JGI Timeline Joint Genome Institute JGI Human Genome












- Slides: 37

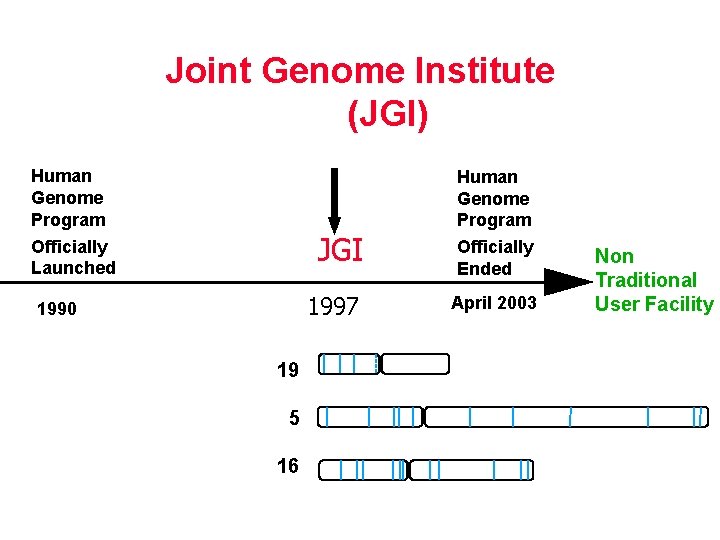
JGI Timeline Joint Genome Institute …………………. (JGI) Human Genome Program JGI Officially Launched 1997 1990 19 5 16 Human Genome Program Officially Ended April 2003 Non Traditional User Facility

The JGI Post Human Genome Project Community Sequencing Program (CSP) Microbial Community Genomics US DOE Joint Genome Institute


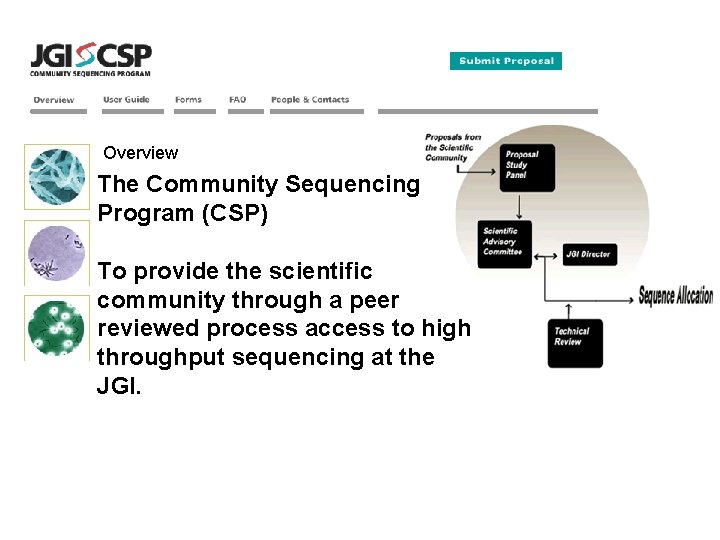
Overview The Community Sequencing Program (CSP) To provide the scientific community through a peer reviewed process access to high throughput sequencing at the JGI.

User Guide > How to Propose a Project What types of projects will the JGI/CSP accept? A wide range of projects. Ultimately, the most important factor in determining if a project will be accepted is its scientific merit.

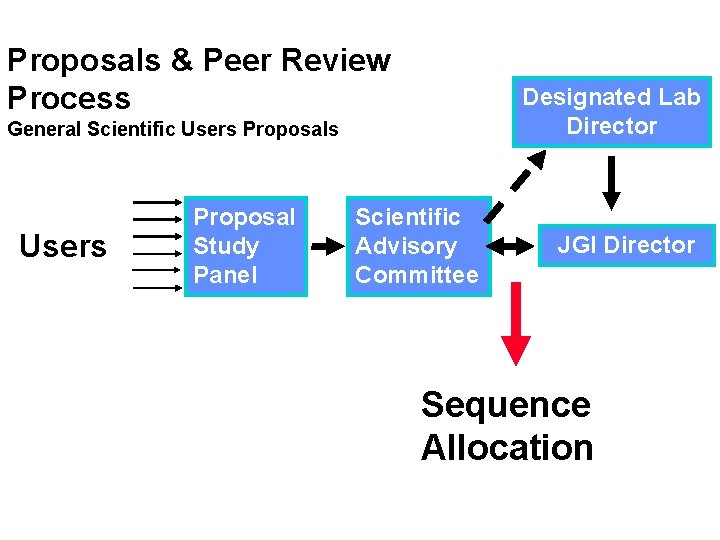
Proposals & Peer Review Process Designated Lab Director General Scientific Users Proposals Users Proposal Study Panel Scientific Advisory Committee JGI Director Sequence Allocation

FAQ What can researchers get from the CSP program? The deliverables can range from raw sequence traces to well-annotated assembled genomes depending on the request in the proposal.

