Genomics Large Genome Sequencing sequencing per partes separated


- Slides: 43

Genomics

Large Genome Sequencing - sequencing per partes (separated chromosomes) - sequencing of non-methylated DNA (= transcriptionally active) - sequencing of ESTs

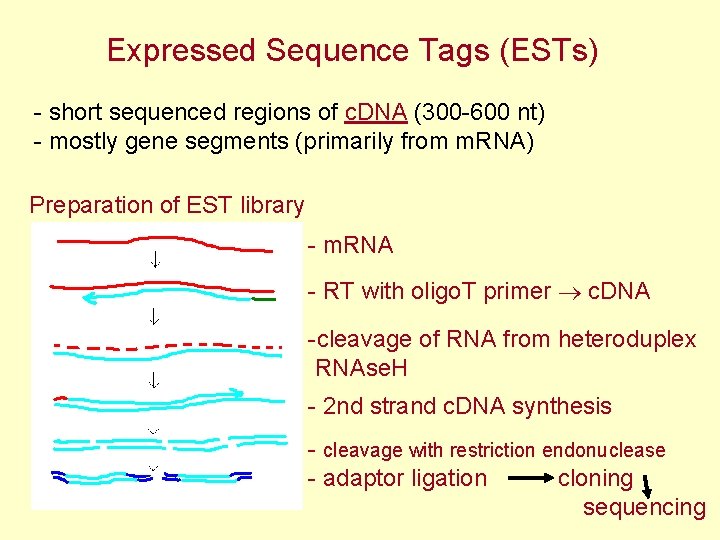
Expressed Sequence Tags (ESTs) - short sequenced regions of c. DNA (300 -600 nt) - mostly gene segments (primarily from m. RNA) Preparation of EST library - m. RNA - RT with oligo. T primer c. DNA -cleavage of RNA from heteroduplex RNAse. H - 2 nd strand c. DNA synthesis - cleavage with restriction endonuclease - adaptor ligation cloning sequencing

Expressed Sequence Tags (ESTs) - short sequenced regions of c. DNA (300 -600 nt) - mostly gene segments (primarily from m. RNA) - alternative sourse of coding sequences for large genomes (rapid and inexpensive) Weak points: - highly redundant, but incomplete as well (!) - problems: various transcript levels - gene expression regulated spatially and temporally, developmentally, environmentally - regulatory sequences not represented (promotors, introns, . . . )

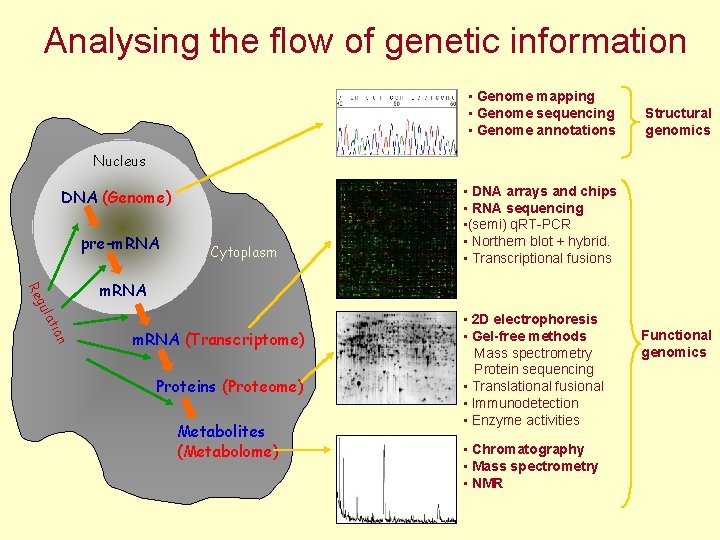
Analysing the flow of genetic information • Genome mapping • Genome sequencing • Genome annotations Structural genomics Nucleus DNA (Genome) pre-m. RNA Cytoplasm • DNA arrays and chips • RNA sequencing • (semi) q. RT-PCR • Northern blot + hybrid. • Transcriptional fusions on ati gul Re m. RNA (Transcriptome) Proteins (Proteome) Metabolites (Metabolome) • 2 D electrophoresis • Gel-free methods Mass spectrometry Protein sequencing • Translational fusional • Immunodetection • Enzyme activities • Chromatography • Mass spectrometry • NMR Functional genomics

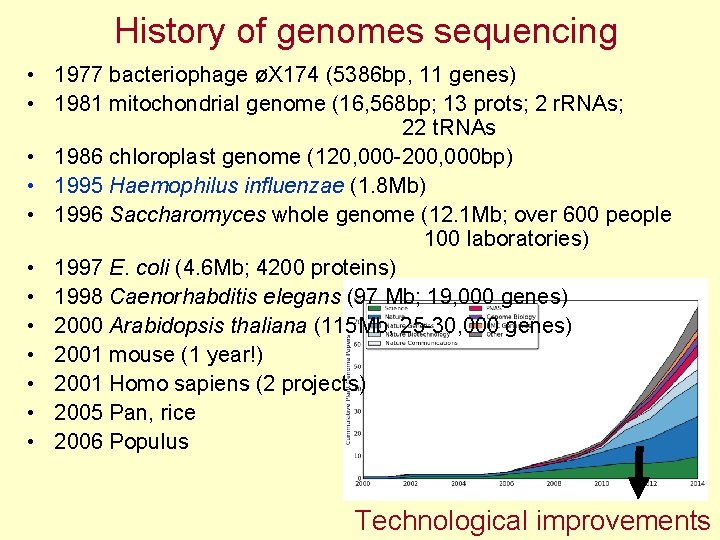
History of genomes sequencing • 1977 bacteriophage øX 174 (5386 bp, 11 genes) • 1981 mitochondrial genome (16, 568 bp; 13 prots; 2 r. RNAs; 22 t. RNAs • 1986 chloroplast genome (120, 000 -200, 000 bp) • 1995 Haemophilus influenzae (1. 8 Mb) • 1996 Saccharomyces whole genome (12. 1 Mb; over 600 people 100 laboratories) • 1997 E. coli (4. 6 Mb; 4200 proteins) • 1998 Caenorhabditis elegans (97 Mb; 19, 000 genes) • 2000 Arabidopsis thaliana (115 Mb, 25 -30, 000 genes) • 2001 mouse (1 year!) • 2001 Homo sapiens (2 projects) • 2005 Pan, rice • 2006 Populus Technological improvements

