Genome Sequence Annotation Server Structural and functional annotation





- Slides: 20

Genome Sequence Annotation Server Structural and functional annotation of model and non-model organisms with Gen. SAS v 5. 0, a web-based annotation platform Jodi L. Humann, Stephen P. Ficklin, Taein Lee, Chun-Huai Cheng, Heidi Hough, Sook Jung, Jill Wegrzyn, David Neale, Dorrie Main jhumann@wsu. edu

What is DNA annotation and why do it? • Getting the DNA sequence is only the first step • Need to know the biological relevance of the DNA sequence • Annotated sequence can be used to find putative genes of interest for study

What scientists want • Current annotation tools: • Many tools available, but run independently of each other • Most of the tools are run via the command line and require server access • Scientists want a platform that: • Is a single location for DNA annotation • Does not require management of computing equipment and software tools • Is easy to use and can be adapted to a variety of DNA sequences

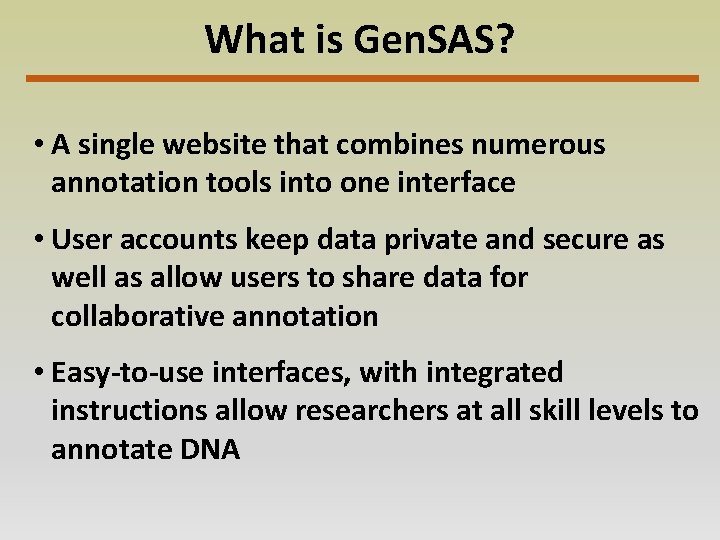
What is Gen. SAS? • A single website that combines numerous annotation tools into one interface • User accounts keep data private and secure as well as allow users to share data for collaborative annotation • Easy-to-use interfaces, with integrated instructions allow researchers at all skill levels to annotate DNA

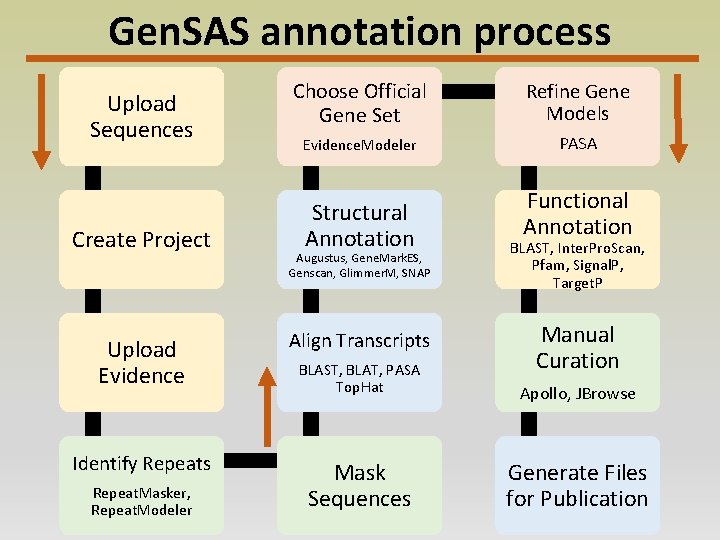
Gen. SAS annotation process Upload Sequences Create Project Choose Official Gene Set Refine Gene Models Evidence. Modeler PASA Structural Annotation Augustus, Gene. Mark. ES, Genscan, Glimmer. M, SNAP Functional Annotation BLAST, Inter. Pro. Scan, Pfam, Signal. P, Target. P Manual Curation Upload Evidence Align Transcripts BLAST, BLAT, PASA Top. Hat Apollo, JBrowse Identify Repeats Mask Sequences Generate Files for Publication Repeat. Masker, Repeat. Modeler