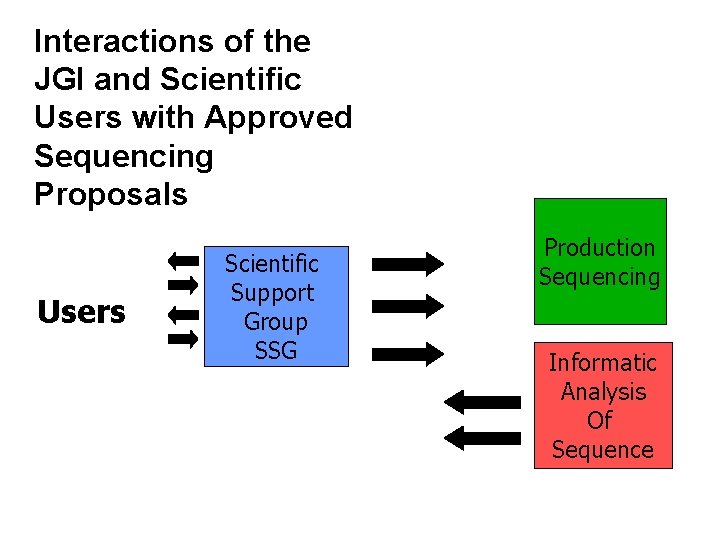
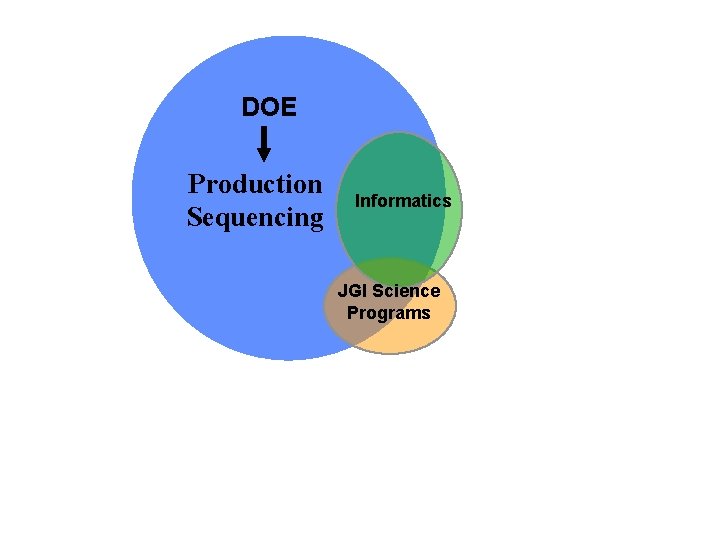
Interactions of the Scientific Support for Approved Projects JGI and Scientific Users with Approved Sequencing Proposals Users Scientific Support Group SSG Production Sequencing Informatic Analysis Of Sequence

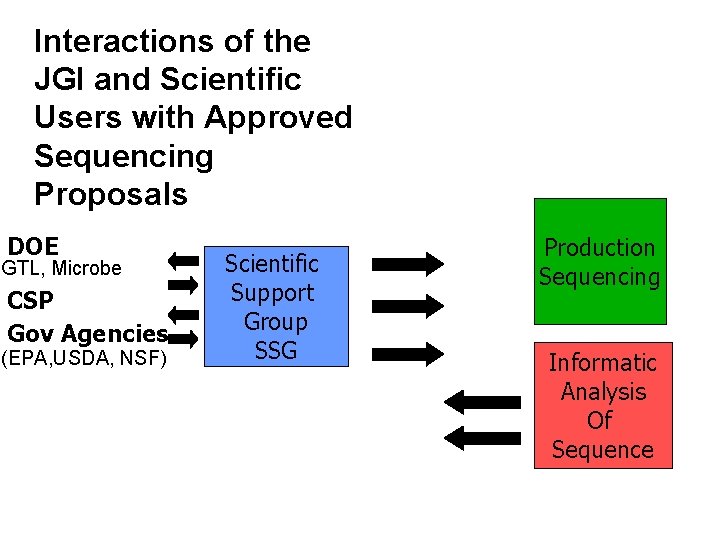
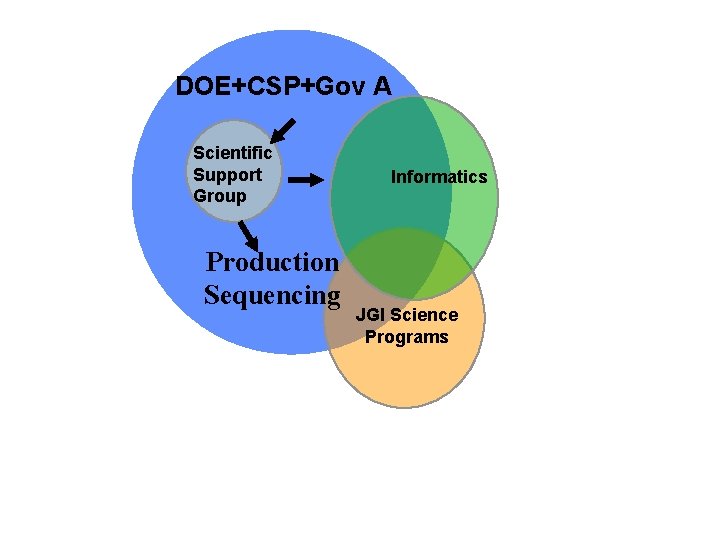
Interactions of the Scientific Support for Approved Projects JGI and Scientific Users with Approved Sequencing Proposals DOE GTL, Microbe CSP Gov Agencies (EPA, USDA, NSF) Scientific Support Group SSG Production Sequencing Informatic Analysis Of Sequence

