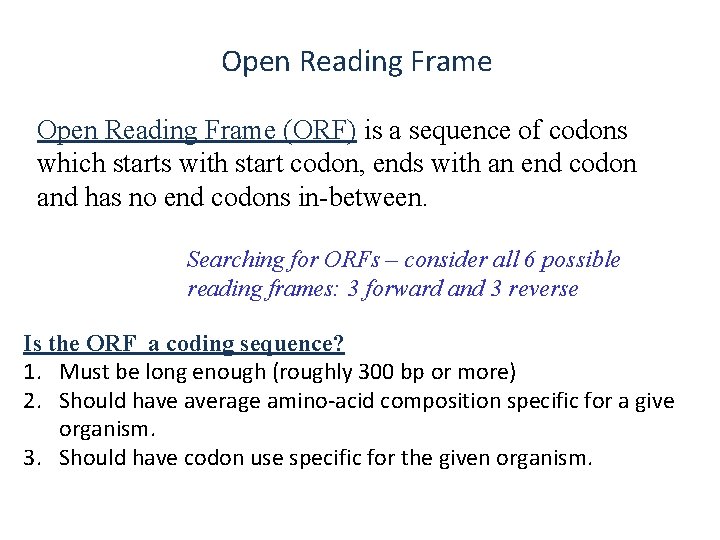
Open Reading Frame (ORF) is a sequence of codons which starts with start codon, ends with an end codon and has no end codons in-between. Searching for ORFs – consider all 6 possible reading frames: 3 forward and 3 reverse Is the ORF a coding sequence? 1. Must be long enough (roughly 300 bp or more) 2. Should have average amino-acid composition specific for a give organism. 3. Should have codon use specific for the given organism.

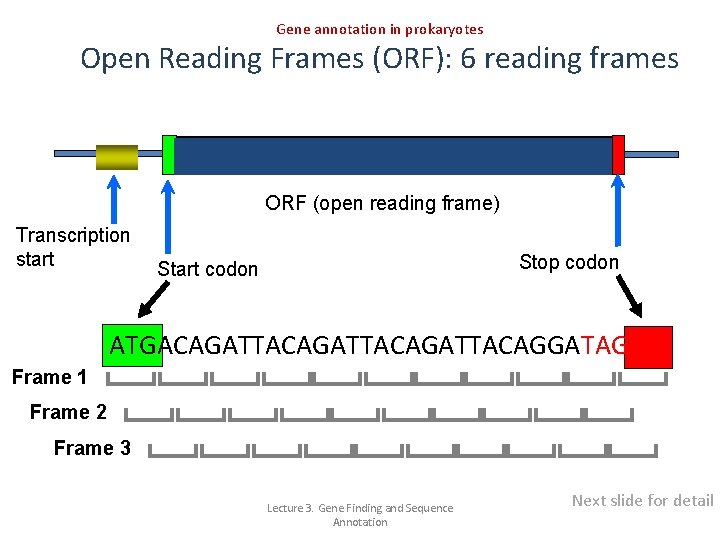
Gene annotation in prokaryotes Open Reading Frames (ORF): 6 reading frames ORF (open reading frame) Transcription start Stop codon Start codon ATGACAGATTACAGATTACAGGATAG Frame 1 Frame 2 Frame 3 Lecture 3. Gene Finding and Sequence Annotation Next slide for detail

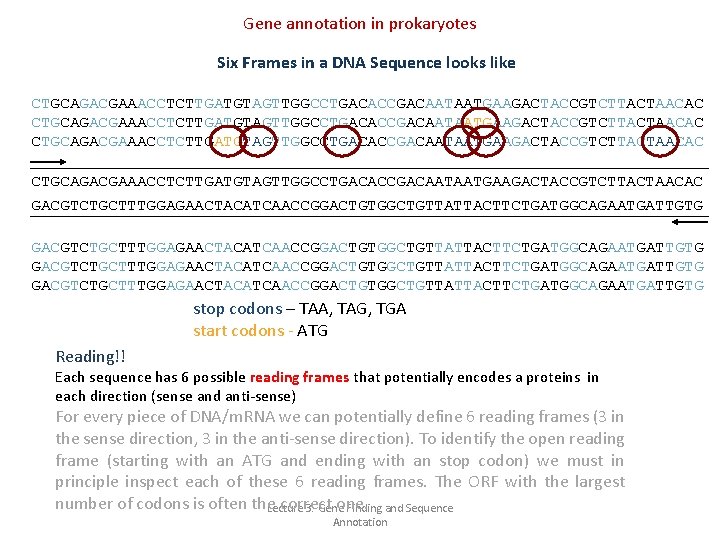
Gene annotation in prokaryotes Six Frames in a DNA Sequence looks like CTGCAGACGAAACCTCTTGATGTAGTTGGCCTGACACCGACAATAATGAAGACTACCGTCTTACTAACAC GACGTCTGCTTTGGAGAACTACATCAACCGGACTGTGGCTGTTATTACTTCTGATGGCAGAATGATTGTG stop codons – TAA, TAG, TGA start codons - ATG Reading!! Each sequence has 6 possible reading frames that potentially encodes a proteins in each direction (sense and anti-sense) For every piece of DNA/m. RNA we can potentially define 6 reading frames (3 in the sense direction, 3 in the anti-sense direction). To identify the open reading frame (starting with an ATG and ending with an stop codon) we must in principle inspect each of these 6 reading frames. The ORF with the largest number of codons is often the correct one. Lecture 3. Gene Finding and Sequence Annotation

