The Artemis Comparison Tool ACT Comparative Genomics It




- Slides: 28

The Artemis Comparison Tool ACT

Comparative Genomics • It might be expected that if two organisms diverged from a common ancestor, they should share: - similar gene content - similar gene organization • However, genome evolution is a highly dynamic process: - gene acquisition and gene loss will change genome content - gene rearrangements will change the order of genes. • Comparative genomics tell us about common and unique features between different organisms at the genome level. • Identification of similarities and differences between genomes may allow us to understand: - How two organisms evolved - Why certain organisms cause diseases while others do not

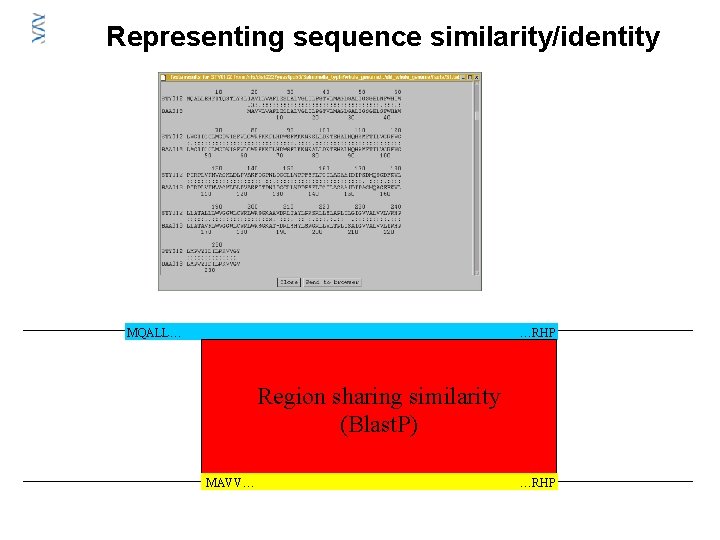
Representing sequence similarity/identity MQALL… …RHP Region sharing similarity (Blast. P) MAVV… …RHP

ACT- Artemis Comparison Tool

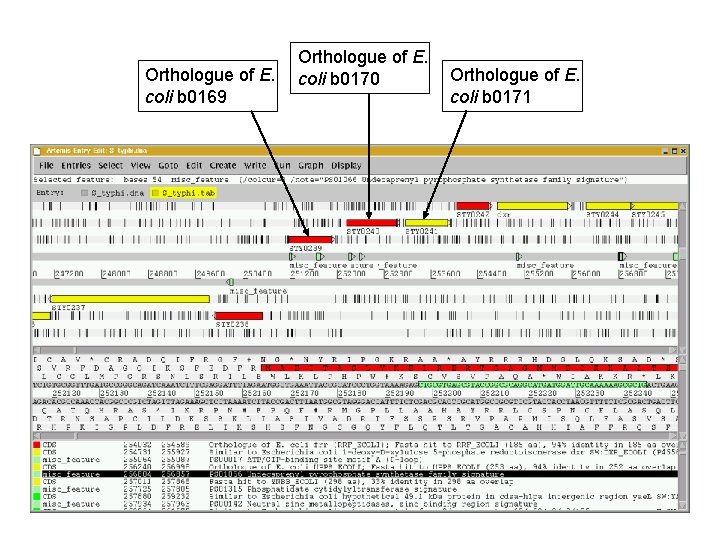
Orthologue of E. coli b 0169 Orthologue of E. coli b 0170 Orthologue of E. coli b 0171

