Sequence Analysis with Artemis Artemis Comparison Tool ACT










- Slides: 36

Sequence Analysis with Artemis & Artemis Comparison Tool (ACT) South East Asian Training Course on Bioinformatics Applied to Tropical Diseases - 2005 (Sponsored by UNDP/World Bank/WHO/TDR) International Centre For Genetic Engineering And Biotechnology , New Delhi, INDIA

Gene finding

atcttttacttttttcatcatctatacaaaaaatcatagaatattcatcatgttgtttaaaataatgtattccattatgaactttattacaaccctcgtt tttaattcacattttatatctttaagtataatatcatttaacattatgttatcttcctcagtgtttttcattattatttgcatgtacagtttatca tttttatgtaccaaactatatcttatattaaatggatctctacttataaagttaaaatctttttttaattttttcacttccaattttatattccg cagtacatcgaattctaaaaaataaataatatataatatataataaataatatataatatataataatatataatatataataaataatatataatatataatactttggaaagattattt atatgaatatatacacctttaataggatacacacatcatatttatatacatataaatattccataaatatttatacaacctcaaataaaca tacatatataaatatatacatatatgtatcattacgtaaaaacatcaaagaaatatactggaaaacatgtcacaaaactaaaaaaggtattagg agatatatttactgattcctcatttttataaatgttaaaattattatccctagtccaaatatccacatttattaaattcacttgaatattgttttttaaa ttgctagatatattaatttgagatttaaaattctgacctatataaacctttcgagaatttataggtagacttaaacttatttcatttgataaactaatat tatcatttatgtccttatcaaaattttctccatttcagttattttaaacatattccaaatattgttattaaacaagggcggacttaaacgaagtaa ttcaatcttaactccttcactcattttatatattccttaatttttactatgtttattaacatataaacaaatatgtcactaa taatatatatatatatattataaatgttttactctattttcacatcttgtcctttttaaaaatcccaattcttattcat taaataataatgtattttttttttttattattatgttactgttttattatatacactcttaatcatatatttatatatatatattattcccttttcatgttttaaacaagaaaactaaaaaaataataaaatatatttttataacatatgt attattaaaatgtataaaaatattccatttattattatttttttatatacattgttataagagtatcttctcccttctggtttatattacta ccatttcactttgaacttttcataaaaattaatagaatatcaaatatgtataatatataacaaaaaaaaaaaaaaaaaaata tatatatatacatatatatttcatctaatcatttaaaattattattatatattttttaaaaaatatatttatgataacataaaaaga atttaattaaatataattacatctaatattattatataataagttttccaaatagaatacttatatatatatattcttccataaaaagaataaaataaaaacaccttaaaagtatttgtaaaaaattccccacattgaatatatagttgtattt ataaaattaaagaaaaagcataaagttaccatttaatagtggagattagtaacattttcttcattatcaaaaatatttcctaattttttg taaaatatatttaaaaatgtaatagattatgtattaaataatatagcaaaatgttcaattttagaaatttgcctctttttgacaaggataattc aaaagatacaggtaaaaaaataaagtaaaacaaaacaaaaaaaaaaaaatgacatgttataatataa taaaaattatgtaatatatcataatcgaagaaacatatatgaaaccaaaaagaaacagatcttgatttattaatacatatataacattcata tctttatttttgtagatgatataaaaaattttataaactcttatgaagggatatatttttcatcatccaataaatttataaatgtatttctagacaaaat tctgatcattgatccgtcttccttaaatgttattacaatacagatctgtagttgatttcctttttaatgagaaaaataagaatcttattgtt ttagggtaatgaaatatagatttatatttttatttattatatattattttttaatttttcttttatatattttatttagtgtataaaa tgatatcctttatatttacatgggatattcaaataataacaaaaatgagtatacacatatatatatatatgtatatttttttatgttcctataggaaagggaagaattcactgatttgtagtgtttacaatattagggaatgcaactttacacttttgaaaaaaattcagtta agcaaaaatattaataacattaaaaagacactgatagcaaaatgtaatgaatatataataacattagaaaataagaaaattactttttatttcttaaata aagattatagtataaatcaaagtgaattaatagaagacggaaaagaacttattgaaaatatctatttgtcaaaaaatcatatcttgttagtaataaaaaa ttcatatgtatataccaattagatattaaaaattcccatattagttatacacttattgatagtttcaatttatcctacctcagagaatct ataataaaagcatataaaataaatgatgtatcaaataatgacccaaaaaaggataataatgaaaaaaatacttcatctaataatataa cacataacaattataatgacatatcaaataataataatattaatggggtgaaagaccatataataacactctggaaaataatga tgaaccaatcttatctataatgaagatcttaatgttttatatgccaaaatatgtataacgtcctttttgaatttaaataacctaagt