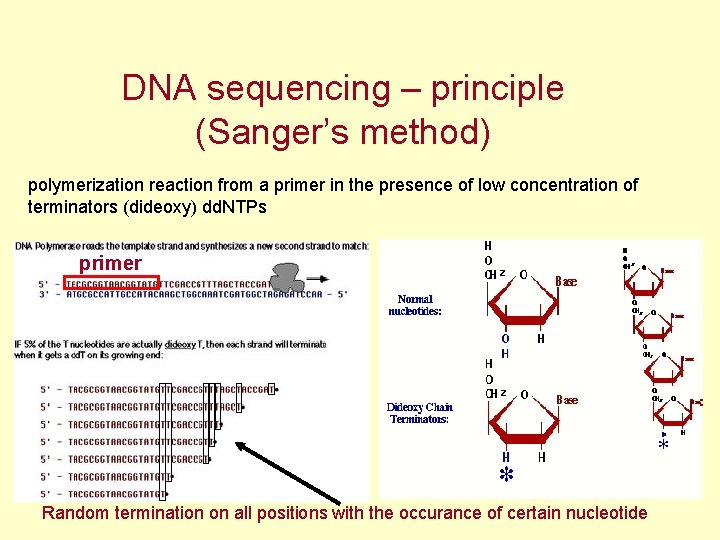
DNA sequencing – principle (Sanger’s method) polymerization reaction from a primer in the presence of low concentration of terminators (dideoxy) dd. NTPs primer Random termination on all positions with the occurance of certain nucleotide

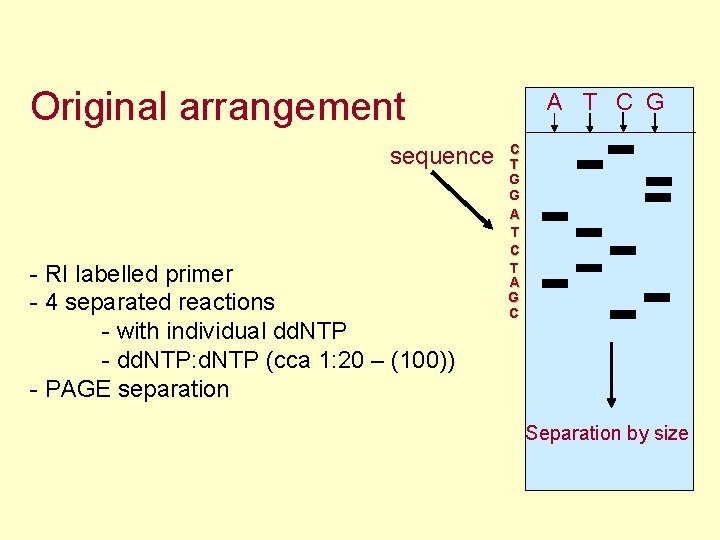
Original arrangement sequence - RI labelled primer - 4 separated reactions - with individual dd. NTP - dd. NTP: d. NTP (cca 1: 20 – (100)) - PAGE separation A T C G C T G G A T C T A G C Separation by size

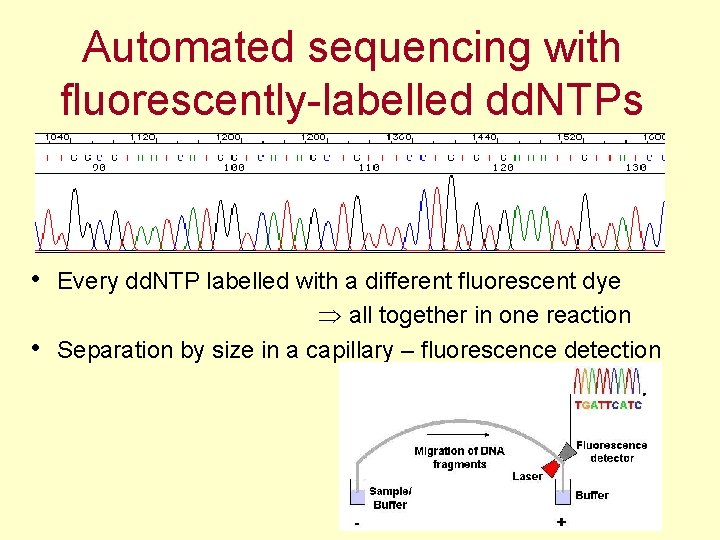
Automated sequencing with fluorescently-labelled dd. NTPs • Every dd. NTP labelled with a different fluorescent dye • all together in one reaction Separation by size in a capillary – fluorescence detection

Next generation sequencing (NGS) - faster and cheaper!!! - parallel sequencing of high numbers of sequences! - no handling with individual sequences! Basic principles: Template - a clone of identical DNA molecules (PCR) - a single DNA molecule Reaction - synthesis of a complementary strand - ligation of oligonucleotides (on the template) - DNA degradation by exonuclease - scanning of ss. DNA strand Detection - optical: substrate incorporation (fluorescent labelled substrates, linked luminiscent reaction) - electronic: products of degradation, NT in ss. DNA

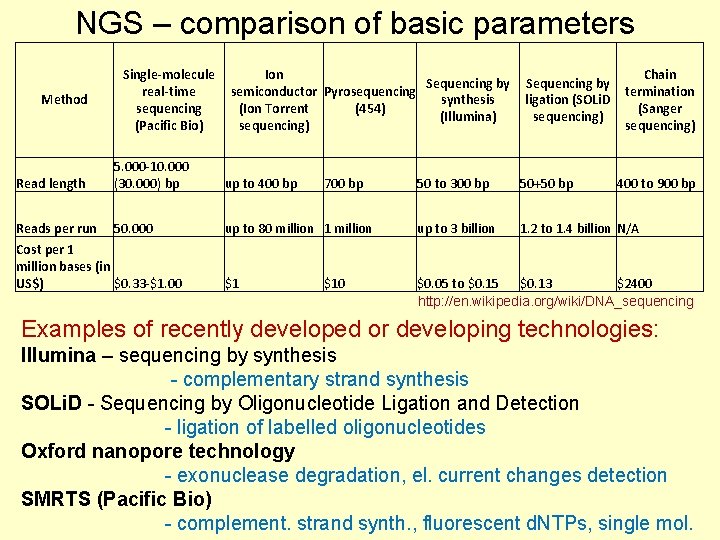
NGS – comparison of basic parameters Method Single-molecule real-time sequencing (Pacific Bio) Ion Sequencing by semiconductor Pyrosequencing synthesis (Ion Torrent (454) (Illumina) sequencing) Read length 5. 000 -10. 000 (30. 000) bp up to 400 bp Reads per run 50. 000 Cost per 1 million bases (in US$) $0. 33 -$1. 00 700 bp Sequencing by ligation (SOLi. D sequencing) Chain termination (Sanger sequencing) 50 to 300 bp 50+50 bp up to 80 million 1 million up to 3 billion 1. 2 to 1. 4 billion N/A $1 $0. 05 to $0. 15 $0. 13 $2400 http: //en. wikipedia. org/wiki/DNA_sequencing $10 400 to 900 bp Examples of recently developed or developing technologies: Illumina – sequencing by synthesis - complementary strand synthesis SOLi. D - Sequencing by Oligonucleotide Ligation and Detection - ligation of labelled oligonucleotides Oxford nanopore technology - exonuclease degradation, el. current changes detection SMRTS (Pacific Bio) - complement. strand synth. , fluorescent d. NTPs, single mol.

