Data analysis of next generation DNA resequencing from





























- Slides: 60

Data analysis of next generation DNA re-sequencing: from raw reads to variants Chuan-Kun Liu 劉傳崑 中央研究院 國家基因體醫學研究中心 2012/09/20

Outlines • • What can we get from NGS DNA Seq Data output of NGS platforms Quality Check and Read Trimming Alignment Variant detection Coverage and qualities Customize data analysis for multi-samples 2

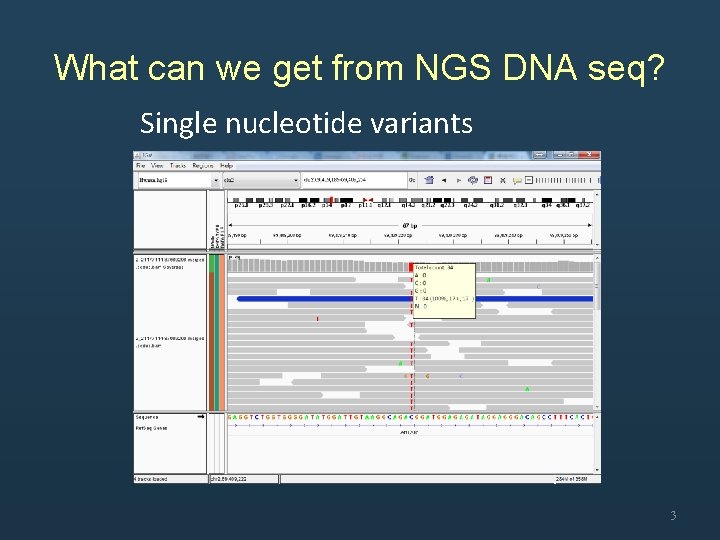
What can we get from NGS DNA seq? Single nucleotide variants 3

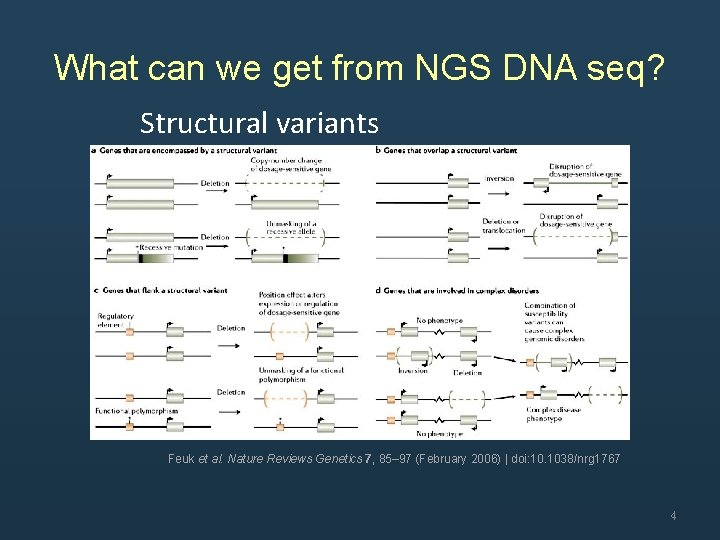
What can we get from NGS DNA seq? Structural variants Feuk et al. Nature Reviews Genetics 7, 85– 97 (February 2006) | doi: 10. 1038/nrg 1767 4

NGS Data processing Hi. Seq 2000 Report Data Backup Data Analysis 5

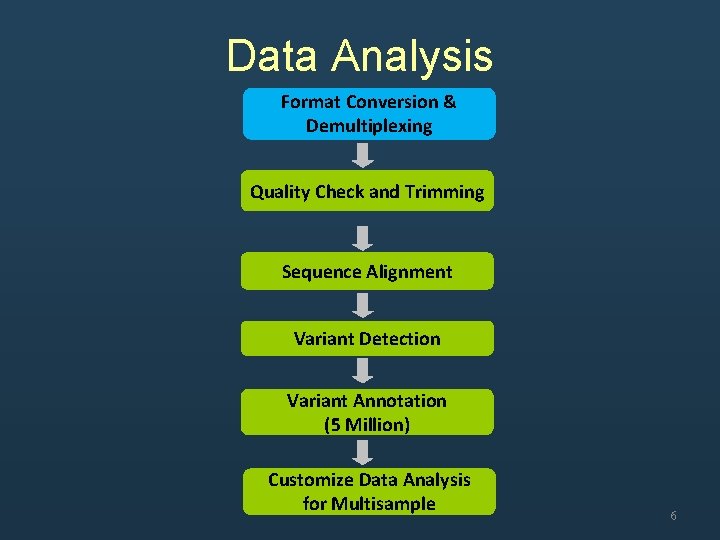
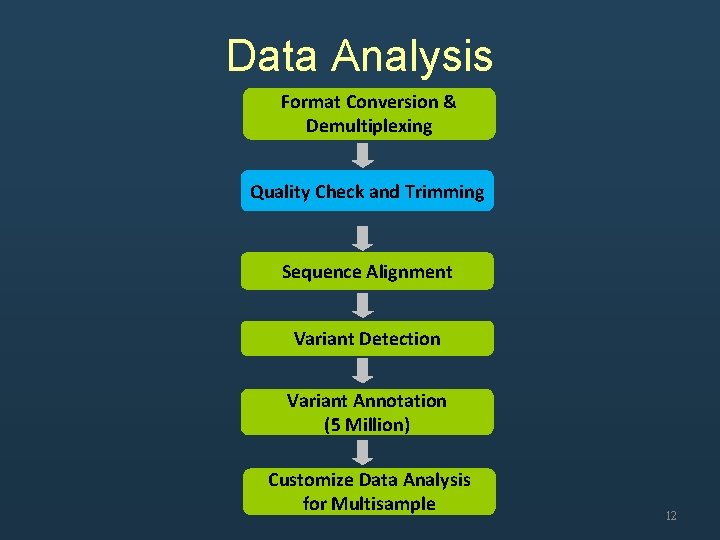
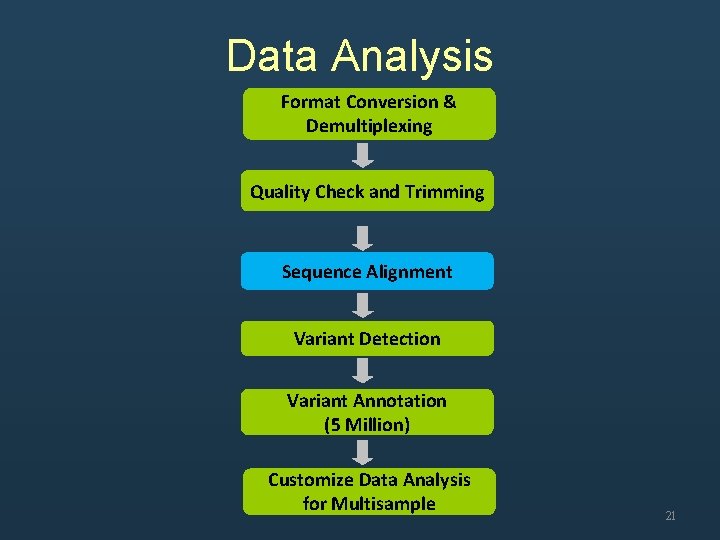
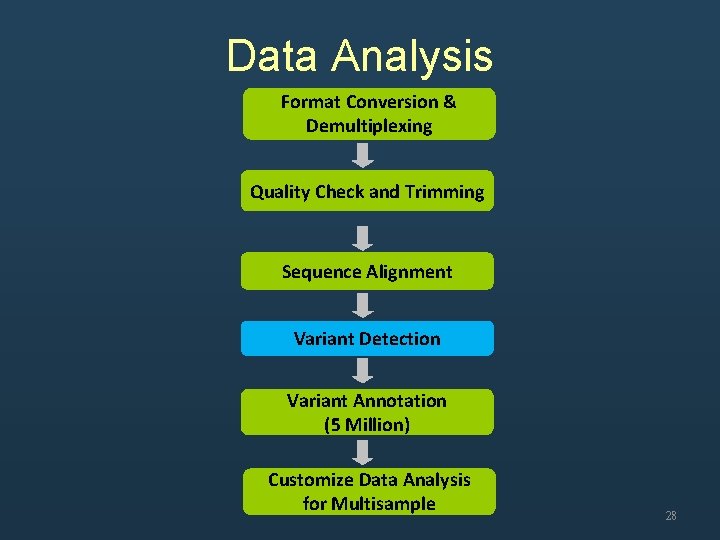
Data Analysis Format Conversion & Demultiplexing Quality Check and Trimming Sequence Alignment Variant Detection Variant Annotation (5 Million) Customize Data Analysis for Multisample 6