atcttttacttttttcatcatctatacaaaaaatcatagaatattcatcatgttgtttaaaataatgtattccattatgaactttattacaaccctcgtt tttaattcacattttatatctttaagtataatatcatttaacattatgttatcttcctcagtgtttttcattattatttgcatgtacagtttatca tttttatgtaccaaactatatcttatattaaatggatctctacttataaagttaaaatctttttttaattttttcacttccaattttatattccg cagtacatcgaattctaaaaaataaataatatataatatataataaataatatataatatataataatatataatatataataaataatatataatatataatactttggaaagattattt atatgaatatatacacctttaataggatacacacatcatatttatatacatataaatattccataaatatttatacaacctcaaataaaca tacatatataaatatatacatatatgtatcattacgtaaaaacatcaaagaaatatactggaaaacat gtcacaaaactaaaaaaggtattagg agatatatttactgattcctcatttttataaatgttaaaattattatccctagtccaaatatccacatttattaaattcacttgaatattgttttttaaa ttgctagatatattaatttgagatttaaaattctgacctatataaacctttcgagaatttataggtagacttaaacttatttcatttgataaactaatat tatcatttatgtccttatcaaaattttctccatttcagttattttaaacatattccaaatattgttattaaacaagggcggacttaaacgaagtaa ttcaatcttaactccttcactcattttatatattccttaatttttactatgtttattaacatataaacaaatatgtcactaa taatatatatatatatattataaatgttttactctattttcacatcttgtcctttttaaaaatcccaattcttattcat taaataataatgtattttttttttttattattatgttactgttttattatatacactcttaatcatatatttatatatatatattattcccttttcatgttttaaacaagaaaactaaaaaaataataaaatatatttttataacag atgt attattaaaatgtataaaaatattccatttattattatttttttatatacattgttataagagtatcttctcccttctggtttatattacta ccatttcactttgaacttttcataaaaattaatagaatatcaaatatgtataatatataacaaaaaaaaaaaaaaaaaaata tatatatatacatatatatttcatctaatcatttaaaattattattatatattttttaaaaaatatatttatgataacataaaaaga atttaattaaatataattacatctaatattattatataataagttttccaaatagaatacttatatatatatattcttccataaaaagaataaaataaaaacaccttaaaagtatttgtaaaaaattccccacattgaatatatagttgtattt ataaaattaaagaaaaagcataaagttaccatttaatagtggagattagtaa gtttttcttcattatcaaaaatatttcctaattttttg taaaatatatttaaaaatgtaatagattatgtattaaataatatagcaaaatgttcaattttagaaatttgcctctttttgacaaggataattc aaaagatacaggtaaaaaaataaagtaaaacaaaacaaaaaaaaaaaaatgacatgttataatataa taaaaattatgtaatatatcataatcgaagaaacatatatgaaaccaaaaagaaacagatcttgatttattaatacatatataacattcata tctttatttttgtagatgatataaaaaattttataaactcttatgaagggatatatttttcatcatccaataaatttataaatgtatttctagacaaaat tctgatcattgatccgtcttccttag gtgttattacaatacagatctgtagttgatttcctttttaatgagaaaaataagaatcttattgtt ttagggtaatgaaatatagatttatatttttatttattatatattattttttaatttttcttttatatattttatttagtgtataaaa tgatatcctttatatttacatgggatattcaaataataacaaaaatgagtatacacatatatatatatatgtatatttttttatgttcctataggaaagggaagaattcactgatttgtagtgtttacaatattagggaatgcaactttacacttttgaaaaaaattcagtta agcaaaaatattaataacattaaaaagacactgatagcaaaatgtaatgaatatataataacattagaaaataagaaaattactttttatttcttaaata aagattatagtataaatcaaagtgaattaatagaagacggaaaagaacttattgaaaatatctatttgtcaaaaaatcatatcttgttagtaataaaaaa ttcatatgtatataccaattag atattaaaaattcccatattagttatacacttattgatagtttcaatttatcctacctcagagaatct ataataaaagcatataaaataaatgatgtatcaaataatgacccaaaaaaggataataatgaaaaaaatacttcatctaataatataa cacataacaattataatgacatatcaaataataataatattaatggggtgaaagaccatataataacactctggaaaataatga tgaaccaatcttatctataatgaagatcttaatgttttatatgccaaaatatgtataacgtcctttttgaatttaaataacctaagt

Gene prediction programs: ORFs and CDSs ORFs are not equivalent to CDSs Not all open reading frames are coding sequences

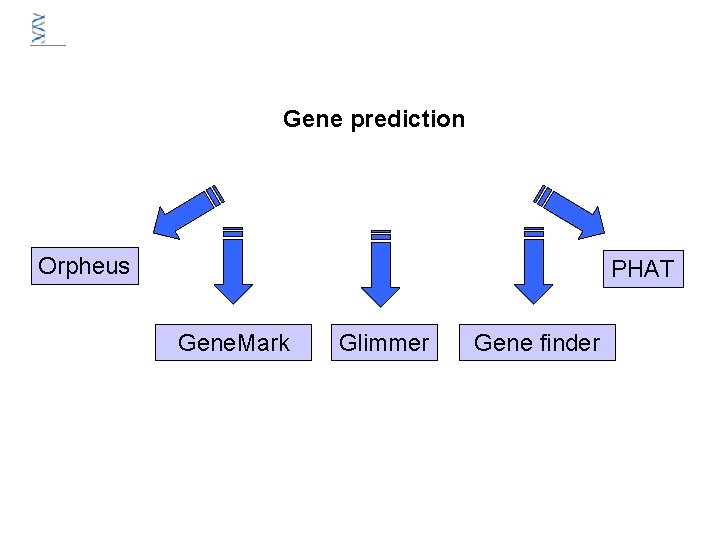
Gene prediction Orpheus PHAT Gene. Mark Glimmer Gene finder

Genefinding programs • Genefinding software packages use Hidden Markov Models. • Predict coding, intergenic and intron sequences • Need to be trained on a specific organism. • Never perfect!