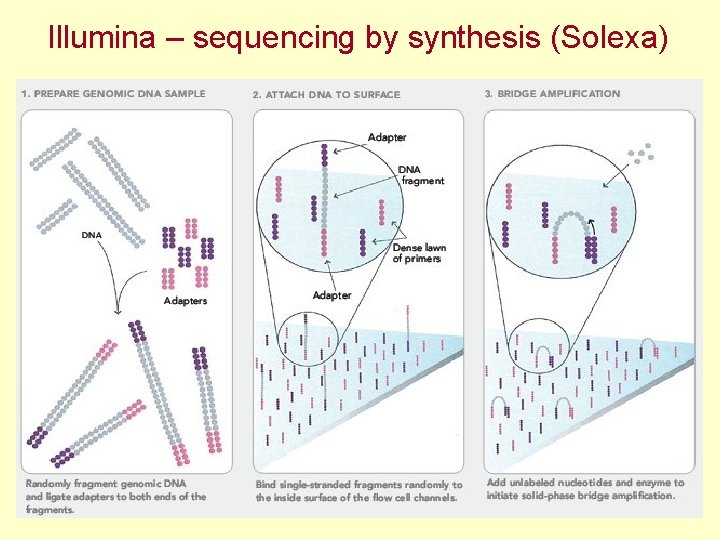
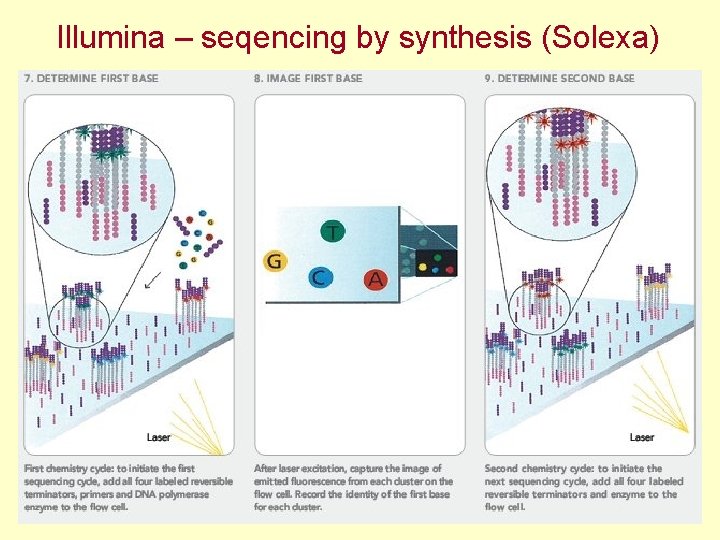
Illumina – sequencing by synthesis (Solexa)

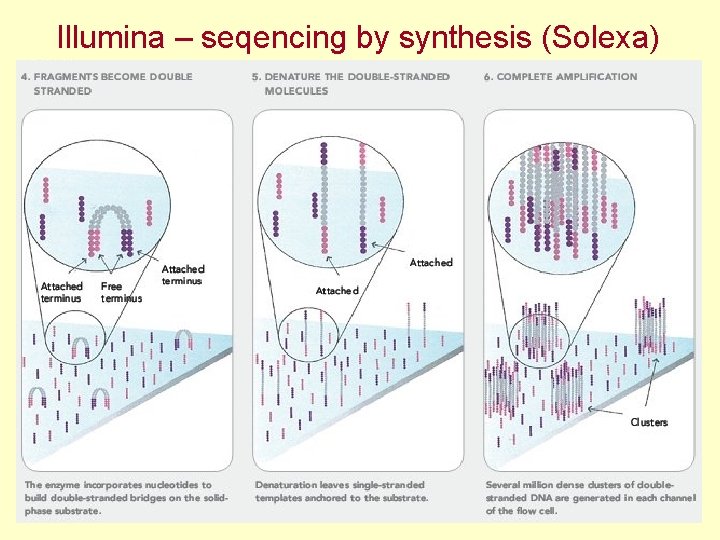
Illumina – seqencing by synthesis (Solexa)

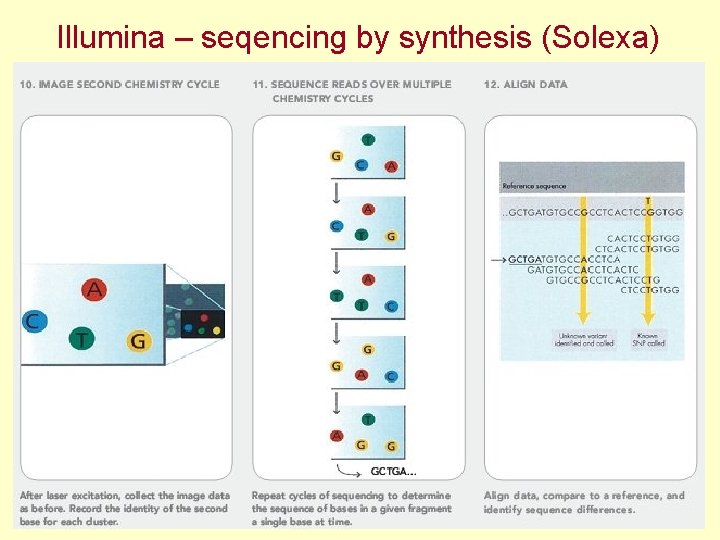
Illumina – seqencing by synthesis (Solexa)

Illumina – seqencing by synthesis (Solexa)

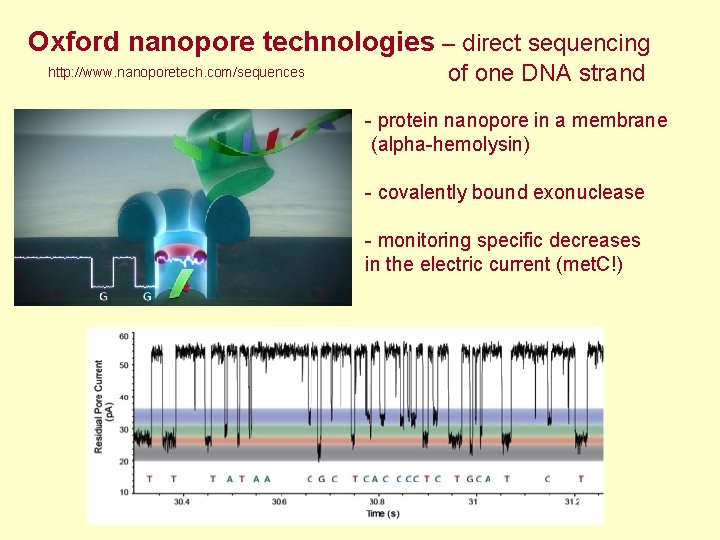
Oxford nanopore technologies – direct sequencing http: //www. nanoporetech. com/sequences of one DNA strand - protein nanopore in a membrane (alpha-hemolysin) - covalently bound exonuclease - monitoring specific decreases in the electric current (met. C!)