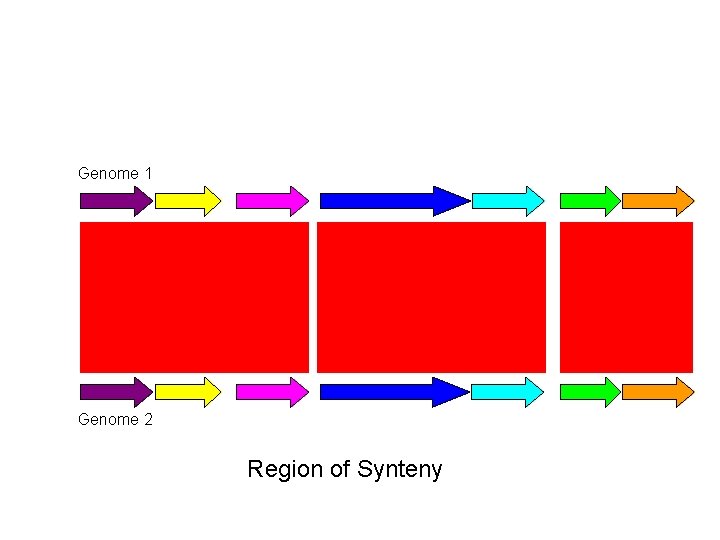
Genome 1 Genome 2 Region of Synteny

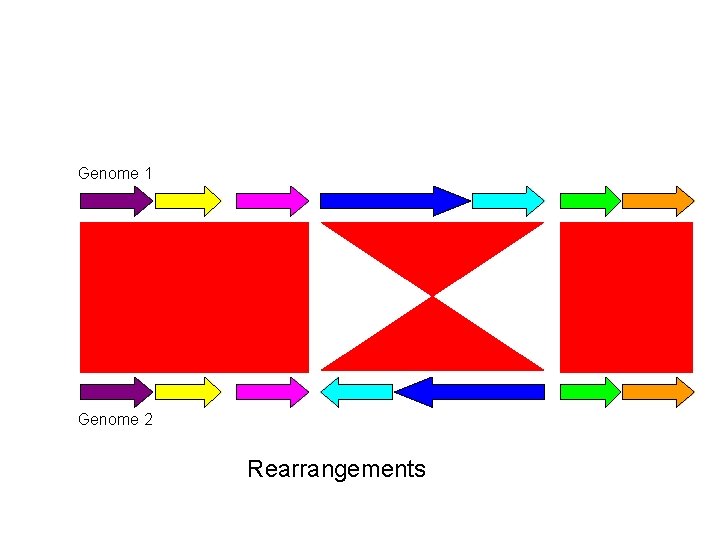
Genome 1 Genome 2 Rearrangements

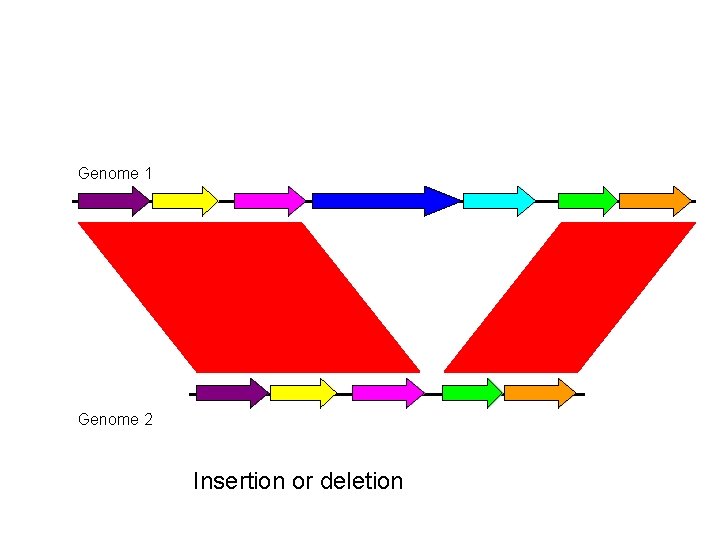
Genome 1 Genome 2 Insertion or deletion

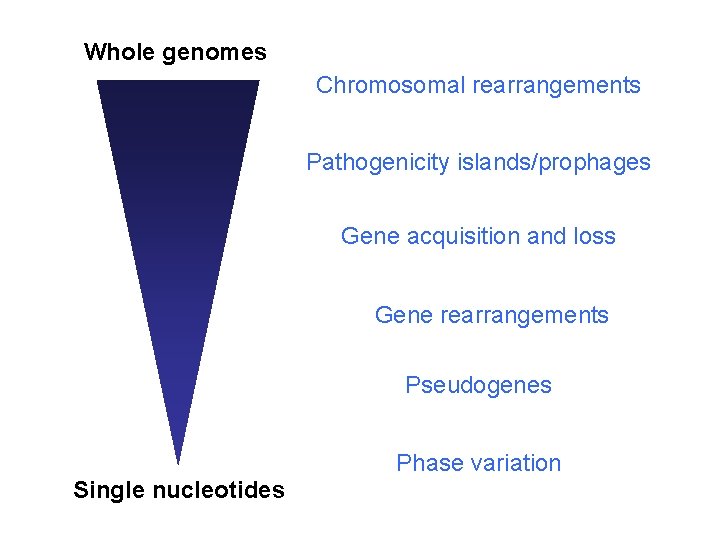
Whole genomes Chromosomal rearrangements Pathogenicity islands/prophages Gene acquisition and loss Gene rearrangements Pseudogenes Phase variation Single nucleotides