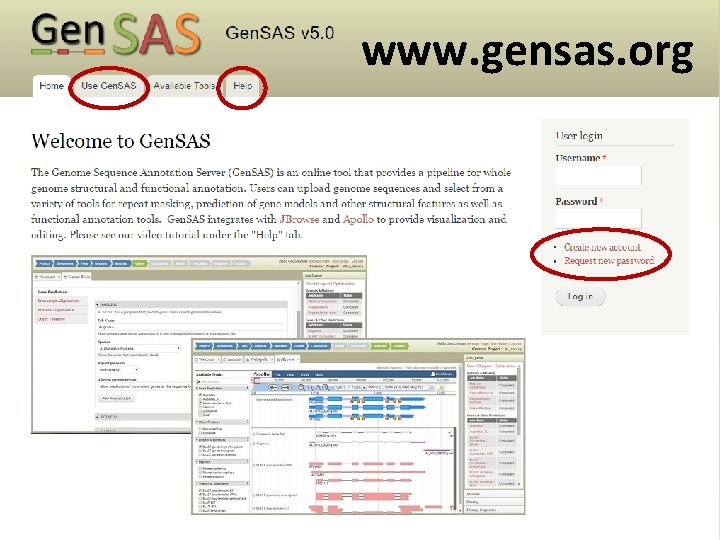
www. gensas. org

Gen. SAS welcome tab provides users with a quick overview of what each of the three screen sections do.

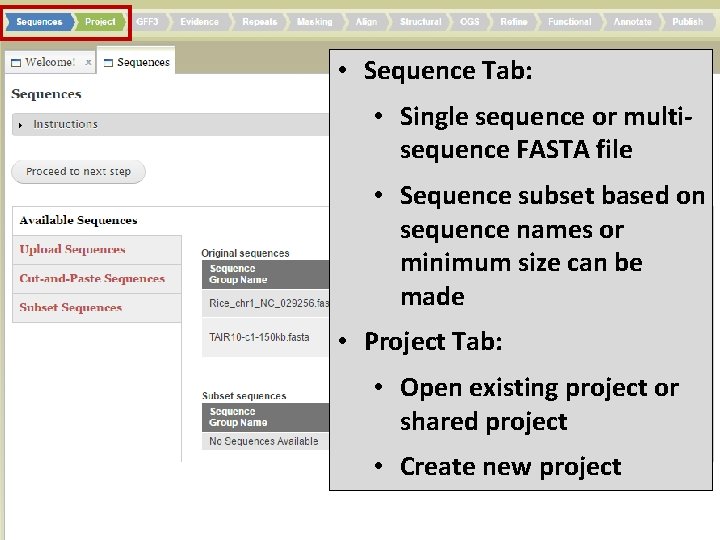
• Sequence Tab: • Single sequence or multisequence FASTA file • Sequence subset based on sequence names or minimum size can be made • Project Tab: • Open existing project or shared project • Create new project

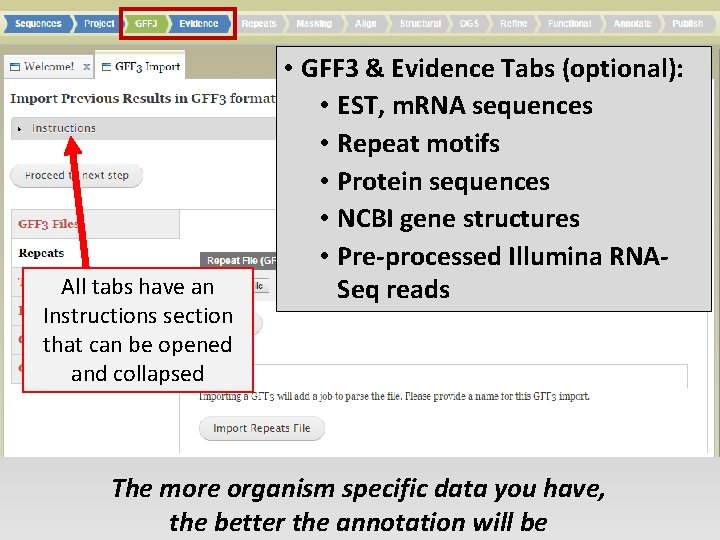
All tabs have an Instructions section that can be opened and collapsed • GFF 3 & Evidence Tabs (optional): • EST, m. RNA sequences • Repeat motifs • Protein sequences • NCBI gene structures • Pre-processed Illumina RNASeq reads The more organism specific data you have, the better the annotation will be

