Mash fast genome and metagenome distance estimation using



























- Slides: 57

Mash fast genome and metagenome distance estimation using Min. Hash Brian D. Ondov, Todd J. Treangen, Páll Melsted, Adam B. Mallonee, Nicholas H. Bergman, Sergey Koren and Adam M. Phillippy, 2016 November 2018 Deep Sequencing Seminar Dvir Ginzburg, Yuri Klayman

Presentation overview • Mash introduction • Min. Hash reminder • Mash distance and p value • Key points, parameters tuning (k, s sizes) and Conceptual issues • Experiments and results • Improvement suggestions • Conclusions * Terminology and derivations appear in the end of the presentation 2

Mash overview • Novel bioinformatics approach for sequences comparison. o Turns large sequences and sequence sets to small, representative sketches. o Provides reliable distance estimation, that can be used for useful features aso Provides clustering and search capabilities for massive sequences collections. o Estimation error depends only on the size of the sketch, and independent of the genome size. • Based on Min. Hash o Reminder in the next slides o Uses “Mash distance” instead of the Jaccard index to address specific domain problems. 3

Mash capabilities • Supports Parallelism • Assembled or unassembled (alignment-free) data • Computationally efficient, addressing tasks unfeasible before • Real-time • Online • Arbitrary alphabets • Free & Open source • Helps with privacy concerns 4

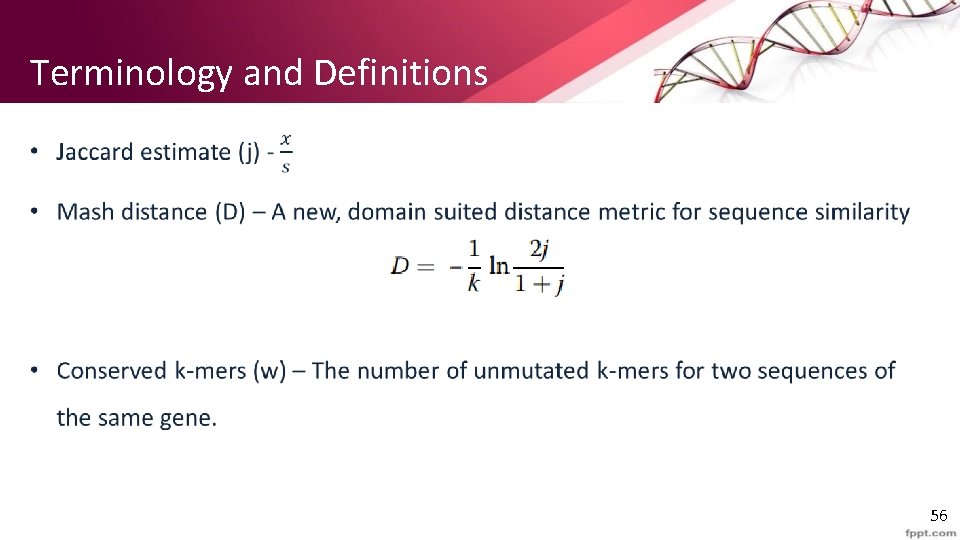
Terminology and Definitions • 5

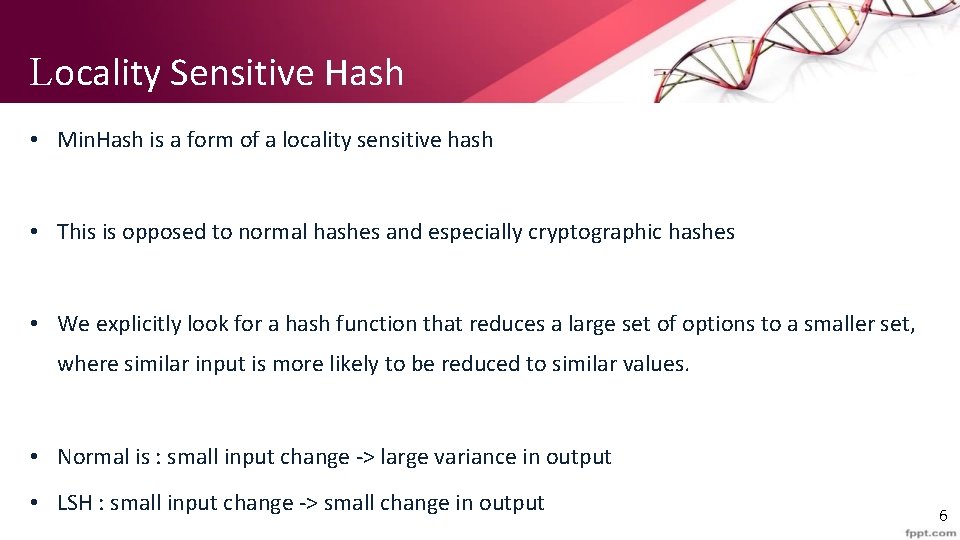
Locality Sensitive Hash • Min. Hash is a form of a locality sensitive hash • This is opposed to normal hashes and especially cryptographic hashes • We explicitly look for a hash function that reduces a large set of options to a smaller set, where similar input is more likely to be reduced to similar values. • Normal is : small input change -> large variance in output • LSH : small input change -> small change in output 6

