Stanford Comprehensive Cancer Center Stanford Genome Technology Center

- Slides: 62

Stanford Comprehensive Cancer Center Stanford Genome Technology Center An Introduction to Next Generation Sequencing Hanlee Ji, M. D. ã Stanford University

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Overview • Principles of next generation DNA sequencing • Analysis of genetic variation and research applications

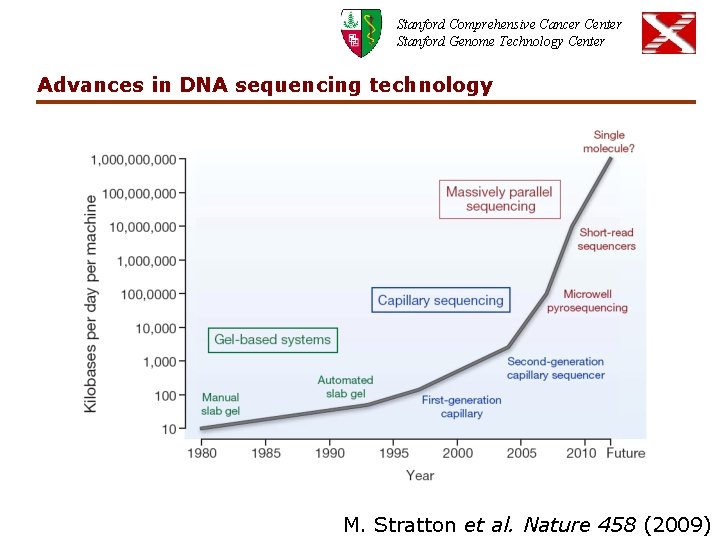
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Advances in DNA sequencing technology M. Stratton et al. Nature 458 (2009)

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Applications • Identifying genetic variants – Whole genome – Exome – Subsets • Transcriptomes (e. g. RNASeq) • Chip-seq • Epigenomes (methylation) • Many others!

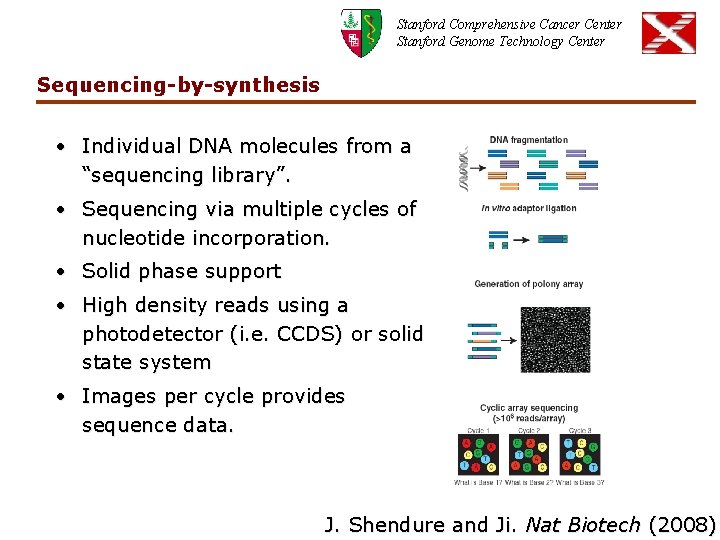
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequencing-by-synthesis • Individual DNA molecules from a “sequencing library”. • Sequencing via multiple cycles of nucleotide incorporation. • Solid phase support • High density reads using a photodetector (i. e. CCDS) or solid state system • Images per cycle provides sequence data. J. Shendure and Ji. Nat Biotech (2008)

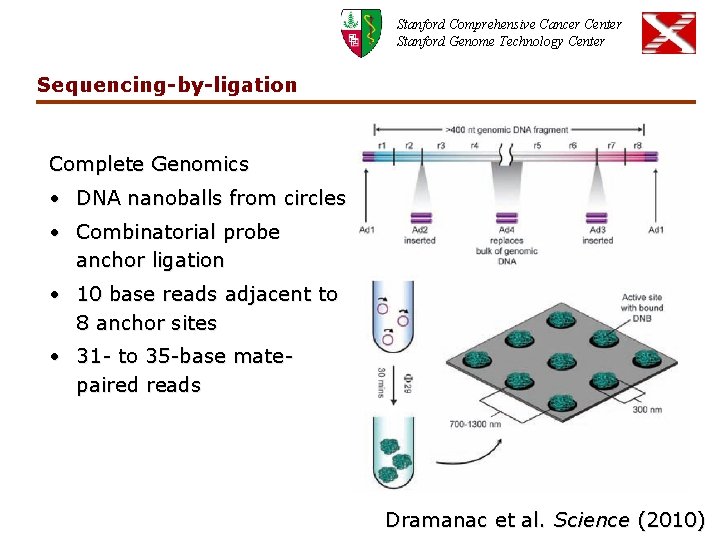
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequencing-by-ligation Complete Genomics • DNA nanoballs from circles • Combinatorial probe anchor ligation • 10 base reads adjacent to 8 anchor sites • 31 - to 35 -base matepaired reads Dramanac et al. Science (2010)

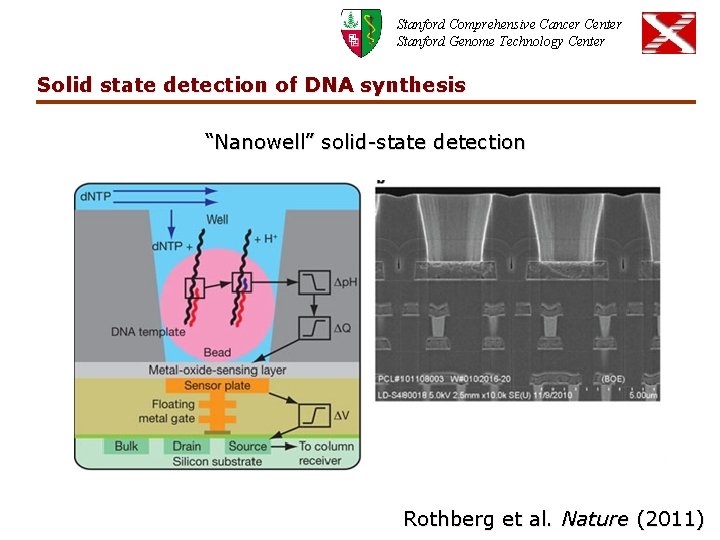
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Solid state detection of DNA synthesis “Nanowell” solid-state detection Rothberg et al. Nature (2011)