Gene prediction programs: Problems • ORFs are not equivalent to CDSs • Gene prediction programs find new genes that share properties with a given set of genes. • They can be confounded by: – – – Sequence constraints (ribosomal proteins etc. ) Sequence biases Different sets of genes Horizontal gene transfer Non-coding DNA

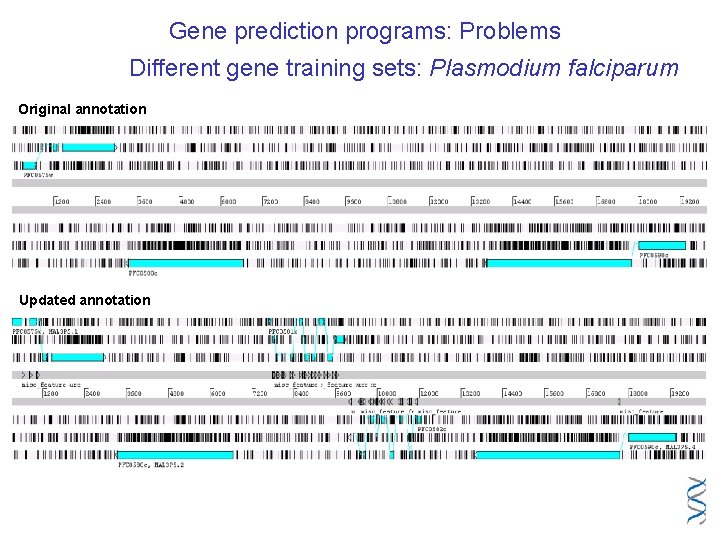
Gene prediction programs: Problems Different gene training sets: Plasmodium falciparum Original annotation Updated annotation

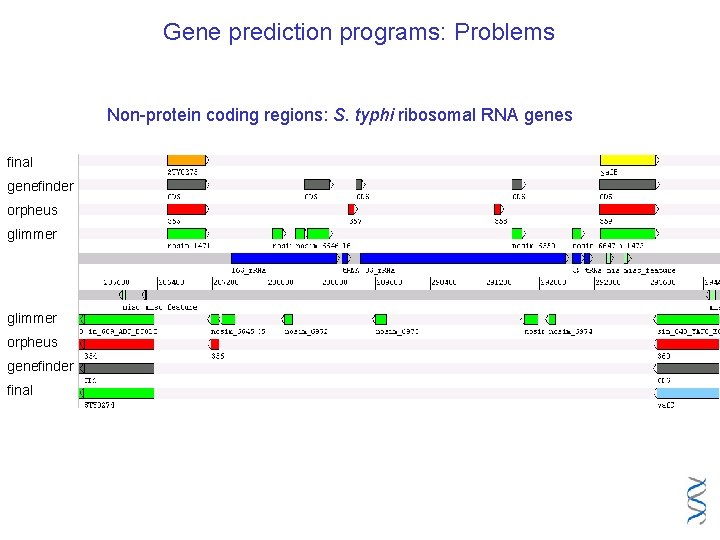
Gene prediction programs: Problems Non-protein coding regions: S. typhi ribosomal RNA genes final genefinder orpheus glimmer orpheus genefinder final

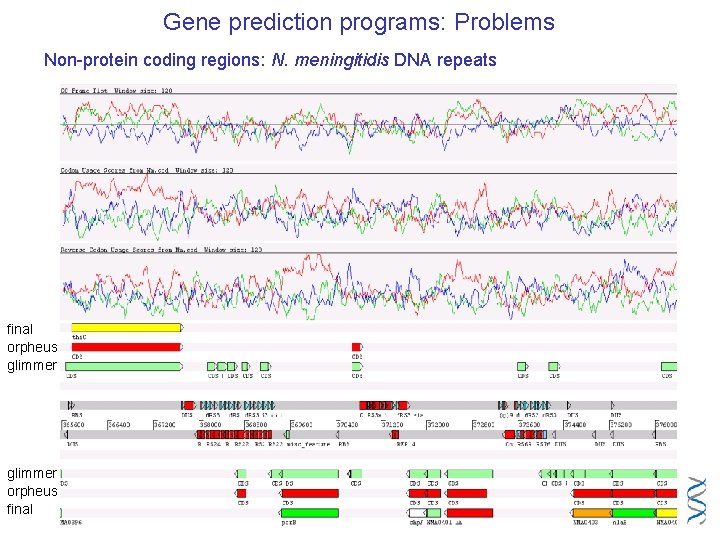
Gene prediction programs: Problems Non-protein coding regions: N. meningitidis DNA repeats final orpheus glimmer orpheus final