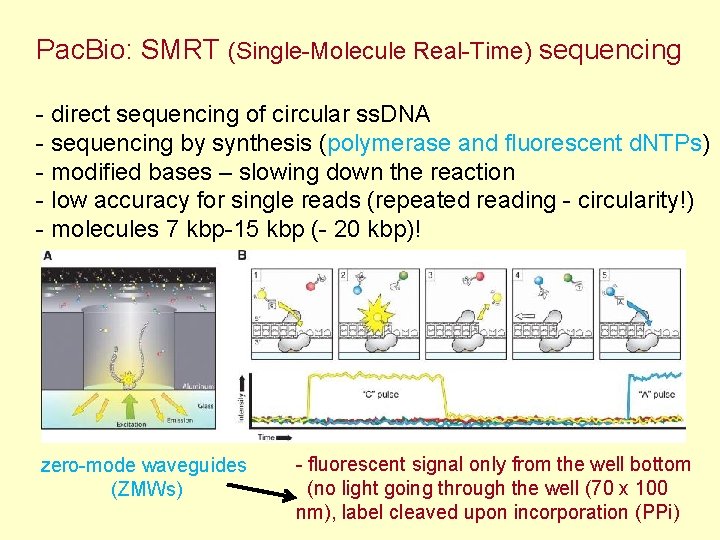
Pac. Bio: SMRT (Single-Molecule Real-Time) sequencing - direct sequencing of circular ss. DNA - sequencing by synthesis (polymerase and fluorescent d. NTPs) - modified bases – slowing down the reaction - low accuracy for single reads (repeated reading - circularity!) - molecules 7 kbp-15 kbp (- 20 kbp)! zero-mode waveguides (ZMWs) - fluorescent signal only from the well bottom (no light going through the well (70 x 100 nm), label cleaved upon incorporation (PPi)

Genome sequencing is not only sequencing of DNA • single sequencing read • 300 – 800 bp (Sanger, 454, Illumina) • 10 s of kbp (Pac. Bio, Oxford nanopore) • Typical genom hunderts of millions to billions bp How to manage?

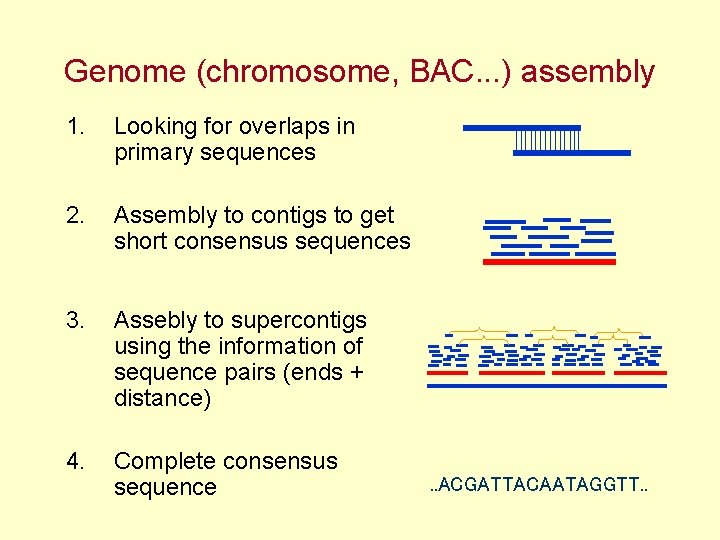
Genome (chromosome, BAC. . . ) assembly 1. Looking for overlaps in primary sequences 2. Assembly to contigs to get short consensus sequences 3. Assebly to supercontigs using the information of sequence pairs (ends + distance) 4. Complete consensus sequence . . ACGATTACAATAGGTT. .

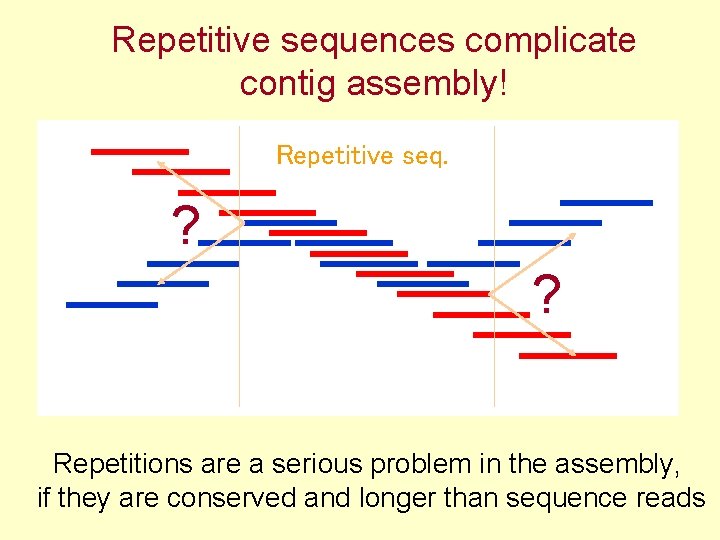
Repetitive sequences complicate contig assembly! Repetitive seq. ? ? Repetitions are a serious problem in the assembly, if they are conserved and longer than sequence reads

Strategies of genome sequencing • Classical strategy (Map-Based Assembly): - minimal quantity of DNA sequencing – sorting of big DNA fragments, successive reading (human genome sequencing – original strategy) - scaffold for genome sequence assemble - time consuming • Whole genome shotgun (WGS) – random (7 -9 x redundant) sequencing – sorting of sequence data (Haemophilus) - problems with repetitive DNA • Combination – „hierarchical shotgun“, „chromosome shotgun“

