DNA Sequencing and the Human Genome Project History


















- Slides: 23

DNA Sequencing and the Human Genome Project • History • Technology • Analysis

History • Timeline – 1953 James Watson and Francis Crick discover the double helical structure of DNA ( Nature ). – 1972 Paul Berg and co-workers create the first recombinant DNA molecule ( PNAS ). – 1977 Allan Maxam and Walter Gilbert (pictured) at Harvard University and Frederick Sanger at the U. K. Medical Research Council (MRC) independently develop methods for sequencing DNA ( PNAS , February; PNAS , December).

History (cont’d) – 1980 David Botstein of the Massachusetts Institute of Technology, Ronald Davis of Stanford University, and Mark Skolnick and Ray White of the University of Utah propose a method to map the entire human genome based on RFLPs (American Journal of Human Genetics ). – 1984 Charles Cantor and David Schwartz of Columbia University develop pulsed field electrophoresis MRC scientists decipher the complete DNA sequence of the Epstein-Barr virus, 170 kb – 1985 Kary Mullis and colleagues at Cetus Corp. develop PCR , a technique to replicate vast amounts of DNA – 1986 Sydney Brenner, DOE, Renato Dulbecco, CSH Symposium all publicly advocate a human genome project. Not everyone convinced!

History (cont’d) – 1986 Leroy Hood and Lloyd Smith of the California Institute of Technology and colleagues announce the first automated DNA sequencing machine – 1987 An advisory panel suggests that DOE should spend $1 billion on mapping and sequencing the human genome over the next 7 years-and that DOE should lead the U. S. effort. DOE's Human Genome Initiative begins. David Burke , Maynard Olson , and George Carle of Washington University in St. Louis develop YACs (left) for cloning, increasing insert size 10 -fold Du. Pont scientists develop a system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides (Marv Caruthers, in Biochem, one of the patent holders) Applied Biosystems Inc. puts the first automated sequencing machine, based on Hood's technology, on the market.

History (cont’d) – 1988 NIH establishes the Office of Human Genome Research and snags Watson (pictured) as its head. Watson declares that 3% of the genome budget should be devoted to studies of social and ethical issues. – 1989 Olson , Hood , Botstein , and Cantor outline a new mapping strategy, using STSs. DOE and NIH start a joint committee on the ethical, legal, and social implications of the HGP. NIH office is elevated to the National Center for Human Genome Research (NCHGR), with grant-awarding authority

History (cont’d) – 1990 NIH and DOE publish a 5 -year plan. Goals include a complete genetic map, a physical map with markers every 100 kb, and sequencing of an aggregate of 20 Mb of DNA in model organisms by 2005 NIH and DOE restart the clock, declaring 1 October the official beginning of the HGP. Cost per base ~$0. 75 David Lipman, Eugene Myers (CU CS Department!), and colleagues at the National Center for Biotechnology Information (NCBI) publish the BLAST algorithm for aligning sequences – 1991 NIH biologist J. Craig Venter announces a strategy to find expressed genes, using ESTs (Science ). A fight erupts at a congressional hearing 1 month later, when Venter reveals that NIH is filing patent applications on thousands of these partial genes.

History (cont’d) – 1992 Watson resigns as head of NCHGR Venter leaves NIH to set up The Institute for Genomic Research (TIGR), William Haseltine heads its sister company, Human Genome Sciences, to commercialize TIGR products. Britain's Wellcome Trust enters the HGP with $95 million Mel Simon of Caltech and colleagues develop BACs for cloning U. S. and French teams complete the first physical maps of chromosomes: David Page of the Whitehead Institute and colleagues map the Y chromosome; Daniel Cohen of the Centre d'Etude du Polymorphisme Humain (CEPH) and Généthon and colleagues map chromosome 21 U. S. and French teams complete genetic maps of mouse and human: mouse, average marker spacing 4. 3 c. M , Eric Lander and colleagues at Whitehead; human, average marker spacing 5 c. M, Jean Weissenbach and colleagues at CEPH

History (cont’d) – 1993 Francis Collins of the University of Michigan is named director of NCHGR. NIH and DOE publish a revised plan for 1993 -98. The goals include sequencing 80 Mb of DNA by the end of 1998 and completing the human genome by 2005. Cost per base target $0. 10/base finished. The Wellcome Trust and MRC open the Sanger Centre at Hinxton Hall, south of Cambridge, U. K. Led by John Sulston The Gen. Bank database officially moves from Los Alamos to NCBI, ending NIH's and DOE's tussle over control – 1994 Jeffrey Murray of the University of Iowa, Cohen of Généthon, and colleagues publish a complete genetic linkage map of the human genome, with an average marker spacing of 0. 7 c. M