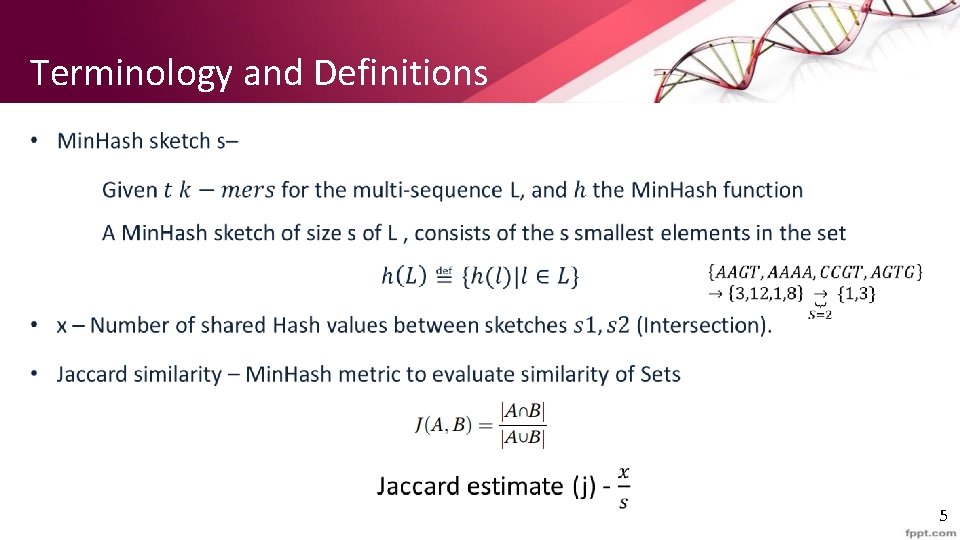
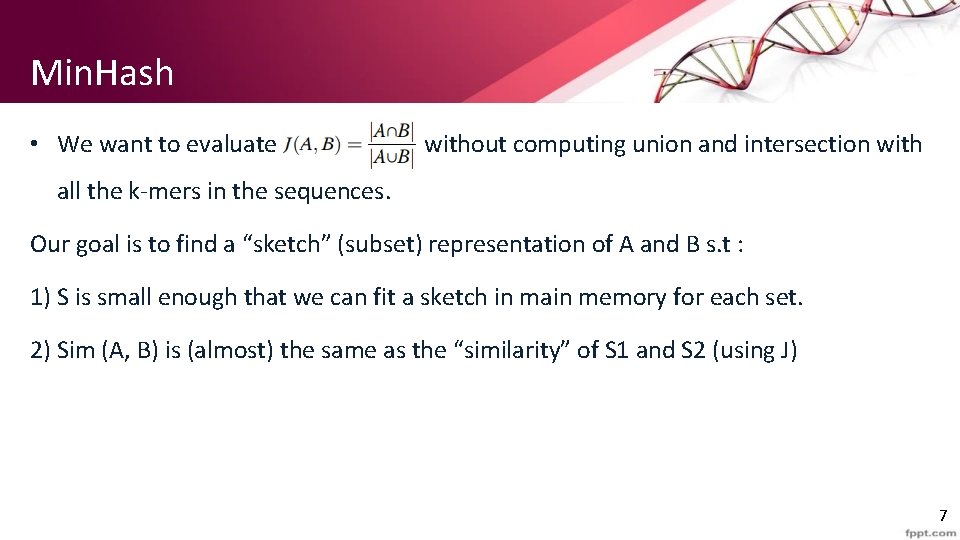
Min. Hash • We want to evaluate without computing union and intersection with all the k-mers in the sequences. Our goal is to find a “sketch” (subset) representation of A and B s. t : 1) S is small enough that we can fit a sketch in main memory for each set. 2) Sim (A, B) is (almost) the same as the “similarity” of S 1 and S 2 (using J) 7

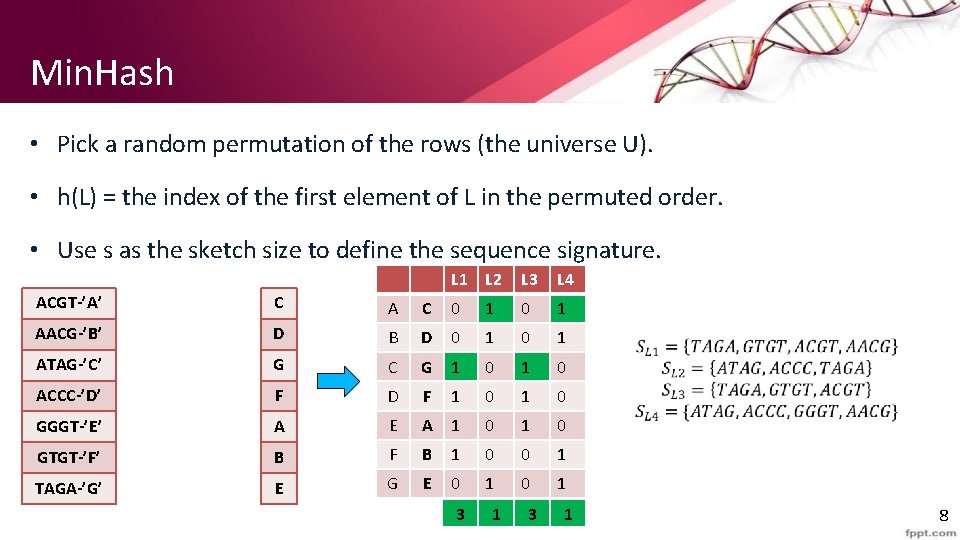
Min. Hash • Pick a random permutation of the rows (the universe U). • h(L) = the index of the first element of L in the permuted order. • Use s as the sketch size to define the sequence signature. L 1 L 2 L 3 L 4 ACGT-’A’ C A C 0 1 AACG-’B’ D B D 0 1 ATAG-’C’ G C G 1 0 ACCC-’D’ F D F 1 0 GGGT-’E’ A E A 1 0 GTGT-’F’ B F B 1 0 0 1 TAGA-’G’ E G E 0 1 3 1 8

Min. Hash • Pr(h(L 1) = h(L 2)) = Sim(L 1, L 2) o The first row where one of the two sets has value 1 belongs to the union. • Recall that union contains rows with at least one 1. o We have equality if both sets have value 1, and this row belongs to the intersection • With multiple signatures we get a good approximation • Input matrix L 1 L 2 L 3 L 4 A 1 0 B 1 0 0 1 C 0 1 D 0 1 E 0 1 F 1 0 G 1 0 Signature matrix ≈ L 1 L 2 L 3 L 4 h 1 1 2 h 2 2 1 3 1 h 3 3 1 Actual Sig (L 1, L 2) 0 0 (L 1, L 3) 3/5 2/3 (L 1, L 4) 1/7 0 (L 2, L 3) 0 0 (L 2, L 4) 3/4 1 (L 3, L 4) 0 0 9