Reading frame A reading frame refers to one of three possible ways of reading a nucleotide sequence. Let's say we have a stretch of 15 DNA base pairs: acttagccgggacta • You can start translating the DNA from the first letter, 'a, ' which would be referred to as the first reading frame. • Or you can start reading from the second letter, 'c, ' which is the second reading frame. • Or you can start reading from the third letter, 't, ' which is the third reading frame. The reading frame affects which protein is made. In the example below, the upper case letters represent amino acids that are coded by the three letters above and to the left of them. The illustration above shows three reading frames. However, there actually six reading frames: three on the positive strand, and three (which are read in the reverse Lecture 3. Gene Finding and Sequence direction) on the negative strand. Annotation

Problems: üThere will be many "ORFs“ occurring by chance üSome will be short - how do we know which are true? üIntrons make this useless in Eukaryotic DNA

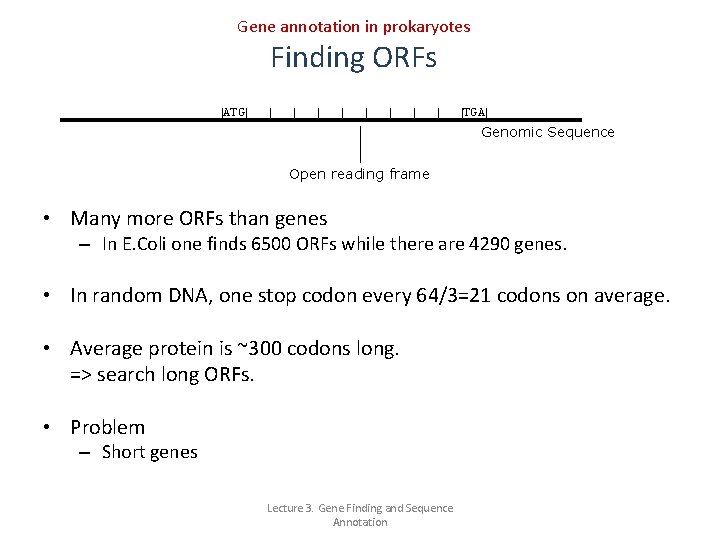
Gene annotation in prokaryotes Finding ORFs ATG TGA Genomic Sequence Open reading frame • Many more ORFs than genes – In E. Coli one finds 6500 ORFs while there are 4290 genes. • In random DNA, one stop codon every 64/3=21 codons on average. • Average protein is ~300 codons long. => search long ORFs. • Problem – Short genes Lecture 3. Gene Finding and Sequence Annotation

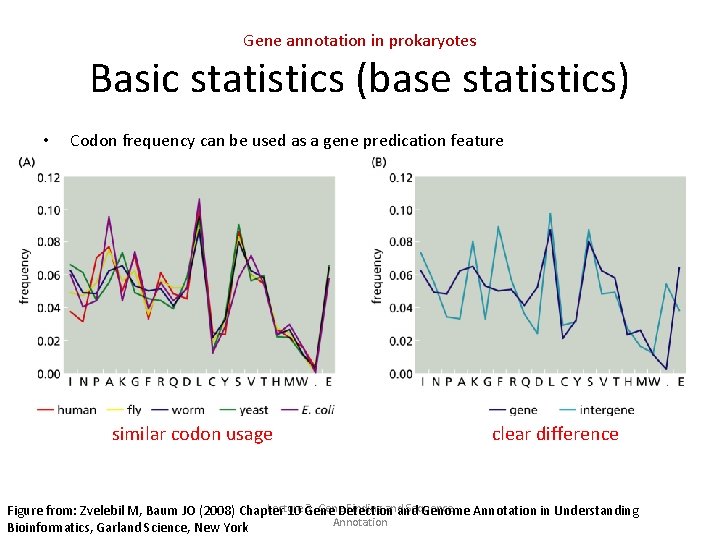
Gene annotation in prokaryotes Basic statistics (base statistics) • Codon frequency can be used as a gene predication feature similar codon usage clear difference Lecture 3. Gene. Detection Finding and Sequence Figure from: Zvelebil M, Baum JO (2008) Chapter 10 Gene and Genome Annotation in Understanding Annotation Bioinformatics, Garland Science, New York