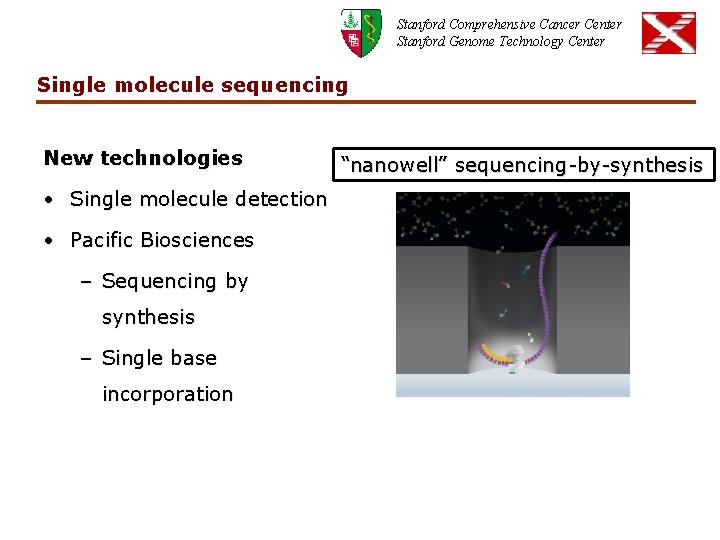
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Single molecule sequencing New technologies • Single molecule detection • Pacific Biosciences – Sequencing by synthesis – Single base incorporation “nanowell” sequencing-by-synthesis

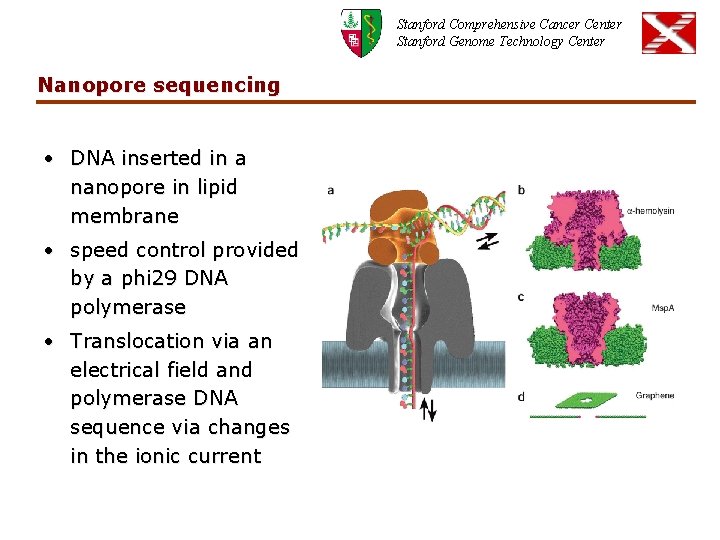
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Nanopore sequencing • DNA inserted in a nanopore in lipid membrane • speed control provided by a phi 29 DNA polymerase • Translocation via an electrical field and polymerase DNA sequence via changes in the ionic current

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Issues with next generation DNA sequencing • Higher sequencing error rates – <0. 1 to 10% or greater depending on sequencing chemistry and configuration • Systematic bias based on approach • Short sequence reads (<250 bases) • Massive data output – Data storage anagement – Variant calling analysis

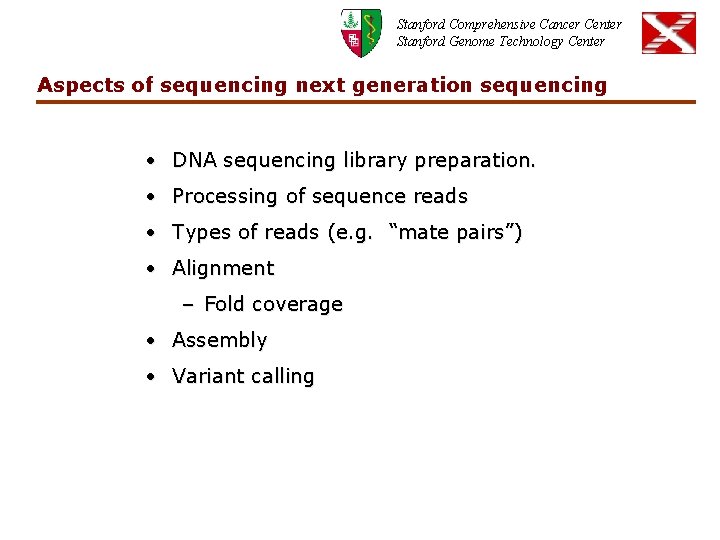
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Aspects of sequencing next generation sequencing • DNA sequencing library preparation. • Processing of sequence reads • Types of reads (e. g. “mate pairs”) • Alignment – Fold coverage • Assembly • Variant calling

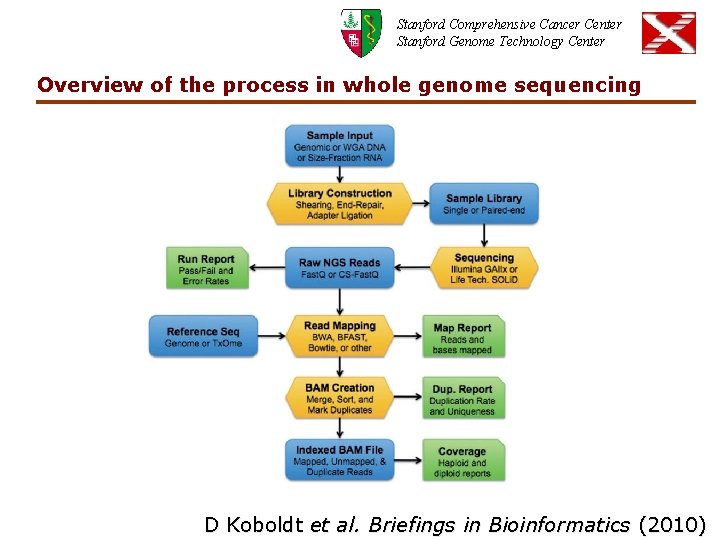
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Overview of the process in whole genome sequencing D Koboldt et al. Briefings in Bioinformatics (2010)