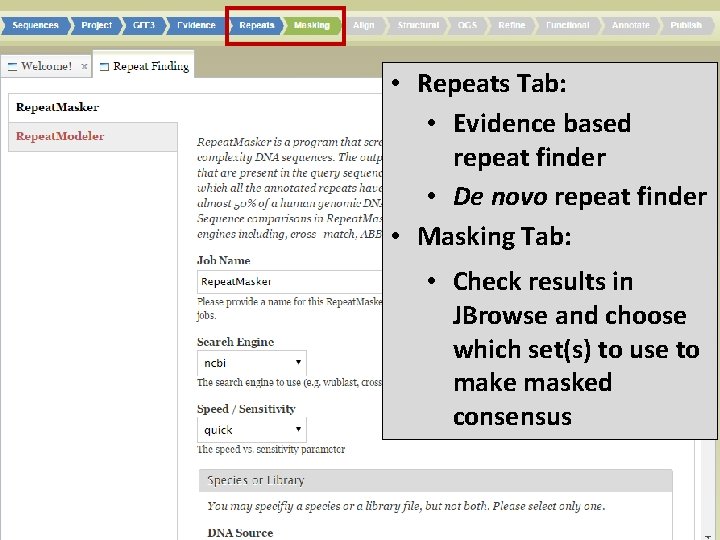
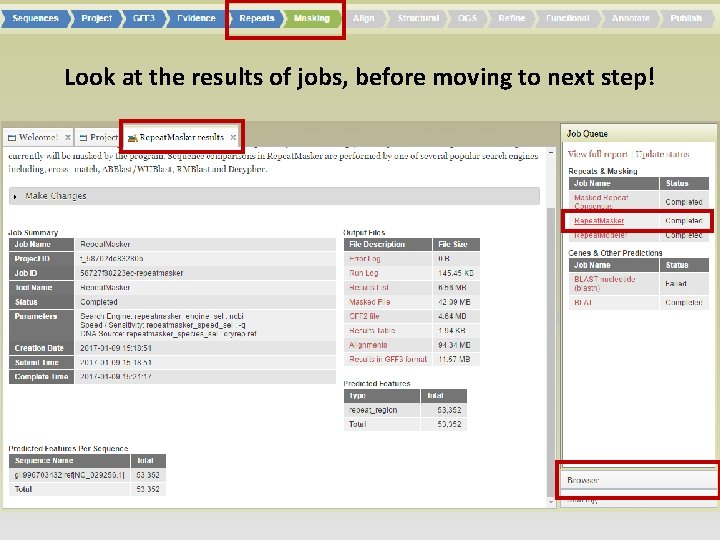
• Repeats Tab: • Evidence based repeat finder • De novo repeat finder • Masking Tab: • Check results in JBrowse and choose which set(s) to use to make masked consensus