Instrument output Illumina Hi. Seq 2000 (BCL files) Life Tech Ion Proton (FASTQ files) Roche GS FLX+ (Standard Flowgram Format (SFF) files) FASTQ Pac. Bio RS (FASTQ files) 7

Sanger FASTQ format @HWI-ST 688: 211: C 0 F 02 ACXX: 2: 2310: 7574: 93175 2: N: 0: GCCAAT ATGCAAATAAACTAGAAAATCTAGAAGAAATGGAGAAATTCCTGGACACAC + CCCFFFFFHHHGHJIIJJIJJIJJJFIIJAJJIJIJJJJI? ? ? 8

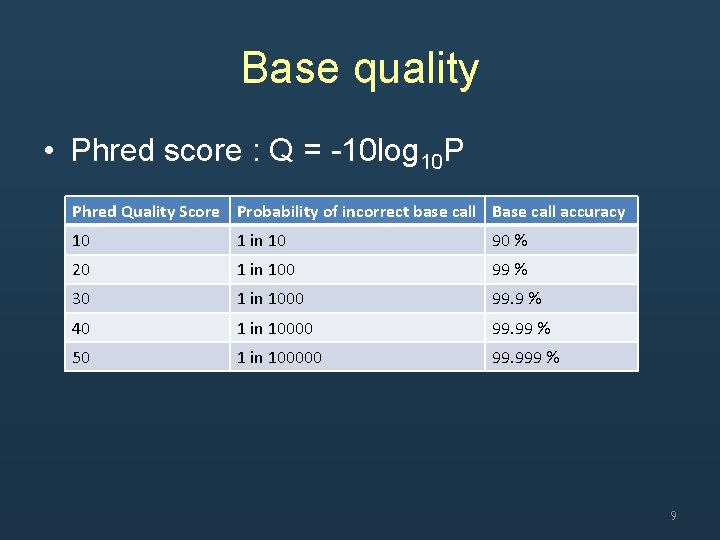
Base quality • Phred score : Q = -10 log 10 P Phred Quality Score Probability of incorrect base call Base call accuracy 10 1 in 10 90 % 20 1 in 100 99 % 30 1 in 1000 99. 9 % 40 1 in 10000 99. 99 % 50 1 in 100000 99. 999 % 9

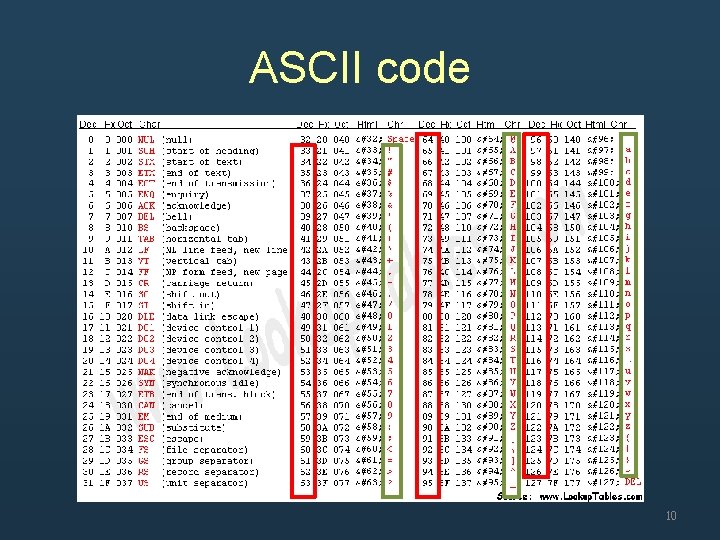
ASCII code 10

Three described FASTQ variants Description, OBF name Sanger standard* fastq-sanger Solexa/early Illumina* fastq-solexa Illumina 1. 3+* fastq-illumina Illumina 1. 8+ fastq-sanger ASCII characters Range Offset 33 -126 33 59 -126 64 64 -126 64 33 -126 33 Quality score Type Range PHRED 0 to 93 Solexa -5 to 62 PHRED 0 to 93 *Nucleic Acids Res. 2010 April; 38(6): 1767– 1771. 11