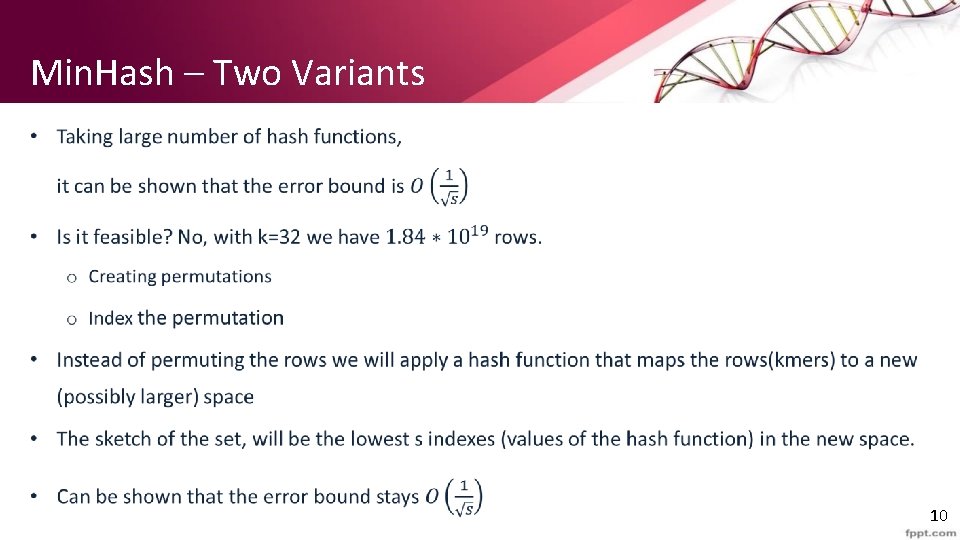
Min. Hash – Two Variants • 10

Mash stages • 11

Mash Distance Jaccard pitfall 12

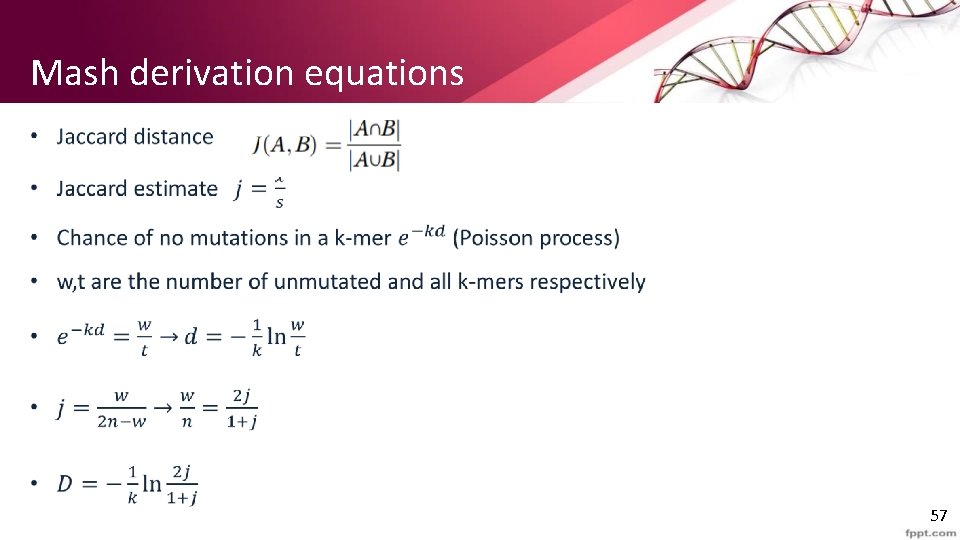
Gene mutations • mutation is the permanent alteration of the nucleotide sequence of the genome. • When comparing sequences of the same genome for “type” matching (i. e. detect virus types etc. ) we seek a metric that would be invariant to mutations. • Jaccard index is sensitive to genome size and simultaneously captures both point mutations and gene content differences • Mash distance D seeks to directly estimate the mutation rate under a simple Poisson process to address this problems. 13