Prokaryotic examples

Whole genomes: S. typhi vs. E. coli SPI-2 SPI-9 SPI-1 SPI-7 Vi SPI-10 S. typhi DNA matches E. coli

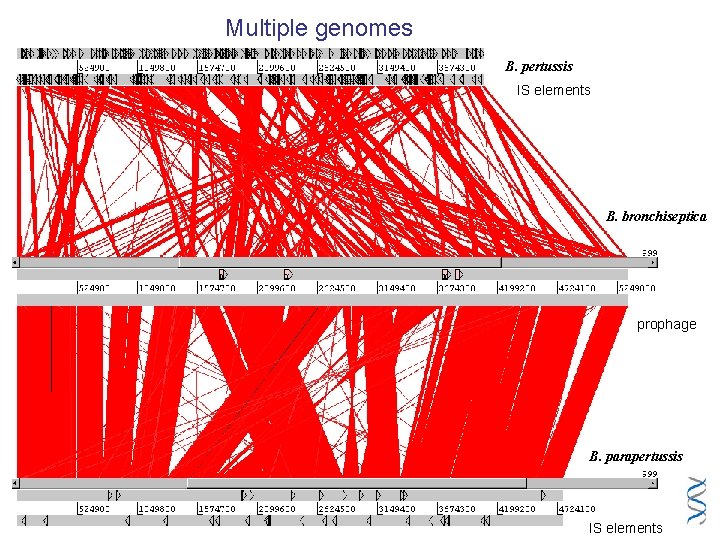
Multiple genomes B. pertussis IS elements B. bronchiseptica prophage B. parapertussis IS elements

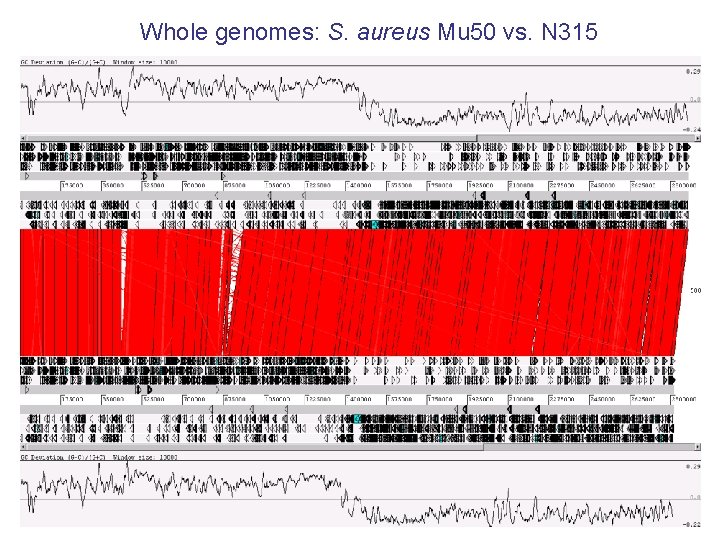
Whole genomes: S. aureus Mu 50 vs. N 315

