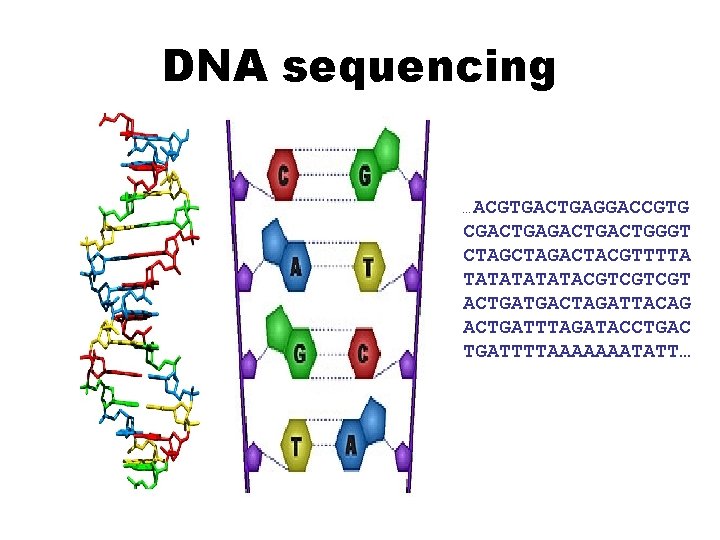
DNA Sequencing DNA sequencing ACGTGACTGAGGACCGTG CGACTGACTGGGT CTAGACTACGTTTTA TATATACGTCGTCGT



- Slides: 24

DNA Sequencing

DNA sequencing …ACGTGACTGAGGACCGTG CGACTGACTGGGT CTAGACTACGTTTTA TATATACGTCGTCGT ACTGATGACTAGATTACAG ACTGATTTAGATACCTGAC TGATTTTAAAAAAATATT…

DNA sequencing ü Determination of nucleotide sequence ü Two similar methods: 1. Maxam and Gilbert method 2. Sanger method ü They depend on the production of a mixture of oligonucleotides labeled either radioactively or fluorescein, with one common end and differing in length by a single nucleotide at the other end ü This mixture of oligonucleotides is separated by high resolution electrophoresis on polyacrilamide gels and the position of the bands determined

Maxam and Gilbert Method üThe single stranded DNA fragment to be sequenced is endlabeled by treatment with alkaline phosphatase to remove the 5’phosphate üIt is then followed by reaction with P-labeled ATP in the presence of polynucleotide kinase, which attaches P labeled to the 5’terminal üThe labeled DNA fragment is then divided into four aliquots, each of which is treated with a reagent which modifies a specific base 1. Aliquot 2. Aliquot 3. Aliquot 4. Aliquot cytosine A + dimethyl sulphate, which methylates guanine residue B + formic acid, which modifies adenine and guanine residues C + Hydrazine, which modifies thymine + cytosine residues D + Hydrazine + 5 mol/l Na. Cl, which makes the reaction specific for üThe four are incubated with piperidine which cleaves the sugar phosphate backbone of DNA next to the residue that has been modified

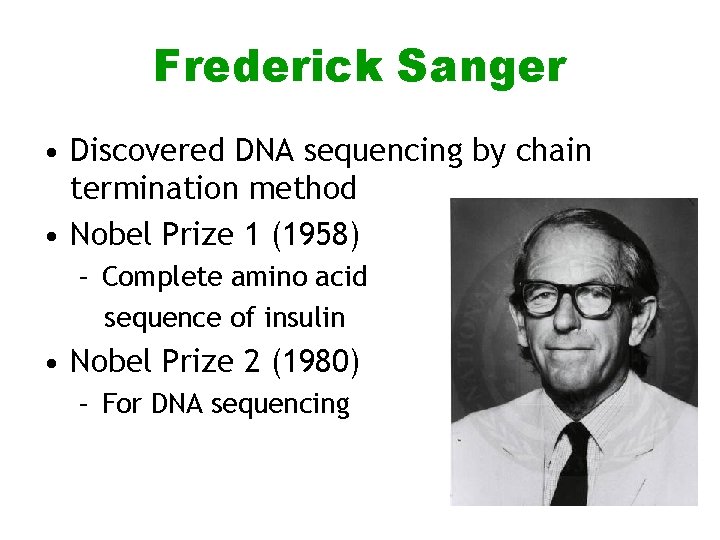
Frederick Sanger • Discovered DNA sequencing by chain termination method • Nobel Prize 1 (1958) – Complete amino acid sequence of insulin • Nobel Prize 2 (1980) – For DNA sequencing