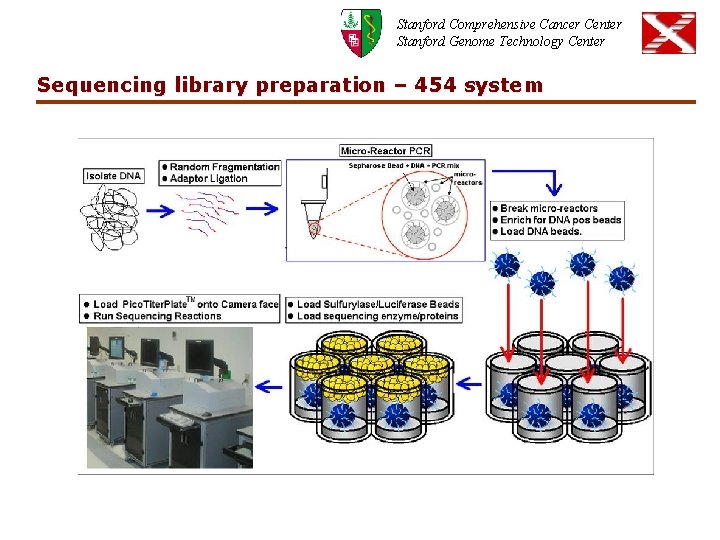
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequencing library preparation – 454 system

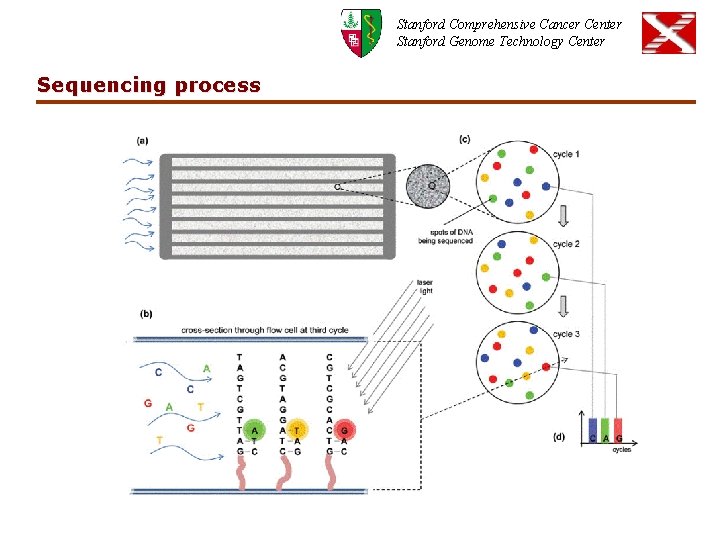
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequencing process

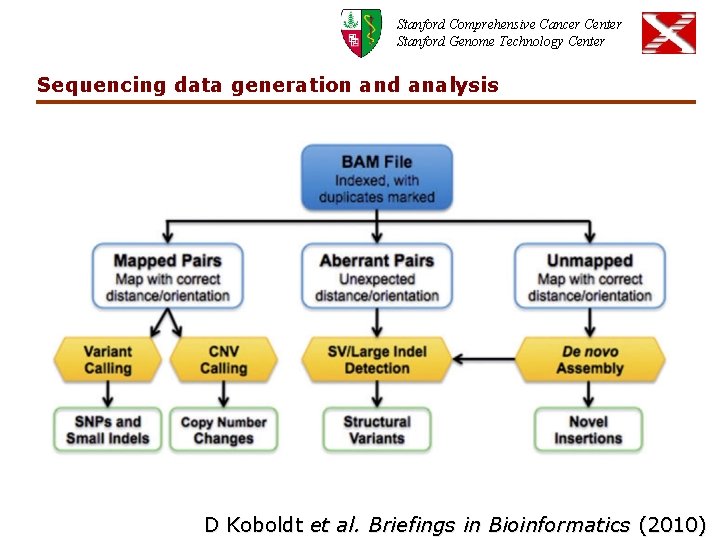
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequencing data generation and analysis D Koboldt et al. Briefings in Bioinformatics (2010)

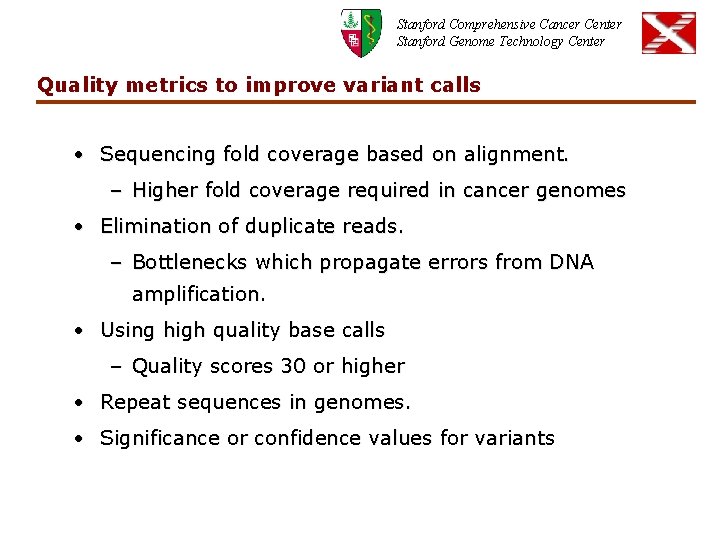
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Quality metrics to improve variant calls • Sequencing fold coverage based on alignment. – Higher fold coverage required in cancer genomes • Elimination of duplicate reads. – Bottlenecks which propagate errors from DNA amplification. • Using high quality base calls – Quality scores 30 or higher • Repeat sequences in genomes. • Significance or confidence values for variants

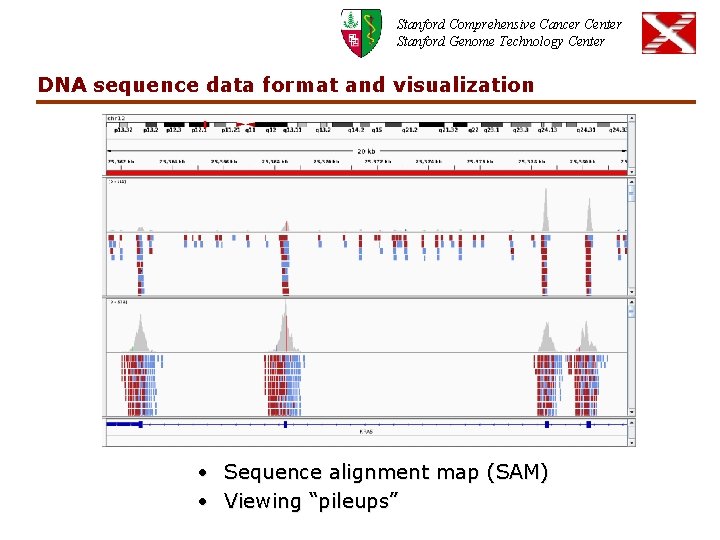
Stanford Comprehensive Cancer Center Stanford Genome Technology Center DNA sequence data format and visualization • Sequence alignment map (SAM) • Viewing “pileups”