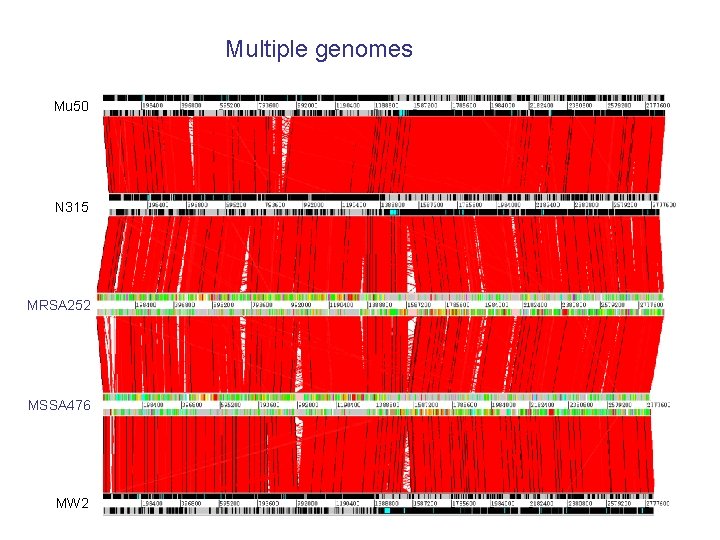
Multiple genomes Mu 50 N 315 MRSA 252 MSSA 476 MW 2

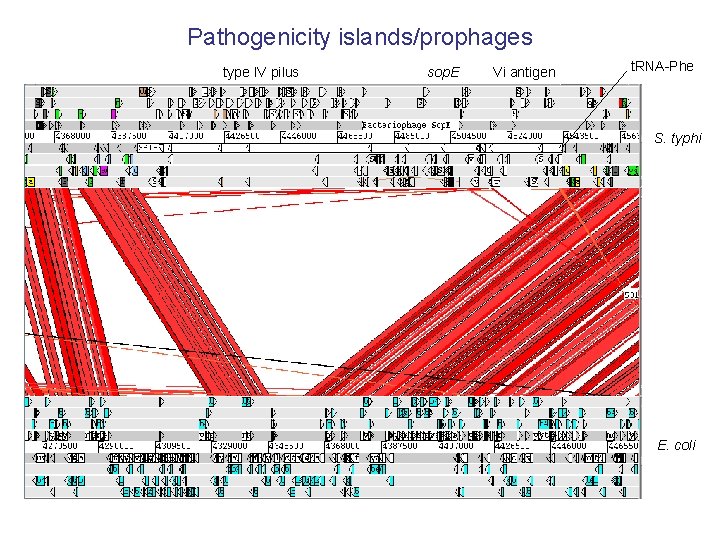
Pathogenicity islands/prophages type IV pilus sop. E Vi antigen t. RNA-Phe S. typhi E. coli

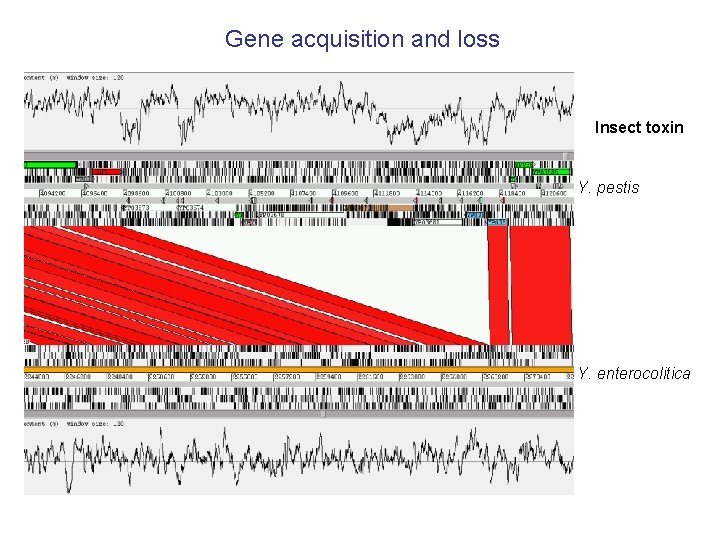
Gene acquisition and loss Insect toxin Y. pestis Y. enterocolitica