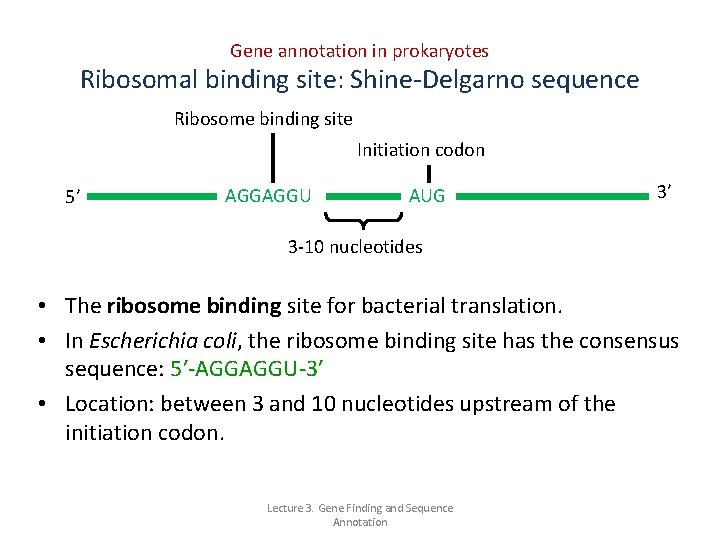
Gene annotation in prokaryotes Ribosomal binding site: Shine-Delgarno sequence Ribosome binding site Initiation codon 5’ AGGAGGU AUG 3’ 3 -10 nucleotides • The ribosome binding site for bacterial translation. • In Escherichia coli, the ribosome binding site has the consensus sequence: 5′-AGGAGGU-3′ • Location: between 3 and 10 nucleotides upstream of the initiation codon. Lecture 3. Gene Finding and Sequence Annotation

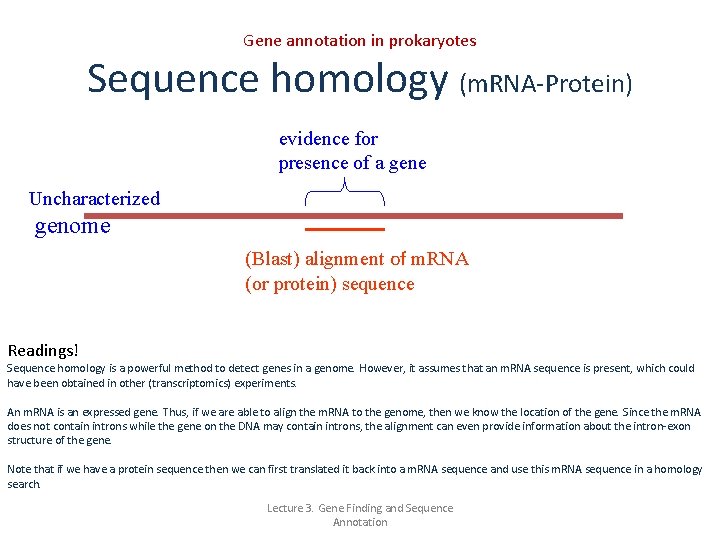
Gene annotation in prokaryotes Sequence homology (m. RNA-Protein) evidence for presence of a gene Uncharacterized genome (Blast) alignment of m. RNA (or protein) sequence Readings! Sequence homology is a powerful method to detect genes in a genome. However, it assumes that an m. RNA sequence is present, which could have been obtained in other (transcriptomics) experiments. An m. RNA is an expressed gene. Thus, if we are able to align the m. RNA to the genome, then we know the location of the gene. Since the m. RNA does not contain introns while the gene on the DNA may contain introns, the alignment can even provide information about the intron-exon structure of the gene. Note that if we have a protein sequence then we can first translated it back into a m. RNA sequence and use this m. RNA sequence in a homology search. Lecture 3. Gene Finding and Sequence Annotation