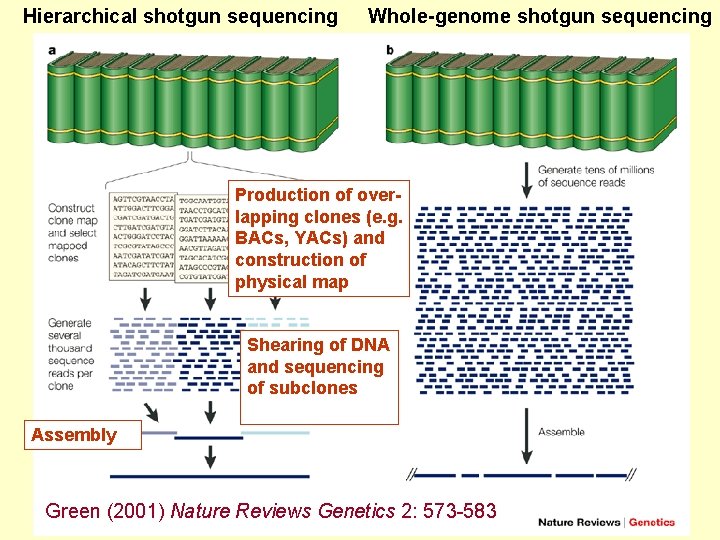
Hierarchical shotgun sequencing Whole-genome shotgun sequencing Production of overlapping clones (e. g. BACs, YACs) and construction of physical map Shearing of DNA and sequencing of subclones Assembly Green (2001) Nature Reviews Genetics 2: 573 -583

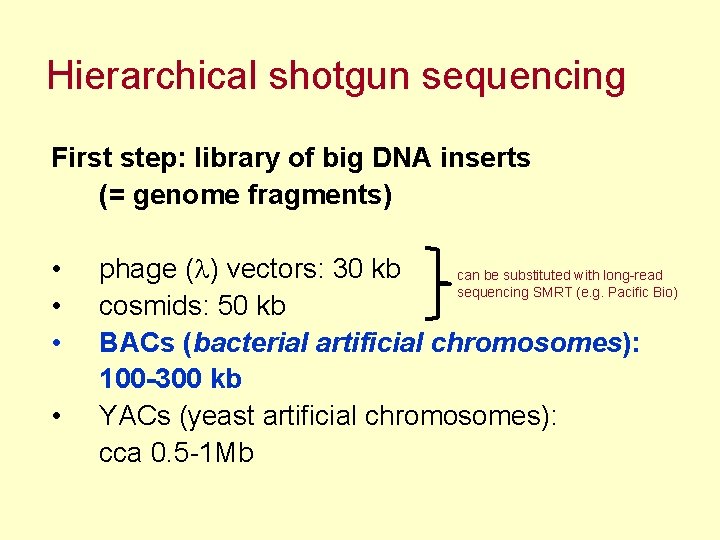
Hierarchical shotgun sequencing First step: library of big DNA inserts (= genome fragments) • • phage (l) vectors: 30 kb can be substituted with long-read sequencing SMRT (e. g. Pacific Bio) cosmids: 50 kb BACs (bacterial artificial chromosomes): 100 -300 kb YACs (yeast artificial chromosomes): cca 0. 5 -1 Mb

Physical „BAC“ map of genome • Arrangement (position, orientation) of individual BAC in the genome • Fundamental for classical sequencing • Very usefull for assembly of „shotgun“ sequences How to make the map from BACs with unknown sequence?

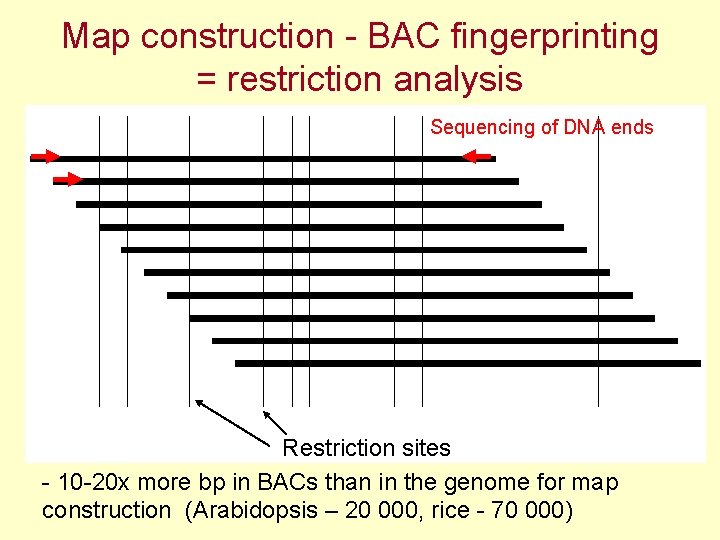
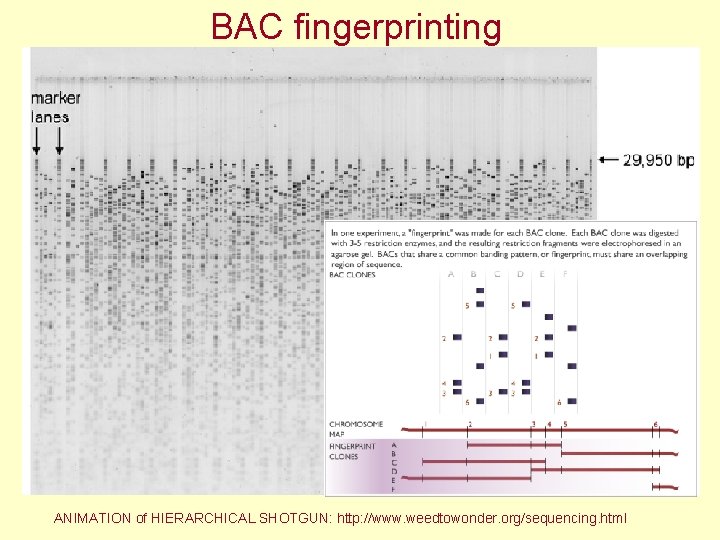
Map construction - BAC fingerprinting = restriction analysis Sequencing of DNA ends Restriction sites - 10 -20 x more bp in BACs than in the genome for map construction (Arabidopsis – 20 000, rice - 70 000)

BAC fingerprinting ANIMATION of HIERARCHICAL SHOTGUN: http: //www. weedtowonder. org/sequencing. html