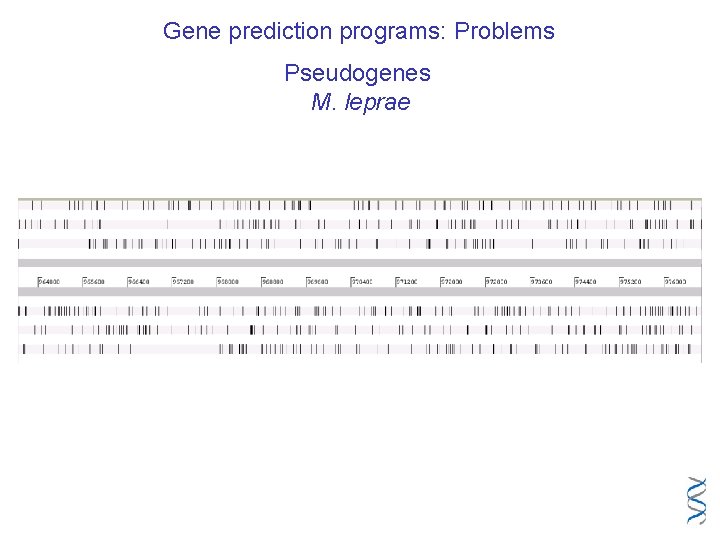
Gene prediction programs: Problems Pseudogenes M. leprae

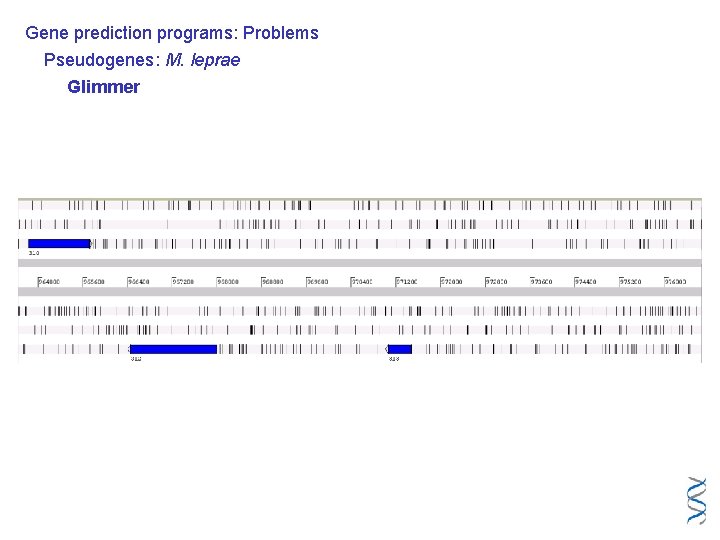
Gene prediction programs: Problems Pseudogenes: M. leprae Glimmer

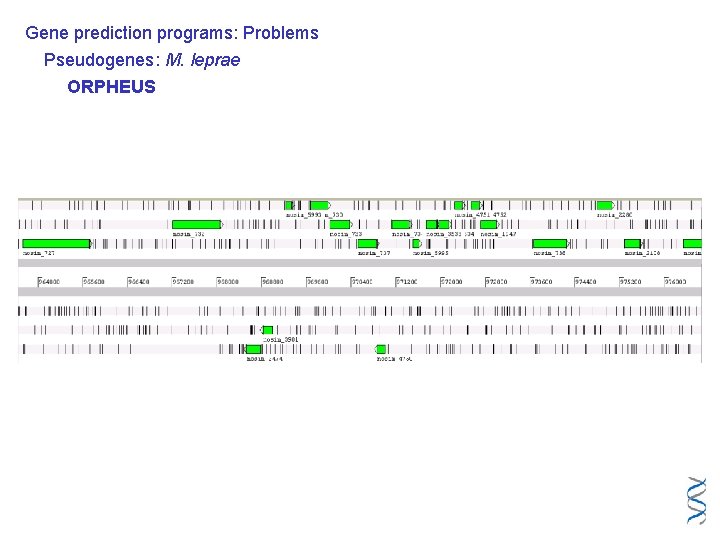
Gene prediction programs: Problems Pseudogenes: M. leprae ORPHEUS

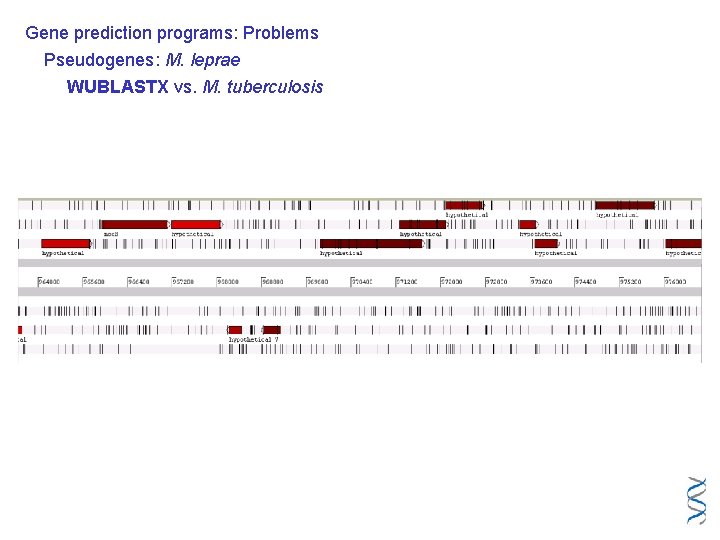
Gene prediction programs: Problems Pseudogenes: M. leprae WUBLASTX vs. M. tuberculosis