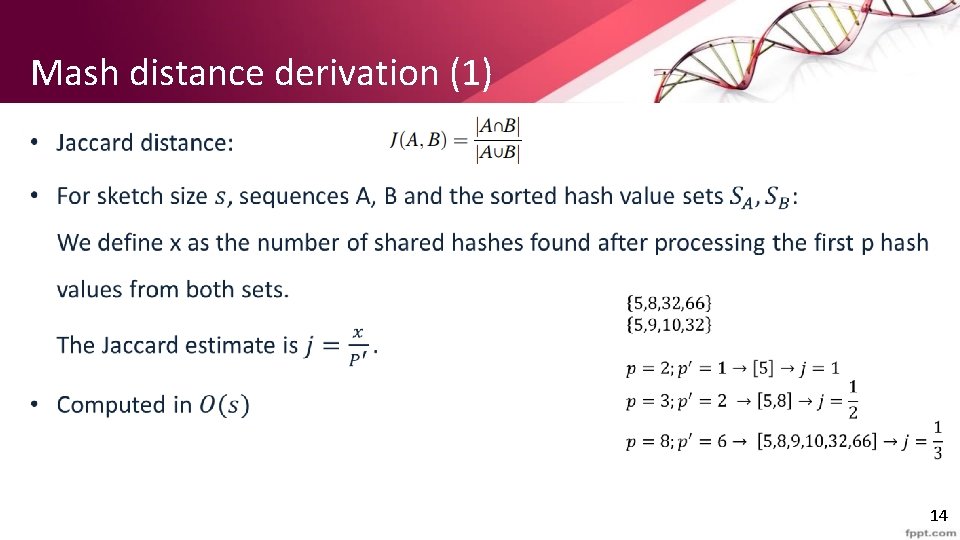
Mash distance derivation (1) • 14

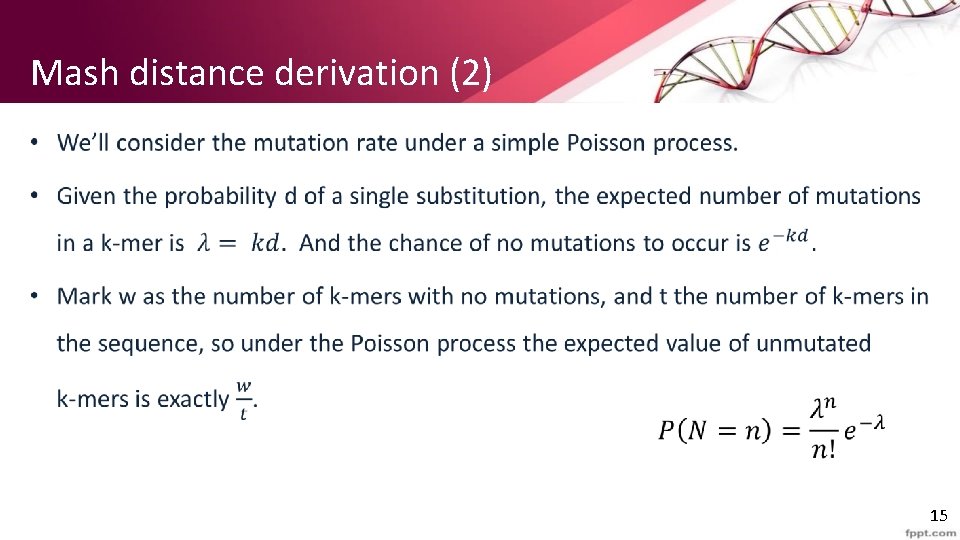
Mash distance derivation (2) • 15

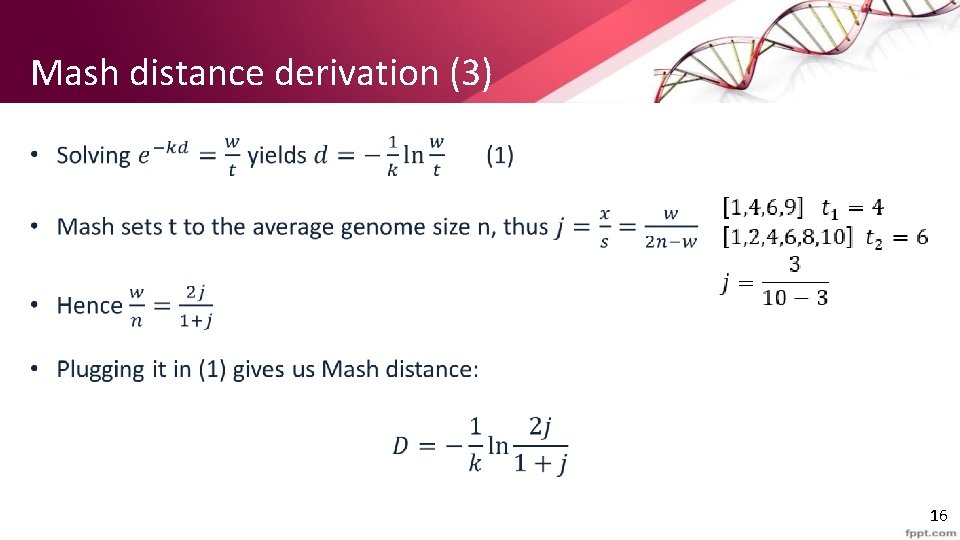
Mash distance derivation (3) • 16

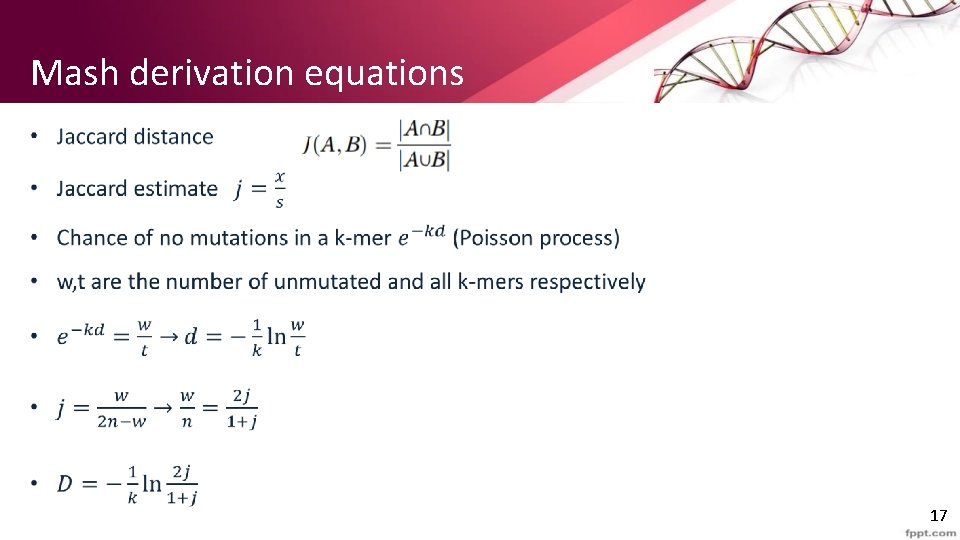
Mash derivation equations • 17

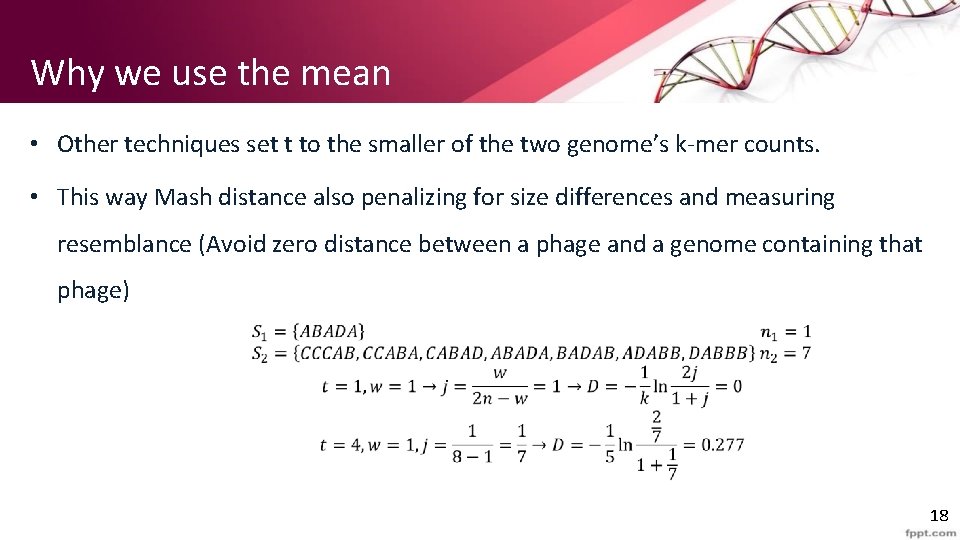
Why we use the mean • Other techniques set t to the smaller of the two genome’s k-mer counts. • This way Mash distance also penalizing for size differences and measuring resemblance (Avoid zero distance between a phage and a genome containing that phage) 18

P-Value We need to know the expected number of x matches or more between two random genomes, to understand if the Jaccard Index and therefore the estimate and the mash distance we calculated is significant 19