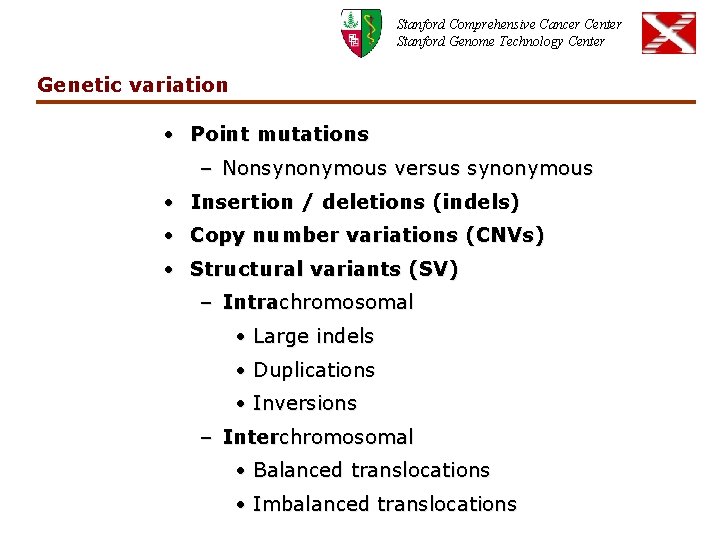
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Genetic variation • Point mutations – Nonsynonymous versus synonymous • Insertion / deletions (indels) • Copy number variations (CNVs) • Structural variants (SV) – Intrachromosomal • Large indels • Duplications • Inversions – Interchromosomal • Balanced translocations • Imbalanced translocations

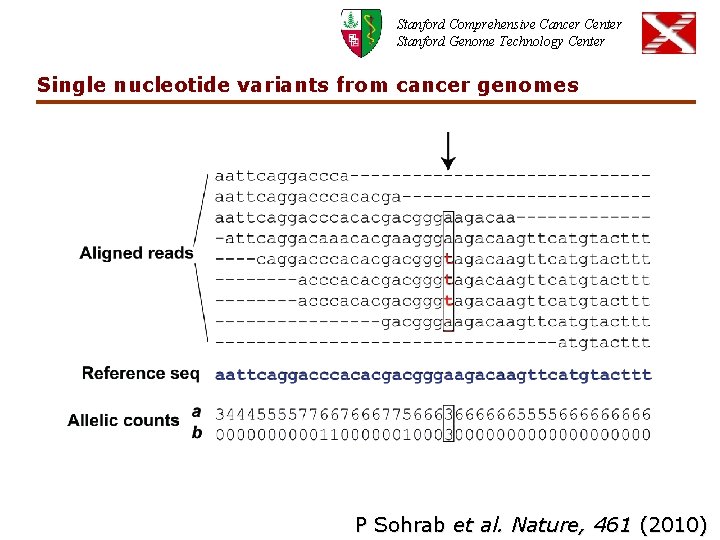
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Single nucleotide variants from cancer genomes P Sohrab et al. Nature, 461 (2010)

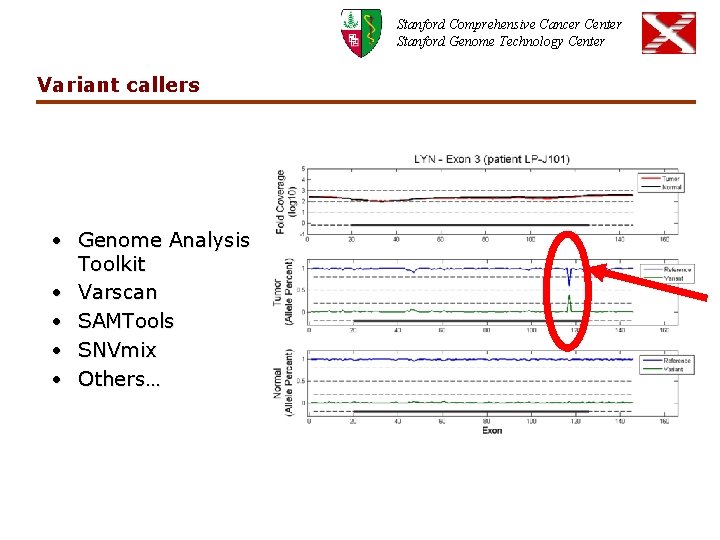
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Variant callers • Genome Analysis Toolkit • Varscan • SAMTools • SNVmix • Others…

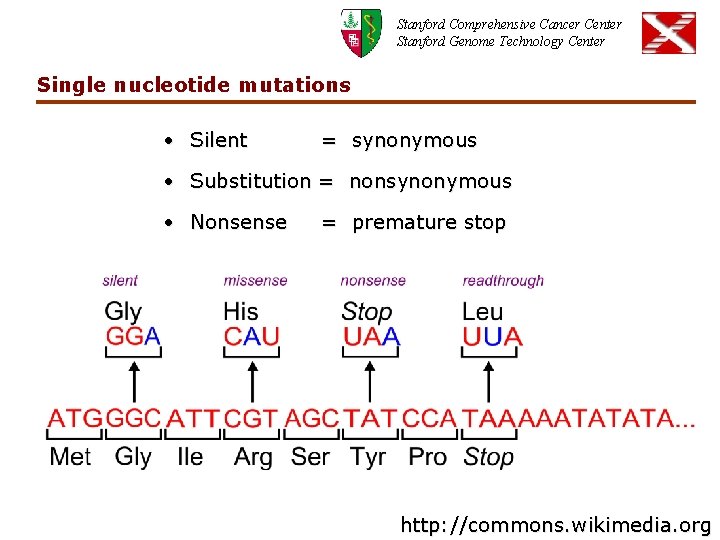
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Single nucleotide mutations • Silent = synonymous • Substitution = nonsynonymous • Nonsense = premature stop http: //commons. wikimedia. org