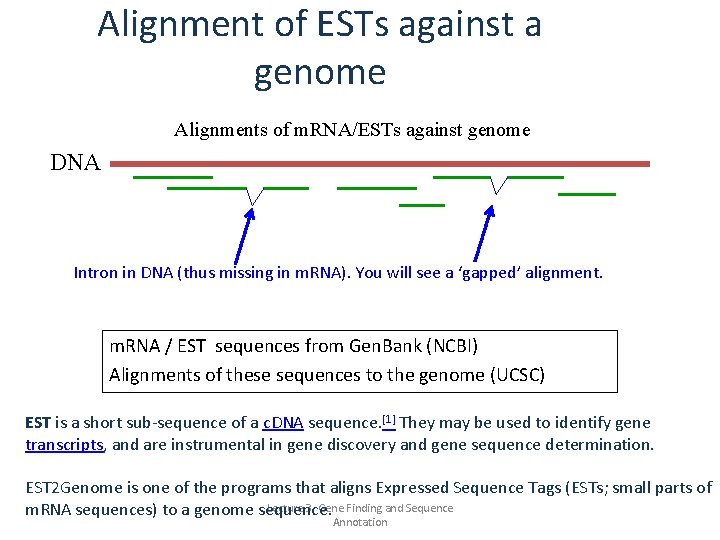
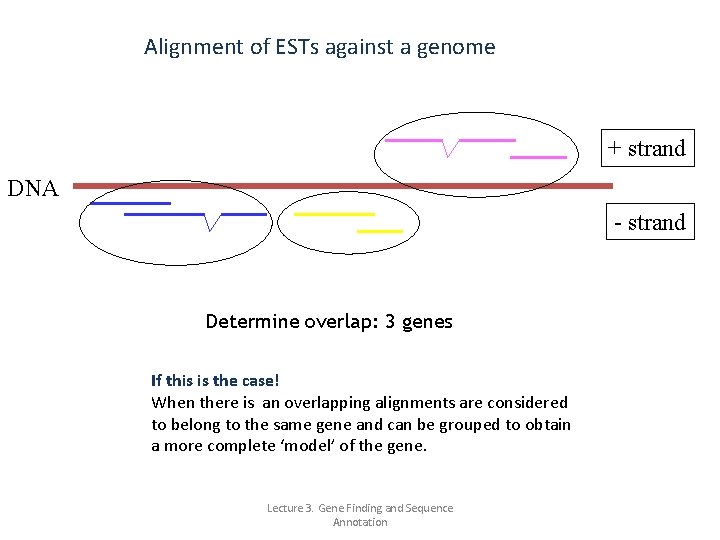
Alignment of ESTs against a genome Alignments of m. RNA/ESTs against genome DNA Intron in DNA (thus missing in m. RNA). You will see a ‘gapped’ alignment. m. RNA / EST sequences from Gen. Bank (NCBI) Alignments of these sequences to the genome (UCSC) EST is a short sub-sequence of a c. DNA sequence. [1] They may be used to identify gene transcripts, and are instrumental in gene discovery and gene sequence determination. EST 2 Genome is one of the programs that aligns Expressed Sequence Tags (ESTs; small parts of Lecture 3. Gene Finding and Sequence m. RNA sequences) to a genome sequence. Annotation

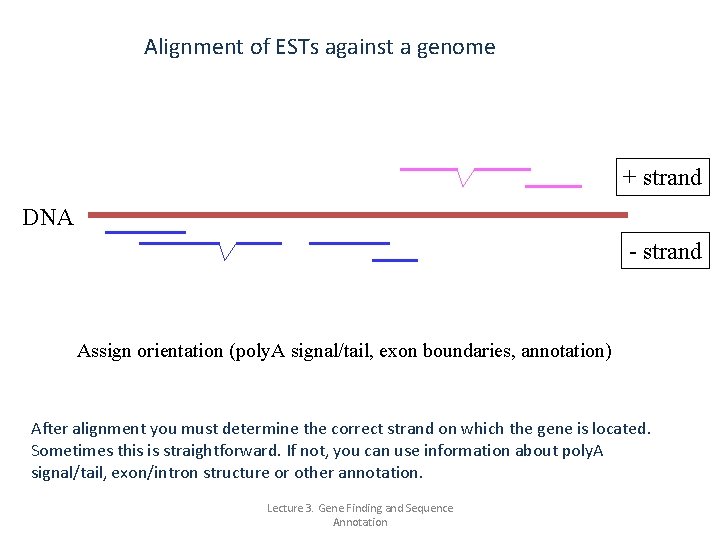
Alignment of ESTs against a genome + strand DNA - strand Assign orientation (poly. A signal/tail, exon boundaries, annotation) After alignment you must determine the correct strand on which the gene is located. Sometimes this is straightforward. If not, you can use information about poly. A signal/tail, exon/intron structure or other annotation. Lecture 3. Gene Finding and Sequence Annotation

Alignment of ESTs against a genome + strand DNA - strand Determine overlap: 3 genes If this is the case! When there is an overlapping alignments are considered to belong to the same gene and can be grouped to obtain a more complete ‘model’ of the gene. Lecture 3. Gene Finding and Sequence Annotation

Gene annotation in prokaryotes Algorithms for Gene Detection in prokaryotes • Some of the programs available • Gene. Mark. hmm • GLIMMER • Eco. Parse • ORPHEUS • Prodigal Many programs for gene identification are available. You don’t have to memorize all these programs for the examination. Lecture 3. Gene Finding and Sequence Annotation

