Gene Annotation Gene Ontology May 28 2020 Gene


















- Slides: 37

Gene Annotation & Gene Ontology May 28, 2020

Gene lists from RNAseq analysis What do you do with a list of 100 s of genes that contain only the following information? • Gene name or symbol • Ratio between groups (UP or DOWN) • One or more database IDs (accession numbers) How do you figure out the role of the genes in the model you are studying?

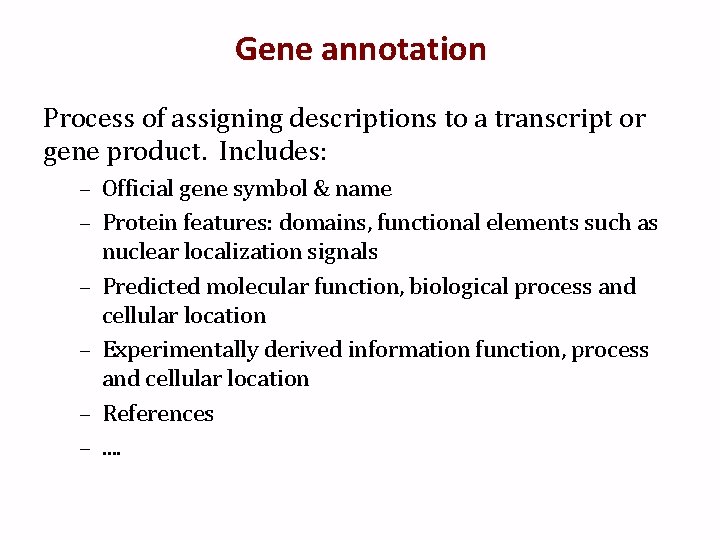
Gene annotation Process of assigning descriptions to a transcript or gene product. Includes: – Official gene symbol & name – Protein features: domains, functional elements such as nuclear localization signals – Predicted molecular function, biological process and cellular location – Experimentally derived information function, process and cellular location – References –. .

Who does the gene annotation? • Refseq & Gene databases – NCBI staff • Ensemble databases – http: //useast. ensembl. org – EMBL & Welcome Trust at Sanger Institute • Uniprot – Staff at European Bioinformatics Institute (EBI), Swiss Institute of Bioinformatics (SIB) and the Protein Information Resource (PIR) • Yeast DB, Fly. Base, Mouse Genome Informatics (MGI) & other organism specific databases

Gene record for BEST 1

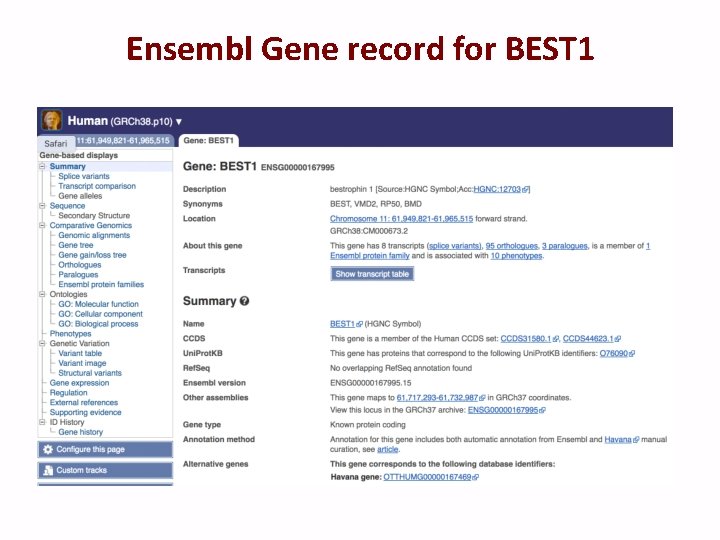
Ensembl Gene record for BEST 1

Uniprot record for BEST 1

Gene, Ensembl or Uniprot? • • What information are you looking for? Comfort level with the interface All have a little to LOTS of information Use as a starting point