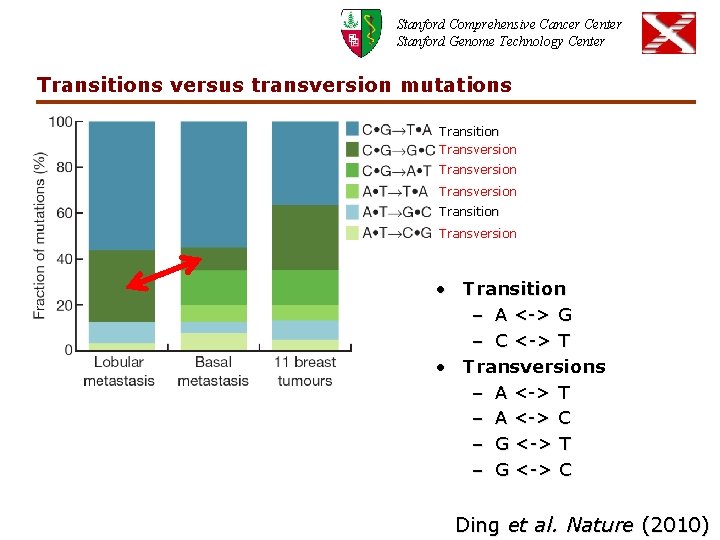
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Transitions versus transversion mutations Transition Transversion Transition Transversion • Transition – A <-> G – C <-> T • Transversions – A <-> T – A <-> C – G <-> T – G <-> C Ding et al. Nature (2010)

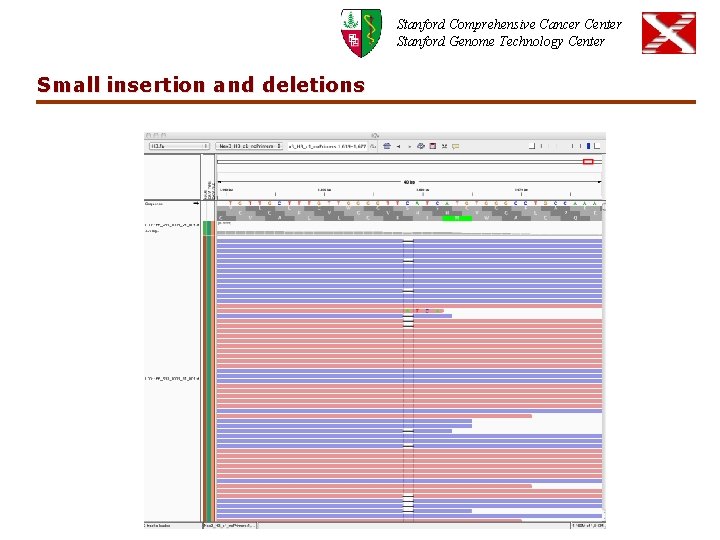
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Small insertion and deletions

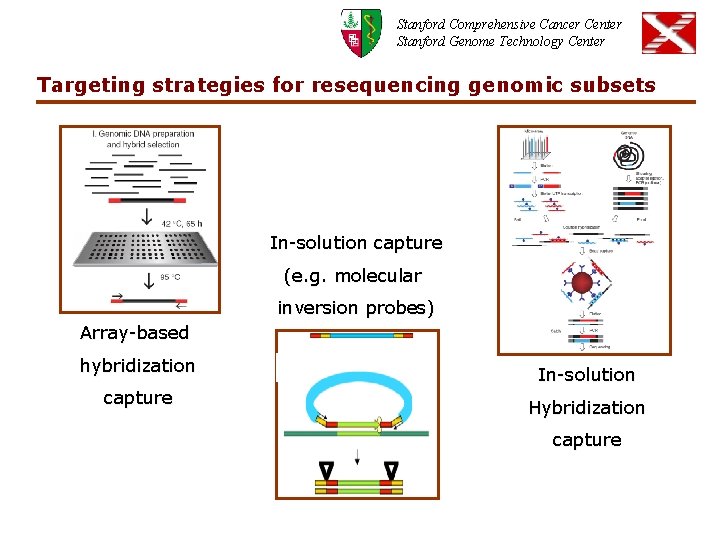
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Targeting strategies for resequencing genomic subsets In-solution capture (e. g. molecular inversion probes) Array-based hybridization capture In-solution Hybridization capture

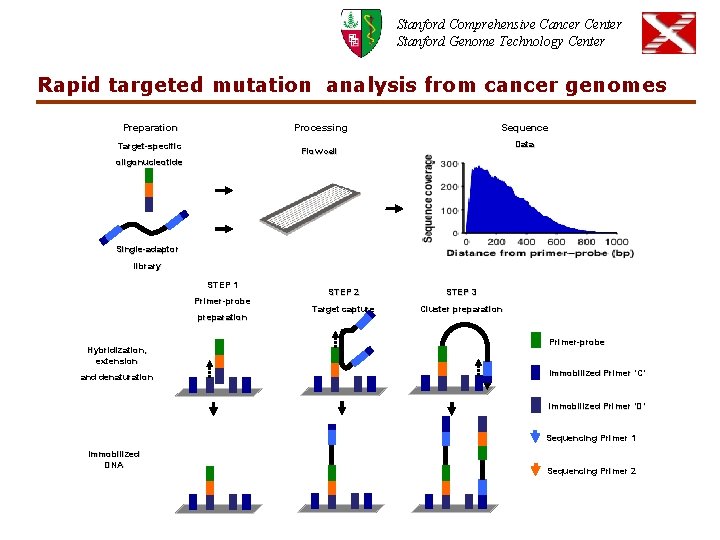
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Rapid targeted mutation analysis from cancer genomes a Preparation Sequence Processing Target-specific Data Flow cell oligonucleotide Single-adaptor b library STEP 1 Primer-probe preparation Hybridization, extension and denaturation STEP 2 STEP 3 Target capture Cluster preparation Primer-probe Immobilized Primer ‘C’ Immobilized Primer ‘D’ Sequencing Primer 1 Immobilized DNA Sequencing Primer 2