DOE Production Sequencing Informatics JGI Science Programs

DOE+CSP+Gov A Scientific Support Group Production Sequencing Informatics JGI Science Programs

Sequence Based Science at the JGI • Gene Regulatory Vocabulary of Animals • Studies of Body Plan Evolution • Microbial Community Genomics

• < 1% of microbes are culturable • Many unculturables live in interdependent consortia of considerable diversity • Aim: to recover genome-scale sequences and reveal metabolic capabilities • What is the structure of natural microbial populations? What is a microbial species? Can we harness their metabolic capabilities

What Enviroments to Study? • Ones with minimal microbial complexity

Iron Mountain Jill Banfield et al. UC Berkeley Jill. Banfield Gene Tyson Phil Hugenholtz UC Berkeley Geology

Iron Mountain Superfund site Discharging >1 ton of toxic metals/day (p. H <1) Fe. S 2

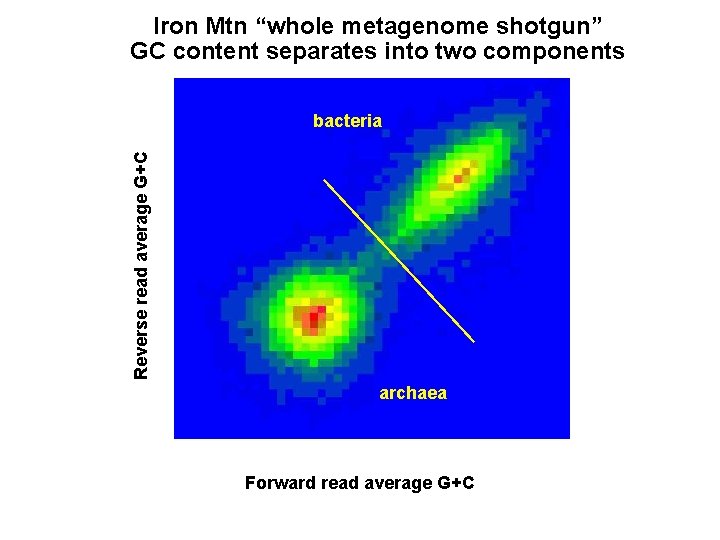