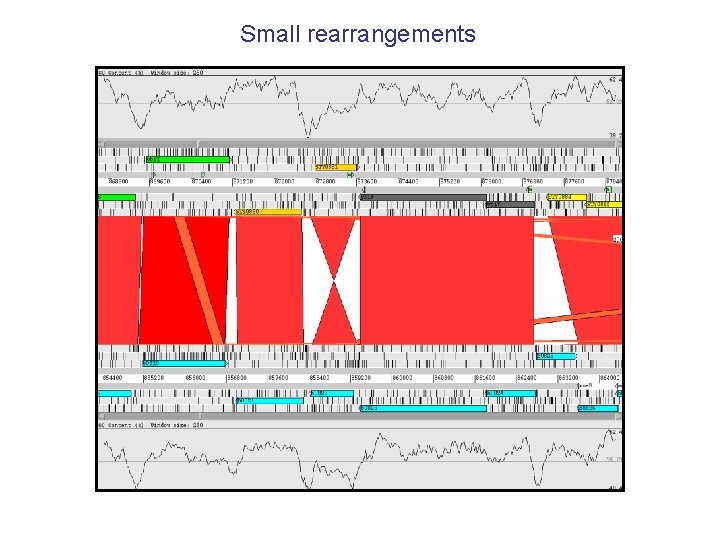
Small rearrangements

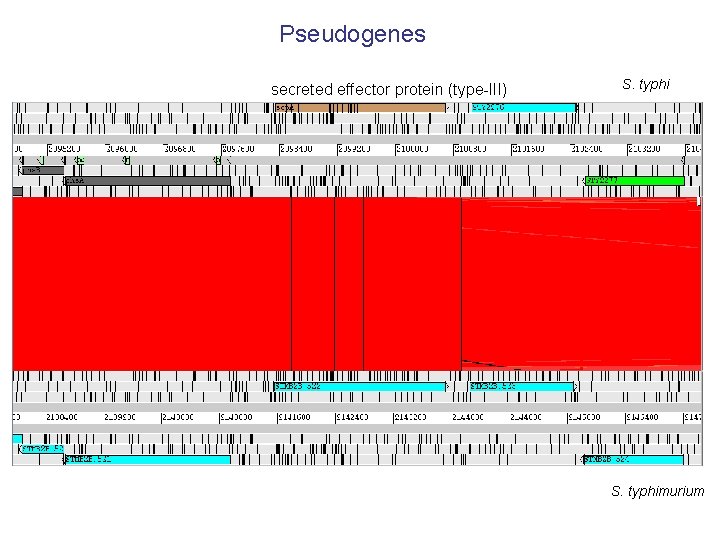
Pseudogenes secreted effector protein (type-III) S. typhimurium

ACT Eukaryotic examples

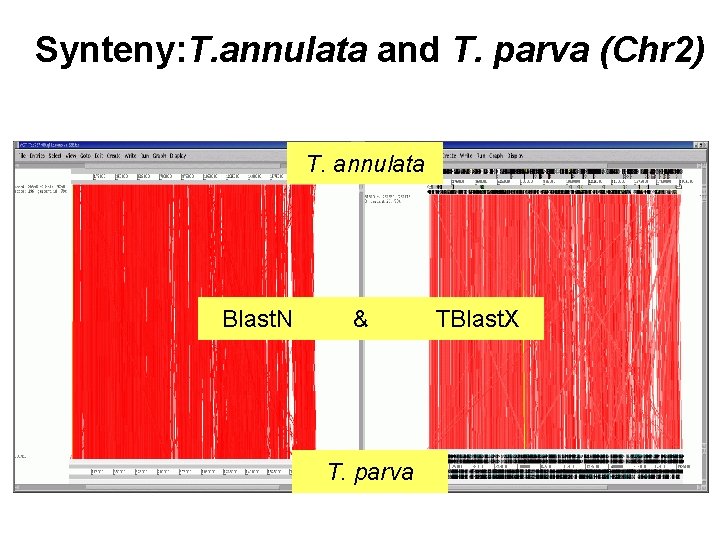
Synteny: T. annulata and T. parva (Chr 2) T. annulata Blast. N & T. parva TBlast. X