History (cont’d) – 1995 Venter and Claire Fraser of TIGR and Hamilton Smith of Johns Hopkins publish the first sequence of a free-living organism, Haemophilus influenzae , 1. 8 Mb Patrick Brown of Stanford and colleagues publish first paper using a printed glass microarray of complementary DNA (c. DNA) probe Researchers at Whitehead and Généthon (led by Lander and Thomas Hudson at Whitehead) publish a physical map of the human genome containing 15, 000 markers – 1996 NIH funds six groups to attempt large-scale sequencing of the human genome. Affymetrix makes DNA chips commercially available. An international consortium publicly releases the complete genome sequence of the yeast S. cerevisiae

History (cont’d) – 1997 Fred Blattner , Guy Plunkett , and University of Wisconsin, Madison, colleagues complete the DNA sequence of E. coli , 5 Mb – 1998 NIH announces a new project to find SNPs Phil Green (pictured) and Brent Ewing of Washington University and colleagues publish a program called phred for automatically interpreting sequencer data. Both phred and its sister program phrap (used for assembling sequences) had been in wide use since 1995. PE Biosystems Inc. introduces the PE Prism 3700 capillary sequencing machine Venter announces a new company named Celera and declares that it will sequence the human genome within 3 years for $300 million In response, the Wellcome Trust doubles its support for the HGP to $330 million, taking on responsibility for one-third of the sequencing

History (cont’d) – 1998 NIH and DOE throw HGP into overdrive with a new goal of creating a "working draft" of the human genome by 2001, and they move the completion date for the finished draft from 2005 to 2003. Sulston of the Sanger Centre and Robert Waterston of Washington University and colleagues complete the genomic sequence of C. elegans (100 mb). – 1999 NIH again moves up the completion date for the rough draft, to spring 2000 Ten companies and the Wellcome Trust launch the SNP consortium, with plans to publicly release data quarterly NIH launches a project to sequence the mouse genome, devoting $130 million over 3 years British, Japanese, and U. S. researchers complete the first sequence of a human chromosome, number 22

History (cont’d) – 2000 Celera and academic collaborators sequence the 180 -Mb genome of the fruit fly Drosophila melanogaster Because of disagreement over a data-release policy, plans for HGP and Celera to collaborate disintegrate amid considerable sniping. HGP consortium led by German and Japanese researchers publishes the complete sequence of chromosome 21 At a White House ceremony, HGP and Celera jointly announce working drafts of the human genome sequence, declare their feud at an end, and promise simultaneous publication An international consortium completes the sequencing of the first plant, Arabidopsis thaliana 125 Mb HGP and Celera's plans for joint publication in Science collapse; HGP sends its paper to Nature

TADA!! • 2001 The HGP consortium publishes its working draft in Nature (15 February), and Celera publishes its draft in Science (16 February).

Technology

Aspects of Sequencing Genomes • Sequencing method • Cloned DNA • Clone/Sequence Assembly

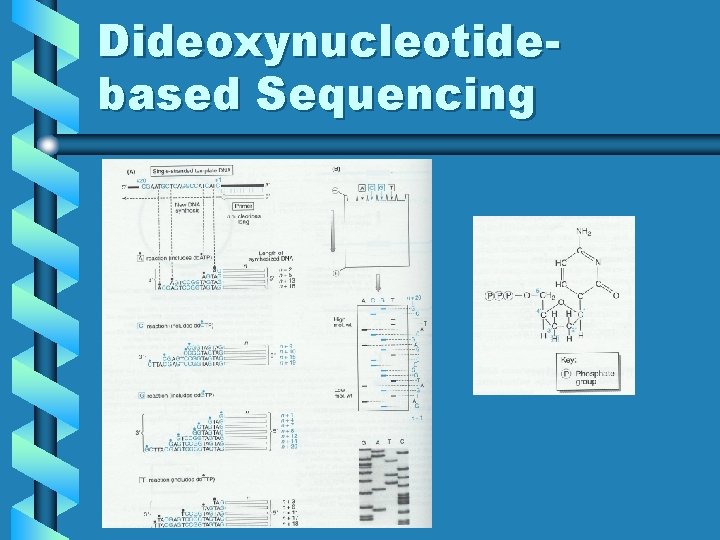
Sequencing Methods • “Sanger” chain termination method; >90% of all sequencing – Relies on ability of DNA polymerase to incorporate nucleotide analogs while synthesizing template driven DNA

