Tomato genome annotation pipeline in Cyrille 2 Erwin

- Slides: 19

Tomato genome annotation pipeline in Cyrille 2 Erwin Datema

Contents of the annotation pipeline n Annotation on the BAC level l n Gene prediction Repeat identification Other features Annotation on the gene level (work in progress) l l blastx vs NCBI’s nr (sequence similarity) Inter. Pro. Scan (domain identifcation)

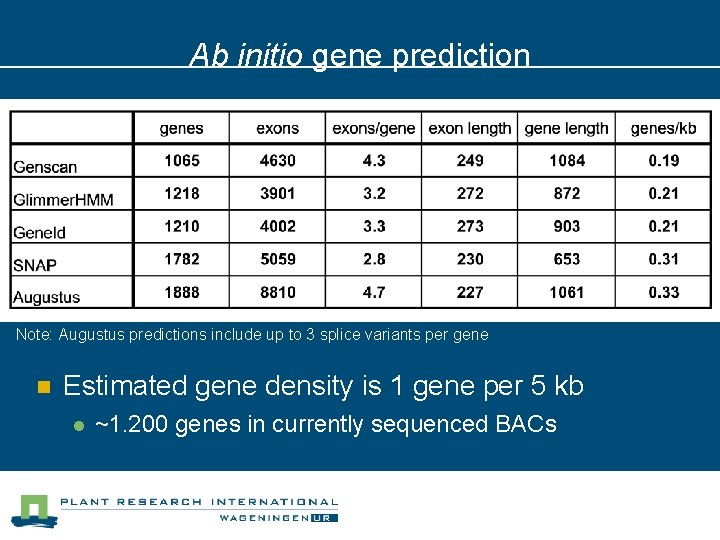
Ab initio gene structure prediction n Ab initio predictors included in the pipeline l l l Genscan Glimmer. HMM Gene. Id SNAP Augustus (trained on tomato!) (has been trained on Solanaceae) (predicts alternative spliced variants)

Alignment-based gene structure prediction (1) n Transcript alignment (blastn + Sim 4) l l l l n SGN tomato Uni. Genes SGN potato Uni. Genes SGN coffee Uni. Genes SGN pepper Uni. Genes SGN petunia Unigenes SGN S. melongena Uni. Genes NCBI full-length tomato c. DNAs (34. 829 Uni. Genes) (31. 072 Uni. Genes) (13. 171 Uni. Genes) (9. 554 Uni. Genes) (5. 135 Uni. Genes) (1. 841 Uni. Genes) (678 c. DNAs) Protein alignment (tblastn + Gene. Wise) l l l TAIR 6 Arabidopsis thaliana proteome TIGR 4 Oryza sativa proteome Uni. Prot Plant division (30. 690 proteins) (62. 827 proteins) (17. 831 proteins)

Additional feature prediction n Repeat Identification l l Tandem Repeats Finder Repeat. Masker • Rep. Base + ‘default’ features (low complexity, etc) • TIGR Solanum lycopersicon repeat library V 2 • SGN Solanum lycopersicon Uni. Repeats n Feature prediction l l t. RNAscan-SE Mar. Scan Gene. Splicer Marker identification (blastn + Sim 4)

Preliminary results n Annotation of chromosome 6 BACs l l l phase 1, 2 and 3 632 contigs Older version of the pipeline • • Glimmer. HMM only trained on Arabidopsis 2 Uni. Gene sets (tomato, potato) 2 protein sets (Arabidopsis, Uni. Prot plant) Protein alignment parameters too strict

The genomic landscape of chromosome 6 n 632 contigs have been annotated l l Length of contigs varies between 348 – 148. 256 nt Average length of 9. 061 nt, median length of 5. 105 nt Total length of 5. 726. 791 nt GC content: 29. 9% min, 34. 1% avg, 42. 2% max (sequences longer than 10. 000 nt)

Ab initio gene prediction Note: Augustus predictions include up to 3 splice variants per gene n Estimated gene density is 1 gene per 5 kb l ~1. 200 genes in currently sequenced BACs