Data Analysis Format Conversion & Demultiplexing Quality Check and Trimming Sequence Alignment Variant Detection Variant Annotation (5 Million) Customize Data Analysis for Multisample 12

Quality Check • Tools for Quality Check – Sequence Analysis Viewer (SAV) – FASTQC – FASTX • Input files – Instrument system files • Inter. Op folder • Run. Info. xml • run. Parameters. xml – FASTQ 13

Quality Check : SAV • Input files – 儀器端的紀錄檔,檔案不易取得 • 可以在還沒有進行 format convert之前了解 reads的 quality狀況 14

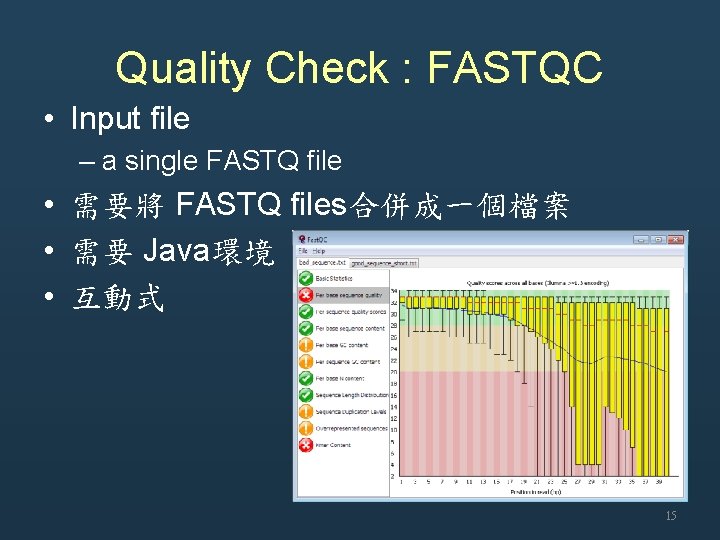
Quality Check : FASTQC • Input file – a single FASTQ file • 需要將 FASTQ files合併成一個檔案 • 需要 Java環境 • 互動式 15

Quality Check : FASTX • Input file – a single FASTQ file • 需要將 FASTQ files合併成一個檔案 • 需要 Linux作業系統 • Command Line column 1 2 3 4 5 6 7 8 9 10 count min max sum mean Q 1 med Q 3 IQR l. W r. W A_Count 953574 2 34 31359668 32. 89 31 34 34 3 27 34 276400 953574 2 34 31597969 33. 14 33 34 34 1 32 34 271486 953574 2 34 31650189 33. 19 33 34 34 1 32 34 261400 953574 2 37 34818060 36. 51 37 37 37 0 37 37 253382 953574 2 37 34763134 36. 46 37 37 37 0 37 37 252777 953574 2 37 34715168 36. 41 37 37 37 0 37 37 252718 953574 2 37 34667721 36. 36 37 37 37 0 37 37 252225 953574 2 37 34698520 36. 39 37 37 37 0 37 37 251982 953574 2 39 36422177 38. 2 39 39 39 0 39 39 246931 953574 2 39 36246082 38. 01 39 39 39 0 39 39 251253 C_Count 201286 186819 204156 217675 220801 231894 224374 228680 230019 226726 G_Count 210089 228290 228743 228607 226468 221480 219947 215144 215002 217366 T_Count N_Count Max_count 265799 0 953574 266975 4 953574 259275 0 953574 253910 0 953574 253528 0 953574 247482 0 953574 257028 0 953574 257768 0 953574 261610 12 953574 258229 0 953574 16

Trimming • Trimming的時機 – Format conversion時 – 將 FASTQ送入 Alignment的 具前 – 使用 Alignment 具內建的 trimming機制 • Trimming的單位 – By flow cell的 Lane為單位 trimming – By read為單位 trimming 17

Trimming • Trimming的方式 – 指定那些 base要使用 (如 n 5 Y*n 5) – 決定 Q值的門檻再透過公式計算出 trimming的 位置 @HWI-ST 688: 211: C 0 F 02 ACXX: 2: 2310: 7574: 93175 2: N: 0: GCCAAT ATGCAAATAAACTAGAAAATCTAGAAGAAATGGAGAAATTCCTGGACACAC + CCCFFFFFHHHGHJIIJJIJJIJJJFIIJAJJIJIJJJJI? ? ? 18

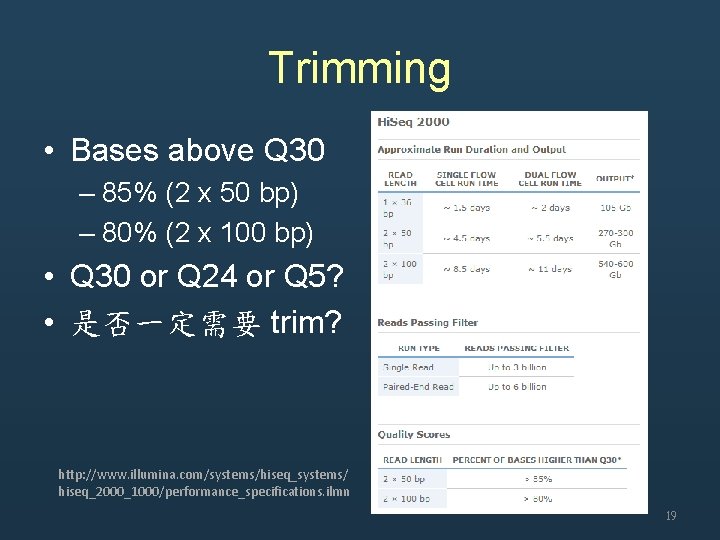
Trimming • Bases above Q 30 – 85% (2 x 50 bp) – 80% (2 x 100 bp) • Q 30 or Q 24 or Q 5? • 是否一定需要 trim? http: //www. illumina. com/systems/hiseq_systems/ hiseq_2000_1000/performance_specifications. ilmn 19