“whole metagenome shotgun” dataset

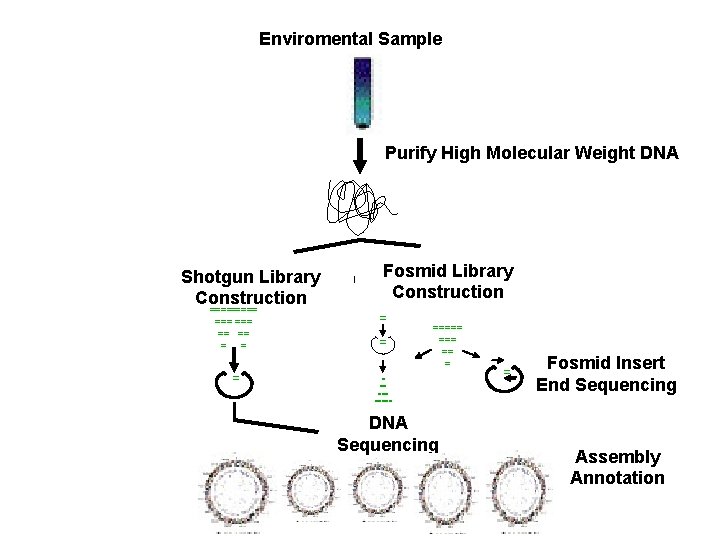
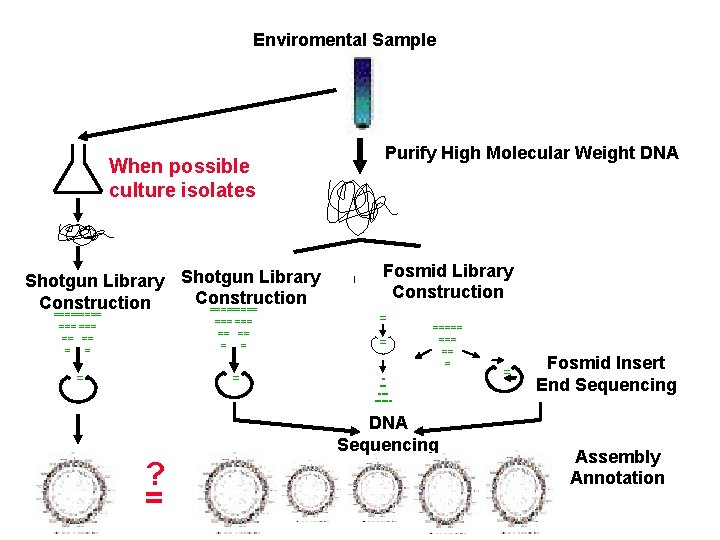
Enviromental Sample Purify High Molecular Weight DNA ===== === == == = Fosmid Library Construction = = ===== === == = Shotgun Library Construction DNA Sequencing = Fosmid Insert End Sequencing Assembly Annotation

Enviromental Sample Shotgun Library Construction ===== === === == == = = = = Fosmid Library Construction = = ===== === == = When possible culture isolates Purify High Molecular Weight DNA Sequencing ? = = Fosmid Insert End Sequencing Assembly Annotation

Iron Mtn “whole metagenome shotgun” GC content separates into two components Reverse read average G+C bacteria archaea Forward read average G+C

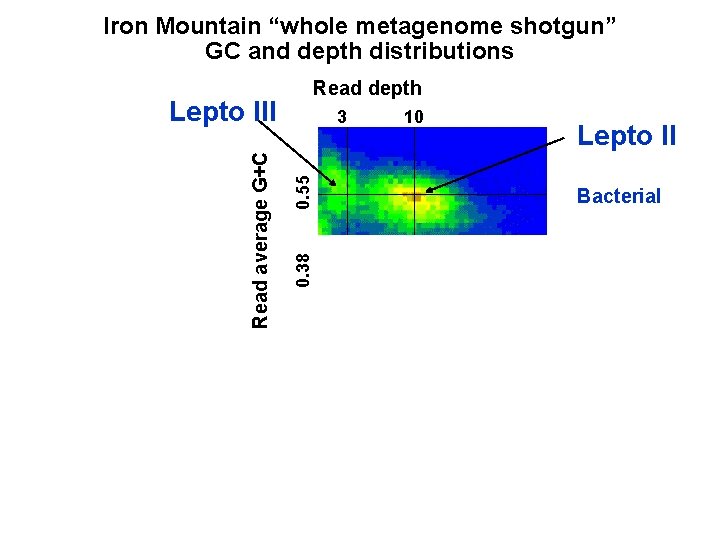
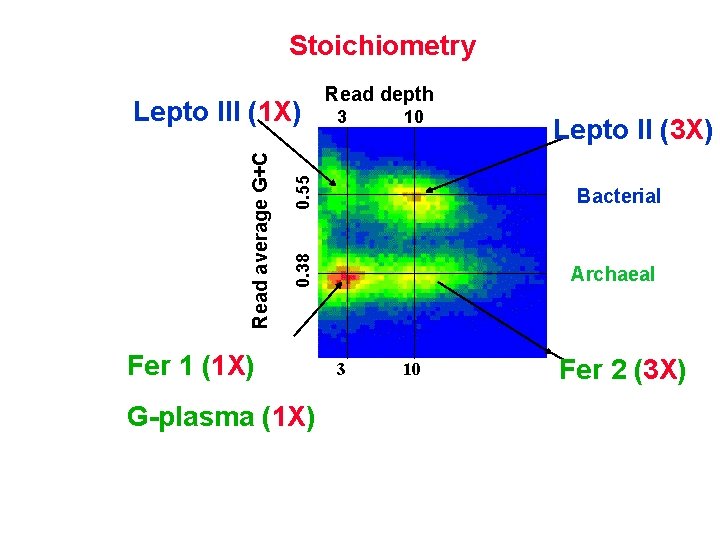
Iron Mountain “whole metagenome shotgun” GC and depth distributions Read depth 0. 55 3 0. 38 Read average G+C Lepto III 10 Lepto II Bacterial