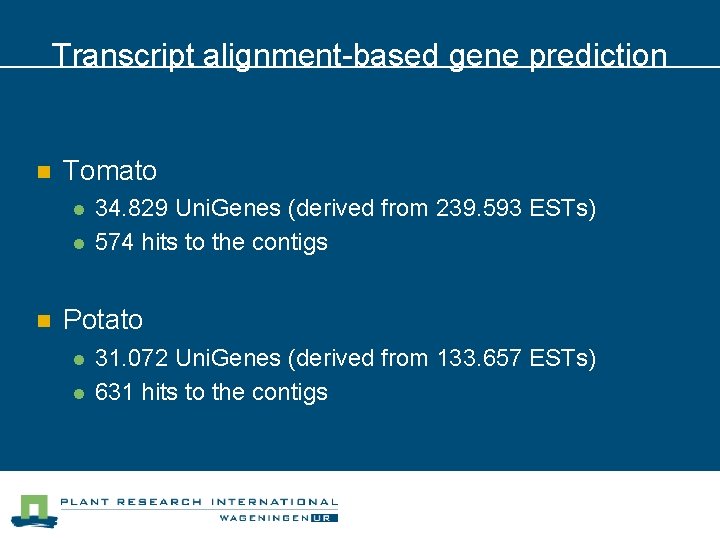
Transcript alignment-based gene prediction n Tomato l l n 34. 829 Uni. Genes (derived from 239. 593 ESTs) 574 hits to the contigs Potato l l 31. 072 Uni. Genes (derived from 133. 657 ESTs) 631 hits to the contigs

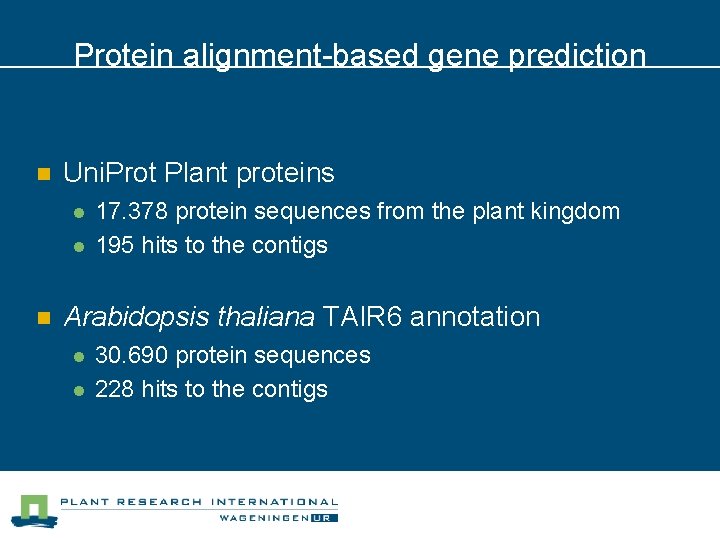
Protein alignment-based gene prediction n Uni. Prot Plant proteins l l n 17. 378 protein sequences from the plant kingdom 195 hits to the contigs Arabidopsis thaliana TAIR 6 annotation l l 30. 690 protein sequences 228 hits to the contigs

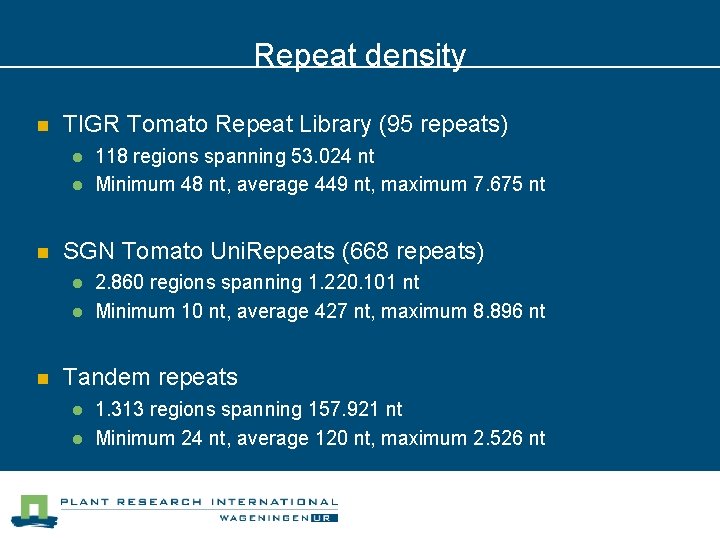
Repeat density n TIGR Tomato Repeat Library (95 repeats) l l n SGN Tomato Uni. Repeats (668 repeats) l l n 118 regions spanning 53. 024 nt Minimum 48 nt, average 449 nt, maximum 7. 675 nt 2. 860 regions spanning 1. 220. 101 nt Minimum 10 nt, average 427 nt, maximum 8. 896 nt Tandem repeats l l 1. 313 regions spanning 157. 921 nt Minimum 24 nt, average 120 nt, maximum 2. 526 nt