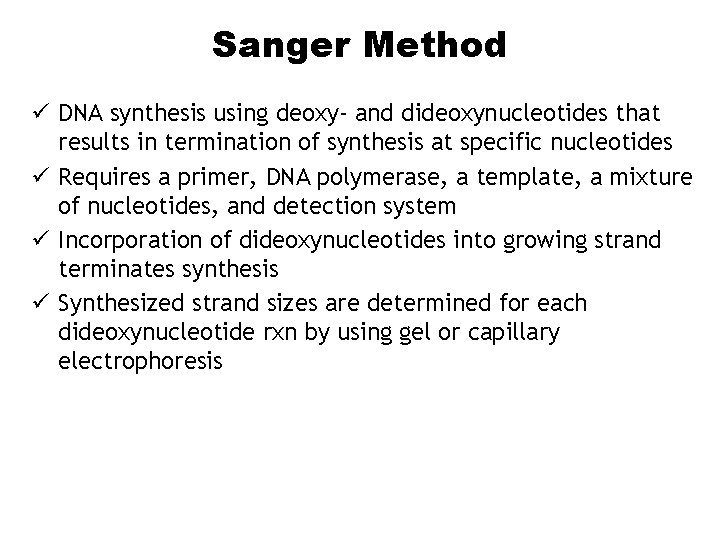
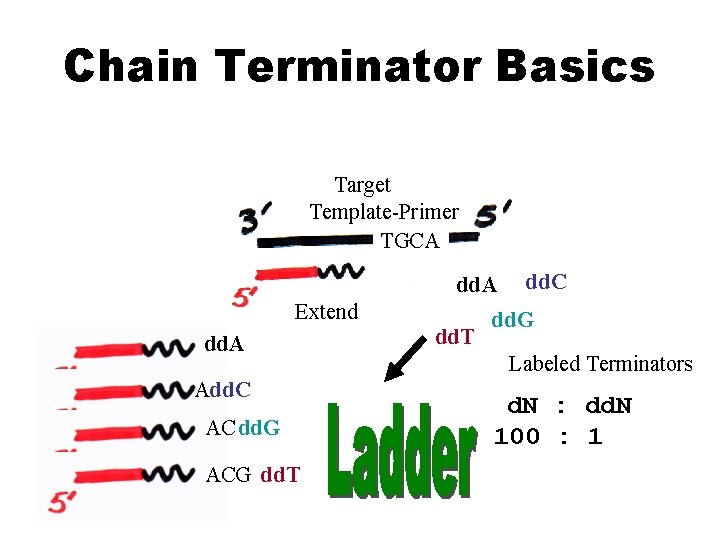
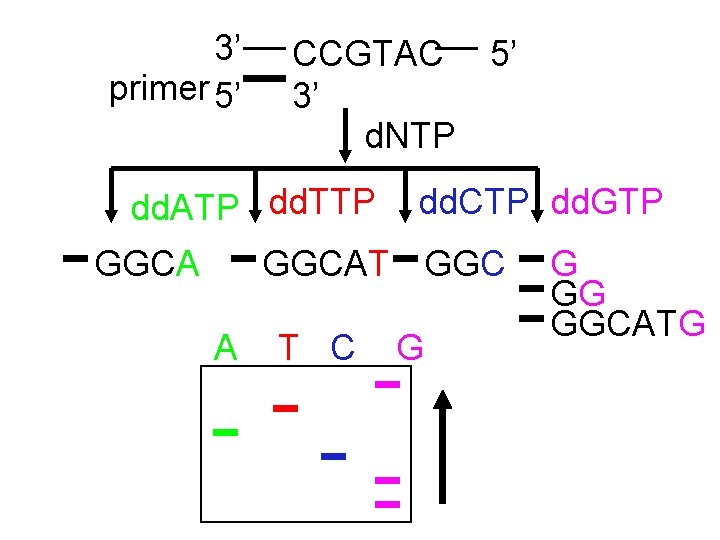
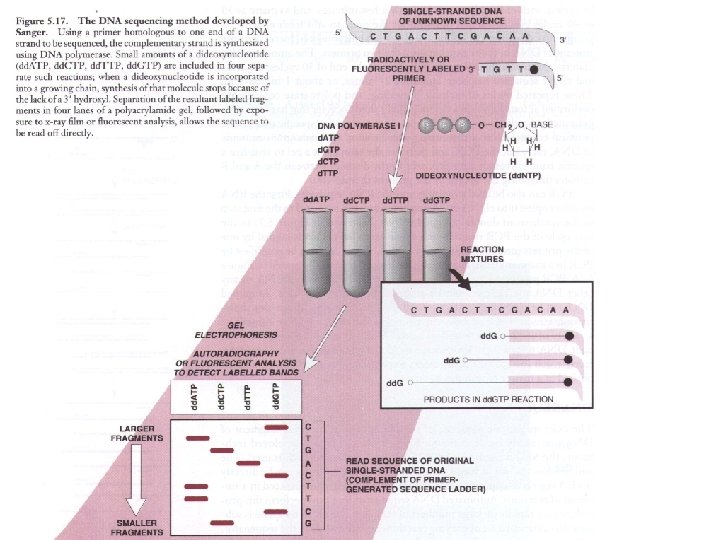
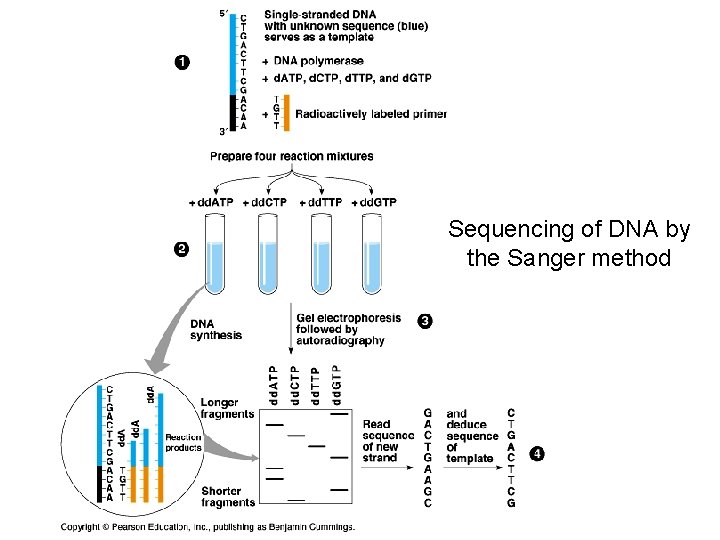
Sanger Method ü DNA synthesis using deoxy- and dideoxynucleotides that results in termination of synthesis at specific nucleotides ü Requires a primer, DNA polymerase, a template, a mixture of nucleotides, and detection system ü Incorporation of dideoxynucleotides into growing strand terminates synthesis ü Synthesized strand sizes are determined for each dideoxynucleotide rxn by using gel or capillary electrophoresis