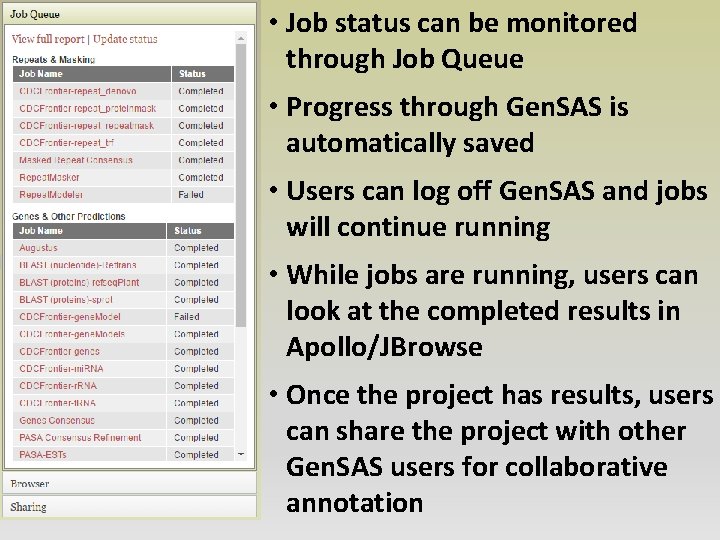
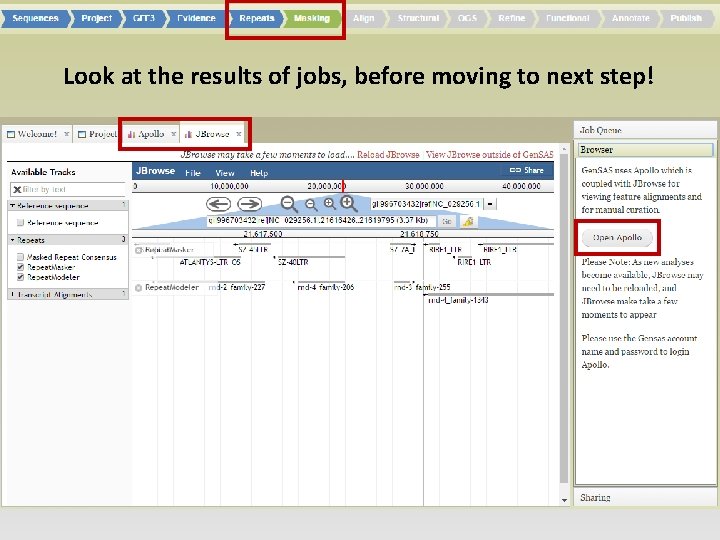
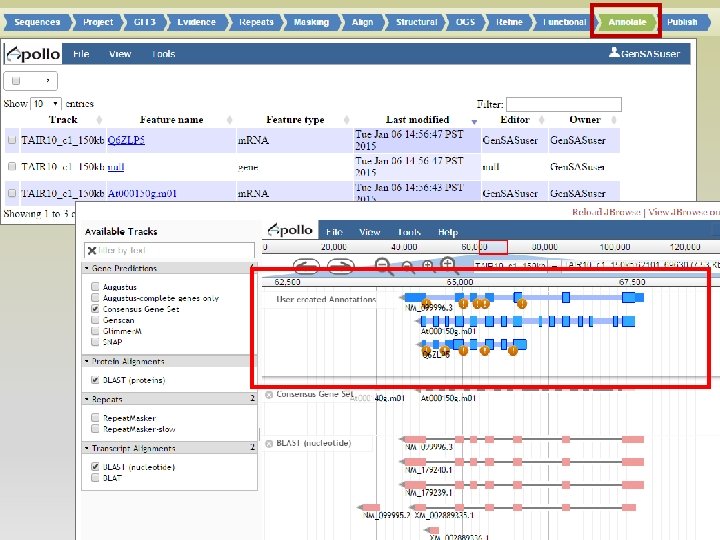
• Job status can be monitored through Job Queue • Progress through Gen. SAS is automatically saved • Users can log off Gen. SAS and jobs will continue running • While jobs are running, users can look at the completed results in Apollo/JBrowse • Once the project has results, users can share the project with other Gen. SAS users for collaborative annotation

Look at the results of jobs, before moving to next step!

Look at the results of jobs, before moving to next step!

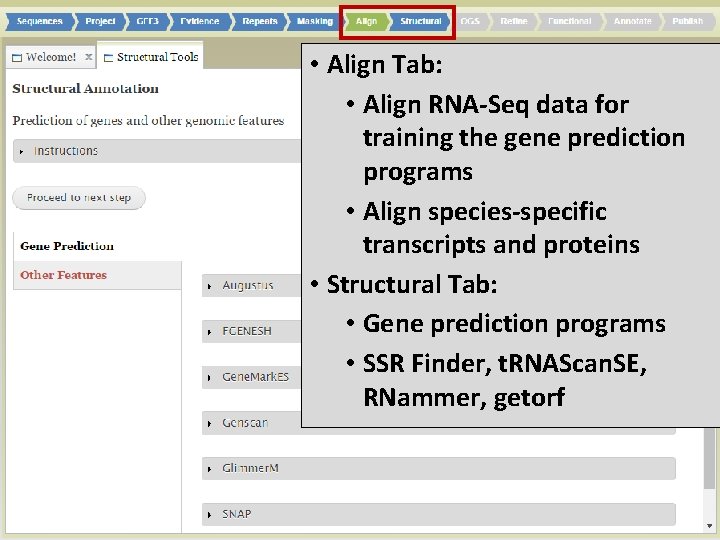
• Align Tab: • Align RNA-Seq data for training the gene prediction programs • Align species-specific transcripts and proteins • Structural Tab: • Gene prediction programs • SSR Finder, t. RNAScan. SE, RNammer, getorf