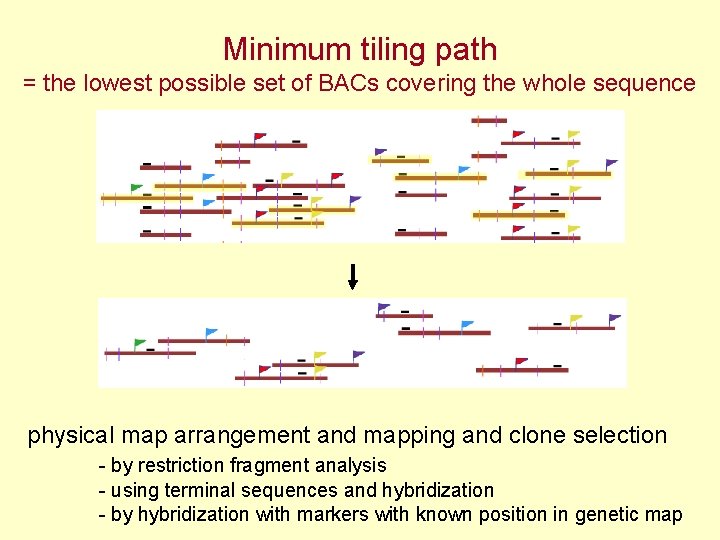
Minimum tiling path = the lowest possible set of BACs covering the whole sequence physical map arrangement and mapping and clone selection - by restriction fragment analysis - using terminal sequences and hybridization - by hybridization with markers with known position in genetic map

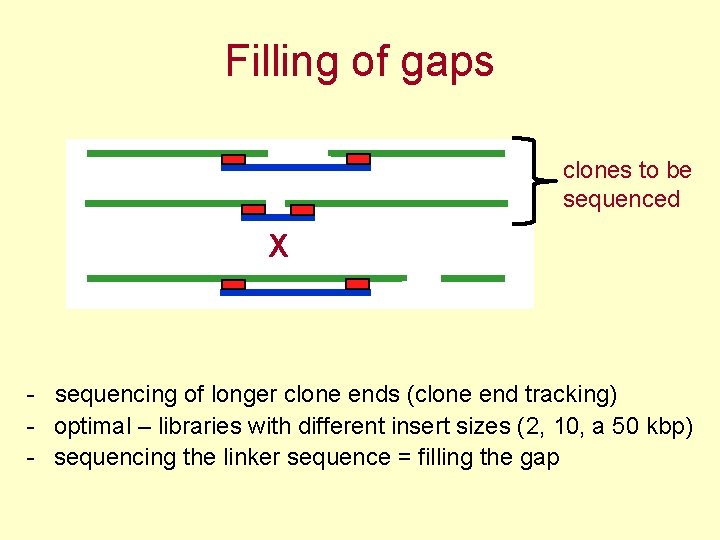
Filling of gaps clones to be sequenced X - sequencing of longer clone ends (clone end tracking) - optimal – libraries with different insert sizes (2, 10, a 50 kbp) - sequencing the linker sequence = filling the gap

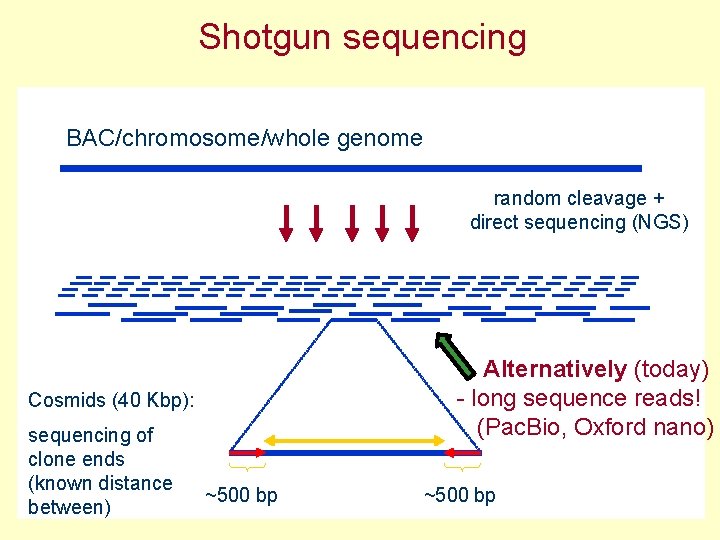
Shotgun sequencing BAC/chromosome/whole genome random cleavage + direct sequencing (NGS) Alternatively (today) - long sequence reads! (Pac. Bio, Oxford nano) Cosmids (40 Kbp): sequencing of clone ends (known distance between) ~500 bp

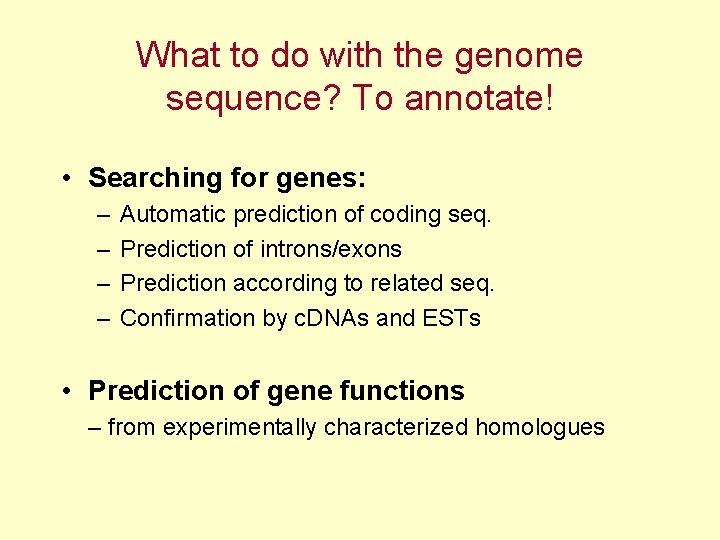
What to do with the genome sequence? To annotate! • Searching for genes: – – Automatic prediction of coding seq. Prediction of introns/exons Prediction according to related seq. Confirmation by c. DNAs and ESTs • Prediction of gene functions – from experimentally characterized homologues

Fragment of Gen. Bank BAC clone annotation

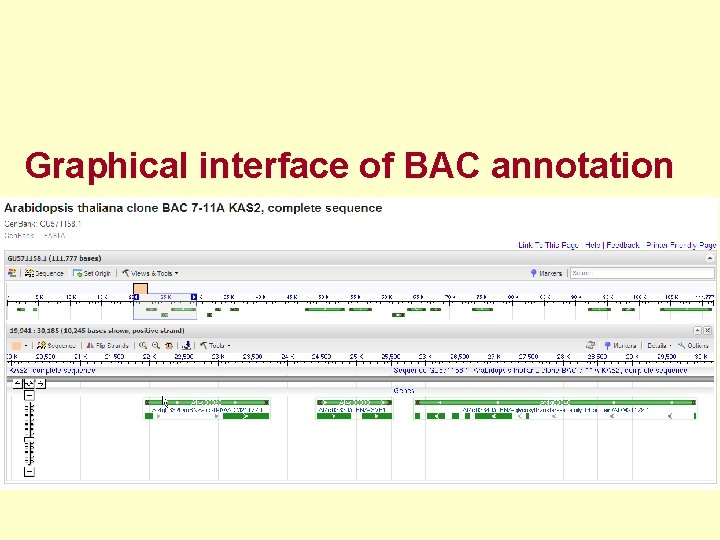
Graphical interface of BAC annotation

Large polyploid genomes alternative strategies for sequencing: - isolation of individual chromosomes e. g. wheat – allows assembly of homeologous chromosomes (allohexaploid) - shotgun sequencing of non-methylated DNA (maize) - sequencing of ESTs (potato)