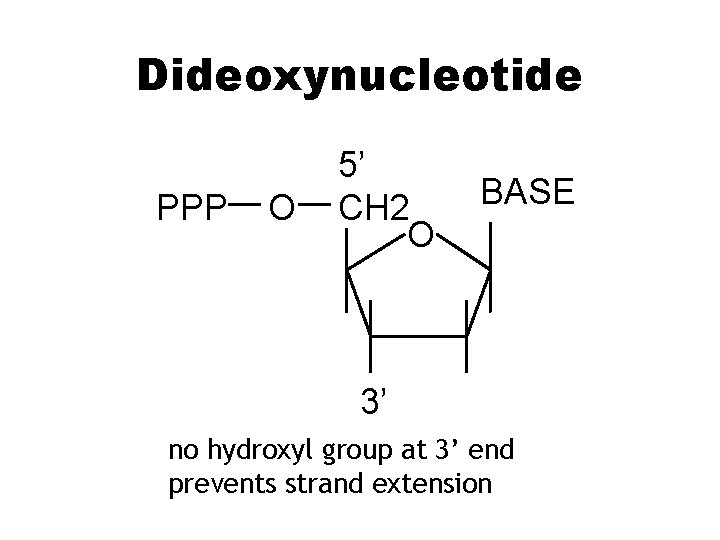
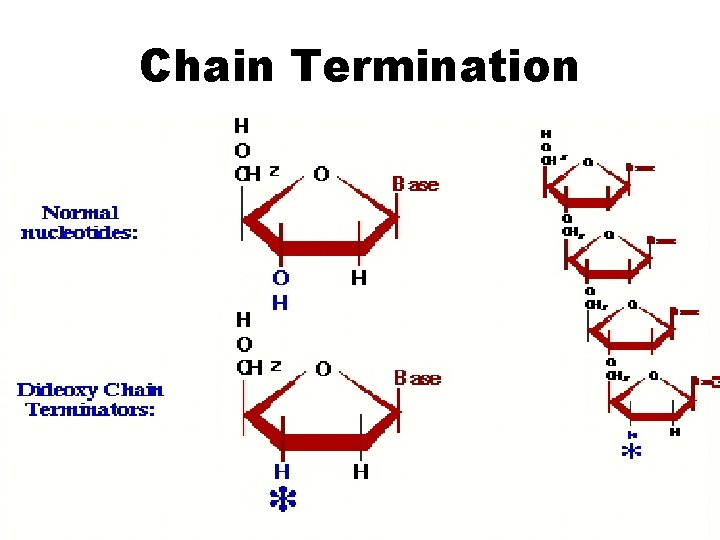
Dideoxynucleotide PPP O 5’ CH 2 O BASE 3’ no hydroxyl group at 3’ end prevents strand extension