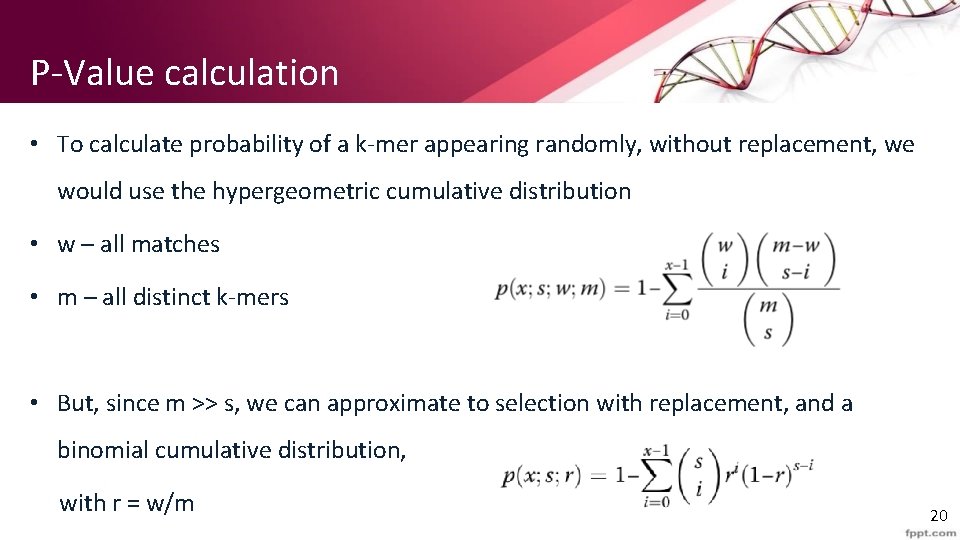
P-Value calculation • To calculate probability of a k-mer appearing randomly, without replacement, we would use the hypergeometric cumulative distribution • w – all matches • m – all distinct k-mers • But, since m >> s, we can approximate to selection with replacement, and a binomial cumulative distribution, with r = w/m 20

How do we get r ? • 21

How do we get m ? • 22

P-Value Significance • The P value tells us the mash distance we calculated has significance, but what does it mean ? • We calculated a significant number, but does that number mean two things are close or far ? How close ? Is it transitive ? 23

ANI - Average Nucleotide Identity • We know the mash distance is a useful measure of global sequence similarity because it correlates with ANI. • The most wildly used genomic relatedness index. [1] • A variation on alignment and scoring, actually simulates “in silico” the previously most wildly used technique, which was in use for 50 years. 24

ANI • Due to the high cost of computing ANI via whole-genome alignment, a subset of 500 Escherichia genomes was selected for comparison • The correlation for ANI of values 90– 100 % is very good • A side note, an ANI of 95% is not too useful for distinguishing between eukaryotes species, but values above 99% can be used. 25

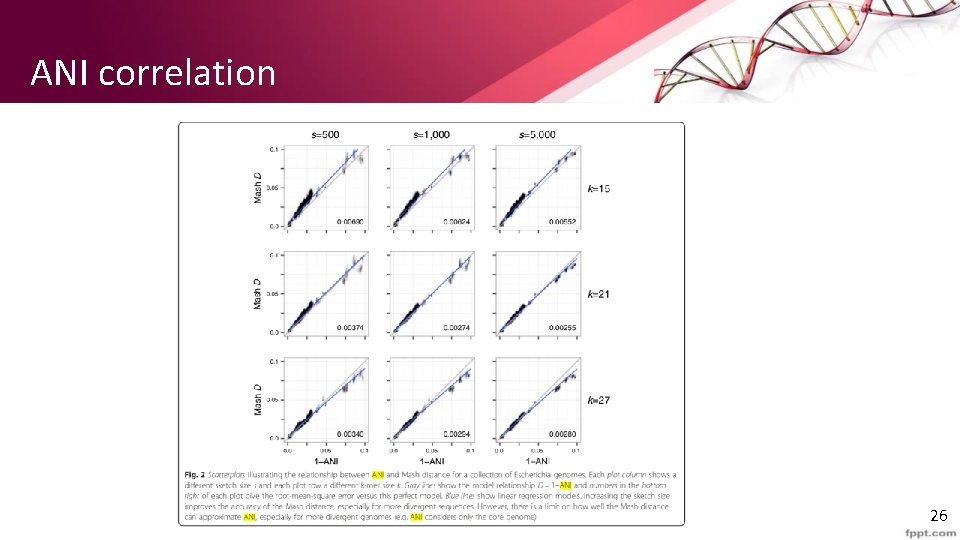
ANI correlation 26

Estimate of Jaccard Index • Note this is still the straight forward Jaccard Index • Each line is a different value of s, increasing from light gray to black : s=100 (light gray), s=1, 000, s=10, 000, and s=100, 000 (black) 27

Impact of S, sketch size • Increasing sketch size helps with divergent sequences. • Increasing s, has negligible impact on time it takes to sketch. Computed in O(n*log(s)), where n is the genome size. • However, it has linear impact on how long it takes to calculate distance, since we store the sketches sorted. • Has serious impact on storage size 28

Jaccard & Mash with different K’s If we look at how we calculate mash distance from the JE, we can see we divide by the k-mer length k = 15, 21, 27 ( top to bottom, red to blue ) Mathematically, the relation is obvious, but what it represents is two things : a. Longer strings, are less likely to match. This is true for the Jaccard Index without Mash. b. Longer strings are more likely to have mutated, and this is something mash D takes into account, while JI doesn’t The paper doesn’t try to distinguish the two. Also, choice of s is unknown. 29

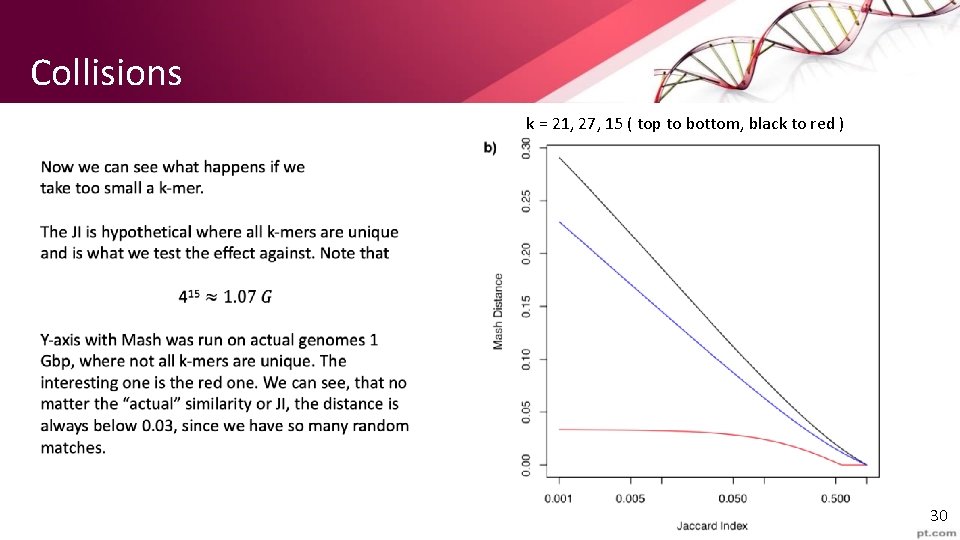
Collisions k = 21, 27, 15 ( top to bottom, black to red ) 30