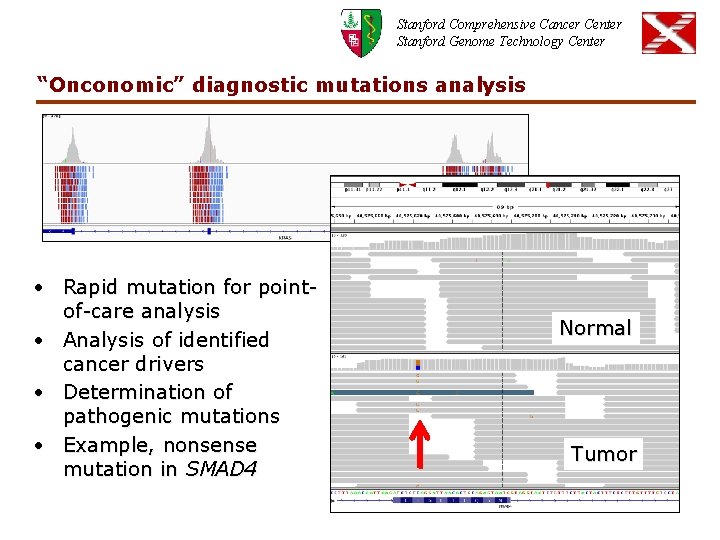
Stanford Comprehensive Cancer Center Stanford Genome Technology Center “Onconomic” diagnostic mutations analysis • Rapid mutation for pointof-care analysis • Analysis of identified cancer drivers • Determination of pathogenic mutations • Example, nonsense mutation in SMAD 4 Normal Tumor

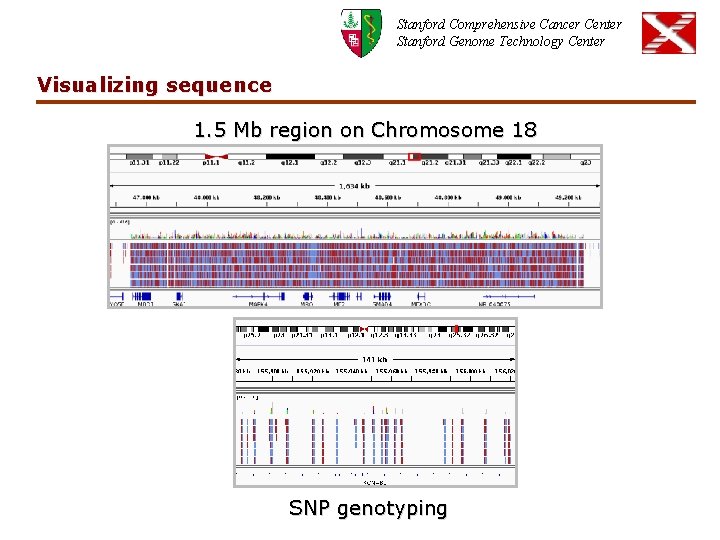
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Visualizing sequence 1. 5 Mb region on Chromosome 18 SNP genotyping

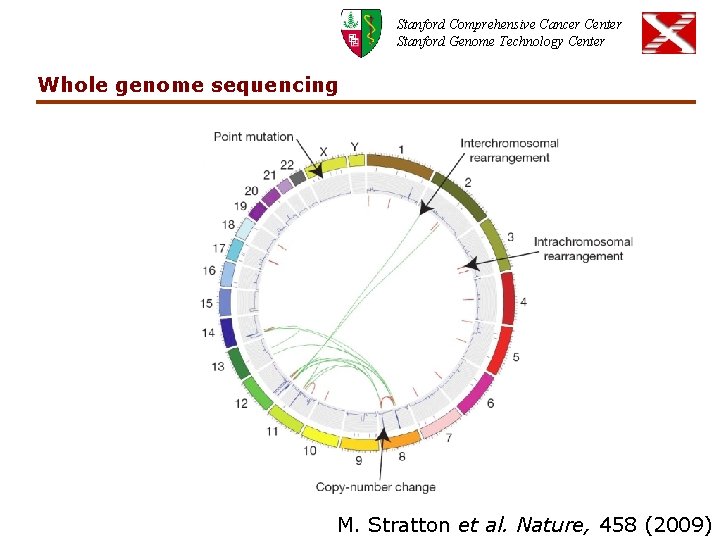
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Whole genome sequencing M. Stratton et al. Nature, 458 (2009)

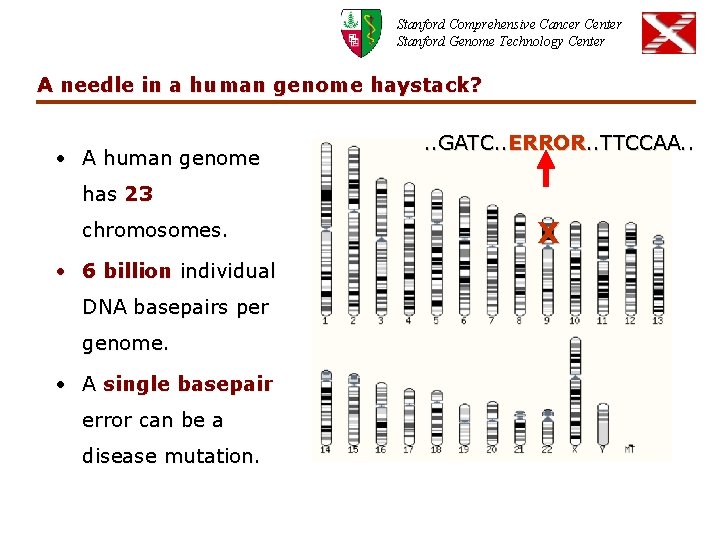
Stanford Comprehensive Cancer Center Stanford Genome Technology Center A needle in a human genome haystack? • A human genome . . GATC. . ERROR. . TTCCAA. . has 23 chromosomes. • 6 billion individual DNA basepairs per genome. • A single basepair error can be a disease mutation. X