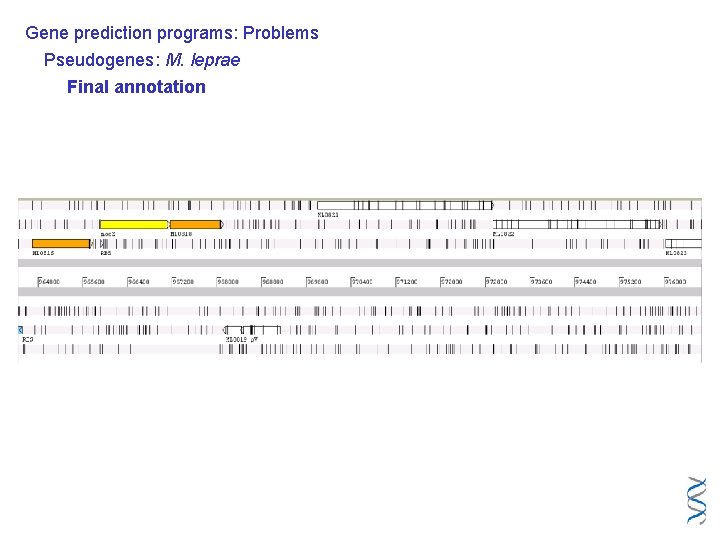
Gene prediction programs: Problems Pseudogenes: M. leprae Final annotation

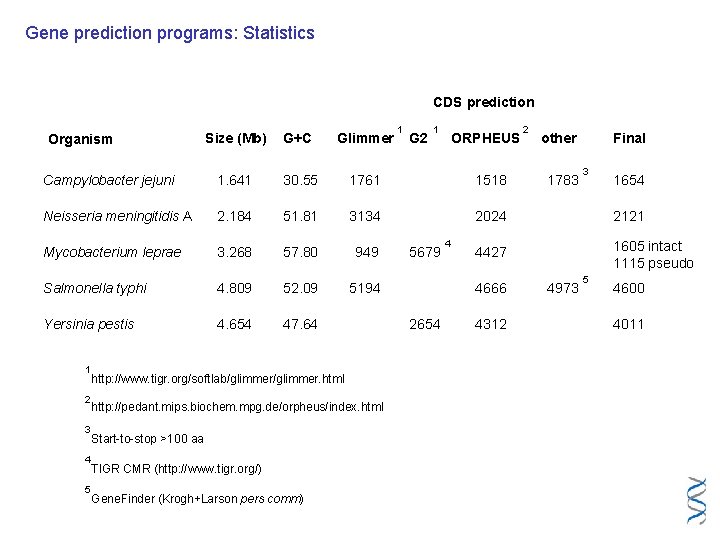
Gene prediction programs: Statistics CDS prediction 1 Glimmer Campylobacter jejuni 1. 641 30. 55 1761 1518 Neisseria meningitidis A 2. 184 51. 81 3134 2024 2121 Mycobacterium leprae 3. 268 57. 80 949 4427 1605 intact 1115 pseudo Salmonella typhi 4. 809 52. 09 5194 Yersinia pestis 4. 654 47. 64 2 3 4 5 http: //www. tigr. org/softlab/glimmer. html http: //pedant. mips. biochem. mpg. de/orpheus/index. html Start-to-stop >100 aa TIGR CMR (http: //www. tigr. org/) Gene. Finder (Krogh+Larson pers comm) 5679 ORPHEUS 2 G+C 1 G 2 1 Size (Mb) Organism 4 4666 2654 4312 other 1783 4973 Final 3 5 1654 4600 4011

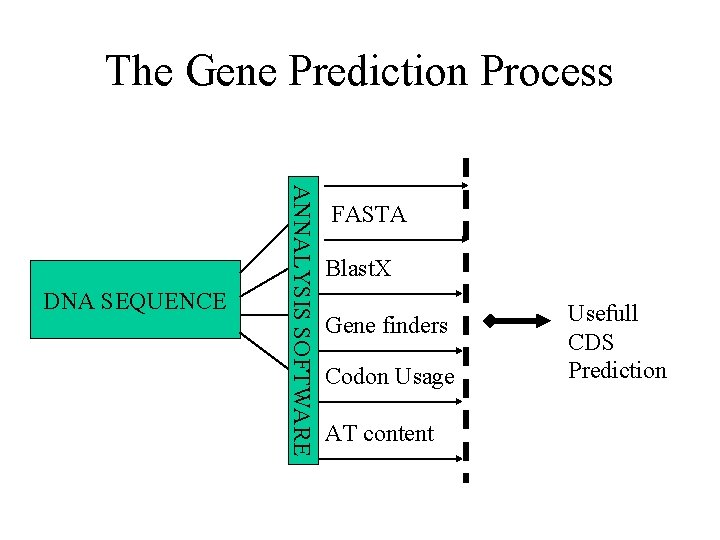
The Gene Prediction Process ESTs ANNALYSIS SOFTWARE DNA SEQUENCE FASTA Blast. X Gene finders Codon Usage AT content Annotator Usefull CDS Prediction