Data Analysis Format Conversion & Demultiplexing Quality Check and Trimming Sequence Alignment Variant Detection Variant Annotation (5 Million) Customize Data Analysis for Multisample 21

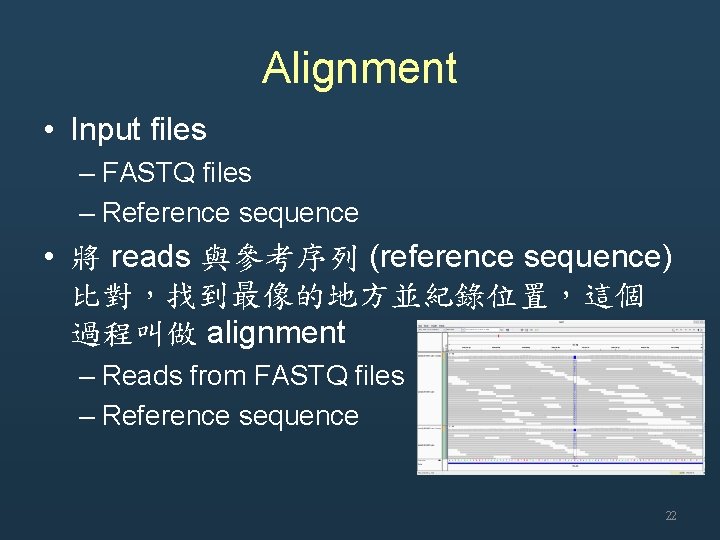
Alignment • Input files – FASTQ files – Reference sequence • 將 reads 與參考序列 (reference sequence) 比對,找到最像的地方並紀錄位置,這個 過程叫做 alignment – Reads from FASTQ files – Reference sequence 22

Software for Alignment • Commercial – CASAVA (Illumina) – Bio. Scope (Life Tech) – CLC Genomics Workbench – GS Reference Mapper (Roche) • Open source – BWA – Bowtie – SOAP 2 23

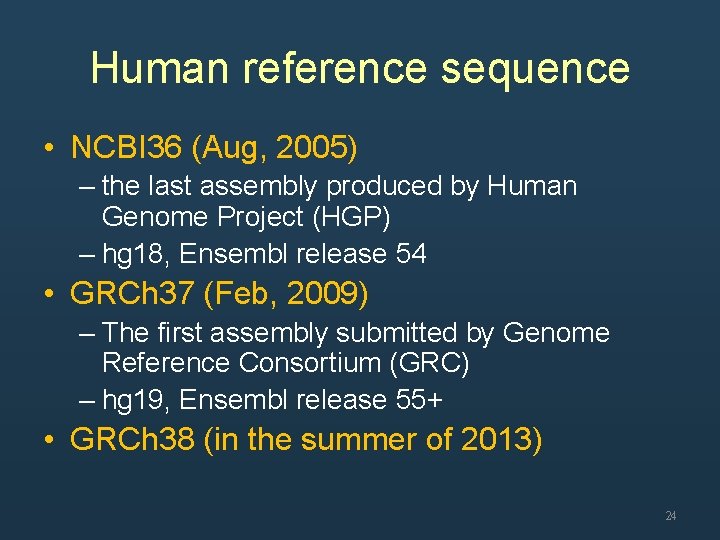
Human reference sequence • NCBI 36 (Aug, 2005) – the last assembly produced by Human Genome Project (HGP) – hg 18, Ensembl release 54 • GRCh 37 (Feb, 2009) – The first assembly submitted by Genome Reference Consortium (GRC) – hg 19, Ensembl release 55+ • GRCh 38 (in the summer of 2013) 24

GRCh 37 • Primary assembly – Chromosome assembly – Unlocalized sequence – Unplaced sequence • Alternate loci – A sequence that provides an alternate representation of a locus found in a largely haploid assembly. (MHC region, UGT 2 B 17, MAPT) • Patches – A contig sequence that is released outside of the full assembly release cycle. 25

Choose your reference genome • Human_g 1 k_v 37 – The reference sequence provided by the 1000 Genomes Project* – Unlocalized and Unplaced sequence are included (Full primary assembly) – Replace the mitochondrial sequence with the revised Cambridge Reference Sequences (r. CRS; AC: NC_012920) (APR, 30, 2010)** • Male or female *ftp: //ftp. ncbi. nih. gov/1000 genomes/ftp/technical/reference/README. human_g 1 k_v 37. fasta. txt **http: //www. ncbi. nlm. nih. gov/nuccore/NC_012920 26

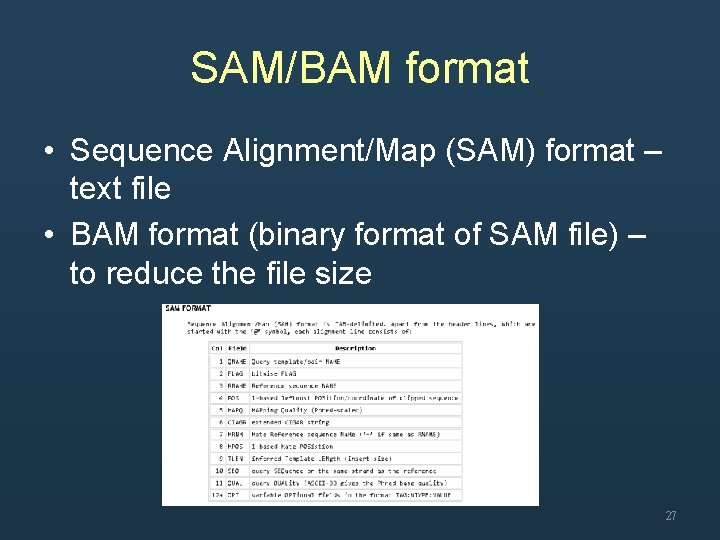
SAM/BAM format • Sequence Alignment/Map (SAM) format – text file • BAM format (binary format of SAM file) – to reduce the file size 27

Data Analysis Format Conversion & Demultiplexing Quality Check and Trimming Sequence Alignment Variant Detection Variant Annotation (5 Million) Customize Data Analysis for Multisample 28