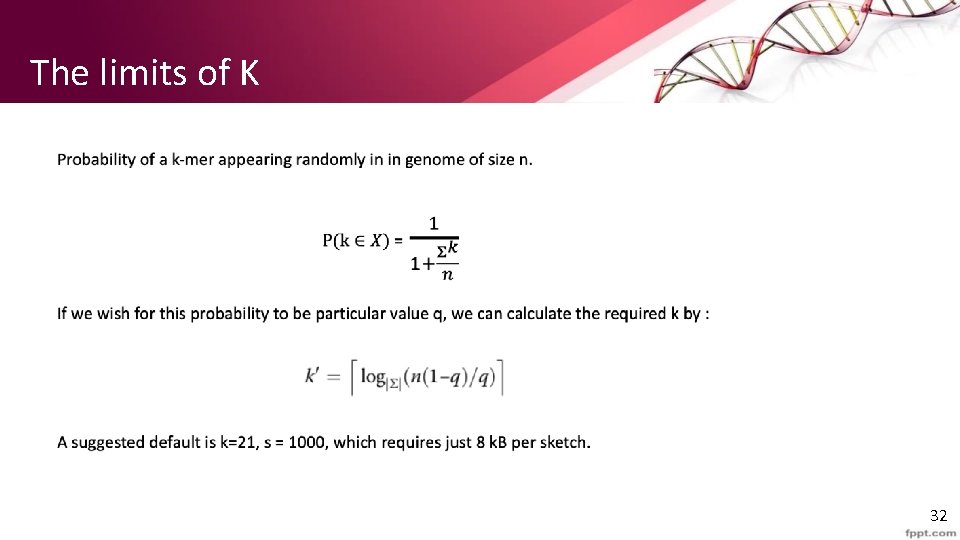
The importance of a just-right K • Tradeoff between sensitivity and specificity. • With a large k, we will miss smaller k matches. In some areas, a sequence match of length 15 is important. • With smaller k, the larger the sequence, the higher the chance of the sequence appearing randomly. • The paper recommends a method that helps to pick a k large enough to avoid random collision, but not larger than needed, to not miss interesting matches. 31

The limits of K 32

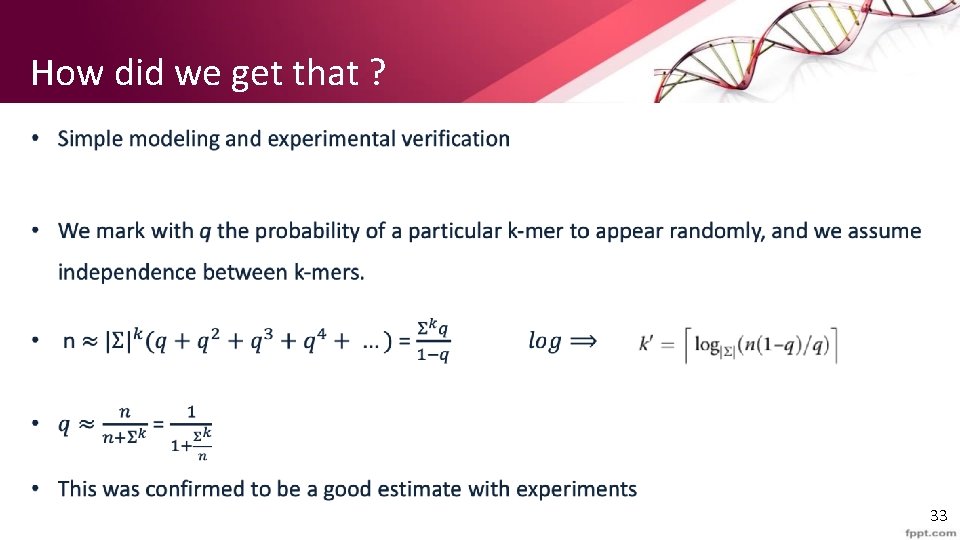
How did we get that ? • 33

Finer points • For DNA sequences, Mash uses canonical k-mers. Meaning we only pick one k-mer from complimentary DNA sequences. • How to handle read errors ? In the next slide. 34

Two Step Extension Ignore low-occurrence k-mers when constructing the sketch, two implementations: o A hash map with a counter, so that only a k-mer that appeared more than c times, is eligible o To avoid large hash-maps, because many k-mers appear less than c times, construct a Bloom Filter. We won’t explore this, because in practice, the authors say the exact method outperforms both extensions in accuracy and memory usage. 35

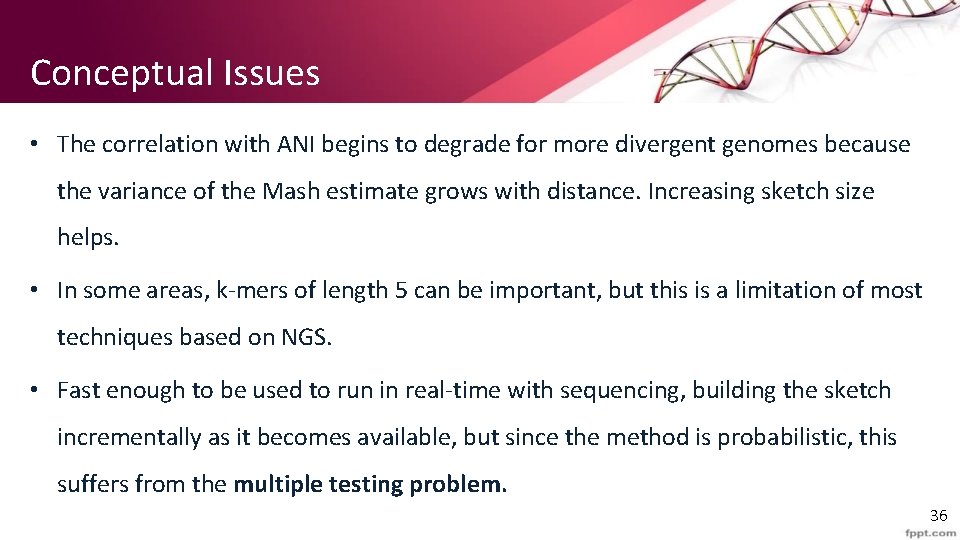
Conceptual Issues • The correlation with ANI begins to degrade for more divergent genomes because the variance of the Mash estimate grows with distance. Increasing sketch size helps. • In some areas, k-mers of length 5 can be important, but this is a limitation of most techniques based on NGS. • Fast enough to be used to run in real-time with sequencing, building the sketch incrementally as it becomes available, but since the method is probabilistic, this suffers from the multiple testing problem. 36