Dealing with gene lists • How can you efficiently categorize the genes in in some biologically meaningful way? • Batch download data from Gene or Uniprot and do a lot of reading? • Pub. Med? • One approach is to use meta-data in the form of terms assigned to each gene that describe its molecular function, participation in a biological process and its location in a cellular component

Gene Ontology • Set of standard biological phrases (terms) which are applied to genes/proteins: – protein kinase – apoptosis – Membrane • Standardize the representation of gene product attributes across species and databases • Maintained by Gene Ontology consortium – http: //geneontology. org/ – Individual groups contribute taxonomic specific terms

Cellular Component Where a gene product acts Mitochondria

Cellular Component Cellular components not all same between organisms

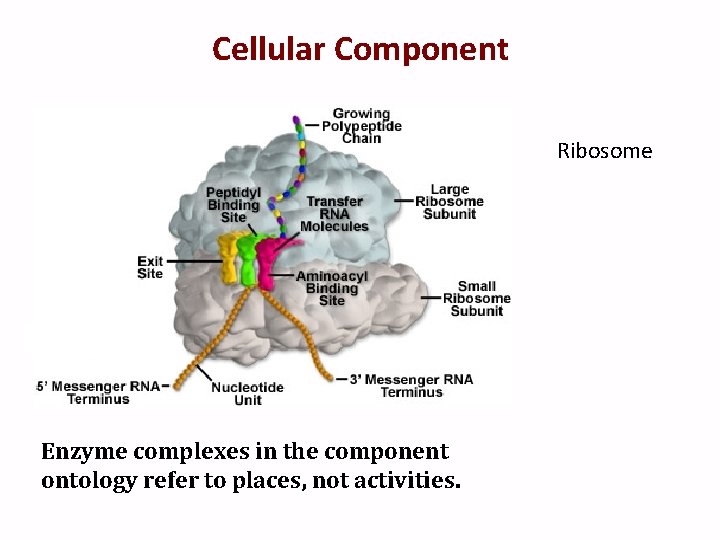
Cellular Component Ribosome Enzyme complexes in the component ontology refer to places, not activities.

Molecular Function Activities or “jobs” of a gene product glucose-6 -phosphate isomerase activity

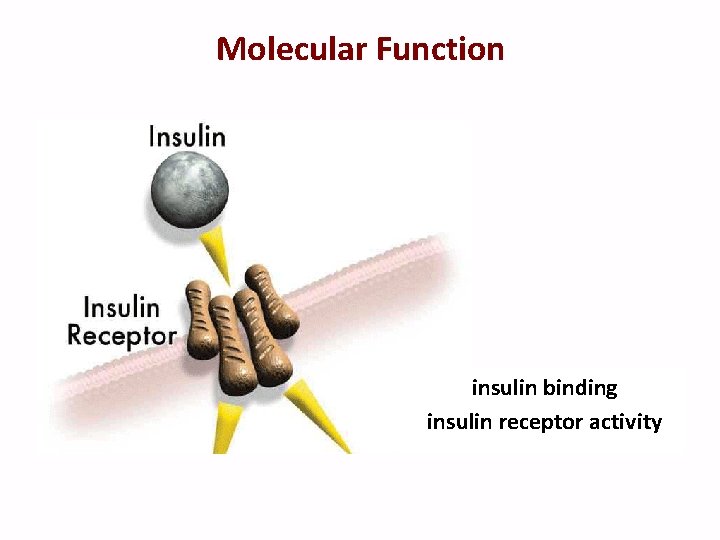
Molecular Function insulin binding insulin receptor activity

Molecular Function • A gene product may have several functions • Sets of functions make up a biological process.