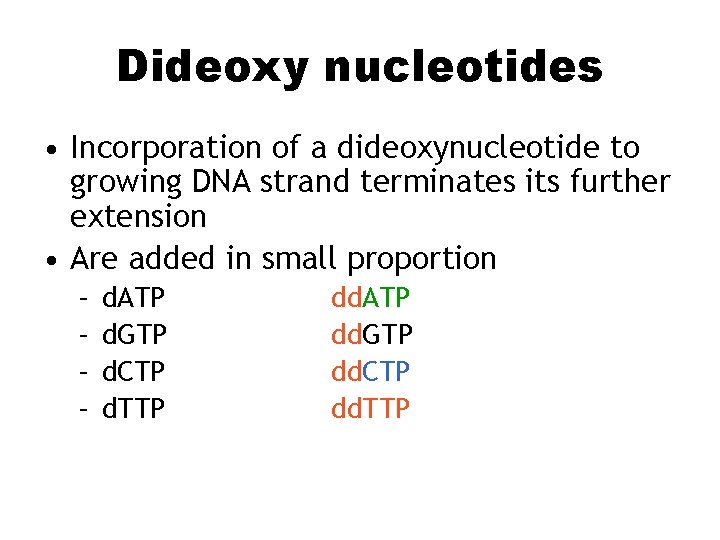
Dideoxy nucleotides • Incorporation of a dideoxynucleotide to growing DNA strand terminates its further extension • Are added in small proportion – – d. ATP d. GTP d. CTP d. TTP dd. ATP dd. GTP dd. CTP dd. TTP