Experiments Results 37

Clustering NCBI Ref. Seq • The Reference Sequence • comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, transcripts, and proteins. • More than 54, 118 organisms, 618 Gbp of genomic sequence 38

Clustering NCBI Ref. Seq • Whole genome sequencing becomes routine • It became impractical to manually assign taxonomic labels for all genomes. • Automated methods are crucial for constructing groups of related genomes • Infeasible with alignment based approaches. • Min. ION - ONT 39

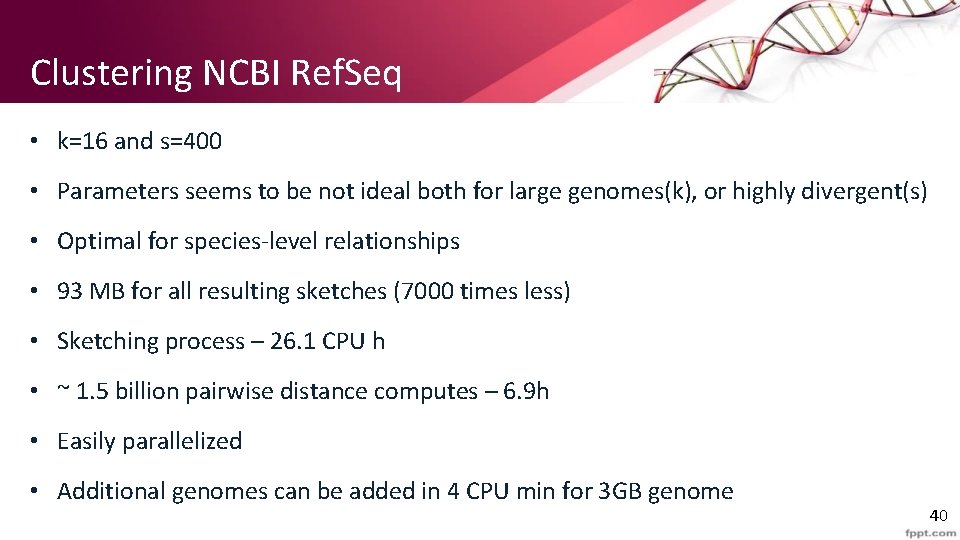
Clustering NCBI Ref. Seq • k=16 and s=400 • Parameters seems to be not ideal both for large genomes(k), or highly divergent(s) • Optimal for species-level relationships • 93 MB for all resulting sketches (7000 times less) • Sketching process – 26. 1 CPU h • ~ 1. 5 billion pairwise distance computes – 6. 9 h • Easily parallelized • Additional genomes can be added in 4 CPU min for 3 GB genome 40

Clustering NCBI Ref. Seq • Unfeasible with prior methods, By using ANI metric, and choosing 500 genomes a comparison has been made. RMSE of 0. 00274 For all pairs with ANI higher than 90%. 41

Hierarchical clustering for mutation rate • 17 Ref. Seq primate genomes were computed in just 2. 5 CPU h (11 min wall clock on 17 cores) with default parameters (s = 1000 and k = 21). • Compared with the UCSC alignment based model. • The trees are topologically consistent for everything except the Homo/Pan split, for which the Mash topology is more similar to past phylogenetic studies 42

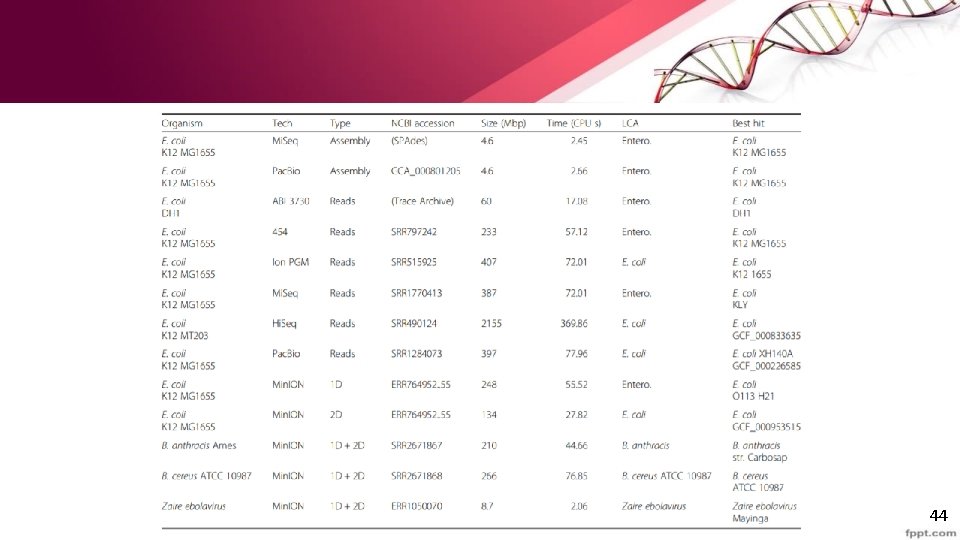
Real-time genome identification from assemblies or reads • Mash is able to rapidly identify genomes from both assemblies and raw sequencing reads. • Generate sketches , compute Mash distances for multiple E coli datasets • compare against the Ref. Seq sketch. • For the assembled genomes the correct strain was identified in a few seconds • For the unassembled genome, even with reads as the E. coli 1 D min. ION consisting of 40% sequencing error rate the correct species were identified 43

44

Clustering massive metagenomic datasets • Metagenomic comparison tool o Fraction of the time previously required. • Current Metagenomic comparison tools as the DSM compute the exact J index using all k-mers. • As we previously see, the J distance (1 -J) rapidly drops with mutation rate, where Mash isn’t. 45