Biological Process a commonly recognized series of events cell division

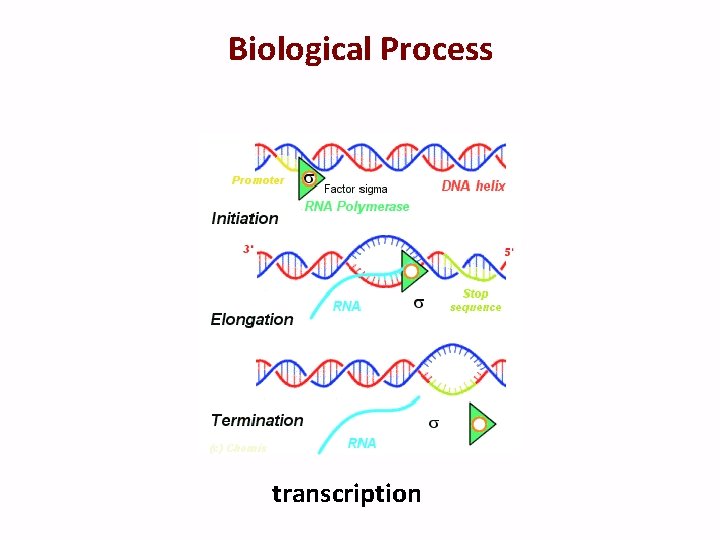
Biological Process transcription

Why use gene ontology? • Allows biologists to make queries across large numbers of genes without researching each one individually • Can find all the PI 3 kinases in a given genome or find all proteins involved in oxidative stress response without prior knowledge of every gene

Gene Ontology for analysis • Biological process terms are more useful for putting gene lists into a context • More GO terms assigned to process than to function or component • Fewest terms assigned to component • Function in the absence of any process information can imply a biological role – i. e. you are looking for transcription factors responsible for some response Ontology annotation is NOT complete

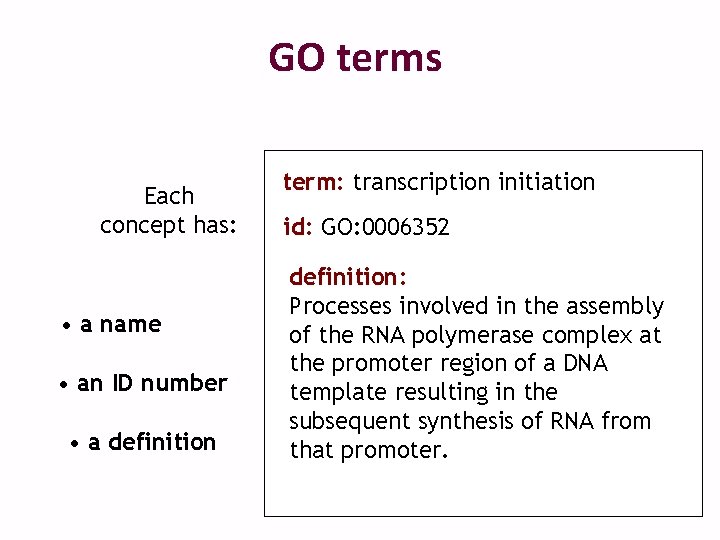
GO terms Each concept has: • a name • an ID number • a definition term: transcription initiation id: GO: 0006352 definition: Processes involved in the assembly of the RNA polymerase complex at the promoter region of a DNA template resulting in the subsequent synthesis of RNA from that promoter.

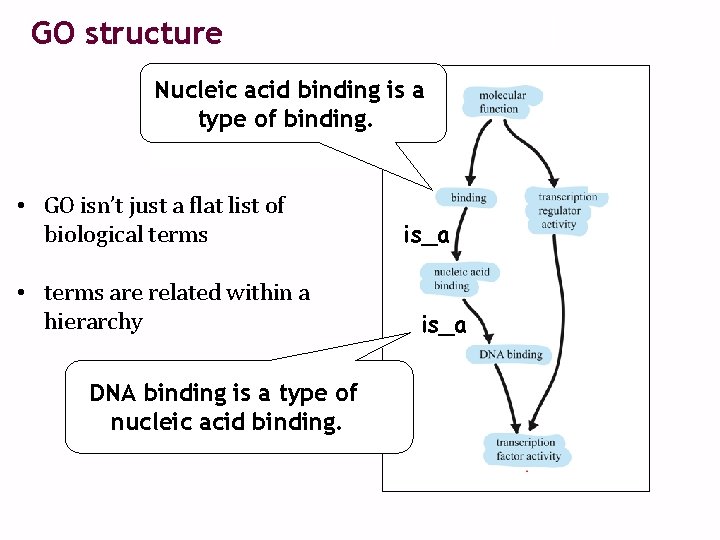
GO structure Nucleic acid binding is a type of binding. • GO isn’t just a flat list of biological terms • terms are related within a hierarchy DNA binding is a type of nucleic acid binding. is_a