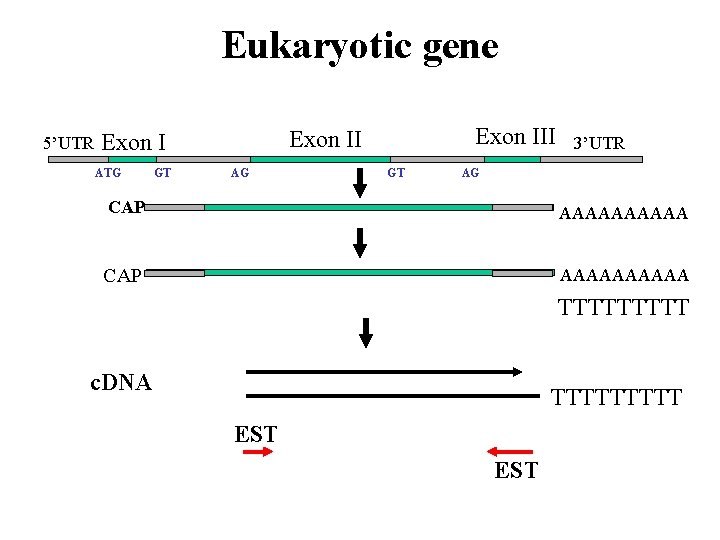
Eukaryotic gene 5’UTR Exon I intron ATG GT AG stop Exon III 3’UTR Exon II GT AG CAP AAAAAAAAAA m. RNA TTTTT c. DNA TTTTT EST

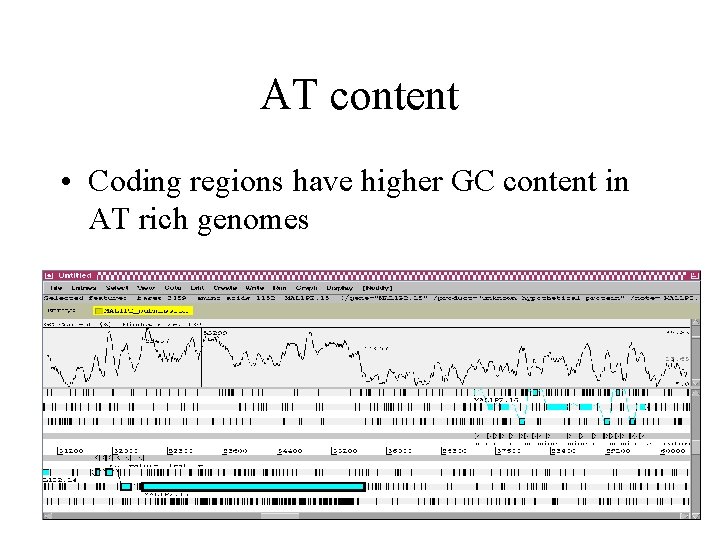
AT content • Coding regions have higher GC content in AT rich genomes

AT content

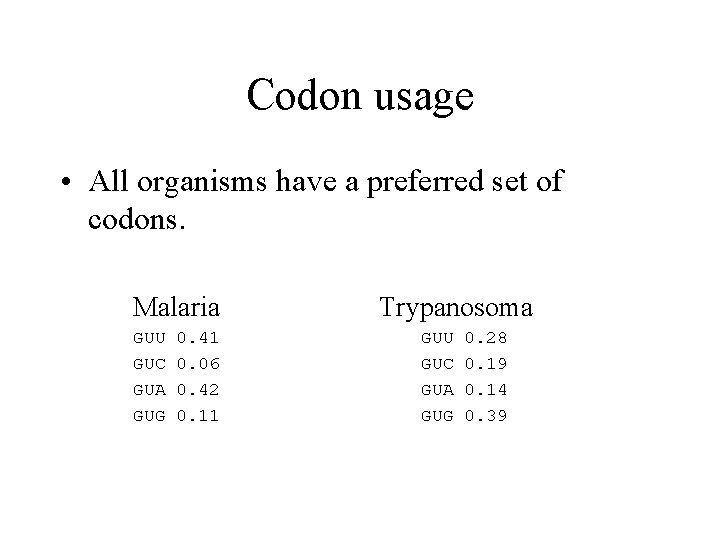
CODON USAGE • Codon bias is different for each organism. • DNA content in coding regions is restricted – but it is not restricted in non coding regions. • The codon usage for any particular gene can influence expression.

Codon usage • All organisms have a preferred set of codons. Malaria GUU GUC GUA GUG 0. 41 0. 06 0. 42 0. 11 Trypanosoma GUU GUC GUA GUG 0. 28 0. 19 0. 14 0. 39

Codon Usage • http: //www. kazusa. or. jp/codon/

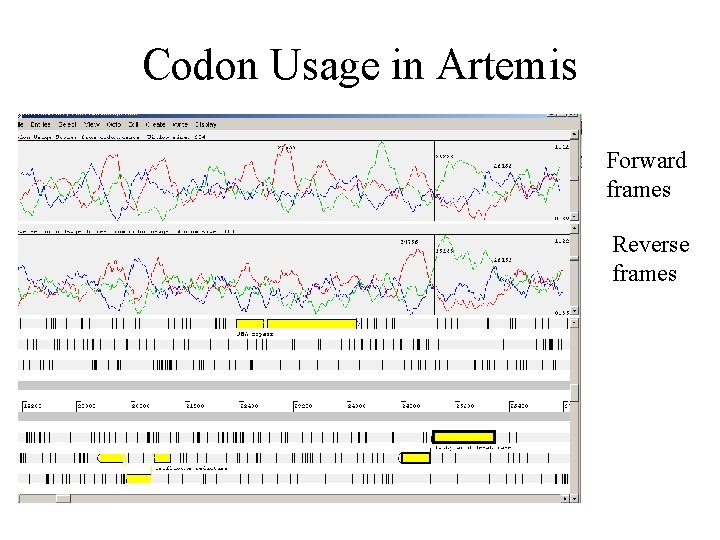
Codon Usage in Artemis Forward frames Reverse frames