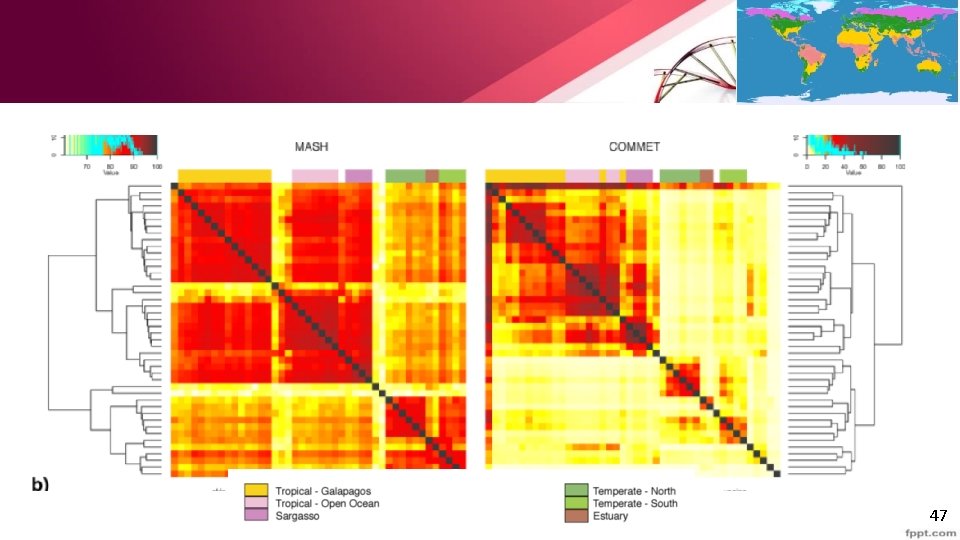
Clustering massive metagenomic datasets • Global Ocen Survey(GOS) data • Mash is tenfold faster, and correlates better to the original GOS study. • Incremetal scalability o Sketching happens only once per sample, and pairwise distances in parallel and instantauous. • Other techniques take 1 h to add a new GOS sample to the cluster • Mash takes less than 1 m. 46

47

Implementation details & Summary 48

Actively developed 49

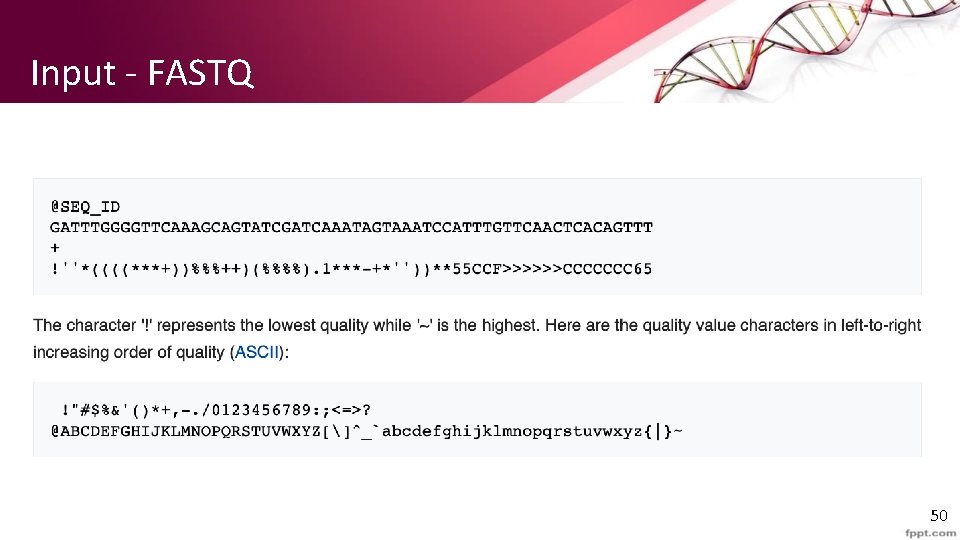
Input - FASTQ 50

Usage Reference-ID, Query-ID, Mash-distance, P-value, Matching hashes 51

Further improvements • Since mash works on FASTQ, and FASTQ comes with quality indicators per nucleotide, we could try to pick k-mers that are of higher quality • The program should probably come with some features to help with the multiple testing problem, like : o Delaying querying and buffering new reads as they are being read, until a reasonable number has been found. o Stability in replacing the tested sketch, so that only significant changes in score replace a value in the sketch. o Warning when the amount of tests performed is high with regards to the p-value • Support for containment testing, which Min. Hash is capable of with some changes. 52

Conclusions • A highly efficient and scalable method of estimating distance between sequences, that is mostly independent of sequence size • Open source and under active development and use 53

54

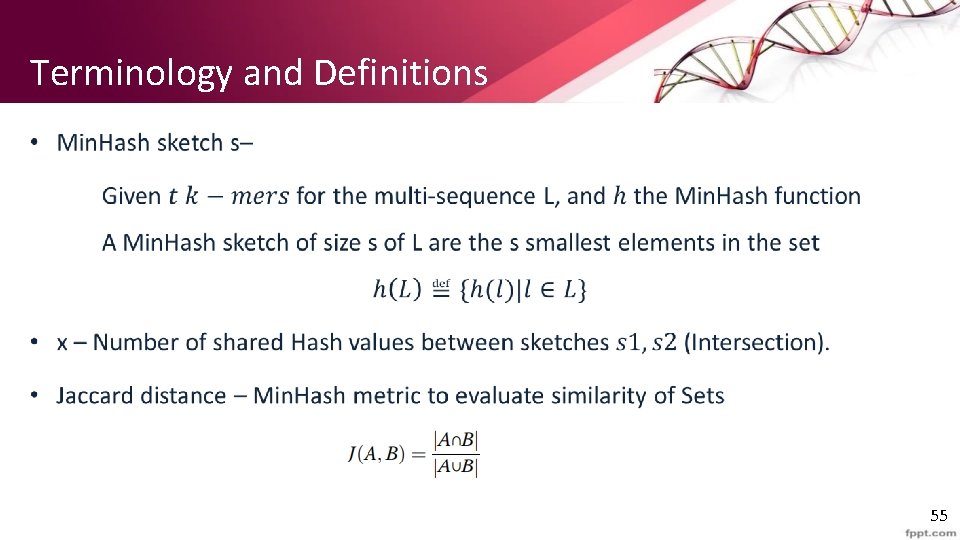
Terminology and Definitions • 55

Terminology and Definitions • 56

Mash derivation equations • 57