GO structure & annotation • A single gene associated with a particular term is automatically annotated to all of the parent terms. • But genes are also annotated with terms from the different categories (BP, MF and CC) The gene product “cytochrome c” can be described by the molecular function oxidoreductase activity, the biological process oxidative phosphorylation, and the cellular component mitochondrial matrix.

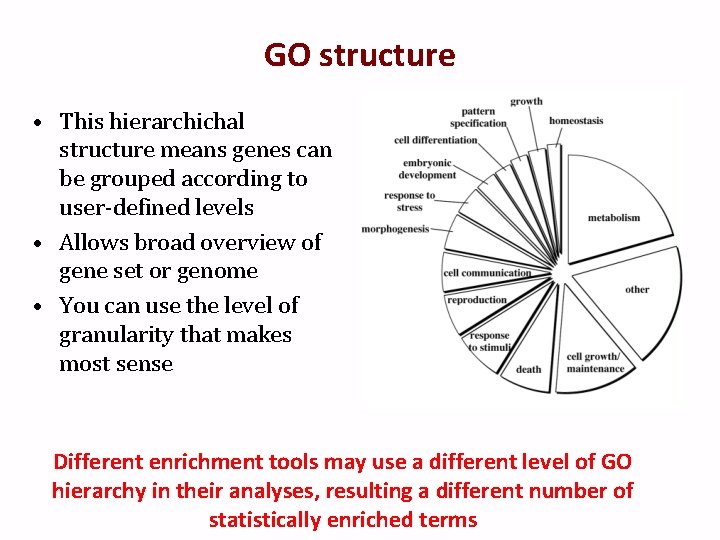
GO structure • This hierarchichal structure means genes can be grouped according to user-defined levels • Allows broad overview of gene set or genome • You can use the level of granularity that makes most sense Different enrichment tools may use a different level of GO hierarchy in their analyses, resulting a different number of statistically enriched terms

TARDBP (TDP-43) • GO biological process: – 3’UTR mediated m. RNA stabilization – RNA splicing – m. RNA processing • GO molecular function: – RNA binding – Double-stranded DNA binding – m. RNA 3’-UTR binding • GO cellular component – Cytoplasm – Interchromatin granule – Nuclear speck

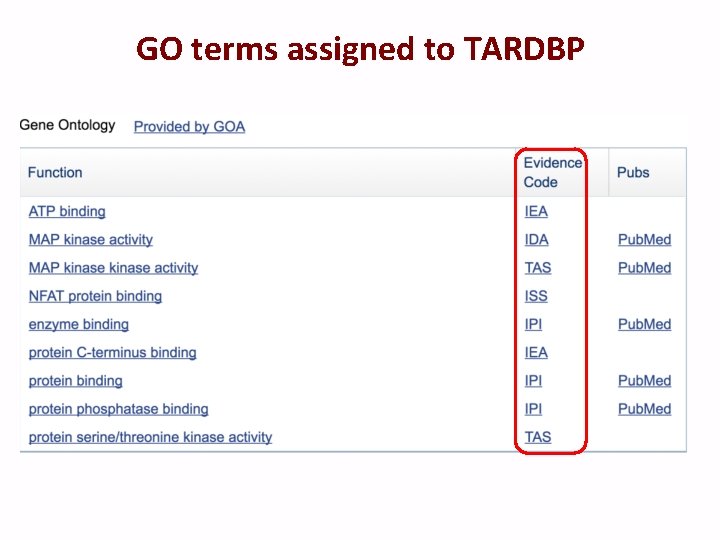
GO terms assigned to TARDBP

Types of evidence codes Experimental: • Inferred from Experiment (EXP) • Inferred from Direct Assay (IDA) • Inferred from Physical Interaction (IPI) • Inferred from Mutant Phenotype (IMP) • Inferred from Genetic Interaction (IGI) • Inferred from Expression Pattern (IEP)