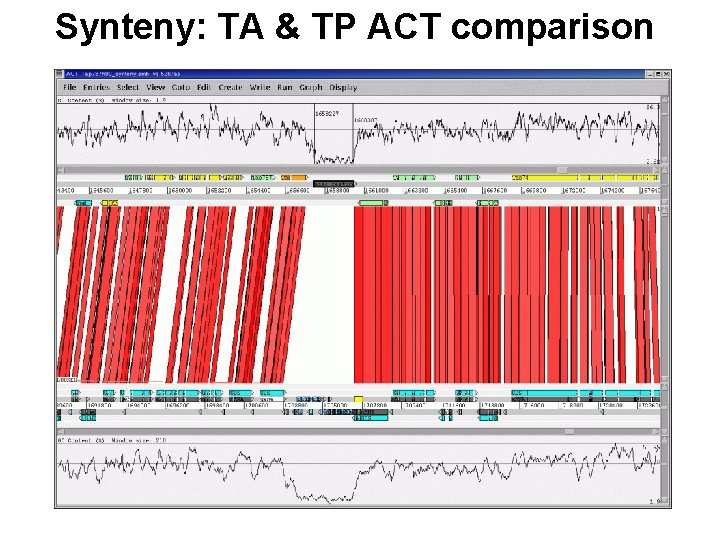
Synteny: TA & TP ACT comparison

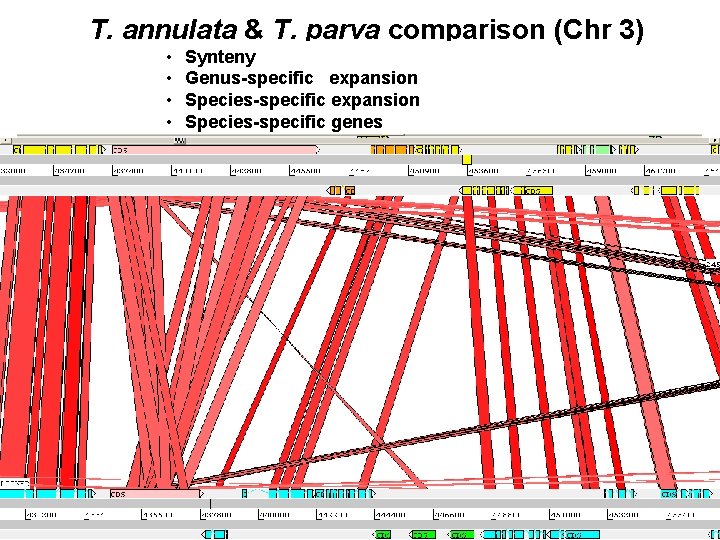
T. annulata & T. parva comparison (Chr 3) • • Synteny Genus-specific expansion Species-specific genes

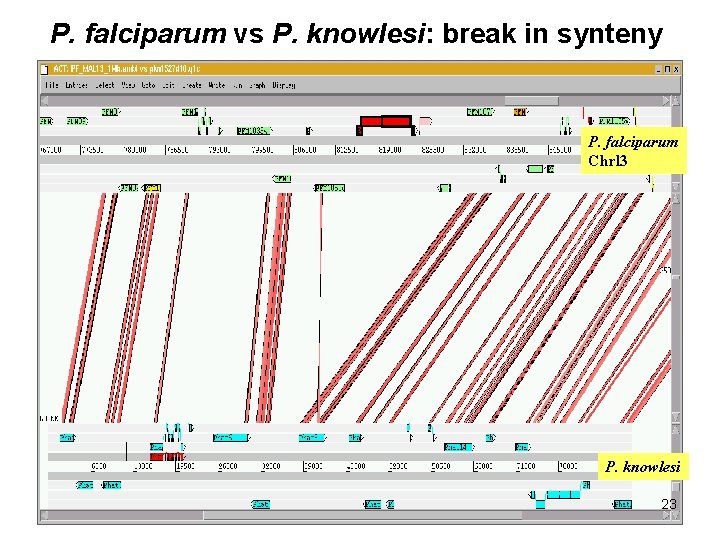
P. falciparum vs P. knowlesi: break in synteny P. falciparum Chrl 3 P. knowlesi 23