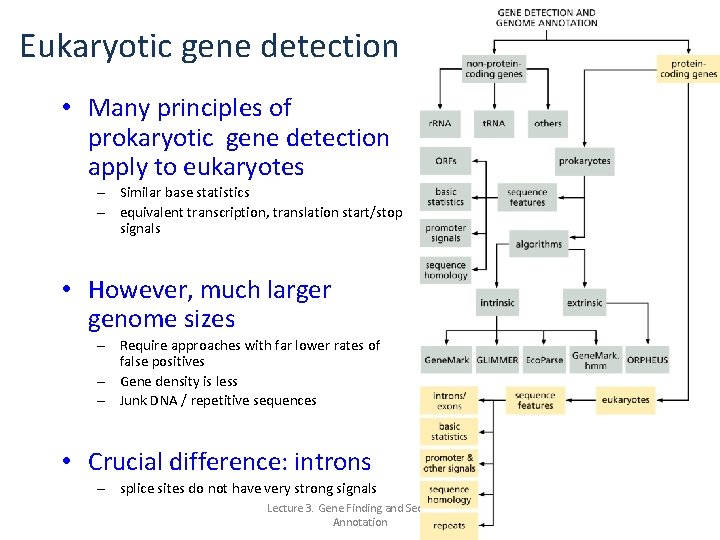
Eukaryotic gene detection • Many principles of prokaryotic gene detection apply to eukaryotes – Similar base statistics – equivalent transcription, translation start/stop signals • However, much larger genome sizes – Require approaches with far lower rates of false positives – Gene density is less – Junk DNA / repetitive sequences • Crucial difference: introns – splice sites do not have very strong signals Lecture 3. Gene Finding and Sequence Annotation

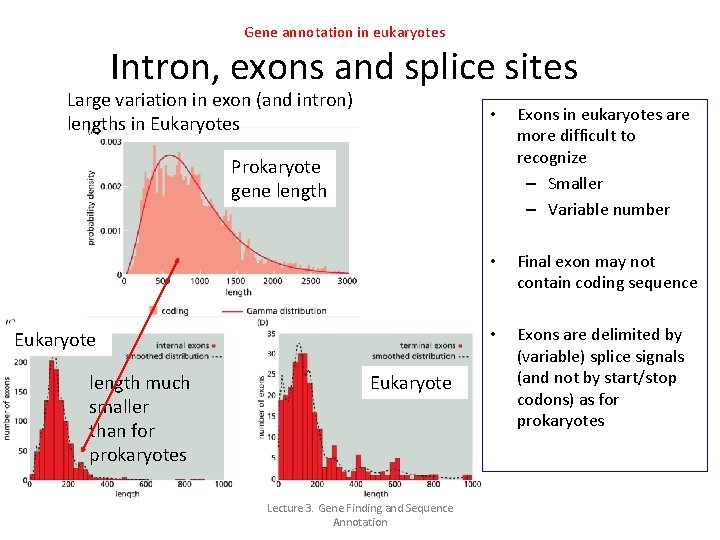
Gene annotation in eukaryotes Intron, exons and splice sites Large variation in exon (and intron) lengths in Eukaryotes • Exons in eukaryotes are more difficult to recognize – Smaller – Variable number • Final exon may not contain coding sequence • Exons are delimited by (variable) splice signals (and not by start/stop codons) as for prokaryotes Prokaryote gene length Eukaryote length much smaller than for prokaryotes Eukaryote Lecture 3. Gene Finding and Sequence Annotation

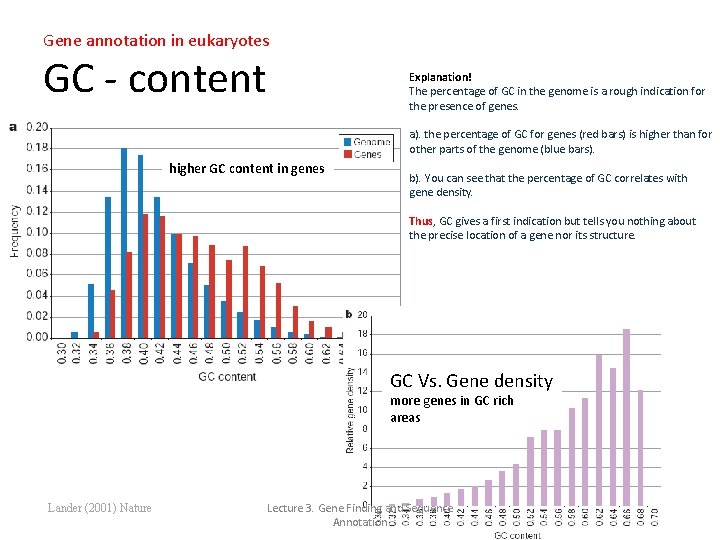
Gene annotation in eukaryotes GC - content Explanation! The percentage of GC in the genome is a rough indication for the presence of genes. a). the percentage of GC for genes (red bars) is higher than for other parts of the genome (blue bars). higher GC content in genes b). You can see that the percentage of GC correlates with gene density. Thus, GC gives a first indication but tells you nothing about the precise location of a gene nor its structure. GC Vs. Gene density more genes in GC rich areas Lander (2001) Nature Lecture 3. Gene Finding and Sequence Annotation