Types of evidence codes Computational: • • • Inferred from Sequence or structural Similarity (ISS) Inferred from Sequence Orthology (ISO) Inferred from Sequence Alignment (ISA) Inferred from Sequence Model (ISM) Inferred from Genomic Context (IGC) Inferred from Biological aspect of Ancestor (IBA) Inferred from Biological aspect of Descendant (IBD) Inferred from Key Residues (IKR) Inferred from Rapid Divergence(IRD) Inferred from Reviewed Computational Analysis (RCA)

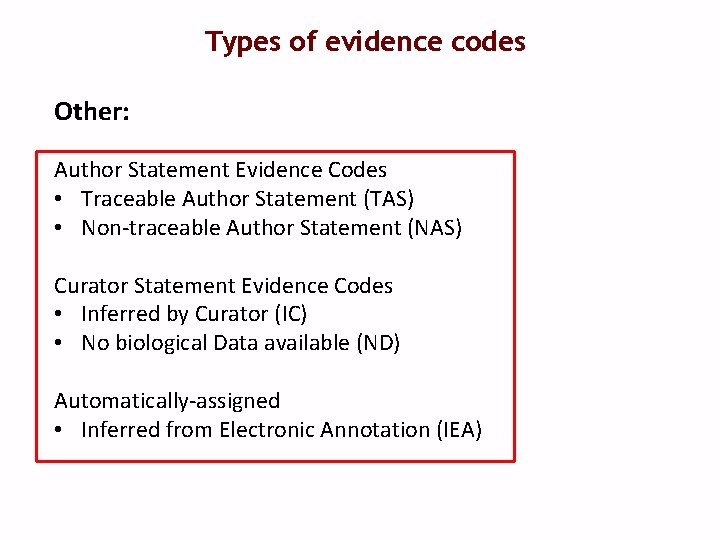
Types of evidence codes Other: Author Statement Evidence Codes • Traceable Author Statement (TAS) • Non-traceable Author Statement (NAS) Curator Statement Evidence Codes • Inferred by Curator (IC) • No biological Data available (ND) Automatically-assigned • Inferred from Electronic Annotation (IEA)

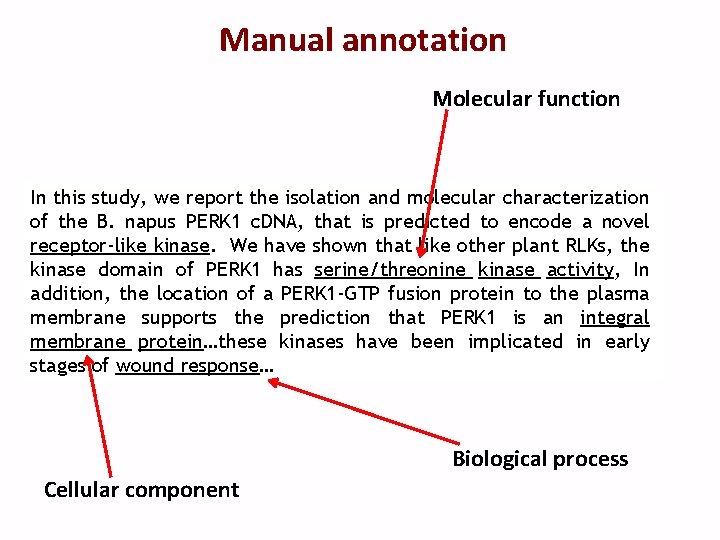
Manual annotation Molecular function In this study, we report the isolation and molecular characterization of the B. napus PERK 1 c. DNA, that is predicted to encode a novel receptor-like kinase. We have shown that like other plant RLKs, the kinase domain of PERK 1 has serine/threonine kinase activity, In addition, the location of a PERK 1 -GTP fusion protein to the plasma membrane supports the prediction that PERK 1 is an integral membrane protein…these kinases have been implicated in early stages of wound response… Biological process Cellular component