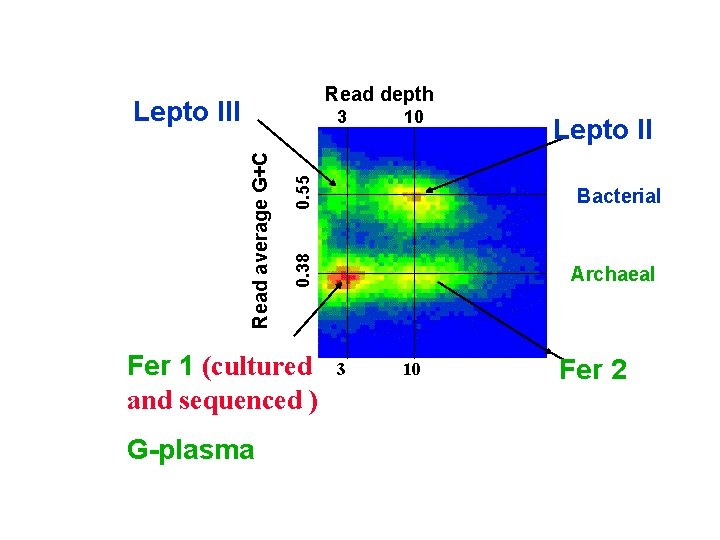
Read depth Lepto III 0. 55 Fer 1 (cultured and sequenced ) G-plasma 10 Lepto II Bacterial 0. 38 Read average G+C 3 Archaeal 3 10 Fer 2

Stoichiometry 10 0. 55 3 Fer 1 (1 X) G-plasma (1 X) Lepto II (3 X) Bacterial 0. 38 Read average G+C Lepto III (1 X) Read depth Archaeal 3 10 Fer 2 (3 X)

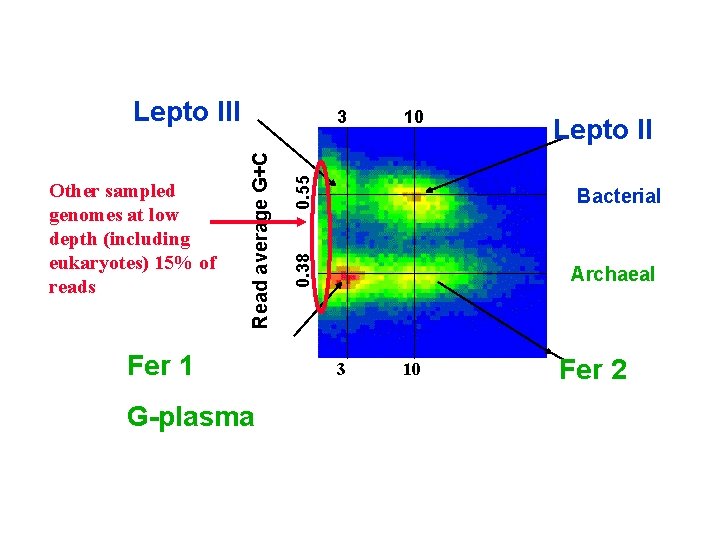
Lepto III Fer 1 G-plasma 0. 55 10 Lepto II Bacterial 0. 38 Read average G+C Other sampled genomes at low depth (including eukaryotes) 15% of reads 3 Archaeal 3 10 Fer 2

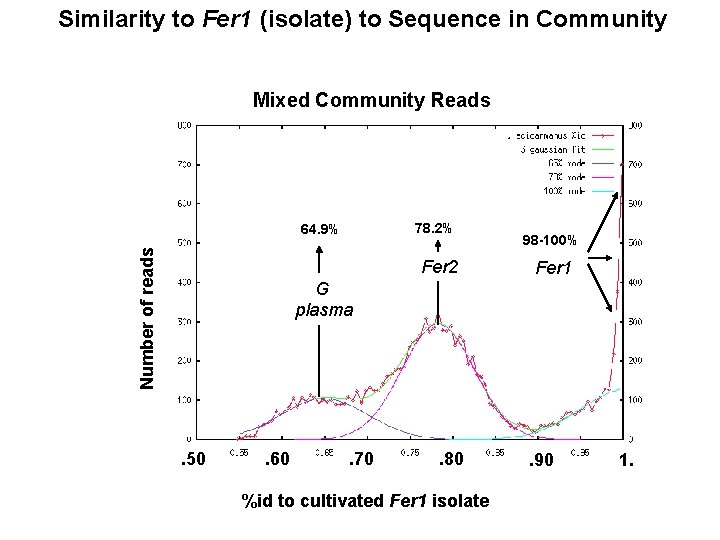
Similarity to Fer 1 (isolate) to Sequence in Community Mixed Community Reads 78. 2% Number of reads 64. 9% Fer 2 98 -100% Fer 1 G plasma . 50 . 60 . 70 . 80 %id to cultivated Fer 1 isolate . 90 1.