Gene annotation in eukaryotes Complexity Eukaryotes • Finding genes in Eukaryotes is difficult due to variation in gene structure – Average vertebrate gene is 30 kb long out of which coding sequence is only about 1 kb – Average coding region consists of 6 exons of about 150 bp BUT – Dystrophin: 2. 4 Mb long – Blood coagulation factor VIII: 26 exons (69 bp to 3106 bp) • Intron 22 produces 2 transcripts unrelated to this gene. Gene finding algorithms are often capable of detecting an ‘average’ gene. However, genes that somehow deviate in length, structure, etc can be missed by gene finding Lecture 3. Gene Finding programs. and Sequence Annotation

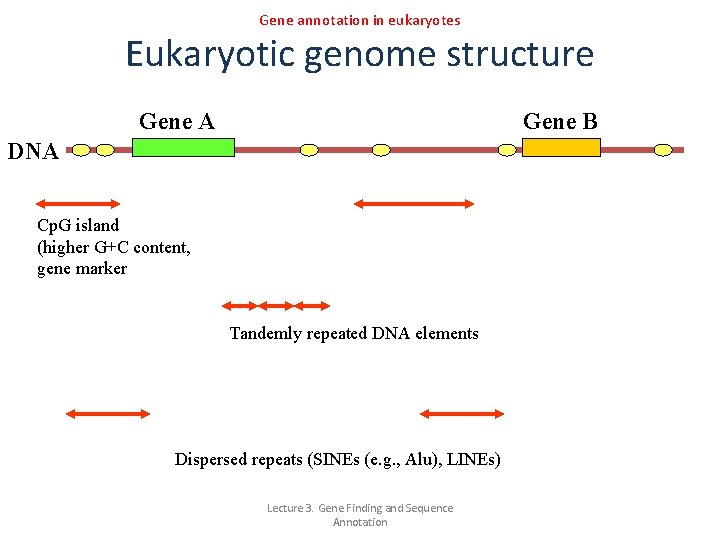
Gene annotation in eukaryotes Eukaryotic genome structure Gene A Gene B DNA Cp. G island (higher G+C content, gene marker Tandemly repeated DNA elements Dispersed repeats (SINEs (e. g. , Alu), LINEs) Lecture 3. Gene Finding and Sequence Annotation

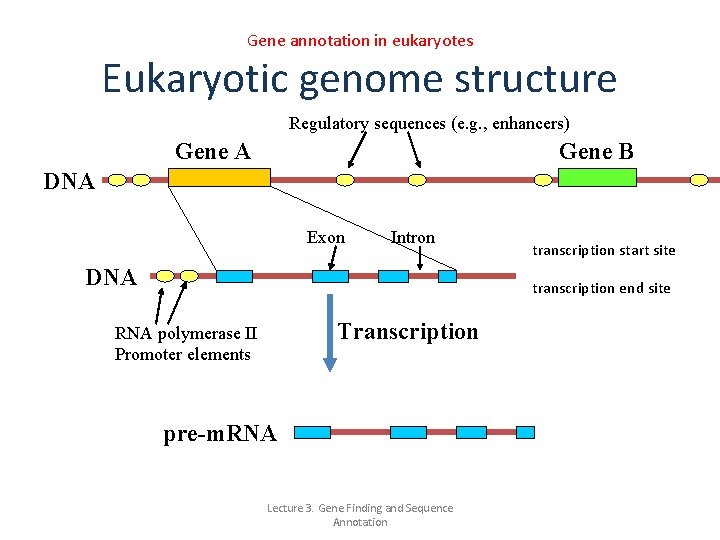
Gene annotation in eukaryotes Eukaryotic genome structure Regulatory sequences (e. g. , enhancers) Gene A Gene B DNA Exon Intron DNA transcription start site transcription end site Transcription RNA polymerase II Promoter elements pre-m. RNA Lecture 3. Gene Finding and Sequence Annotation