Chain Termination

Chain Terminator Basics Target Template-Primer TGCA dd. A Extend dd. A Add. C AC dd. G ACG dd. T dd. C dd. G Labeled Terminators d. N : dd. N 100 : 1

3’ primer 5’ CCGTAC 5’ 3’ d. NTP dd. ATP dd. TTP GGCA dd. CTP dd. GTP GGCAT GGC A T C G G GG GGCATG

All Possible Terminations

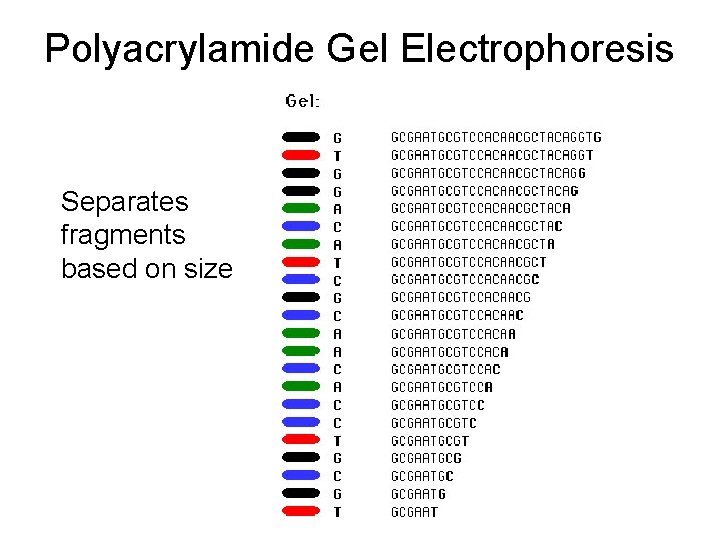
Polyacrylamide Gel Electrophoresis Separates fragments based on size