Software for variant detection • Commercial – CASAVA (Illumina) – Bio. Scope (Life Tech) – CLC Genomics Workbench – GS Reference Mapper (Roche) • Open source – GATK 29

Variant detection • Input files – bam files – Reference sequence – db. SNP • 不是 alignment完成就可以立即進行 variant detection,建議還需要進行進一步處理 – Local realignment around indels – Mark. Duplicates – Base quality score recalibration • XXX. merged. clean. dedup. recal. bam 30

Realignment AGGGAAACCACACAGGCTTCTTAGGCCATTGGAAT GGAAACCACACAGGCTT---AGGCCATTGGAA GAAACCACACAGGC---TTAGGCCATTGGAAT AAACCACACAGGCT---TAGGCCATTGG AAACCACACAGGCTT---AGGCCATTGG CCACACAGGC---TTAGGCCATTGGAA CACACAGG---CTTAGGCCATTGGAAT 31

Realignment AGGGAAACCACACAGGCTTCTTAGGCCATTGGAAT GGAAACCACACAGGCTT---AGGCCATTGGAAT AAACCACACAGGCTT---AGGCCATTGGAA CACACAGGCTT---AGGCCATTGGAAT 32

Realignment • for contrastive calling projects -- such as cancer tumor/normals -- we recommend realigning both the tumor and the normal together in general to avoid slight alignment differences between the two tissue types* • Known indels data (ex. db. SNP 137) *http: //www. broadinstitute. org/gatk/guide/topic? name=intro 33

Mark. Duplicate AGGGAAACCACACAGGCTTCTTAGGCCATTGGAAT GGAAACCACACAGGCTT---AGGCCATTGGAAT AAACCACACAGGCTT---AGGCCATTGGAA CACACAGGCTT---AGGCCATTGGAAT 34

Variant discovery (GATK) • • Unified. Genotyper (standard protocol) Haplotype. Caller (experimental protocol) Multi-sample SNP and indel calling Output file – VCF format 35

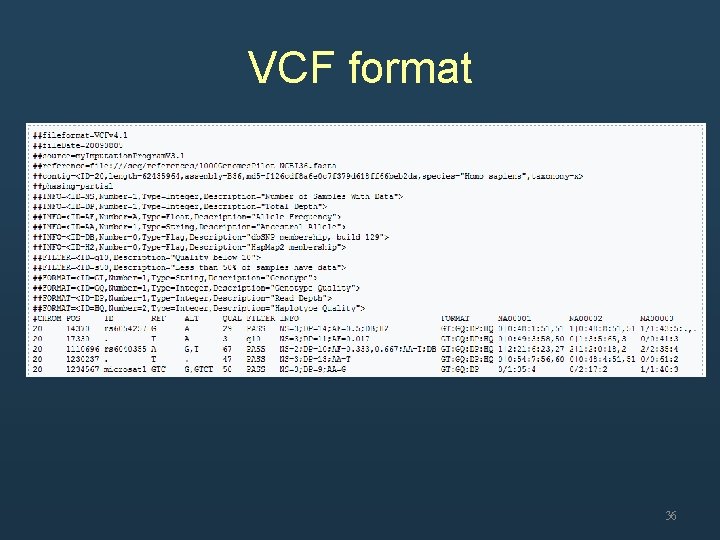
VCF format 36

Variant Detection Output (CASAVA) • Sorted. bam • not. Mapped directory (with the option –sort. Keep. All. Reads) – no. Match – qc. Fail – non. Unique – repeat. Masked – mixed 37

Variant Detection (CASAVA) • • • snps. txt indels. txt 0000/sites. txt 0000/snps. removed. txt 0000/indels. removed. txt 38

Variant Detection Output chr 21. fa. snp. txt (CASAVA 1. 7) position 16050159 16050252 16052239 16052513 16052618 16053659 16054667 16054740 16055942 16058070 16058415 16058766 16058767 16058852 A 0 3 0 0 24 0 0 0 25 2 0 C 2 0 0 27 0 9 1 0 0 0 0 G 0 0 8 0 1 0 37 34 0 8 21 2 25 0 T modified_call total 5 TC 7 6 TA 9 0 G 8 0 C 27 0 A 25 0 C 21 0 G 39 0 G 35 25 T 28 0 G 9 0 G 21 0 A 28 0 G 28 16 T 16 used score reference 7 18. 20: 7. 60 C 9 20. 60: 11. 20 A 8 29. 4 A 27 94. 49 G 25 80. 29: 3. 00 G 9 12. 29 A 38 125. 59 C 34 112. 25 A 25 77. 49 C 8 25. 1 A 21 71. 7 A 27 85. 20: 7. 60 G 27 85. 10: 7. 60 A 16 50. 8 A type SNP_het 2 SNP_diff SNP_diff SNP_diff 39

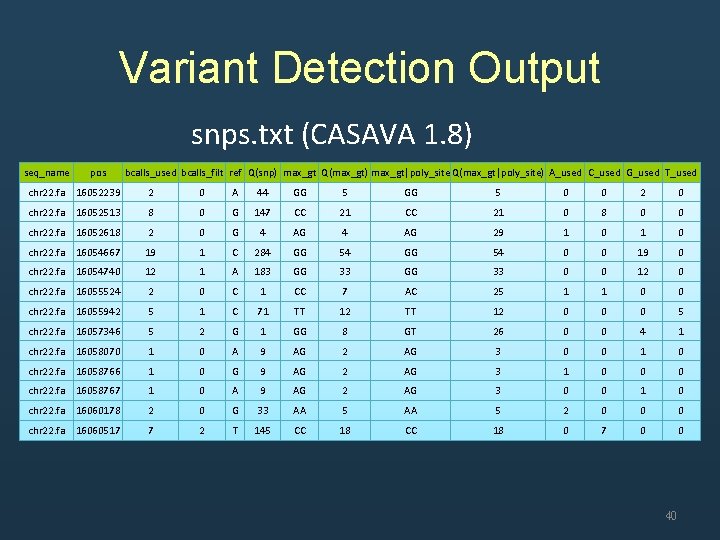
Variant Detection Output snps. txt (CASAVA 1. 8) seq_name pos bcalls_used bcalls_filt ref Q(snp) max_gt Q(max_gt) max_gt|poly_site Q(max_gt|poly_site) A_used C_used G_used T_used chr 22. fa 16052239 2 0 A 44 GG 5 0 0 2 0 chr 22. fa 16052513 8 0 G 147 CC 21 0 8 0 0 chr 22. fa 16052618 2 0 G 4 AG 29 1 0 chr 22. fa 16054667 19 1 C 284 GG 54 0 0 19 0 chr 22. fa 16054740 12 1 A 183 GG 33 0 0 12 0 chr 22. fa 16055524 2 0 C 1 CC 7 AC 25 1 1 0 0 chr 22. fa 16055942 5 1 C 71 TT 12 0 0 0 5 chr 22. fa 16057346 5 2 G 1 GG 8 GT 26 0 0 4 1 chr 22. fa 16058070 1 0 A 9 AG 2 AG 3 0 0 1 0 chr 22. fa 16058766 1 0 G 9 AG 2 AG 3 1 0 0 0 chr 22. fa 16058767 1 0 A 9 AG 2 AG 3 0 0 1 0 chr 22. fa 16060178 2 0 G 33 AA 5 2 0 0 0 chr 22. fa 16060517 7 2 T 145 CC 18 0 7 0 0 40