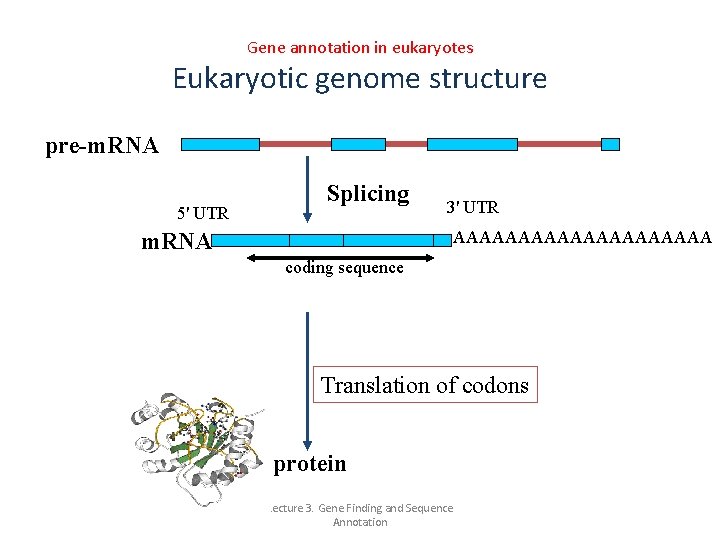
Gene annotation in eukaryotes Eukaryotic genome structure pre-m. RNA 5' UTR Splicing 3' UTR AAAAAAAAAA m. RNA coding sequence Translation of codons protein Lecture 3. Gene Finding and Sequence Annotation

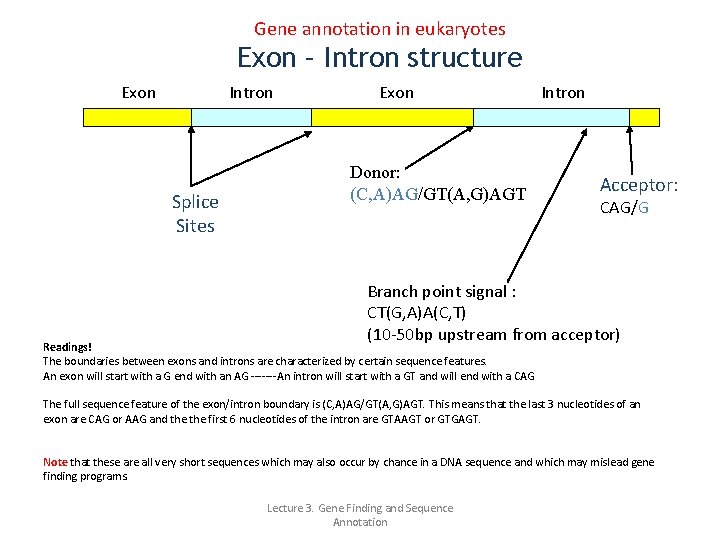
Gene annotation in eukaryotes Exon – Intron structure Exon Intron Splice Sites Exon Donor: (C, A)AG/GT(A, G)AGT Intron Acceptor: CAG/G Branch point signal : CT(G, A)A(C, T) (10 -50 bp upstream from acceptor) Readings! The boundaries between exons and introns are characterized by certain sequence features. An exon will start with a G end with an AG -------An intron will start with a GT and will end with a CAG The full sequence feature of the exon/intron boundary is (C, A)AG/GT(A, G)AGT. This means that the last 3 nucleotides of an exon are CAG or AAG and the first 6 nucleotides of the intron are GTAAGT or GTGAGT. Note that these are all very short sequences which may also occur by chance in a DNA sequence and which may mislead gene finding programs. Lecture 3. Gene Finding and Sequence Annotation

Gene annotation in eukaryotes Polyadenylation signal Eukaryotic m. RNAs are polyadenylated, i. e. , have up to 250 A’s added to their 3’ end after transcription terminates (T) Signals: The poly. A signal is another example of a signal (sequence feature) that signals the end of transcription. For Detail: http: //themedicalbiochemistrypage. org/rna. php#processing Lecture 3. Gene Finding and Sequence Annotation

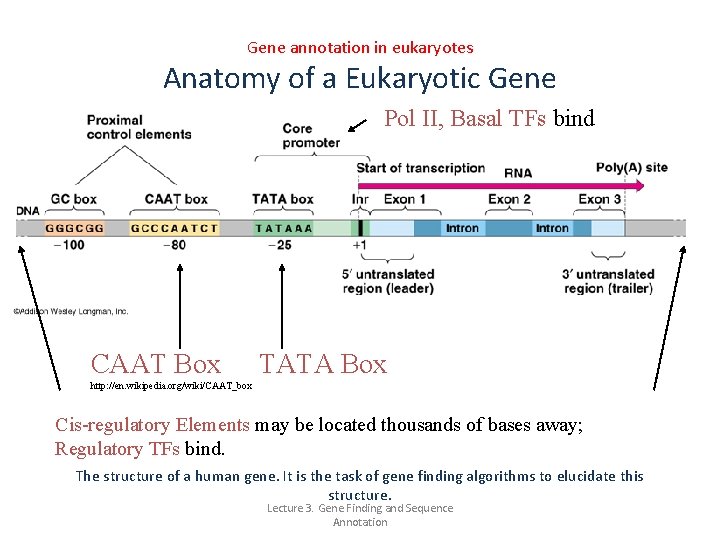
Gene annotation in eukaryotes Anatomy of a Eukaryotic Gene Pol II, Basal TFs bind CAAT Box TATA Box http: //en. wikipedia. org/wiki/CAAT_box Cis-regulatory Elements may be located thousands of bases away; Regulatory TFs bind. The structure of a human gene. It is the task of gene finding algorithms to elucidate this structure. Lecture 3. Gene Finding and Sequence Annotation