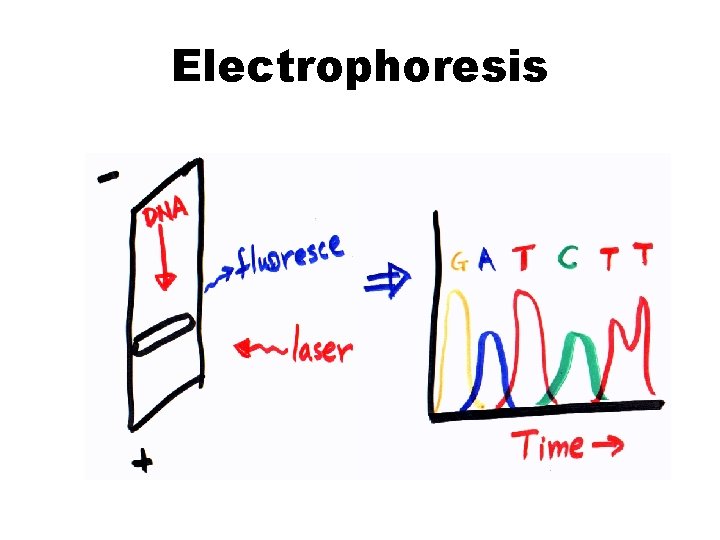
Electrophoresis

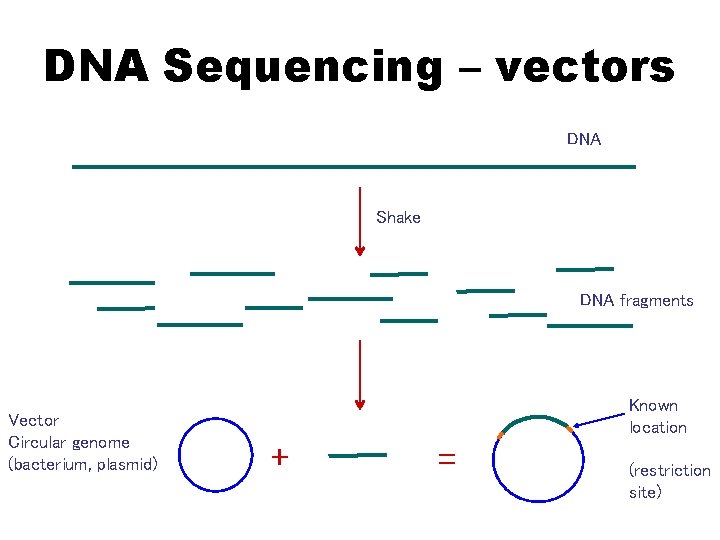
DNA Sequencing – vectors DNA Shake DNA fragments Vector Circular genome (bacterium, plasmid) Known location + = (restriction site)

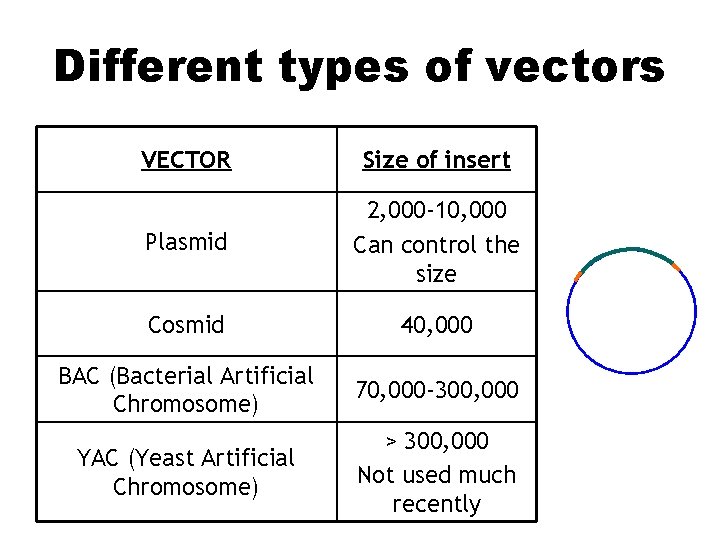
Different types of vectors VECTOR Size of insert Plasmid 2, 000 -10, 000 Can control the size Cosmid 40, 000 BAC (Bacterial Artificial Chromosome) 70, 000 -300, 000 YAC (Yeast Artificial Chromosome) > 300, 000 Not used much recently

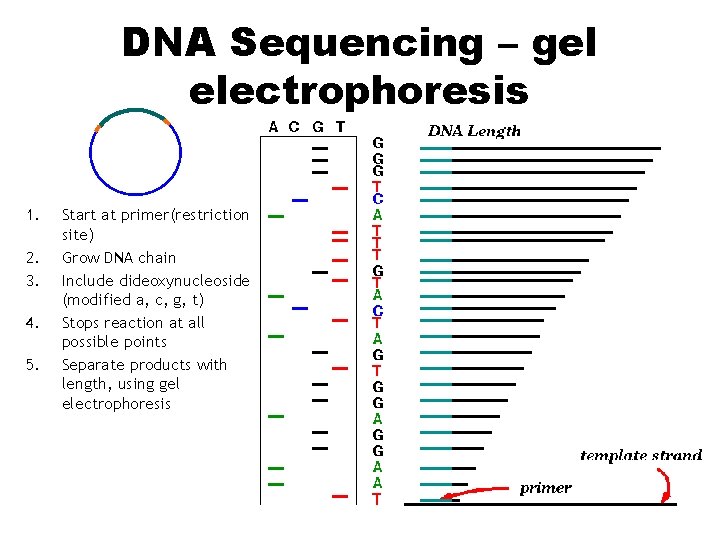
DNA Sequencing – gel electrophoresis 1. 2. 3. 4. 5. Start at primer(restriction site) Grow DNA chain Include dideoxynucleoside (modified a, c, g, t) Stops reaction at all possible points Separate products with length, using gel electrophoresis