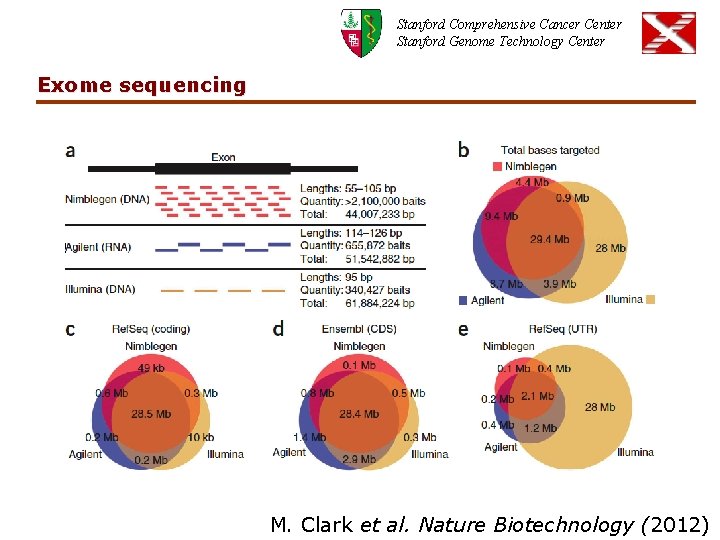
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Exome sequencing M. Clark et al. Nature Biotechnology (2012)

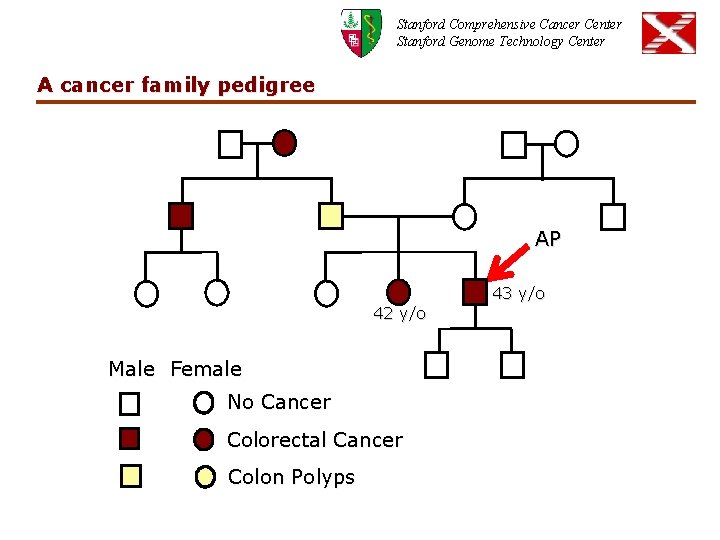
Stanford Comprehensive Cancer Center Stanford Genome Technology Center A cancer family pedigree AP 43 y/o 42 y/o Male Female No Cancer Colorectal Cancer Colon Polyps

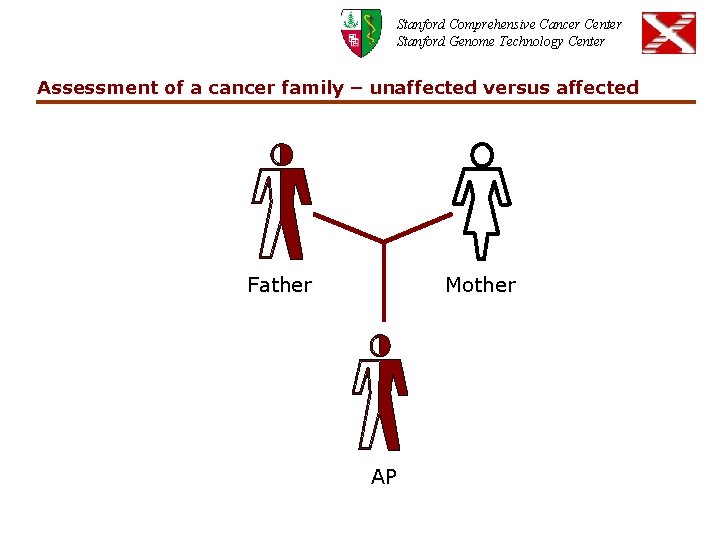
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Assessment of a cancer family – unaffected versus affected Father Mother AP

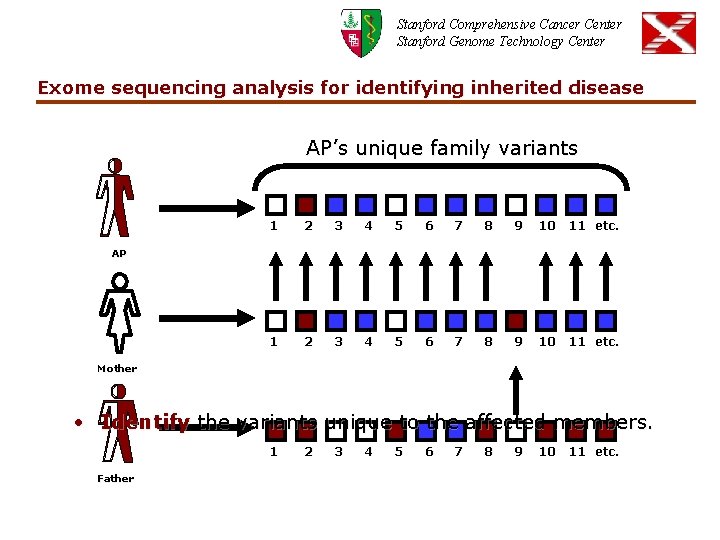
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Exome sequencing analysis for identifying inherited disease AP’s unique family variants 1 2 3 4 5 6 7 8 9 10 11 etc. AP Mother • Identify the variants unique to the affected members. 1 Father 2 3 4 5 6 7 8 9 10 11 etc.

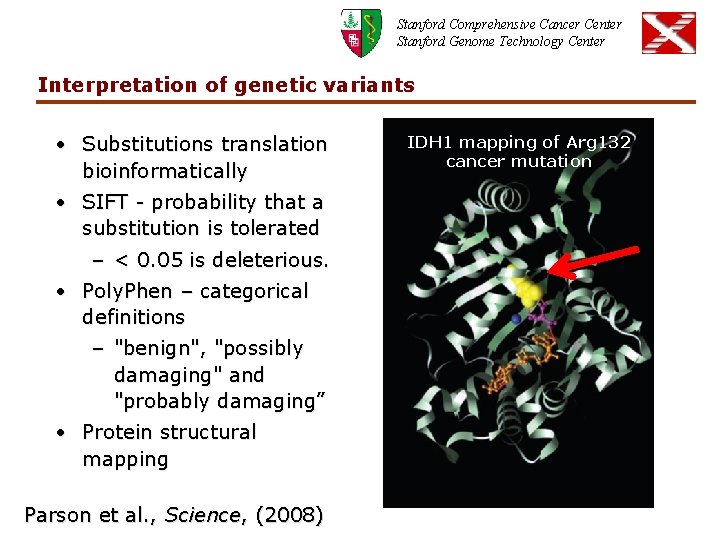
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Interpretation of genetic variants • Substitutions translation bioinformatically • SIFT - probability that a substitution is tolerated – < 0. 05 is deleterious. • Poly. Phen – categorical definitions – "benign", "possibly damaging" and "probably damaging” • Protein structural mapping Parson et al. , Science, (2008) IDH 1 mapping of Arg 132 cancer mutation