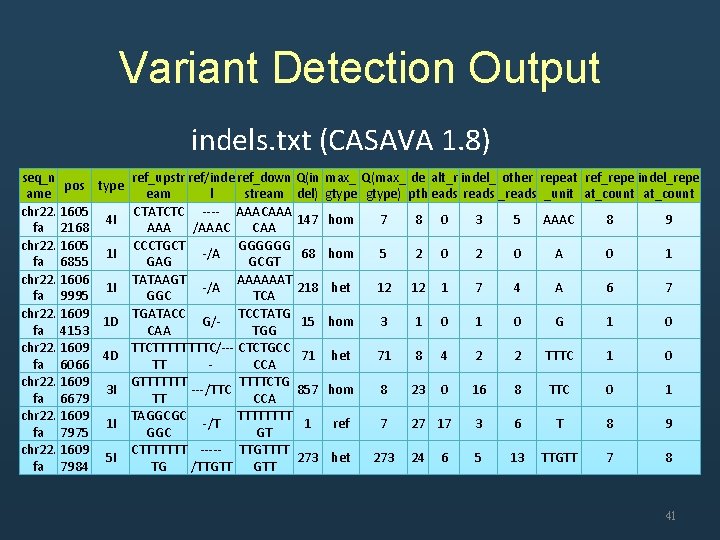
Variant Detection Output indels. txt (CASAVA 1. 8) seq_n ref_upstr ref/inde ref_down Q(in max_ Q(max_ de alt_r indel_ other repeat ref_repe indel_repe pos type ame eam l stream del) gtype) pth eads reads _unit at_count chr 22. 1605 CTATCTC ---- AAACAAA 4 I 147 hom 7 8 0 3 5 AAAC 8 9 fa 2168 AAA /AAAC CAA chr 22. 1605 CCCTGCT GGGGGG 1 I -/A 68 hom 5 2 0 A 0 1 fa 6855 GAG GCGT chr 22. 1606 TATAAGT AAAAAAT 1 I -/A 218 het 12 12 1 7 4 A 6 7 fa 9995 GGC TCA chr 22. 1609 TGATACC TCCTATG 1 D G/15 hom 3 1 0 G 1 0 fa 4153 CAA TGG chr 22. 1609 TTCTTTTT TTTC/--- CTCTGCC 4 D 71 het 71 8 4 2 2 TTTC 1 0 fa 6066 TT CCA chr 22. 1609 GTTTTTTTCTG 3 I ---/TTC 857 hom 8 23 0 16 8 TTC 0 1 fa 6679 TT CCA chr 22. 1609 TAGGCGC TTTT 1 I -/T 1 ref 7 27 17 3 6 T 8 9 fa 7975 GGC GT chr 22. 1609 CTTTTTTT ----- TTGTTTT 5 I 273 het 273 24 6 5 13 TTGTT 7 8 fa 7984 TG /TTGTT 41

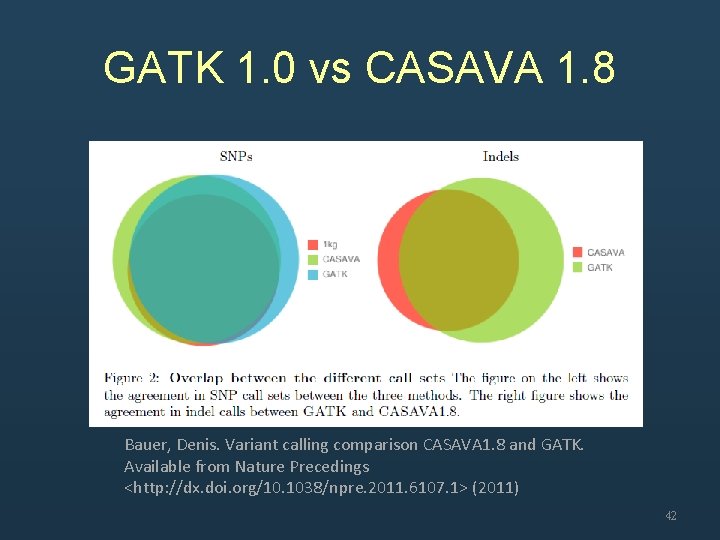
GATK 1. 0 vs CASAVA 1. 8 Bauer, Denis. Variant calling comparison CASAVA 1. 8 and GATK. Available from Nature Precedings <http: //dx. doi. org/10. 1038/npre. 2011. 6107. 1> (2011) 42