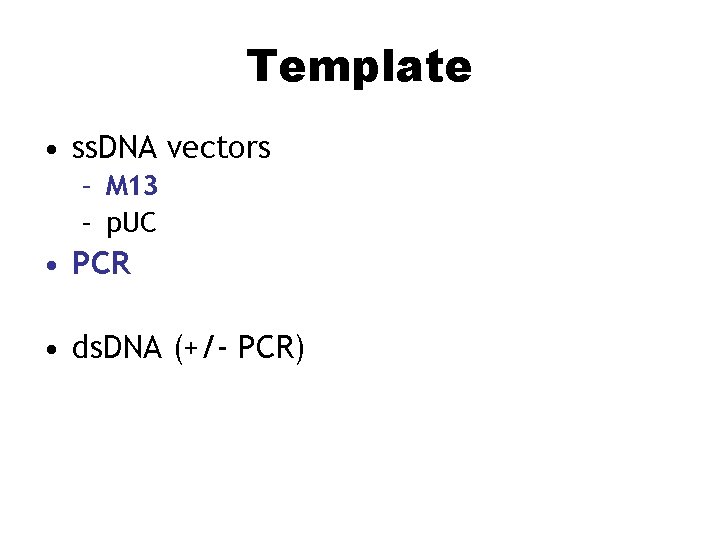
Template • ss. DNA vectors – M 13 – p. UC • PCR • ds. DNA (+/- PCR)

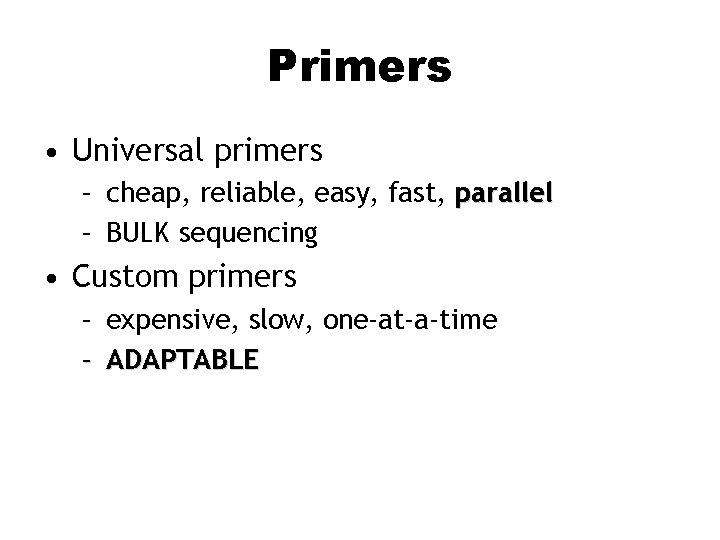
Primers • Universal primers – cheap, reliable, easy, fast, parallel – BULK sequencing • Custom primers – expensive, slow, one-at-a-time – ADAPTABLE

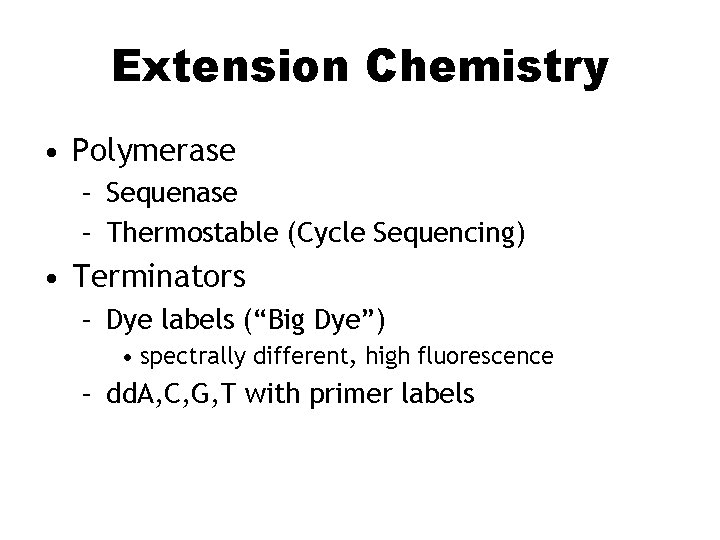
Extension Chemistry • Polymerase – Sequenase – Thermostable (Cycle Sequencing) • Terminators – Dye labels (“Big Dye”) • spectrally different, high fluorescence – dd. A, C, G, T with primer labels