Gene annotation in eukaryotes Promotor sequences and binding sites for transcription factors • Further differences between prokaryotic and eukaryotic gene structures: – Sequence signals in upstream regions are much more variable in eukaryotes • Both in position and compositions – Control of gene expression is more complex in eukaryotes • Can be affected by many molecules binding the DNA in the gene region • This leads to many more potential promotor binding sites • These binding sites may be spread over a much larger region (several thousand bases) • Strict control of gene expression – Some genes are known to be poorly expressed because high levels would be damaging (e. g. , genes for growth factors) – Such genes sometimes lack the TATA box characteristic for promotors. – This complicates the identification of such genes Lecture 3. Gene Finding and Sequence Annotation

Methods to detect eukaryotic gene signals • Promotors • Transcription start/stop signals – e. g. TATA box (30% of genes don’t have TATA box) – e. g. poly. A signal • Translation start/stop signals – no defined ribosome-binding site in eukaryotic genes Lecture 3. Gene Finding and Sequence Annotation

Methods to predict the intron/exon structure • ORF identification methods for prokaryotes don’t work • If exons are long enough then base statistics can be used. • Signals for splice sites are not well defined • Initial/terminal exons also contain non-coding sequence Lecture 3. Gene Finding and Sequence Annotation

Complete Eukaryotic gene models • Programs that use and combine all features of a gene to make a prediction about the complete gene structure (=model) • E. g. , Gen. Scan Lecture 3. Gene Finding and Sequence Annotation

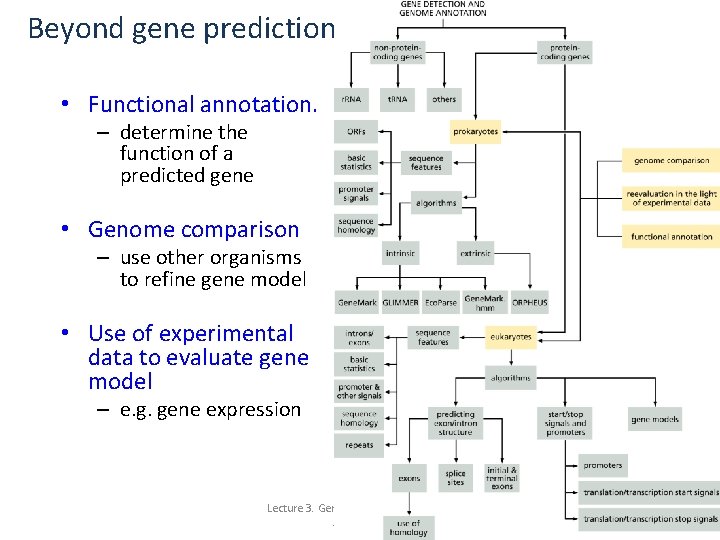
Beyond gene prediction • Functional annotation. – determine the function of a predicted gene • Genome comparison – use other organisms to refine gene model • Use of experimental data to evaluate gene model – e. g. gene expression Lecture 3. Gene Finding and Sequence Annotation

Gene identification programs based on comparison with related genome sequences: TWAIN TWINSCAN Ab initio gene identification programs including those which use homologous gene sequences: GAZE The Gene. Mark set of programs Genie Genome. Scan Gen. Scan GLIMMER, Glimmer. M and Glimmer. HMM Grail. EXP ORPHEUS Wise 2 including Gene. Wise Lecture 3. Gene Finding and Sequence Annotation

Identifying t. RNA genes: t. RNAscan-SE program and web server Promoter prediction programs: Core. Promoter Exon prediction programs: First. EF JTEF MZEF Splice site prediction programs: Gene. Splicer Splice. Predictor Genome annotation visualization programs: Apollo Artemis and Artemis Comparison Tool (ACT) VISTA Lecture 3. Gene Finding and Sequence Annotation

Web Servers: The following web sites provide on-line access to gene annotation tools: Analysis and annotation tool (AAT) First. EF FGENES family of programs Fun. Site. P GAP 2, NAP and other DNA alignment programs Gene. Builder Gene. Splicer Gene. Walker Gene. Wise is part of the Wise 2 suite Gen. Scan Grail. EXP HMMGene Mc. Promoter Net. Plant. Gene NNPP Pro. Scan Lecture 3. Gene Finding and Sequence Annotation