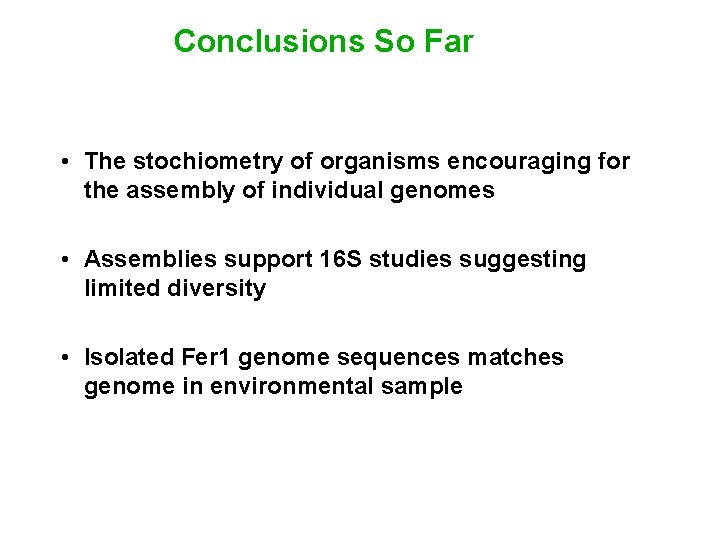
Conclusions So Far • The stochiometry of organisms encouraging for the assembly of individual genomes • Assemblies support 16 S studies suggesting limited diversity • Isolated Fer 1 genome sequences matches genome in environmental sample

How do we know that our assembly is correct?

How do you know you’ve done it right? Check pair ends against scaffold How do we know that our assembly is correct? At the gross level: check pairs (expect few % due to failing/chimeric clones) Align all reads back against assembled scaffolds end where there is no clone coverage in 3 kb plasmids Identifies potentially repetitive areas and/or rearrangements

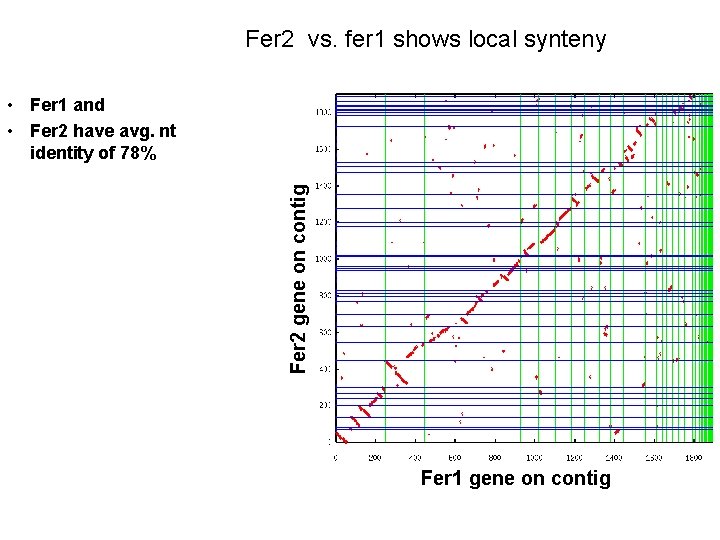
Fer 2 vs. fer 1 shows local synteny Fer 2 gene on contig • Fer 1 and • Fer 2 have avg. nt identity of 78% Fer 1 gene on contig

What does it mean to assemble a community genome? Sample derived from millions of genomes. ? What is a “species” in the enviroment? Members of the same species a) significantly different (many lineages survive and diverge) b) highly similar (selective sweeps)

What does it mean to assemble a community genome? Lepto II : 1 nucleotide variation / 3, 000 bp Fer II: 2. 2 nucleotide variation / 100 bp