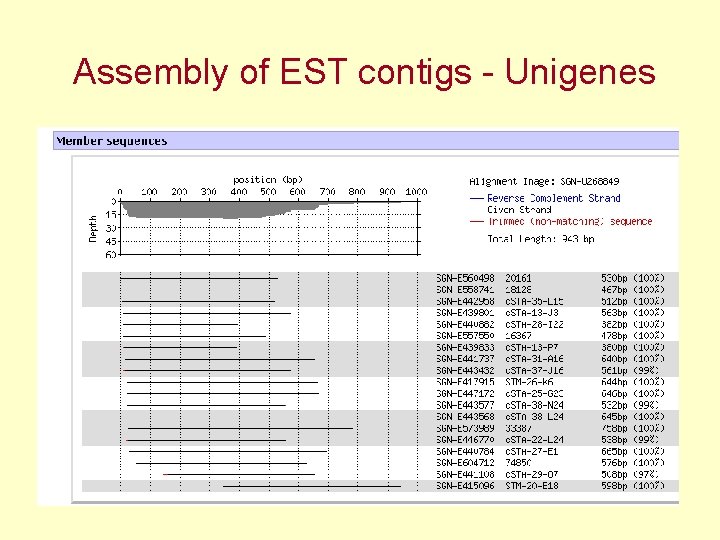
Assembly of EST contigs - Unigenes

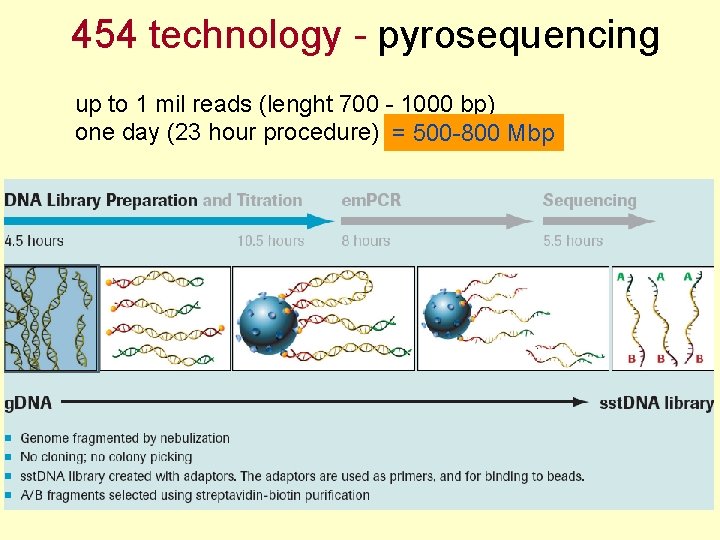
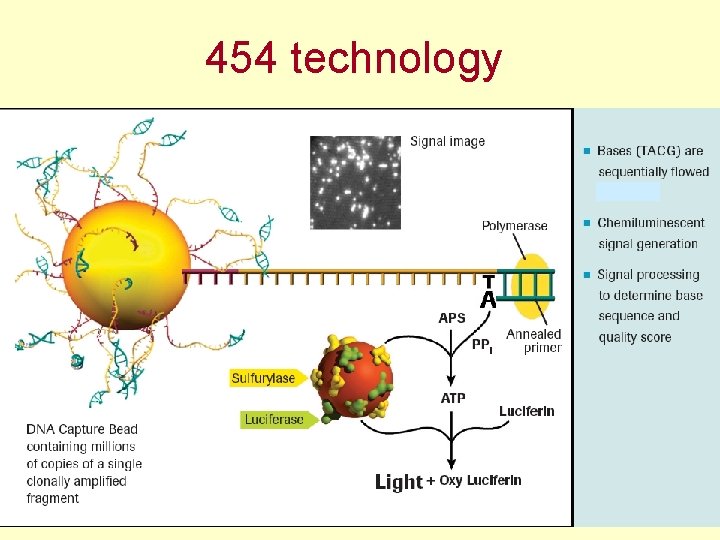
454 technology - pyrosequencing up to 1 mil reads (lenght 700 - 1000 bp) one day (23 hour procedure) = 500 -800 Mbp