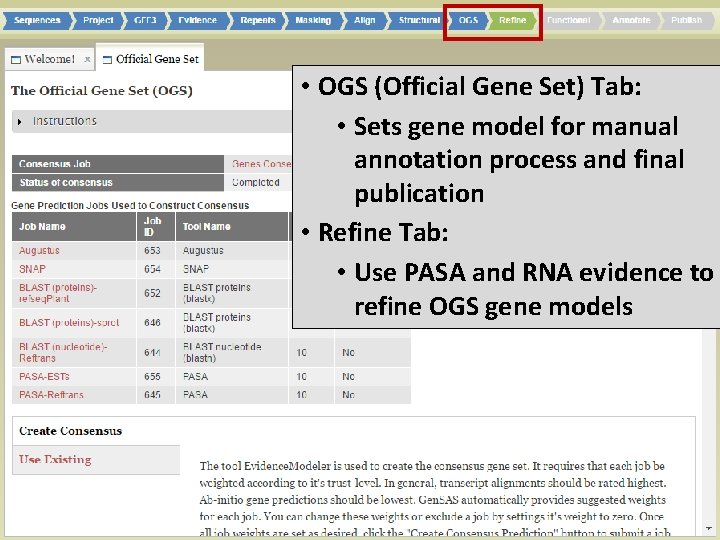
• OGS (Official Gene Set) Tab: • Sets gene model for manual annotation process and final publication • Refine Tab: • Use PASA and RNA evidence to refine OGS gene models

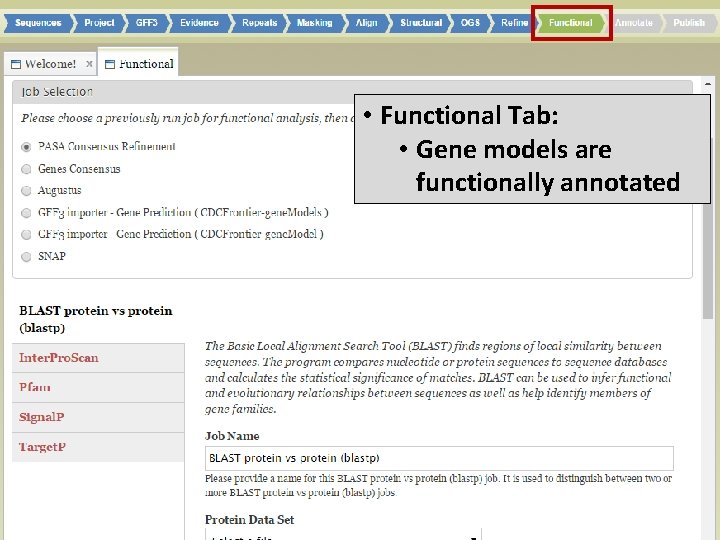
• Functional Tab: • Gene models are functionally annotated


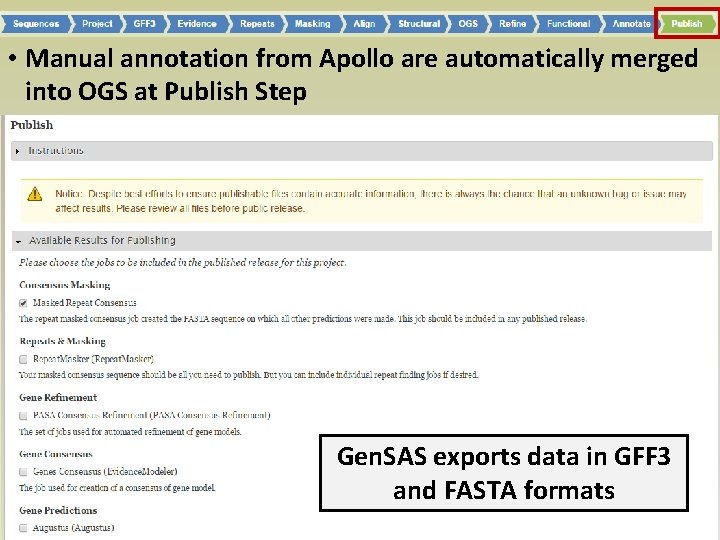
• Manual annotation from Apollo are automatically merged into OGS at Publish Step Gen. SAS exports data in GFF 3 and FASTA formats

Future development • Integrate Apollo 2 and newest JBrowse • Add option to create single merged GFF 3 of all annotation data under Publish step • Improve how BLAST jobs are submitted to cluster to reduce run time

Supported by