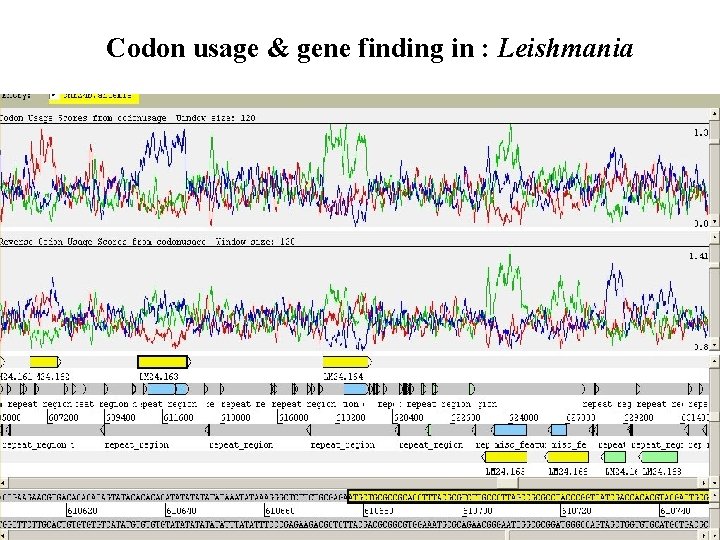
Codon usage & gene finding in : Leishmania

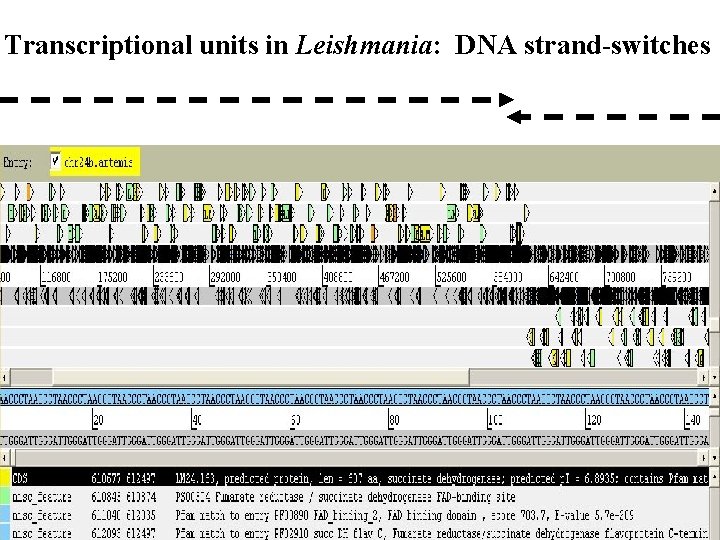
Transcriptional units in Leishmania: DNA strand-switches

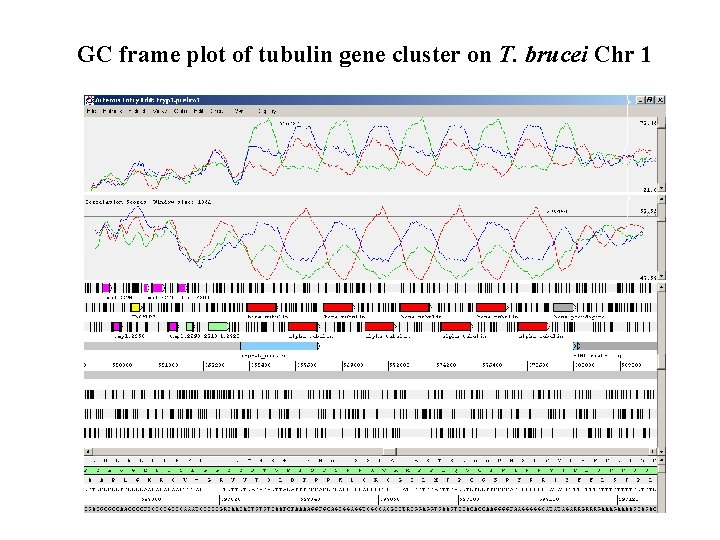
GC frame plot • Plots the third position GC content of each frame of a DNA sequence. • In coding DNA the GC content of the 3 rd base is often higher. • Good prediction of coding in malaria and trypanosomes.