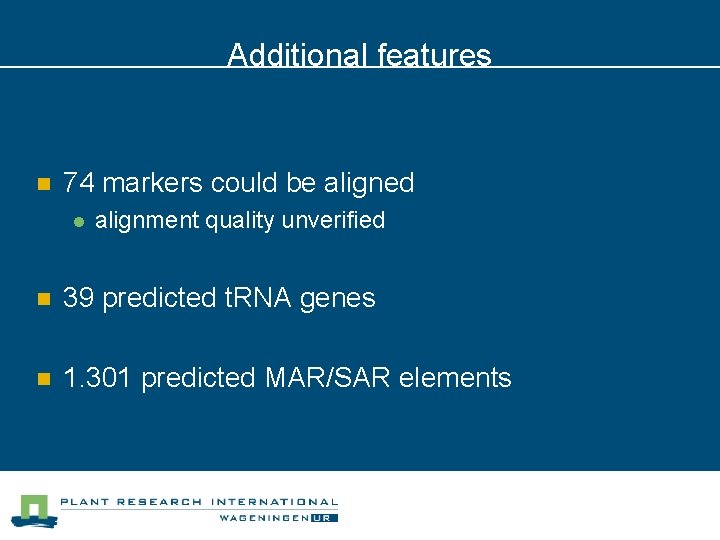
Additional features n 74 markers could be aligned l alignment quality unverified n 39 predicted t. RNA genes n 1. 301 predicted MAR/SAR elements

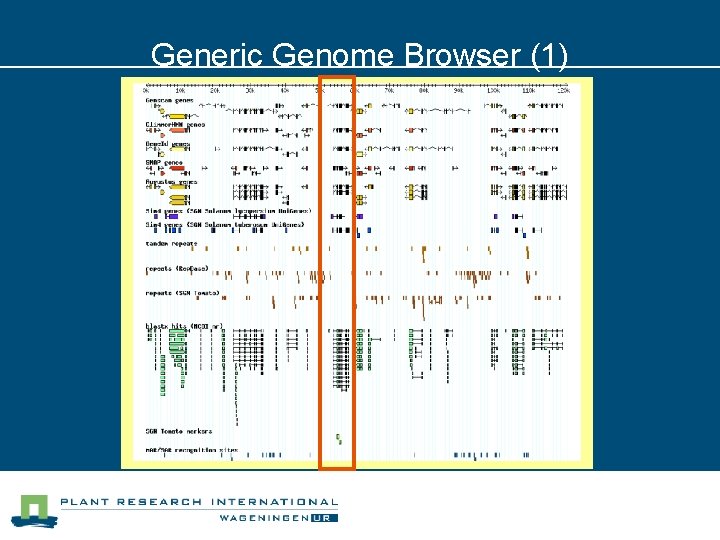
Generic Genome Browser (1)