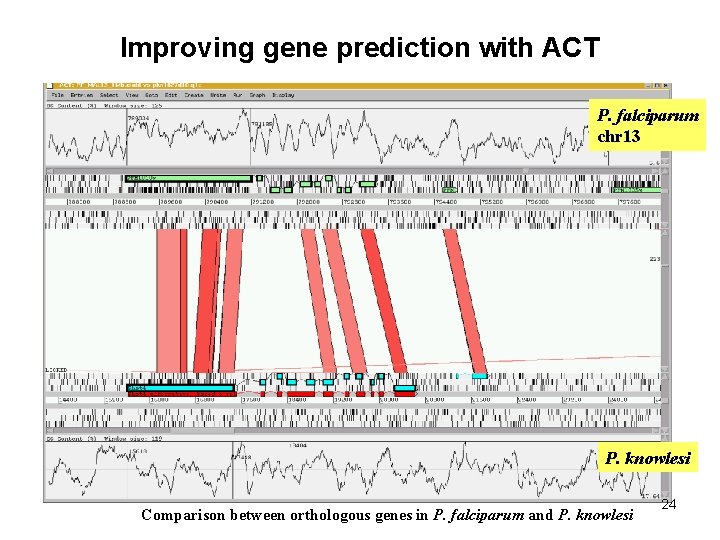
Improving gene prediction with ACT P. falciparum chr 13 P. knowlesi Comparison between orthologous genes in P. falciparum and P. knowlesi 24

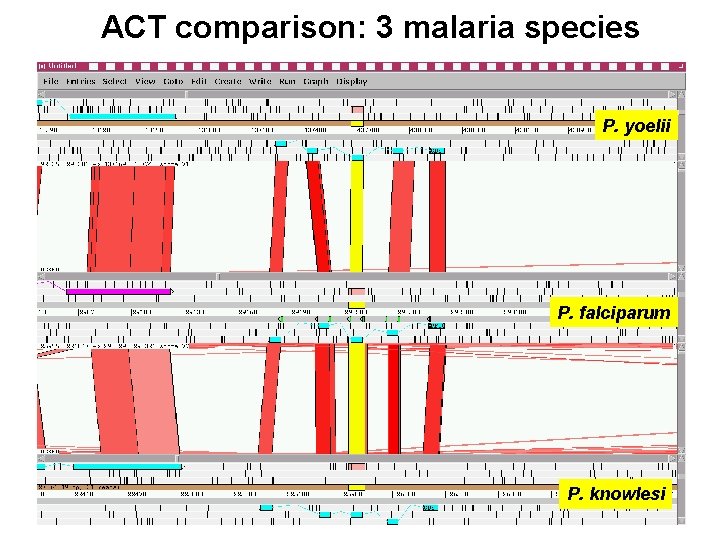
ACT comparison: 3 malaria species P. yoelii P. falciparum P. knowlesi

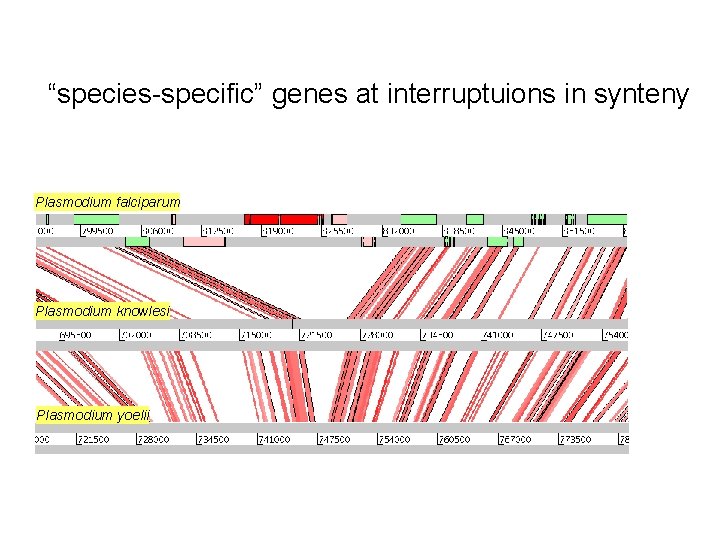
“species-specific” genes at interruptuions in synteny Plasmodium falciparum Plasmodium knowlesi Plasmodium yoelii