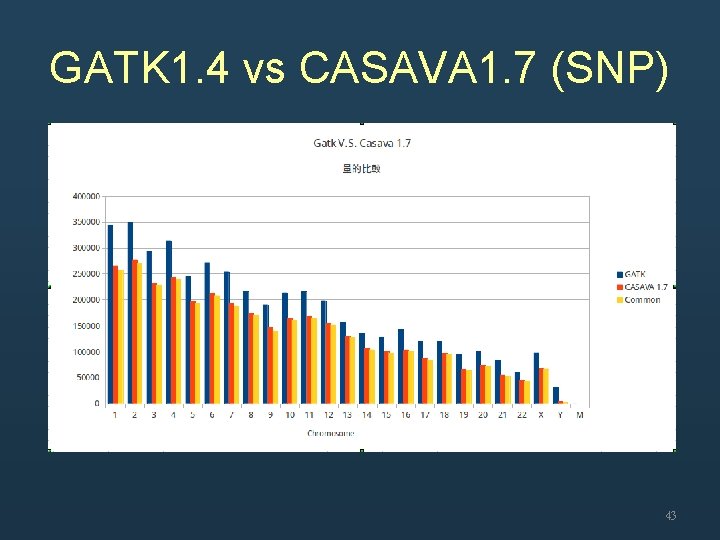
GATK 1. 4 vs CASAVA 1. 7 (SNP) 43

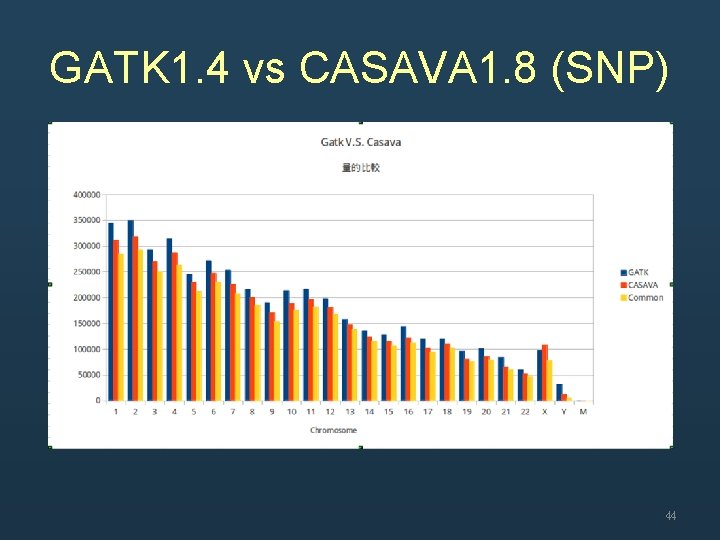
GATK 1. 4 vs CASAVA 1. 8 (SNP) 44

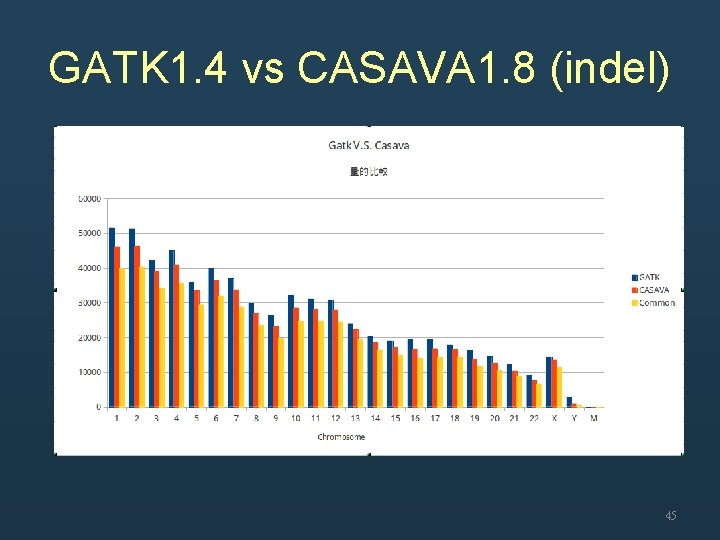
GATK 1. 4 vs CASAVA 1. 8 (indel) 45

Data Analysis r o f s r h <8 n a m u h a ) X 0 3 ( e m o n ge paired-end reads by Hi. Seq 2000 Format Conversion & Demultiplexing Quality Check and Trimming Sequence Alignment Variant Detection Variant Annotation (5 Million) Customize Data Analysis for Multisample 46

提供分析服務 • 分析一個 30 X human whole genome alignment and variant discovery時間已經 從 10天減少至 8小時 • 分析一個 100 X human whole exome的時 間可以在 3小時內完成 • 目前中心已經完成超過 250個 exome seq, 30 whole genome之分析 47

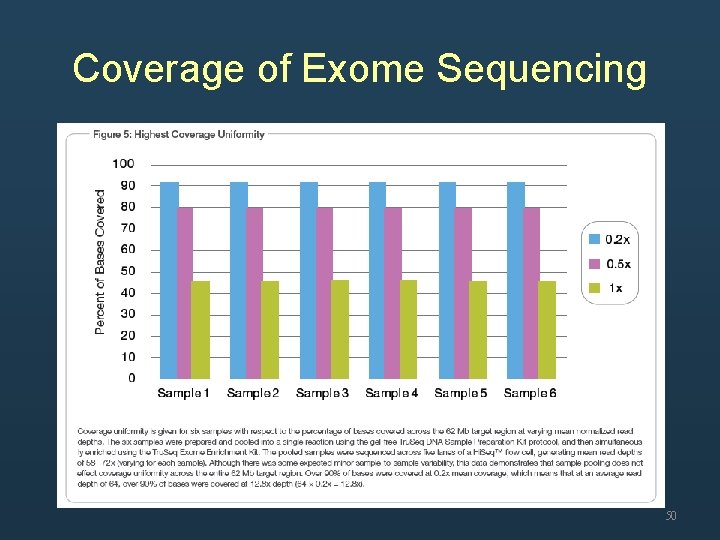
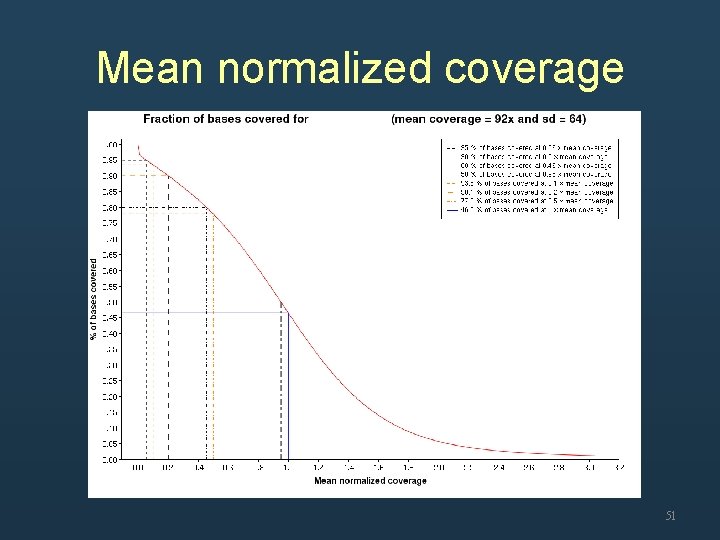
Coverage • Whole genome sequencing – Mean coverage • Exome sequencing – Over 90% of target region were covered at 0. 2 x mean coverage 48