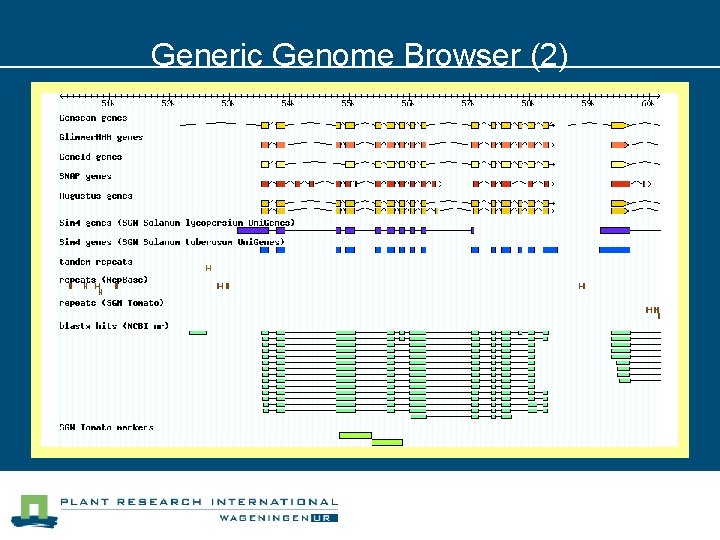
Generic Genome Browser (2)

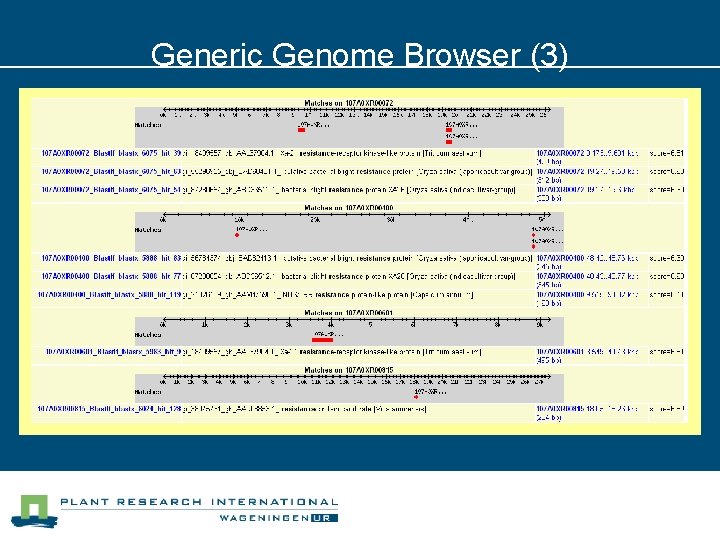
Generic Genome Browser (3)

Recent work n Gene. Model. Collector l l l n Tries to find ‘full’ open reading frames in aligned Uni. Genes Automatic generation of gene predictor training set Parameters? JIGSAW l l Appears not to provide a prediction for every region which contains annotations Training?

Future Work – Tomato Annotation Pipeline n Gene prediction l l n Combining predictions into a single consensus model Train individual predictors with recently curated tomato gene set Automated functional annotation of genes l l l “Giving a biological meaning to the nicely colored bars” blastx Inter. Pro. Scan

Future Work – Tomato Genome Browser n Annotation of features l l n Meaningful names for features such as genes, marker alignments, blast hits More detailed and better readable data when clicking on a feature Links to external data sources l l NCBI Gen. Bank SGN

Acknowledgements n Cyrille 2 development l l l n Tomato BAC sequencing (chromosome 6) l n Mark Fiers Ate van der Burgt Joost de Groot Greenomics Supervision l l Willem Stiekema Roeland van Ham
 Tomato genome browser
Tomato genome browser Sequence assembly ppt
Sequence assembly ppt Maker genome annotation
Maker genome annotation Semi-global alignment
Semi-global alignment Which pipeline is a linear
Which pipeline is a linear Paralleism
Paralleism Cyrille marquet
Cyrille marquet Organigramme dgt
Organigramme dgt Las chicas de alambre
Las chicas de alambre Cyrille marquet
Cyrille marquet Erwin vs powerdesigner
Erwin vs powerdesigner Erwin chargaff
Erwin chargaff Erwin setyo kriswanto
Erwin setyo kriswanto Mg modelo de dalton
Mg modelo de dalton Erwin honkoop
Erwin honkoop Erwin sitompul
Erwin sitompul Erwin sitompul
Erwin sitompul Regla de erwin chargaff 1940
Regla de erwin chargaff 1940 Electromagnetics
Electromagnetics Vertical
Vertical