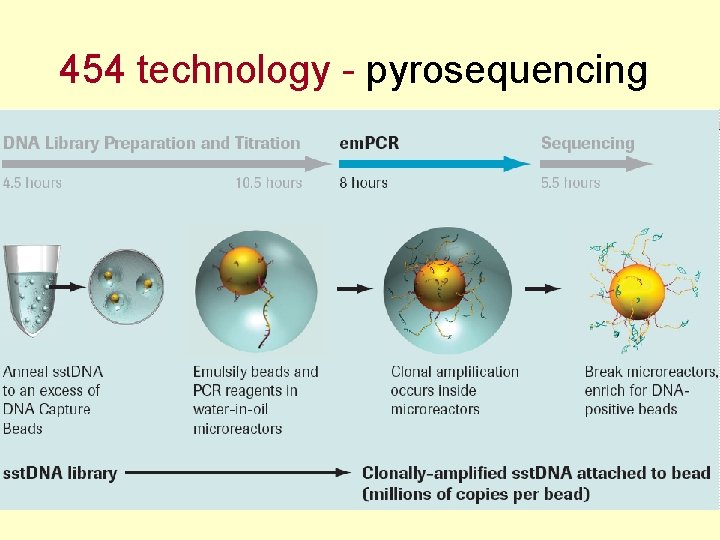
454 technology - pyrosequencing

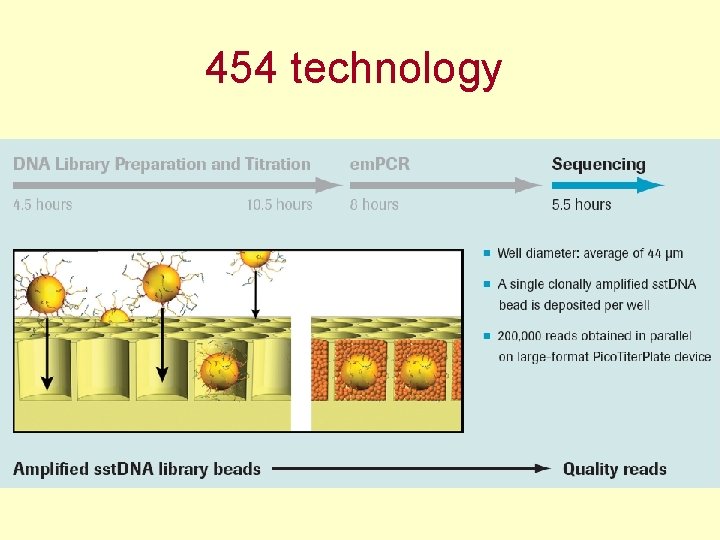
454 technology

454 technology

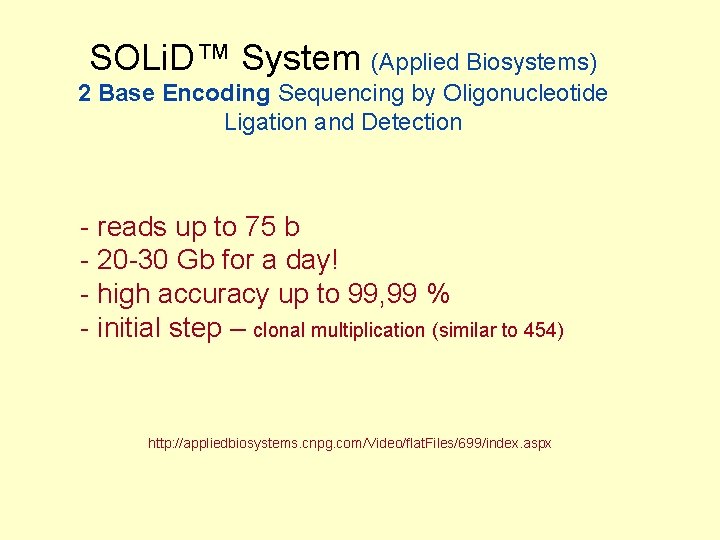
SOLi. D™ System (Applied Biosystems) 2 Base Encoding Sequencing by Oligonucleotide Ligation and Detection - reads up to 75 b - 20 -30 Gb for a day! - high accuracy up to 99, 99 % - initial step – clonal multiplication (similar to 454) http: //appliedbiosystems. cnpg. com/Video/flat. Files/699/index. aspx

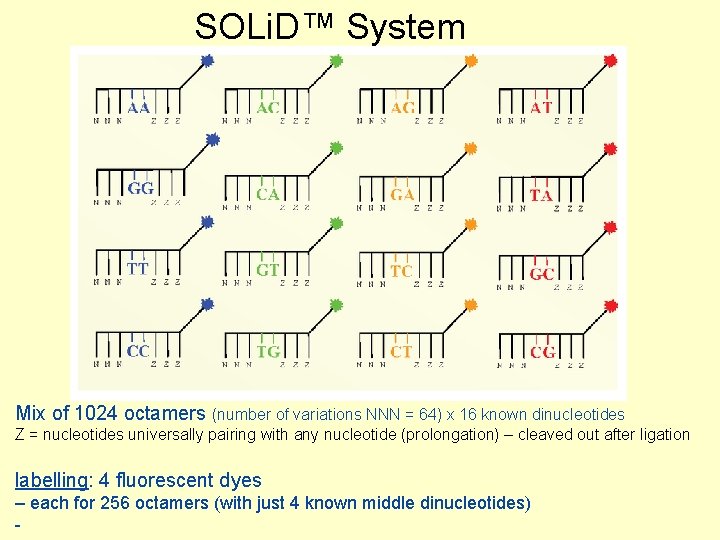
SOLi. D™ System Mix of 1024 octamers (number of variations NNN = 64) x 16 known dinucleotides Z = nucleotides universally pairing with any nucleotide (prolongation) – cleaved out after ligation labelling: 4 fluorescent dyes – each for 256 octamers (with just 4 known middle dinucleotides) -

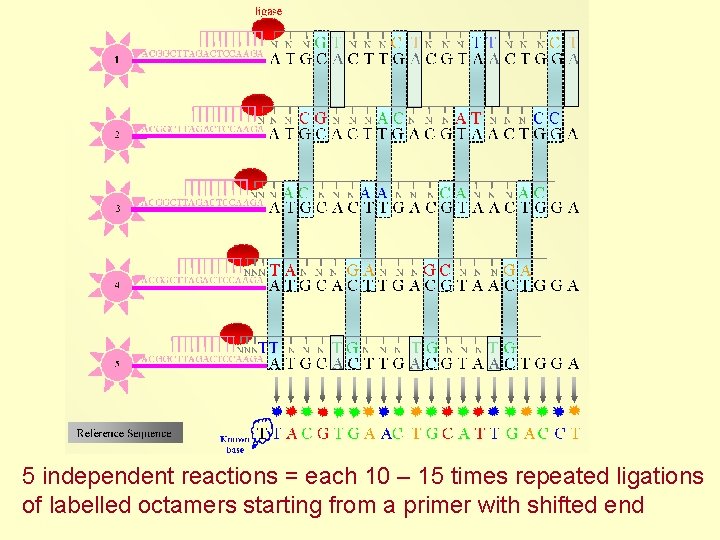
5 independent reactions = each 10 – 15 times repeated ligations of labelled octamers starting from a primer with shifted end

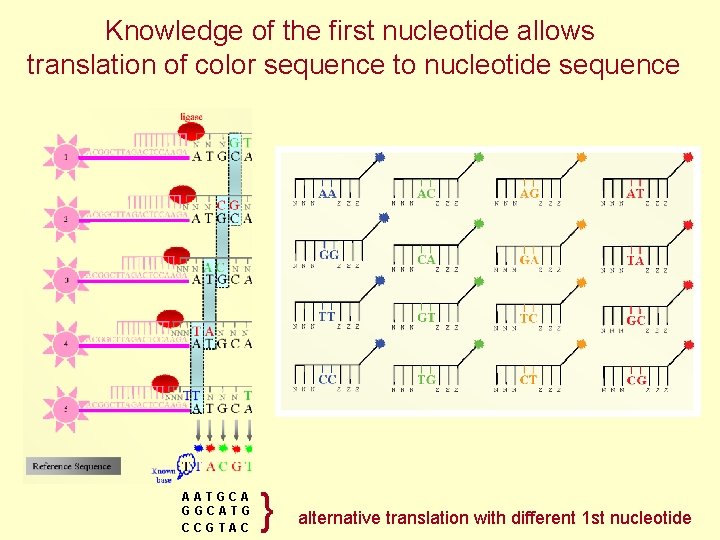
Knowledge of the first nucleotide allows translation of color sequence to nucleotide sequence AATGCA GGCATG CCGTAC } alternative translation with different 1 st nucleotide