Dideoxynucleotidebased Sequencing

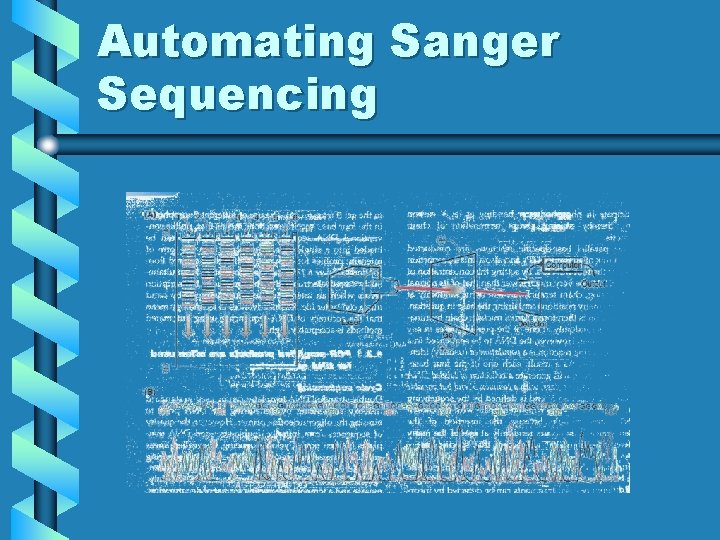
Automating Sanger Sequencing

Other Automated Methods • Hybridization method – Hybridize to oligos on a chip • Affymetrix can do 30 K resequence • Limited by number of features and hybridization specificity • Single molecule methods – Pore-base - threads DNA through molecular pore in membrane - bases determined by changes in conductance – Mass spec - best for small molecules now like SNPs

Most-used hardware • ABI 377 - gel based - 96 lanes a pop - read length ~500 bp - run time ~4 -16 h => ~40, 000 bases/run X 3 runs/day = 120, 000 • ABI 3700 - Capillary based - 48 capillaries - read length ~500 bp run time ~40 minutes => 950,

Whaddaya determine the sequence of? Given chemistry and hardware to read ~500 bp in a row what gets sequenced?

Clones • Large insert clones – YACs (Yeast Artificial Chromosomes • • • Useful for mapping ~1 mb inserts Unstable during construction and propagation Not useful for sequencing – BACs (Bacterial Artificial Chromosomes) • ~150 kb insert • Extremely stable and easy to propagate • Gold standard for sequencing targets and chromosomescale maps – Cosmids • ~50 kb insert • Extremely stable and easy to propagate • Useful for sequencing but too small for chromosome maps

Sequence-ready clones • Plasmids – – – 1 -10 kb insert capacity High copy number Easy to sequence bi-directionally Automated clone picking/DNA isolation possible Examples: p. UC 18, p. BR 322 • Single-stranded Bacteriophage – 1 -5 kb insert capacity – Grows at high copy as plasmid and is shed into medium as single stranded DNA phage – Easy to isolate, pick, sequence – Easy to automate – M 13 is used almost exclusively
 Human genome project
Human genome project History of sequencing
History of sequencing Genome-to-genome distance calculator
Genome-to-genome distance calculator Per partes
Per partes Shotgun sequencing
Shotgun sequencing Shotgun sequencing
Shotgun sequencing Genome sequencing
Genome sequencing Genome sequencing
Genome sequencing Difference between bac and yac
Difference between bac and yac Human genome project source code
Human genome project source code Human genome project
Human genome project Human genome project
Human genome project Importance of dna sequencing
Importance of dna sequencing 3rd generation dna sequencing
3rd generation dna sequencing Helioscope sequencing
Helioscope sequencing Dna sequencing gel
Dna sequencing gel Dna sequencing
Dna sequencing Dna sequencing applications
Dna sequencing applications Human genome consists of
Human genome consists of Human genome size
Human genome size Human genome size
Human genome size Strs and vntrs
Strs and vntrs Human genome structure
Human genome structure Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key