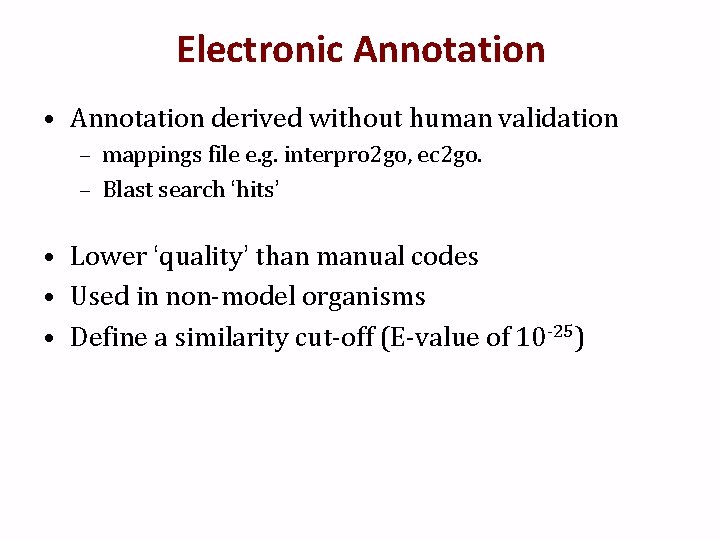
Electronic Annotation • Annotation derived without human validation – mappings file e. g. interpro 2 go, ec 2 go. – Blast search ‘hits’ • Lower ‘quality’ than manual codes • Used in non-model organisms • Define a similarity cut-off (E-value of 10 -25)

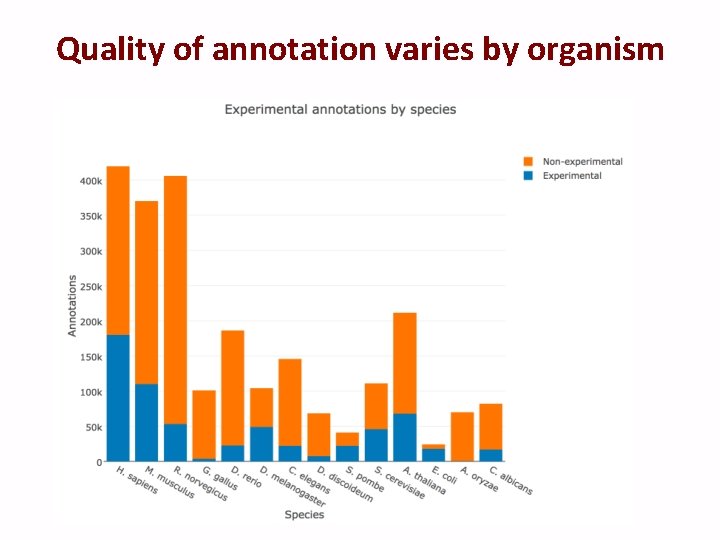
Quality of annotation varies by organism

GO & analysis of gene lists • www. geneontology. org – Maintains the databases of GO terms, serves a clearing house for terms as they are assigned in new organisms • Tools for exploring gene lists using GO: – Web. Gestalt, g. Profiler, Gorilla, Panther – DAVID is a suite of tools for gene enrichment analysis that also includes GO. – We’ll use both DAVID and Web. Gestalt to explore our gene list – DAVID does not reduce the redundancy of terms as well as Web. Gestalt does