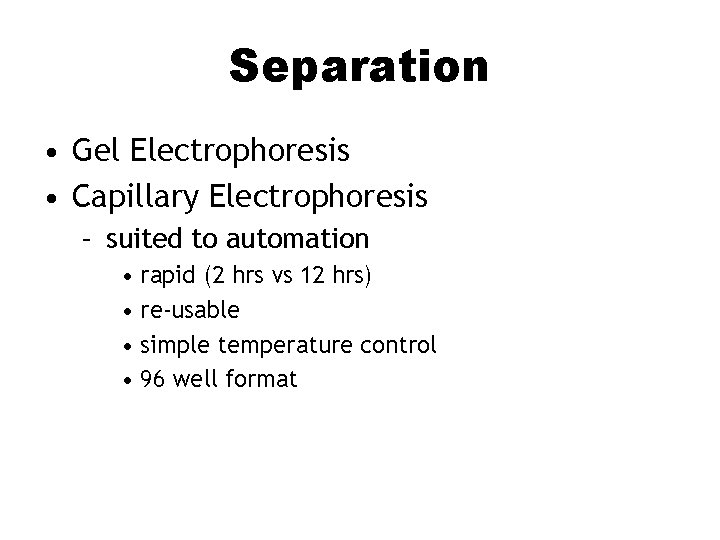
Separation • Gel Electrophoresis • Capillary Electrophoresis – suited to automation • rapid (2 hrs vs 12 hrs) • re-usable • simple temperature control • 96 well format

Sequencing Strategies • Ordered – Divide and Conquer • Random Sequence – Brute Force

DNA Sequencing 5. 17)

Sequencing of DNA by the Sanger method
 Dna sequencing importance
Dna sequencing importance Dna sequencing
Dna sequencing 3rd generation dna sequencing
3rd generation dna sequencing Dna sequencing applications
Dna sequencing applications Dna sequencing methods
Dna sequencing methods Dna sequencing gel
Dna sequencing gel Bioflix activity dna replication nucleotide pairing
Bioflix activity dna replication nucleotide pairing Coding dna and non coding dna
Coding dna and non coding dna Replication process
Replication process Function of dna polymerase 3
Function of dna polymerase 3 Dna rna protein synthesis homework #2 dna replication
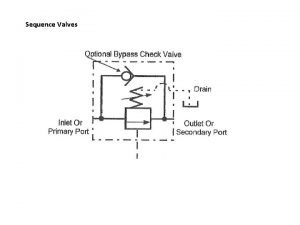
Dna rna protein synthesis homework #2 dna replication Symbol of sequence valve
Symbol of sequence valve Wilkes control unit
Wilkes control unit Per partes
Per partes Sequencing analysis viewer
Sequencing analysis viewer Microprogram sequencer diagram
Microprogram sequencer diagram Next generation sequencing methods
Next generation sequencing methods Sequencing computer science
Sequencing computer science Cloning and sequencing explorer series
Cloning and sequencing explorer series Panfacial fractures sequencing
Panfacial fractures sequencing Ap csp sequencing
Ap csp sequencing Scheduling loading sequencing and monitoring
Scheduling loading sequencing and monitoring Sequencing batch reactor advantages and disadvantages
Sequencing batch reactor advantages and disadvantages The cask of amontillado quiz
The cask of amontillado quiz Scheduling decisions
Scheduling decisions