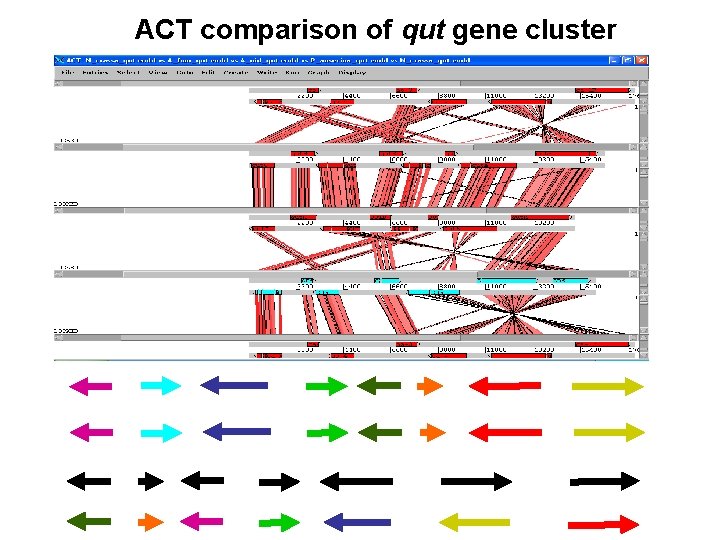
ACT comparison of qut gene cluster

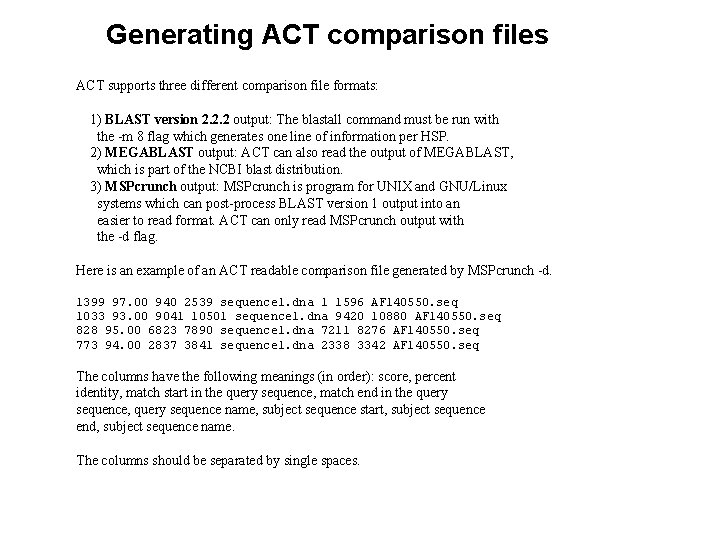
Generating ACT comparison files ACT supports three different comparison file formats: 1) BLAST version 2. 2. 2 output: The blastall command must be run with the -m 8 flag which generates one line of information per HSP. 2) MEGABLAST output: ACT can also read the output of MEGABLAST, which is part of the NCBI blast distribution. 3) MSPcrunch output: MSPcrunch is program for UNIX and GNU/Linux systems which can post-process BLAST version 1 output into an easier to read format. ACT can only read MSPcrunch output with the -d flag. Here is an example of an ACT readable comparison file generated by MSPcrunch -d. 1399 97. 00 940 2539 sequence 1. dna 1 1596 AF 140550. seq 1033 93. 00 9041 10501 sequence 1. dna 9420 10880 AF 140550. seq 828 95. 00 6823 7890 sequence 1. dna 7211 8276 AF 140550. seq 773 94. 00 2837 3841 sequence 1. dna 2338 3342 AF 140550. seq The columns have the following meanings (in order): score, percent identity, match start in the query sequence, match end in the query sequence, query sequence name, subject sequence start, subject sequence end, subject sequence name. The columns should be separated by single spaces.