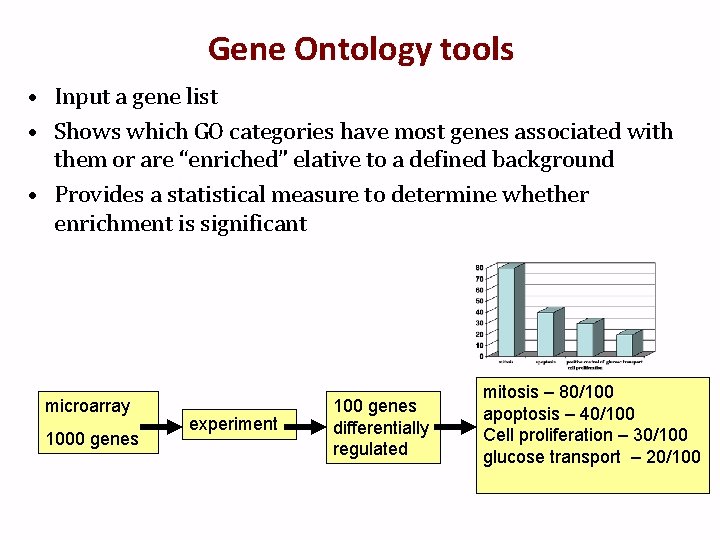
Gene Ontology tools • Input a gene list • Shows which GO categories have most genes associated with them or are “enriched” elative to a defined background • Provides a statistical measure to determine whether enrichment is significant microarray 1000 genes experiment 100 genes differentially regulated mitosis – 80/100 apoptosis – 40/100 Cell proliferation – 30/100 glucose transport – 20/100

Using GO in practice • The distribution of all genes on the microarray and their GO assignment serves as the background: Process Genes on array # genes expected (out of 100) # genes observed Mitosis 800/1000 80 80 Apoptosis 400/1000 40 40 Cell proliferation 100/1000 10 30 Glucose transport 50/1000 5 20 • Proportions analysis – More sophisticated version of a Chi-squared or Fisher’s exact test • Result is terms with p-values and adjusted p-values. You decide what p-value is significant

Other sources of annotation • Uniprot (Swiss-Prot) keywords • Protein domain databases – PFAM, Panther, PDB, PROSITE, ect • Gene. DB summaries from NCBI • Protein-protein interactions databases • Pathway databases – KEGG, Bio. Carta, BBID, Reactome DAVID incorporates annotation from all of these and clusters the redundant terms

Today in computer lab • Tutorial on using DAVID & Web. Gestalt • Analysis of gene lists using DAVID and Web. Gestalt and comparing the results from the two different tools
 Gene ontology project
Gene ontology project Gene ontology project
Gene ontology project What is meant by gene ontology?
What is meant by gene ontology? Gene ontology
Gene ontology David gene functional classification tool
David gene functional classification tool Open reading frame
Open reading frame Gene by gene test results
Gene by gene test results Chapter 17 from gene to protein
Chapter 17 from gene to protein Ontology, epistemology, axiology
Ontology, epistemology, axiology Ontological definition
Ontological definition Ontology research methods
Ontology research methods Ontology in biology
Ontology in biology Ontology 101
Ontology 101 Suggested upper merged ontology
Suggested upper merged ontology Resources events agents
Resources events agents Pizza ontology
Pizza ontology Basic formal ontology
Basic formal ontology Business model ontology
Business model ontology Epistemology vs ontology
Epistemology vs ontology Business ontology
Business ontology Ontology editors
Ontology editors Financial industry business ontology
Financial industry business ontology Ontology schema
Ontology schema Research ontology and epistemology
Research ontology and epistemology Provi tutorial
Provi tutorial Dolce ontology
Dolce ontology Ontology kurssi
Ontology kurssi Ontology alignment
Ontology alignment Analyticism
Analyticism Twinkle helicase
Twinkle helicase Ontology creation
Ontology creation Barry smith buffalo
Barry smith buffalo Ontology alignment
Ontology alignment Napolen pizza
Napolen pizza Sims position
Sims position Business ontology
Business ontology Types of ontology
Types of ontology Ontology
Ontology