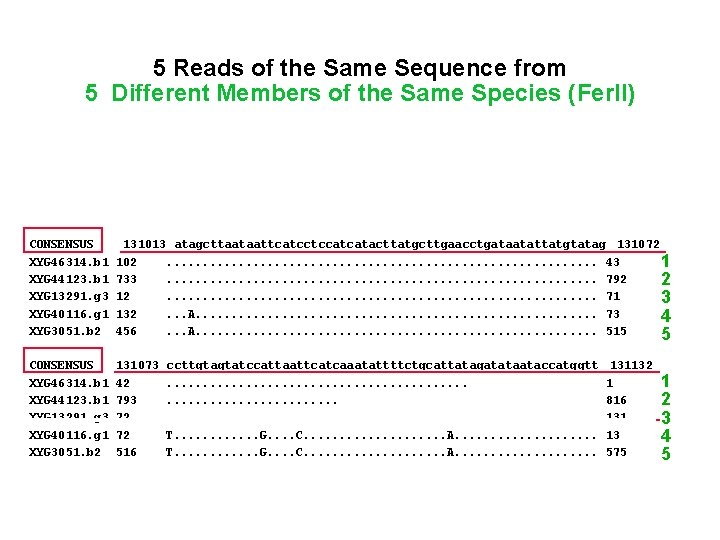
• • 5 Reads of the Same Sequence from 5 Different Members of the Same Species (Fer. II) CONSENSUS 130953 gtttatattaaatccattgatttctaagcttccggttcttcttccgtataatggagattt 131012 XYG 46314. b 1 XYG 44123. b 1 XYG 44918. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 162 673 48 2 192 396 • • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 131013 atagcttaataattcatcctccatcatacttatgcttgaacctgataatattatgtatag 131072 102. . . . 43 11 733. . . . 792 2 12. . . . 71 3 132. . . A. . . . 73 4 456. . . A. . . . 515 5 • • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 131073 42 793 72 72 516 1 A. . . C. . . . . . A. . . . C. . . A. . . . G. . . . . . ccttgtagtatccattaattcatcaaatattttctgcattatagatataataccatggtt. . . . . . . . T. . . G. . . . C. . . . . A. . . . . 103 732 4 11 133 455 131132 1 816 131 13 575 1 2 3 4 5

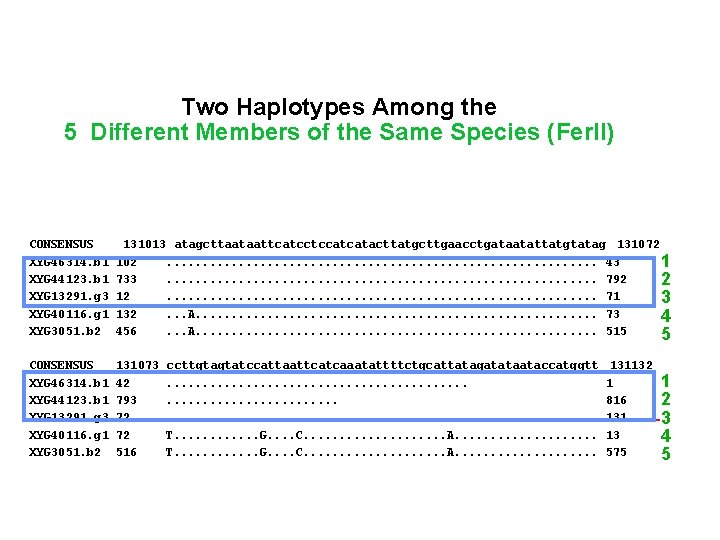
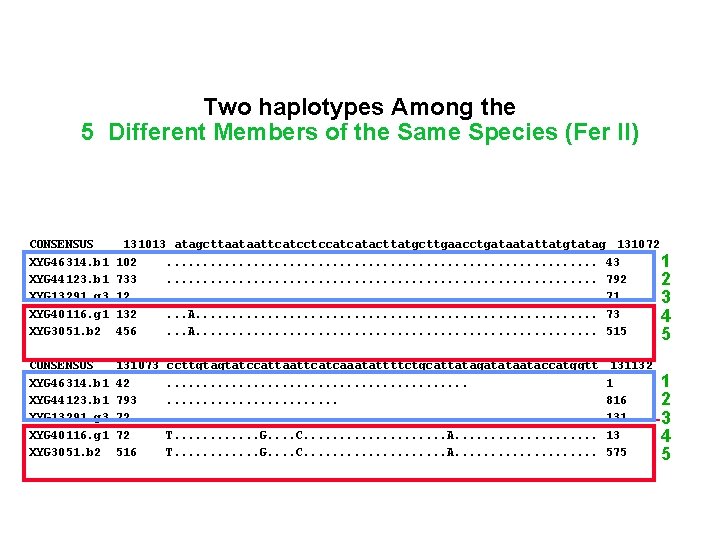
131012 Two Haplotypes Among the 103 5 Different Members of the Same Species (Fer. II)732 • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 44918. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 130953 162 673 48 2 192 396 • • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 131013 atagcttaataattcatcctccatcatacttatgcttgaacctgataatattatgtatag 131072 102. . . . 43 11 733. . . . 792 2 12. . . . 71 3 132. . . A. . . . 73 4 456. . . A. . . . 515 5 • • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 131073 42 793 72 72 516 1 gtttatattaaatccattgatttctaagcttccggttcttcttccgtataatggagattt A. . . C. . . . . . A. . . . C. . . A. . . . G. . . . . . ccttgtagtatccattaattcatcaaatattttctgcattatagatataataccatggtt. . . . . . . . T. . . G. . . . C. . . . . A. . . . . 4 11 133 455 131132 1 816 131 13 575 1 2 3 4 5