GC frame plot of tubulin gene cluster on T. brucei Chr 1

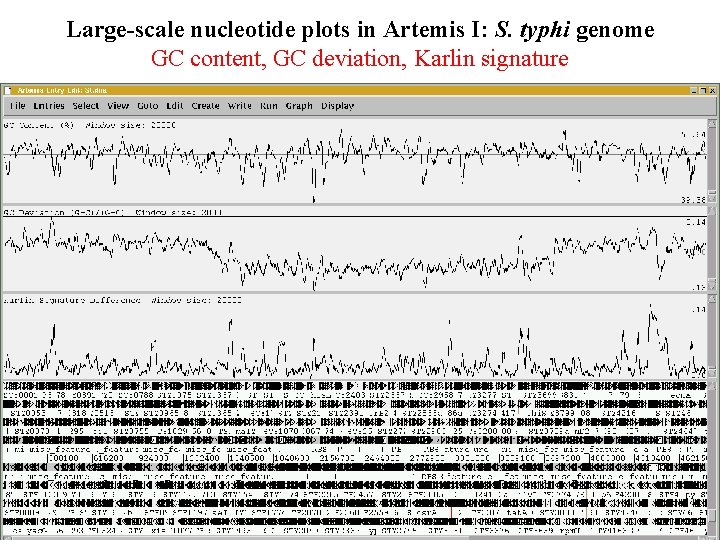
Large-scale nucleotide plots in Artemis I: S. typhi genome GC content, GC deviation, Karlin signature

Homology Data • Coding regions are more conserved than non coding regions due to selective pressure. • Comparing all possible translations against all known proteins will give clues to known genes. • Blastx

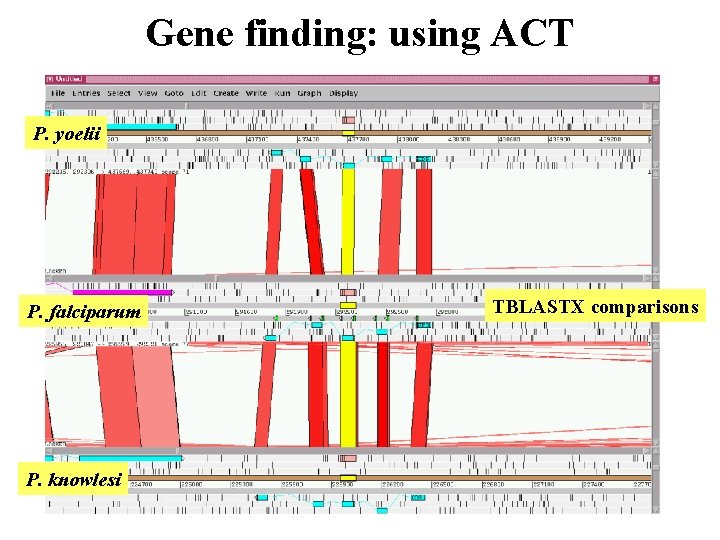
Gene finding: using ACT P. yoelii P. falciparum P. knowlesi TBLASTX comparisons

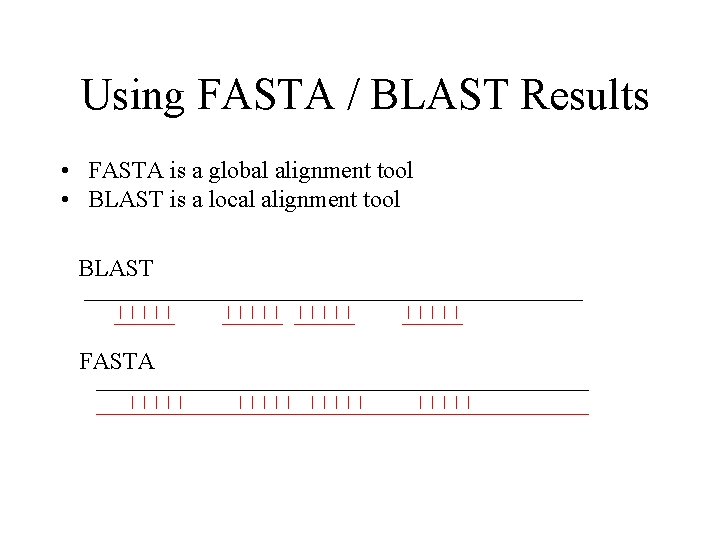
Using FASTA / BLAST Results • FASTA is a global alignment tool • BLAST is a local alignment tool BLAST FASTA

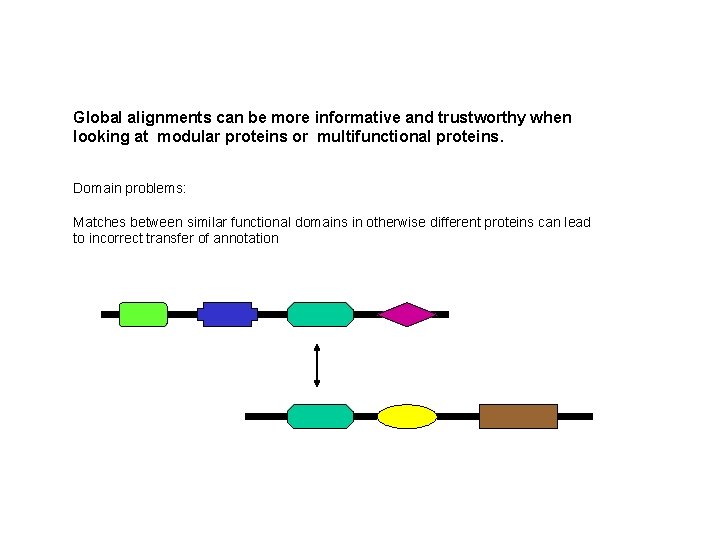
Global alignments can be more informative and trustworthy when looking at modular proteins or multifunctional proteins. Domain problems: Matches between similar functional domains in otherwise different proteins can lead to incorrect transfer of annotation