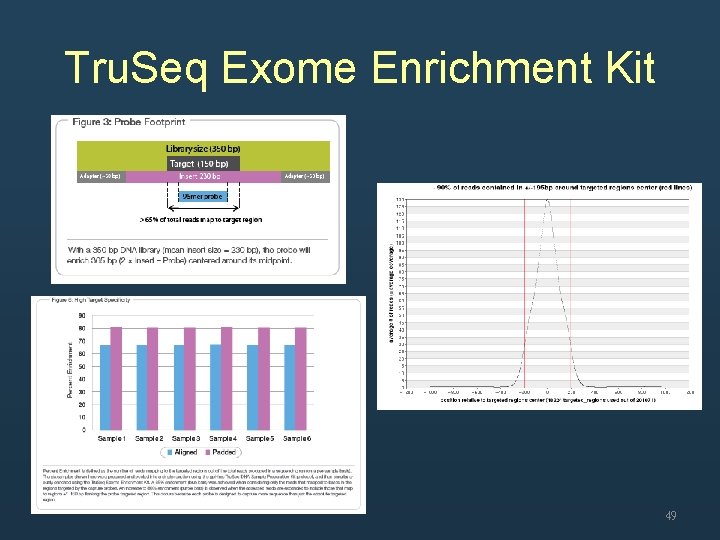
Tru. Seq Exome Enrichment Kit 49

Coverage of Exome Sequencing 50

Mean normalized coverage 51

Qualities • Base Quality – In FASTQ files • Alignment Quality – In BAM files • Quality for the assertion made in alternative allele – In VCF file, snps. txt, or indels. txt 52

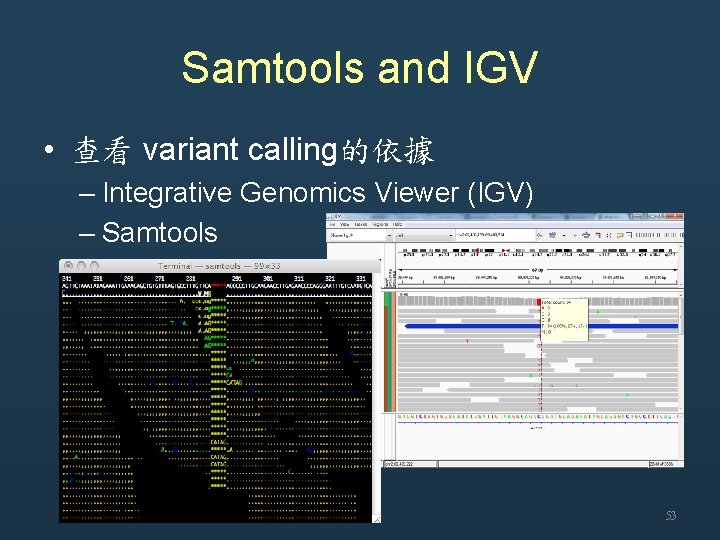
Samtools and IGV • 查看 variant calling的依據 – Integrative Genomics Viewer (IGV) – Samtools 53


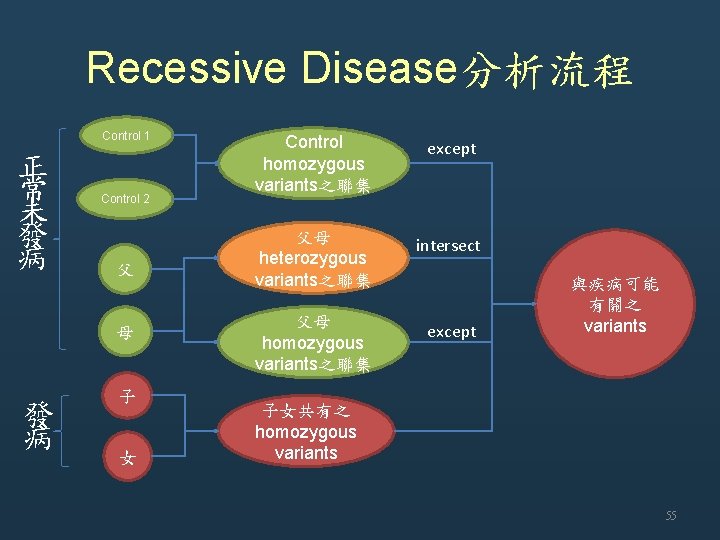
Recessive Disease分析流程 Control 1 正 常 未 發 病 Control 2 父 母 發 病 子 女 Control homozygous variants之聯集 except 父母 heterozygous variants之聯集 intersect 父母 homozygous variants之聯集 except 與疾病可能 有關之 variants 子女共有之 homozygous variants 55

SQL 語法 ((select chr, pos, variant from case_variant_homo where sample='case_2' intersect select chr, pos, variant from case_variant_homo where sample='case_3' except select chr, pos, variant from case_variant_homo where sample='case_1') intersect select chr, pos, variant from case_variant_het where sample='case_1') except select chr, pos, variant from control_variant_homo 56

分析前需決定的事項 • Trimming 的 Q值 (Q 30 ? ) • 分析的 pipeline – BWA/GATK – CASAVA • Reference Genome • db. SNP version • 測序與分析結果儲存的方式 – FASTQ files (120 GB / 30 X human whole genome) – BAM files and VCF files (380 GB / 30 X human whole genome) 57


59

Thanks for your attention chuankun@ibms. sinica. edu. tw