131012 Two haplotypes Among the 103 5 Different Members of the Same Species (Fer II) 732 • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 44918. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 130953 162 673 48 2 192 396 • • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 131013 atagcttaataattcatcctccatcatacttatgcttgaacctgataatattatgtatag 131072 102. . . . 43 11 733. . . . 792 2 12. . . . 71 3 132. . . A. . . . 73 4 456. . . A. . . . 515 5 • • • CONSENSUS XYG 46314. b 1 XYG 44123. b 1 XYG 13291. g 3 XYG 40116. g 1 XYG 3051. b 2 131073 42 793 72 72 516 1 gtttatattaaatccattgatttctaagcttccggttcttcttccgtataatggagattt A. . . C. . . . . . A. . . . C. . . A. . . . G. . . . . . ccttgtagtatccattaattcatcaaatattttctgcattatagatataataccatggtt. . . . . . . . T. . . G. . . . C. . . . . A. . . . . 4 11 133 455 131132 1 816 131 13 575 1 2 3 4 5

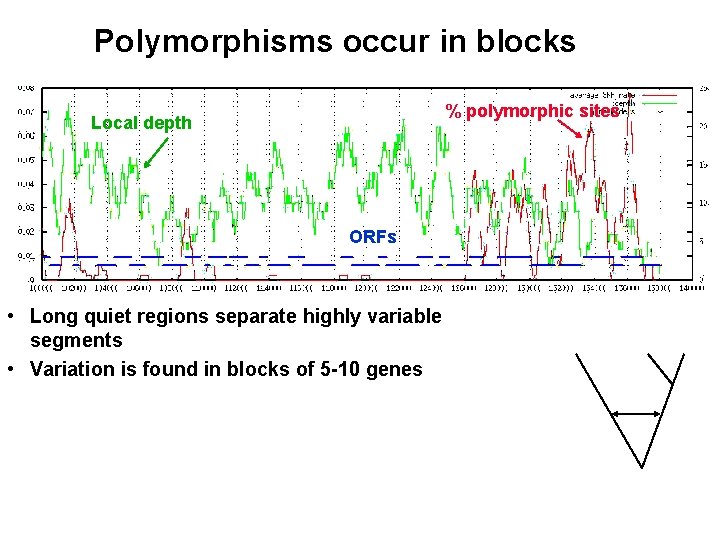
Polymorphisms occur in blocks % polymorphic sites Local depth ORFs • Long quiet regions separate highly variable segments • Variation is found in blocks of 5 -10 genes

Summary of Iron Mountain Biofilm • Limited number of predominant species present in biofilm the majority have never been cultured • Several lines of evidence suggest that we can assemble genomes of these organisms • Simplicity of community suggests removal of most variants by natural selection • Now studying the metabolic capabilities of microbes

 Jgi csp
Jgi csp National human genome research institute
National human genome research institute National human genome research institute
National human genome research institute National human genome research institute
National human genome research institute Encode
Encode Genome-to-genome distance calculator
Genome-to-genome distance calculator The human genome consists of
The human genome consists of Human genome size
Human genome size Human genome size
Human genome size Human genome project
Human genome project Human genome structure
Human genome structure Repeated sequences
Repeated sequences Human genome project source code
Human genome project source code Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key Sequencing human genome
Sequencing human genome History of human genome project
History of human genome project Human genome project
Human genome project Chapter 14 the human genome
Chapter 14 the human genome Human genome project
Human genome project 14-3 human molecular genetics answer key
14-3 human molecular genetics answer key Who discovered the structure of dna
Who discovered the structure of dna Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Break joint lamb
Break joint lamb Ligamentum nuchae
Ligamentum nuchae Difference between permanent joint and temporary joint
Difference between permanent joint and temporary joint Memorandum joint venture account format
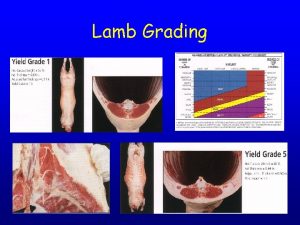
Memorandum joint venture account format Lamb carcass grading
Lamb carcass grading Hinge joint
Hinge joint Duke human vaccine institute
Duke human vaccine institute What is genome
What is genome Plant genome research program
Plant genome research program Stanford
Stanford Mash distance
Mash distance Perpartes
Perpartes Shotgun sequencing
Shotgun sequencing Shotgun sequencing
Shotgun sequencing Genome sequencing
Genome sequencing Patric genome
Patric genome