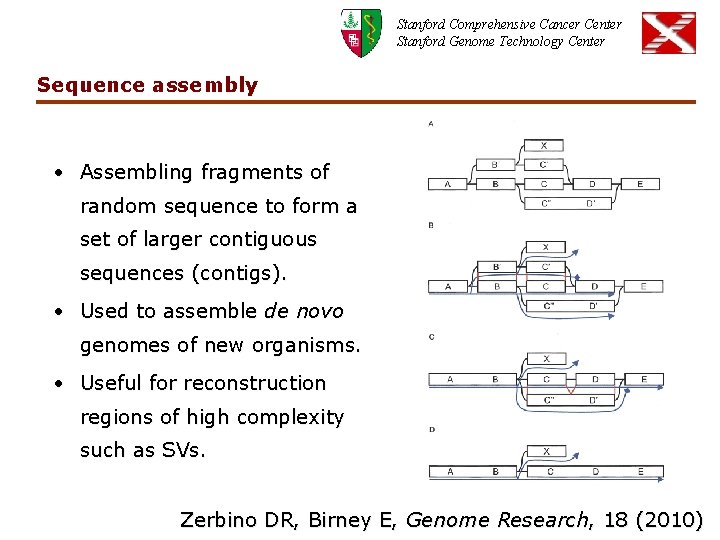
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequence assembly • Assembling fragments of random sequence to form a set of larger contiguous sequences (contigs). • Used to assemble de novo genomes of new organisms. • Useful for reconstruction regions of high complexity such as SVs. Zerbino DR, Birney E, Genome Research, 18 (2010)

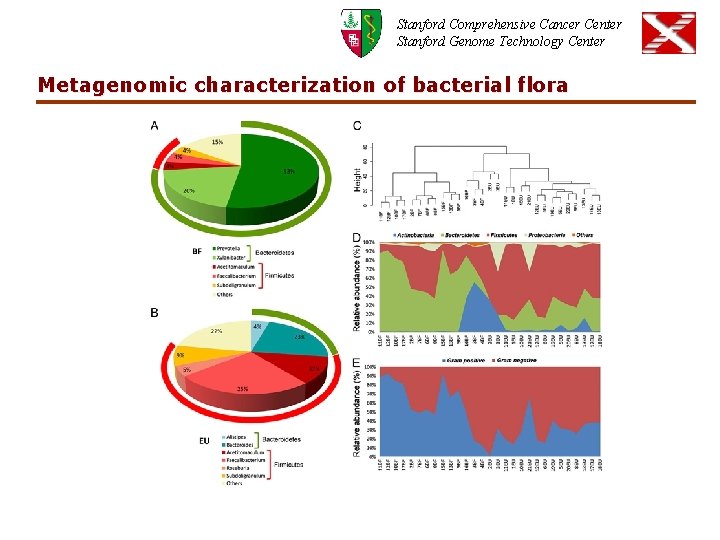
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Metagenomic characterization of bacterial flora

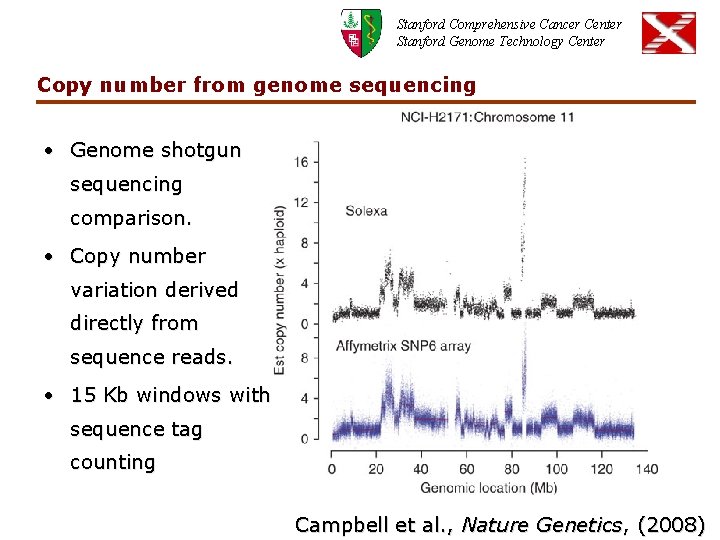
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Copy number from genome sequencing • Genome shotgun sequencing comparison. • Copy number variation derived directly from sequence reads. • 15 Kb windows with sequence tag counting Campbell et al. , Nature Genetics, (2008)

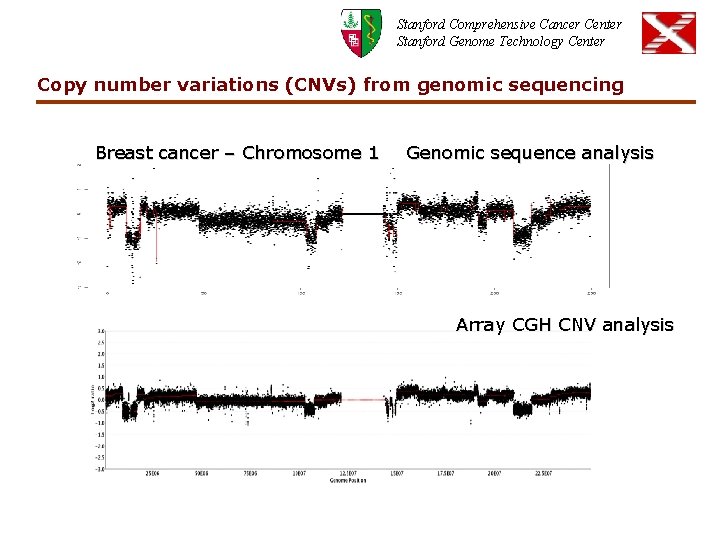
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Copy number variations (CNVs) from genomic sequencing Breast cancer – Chromosome 1 Genomic sequence analysis Array CGH CNV analysis

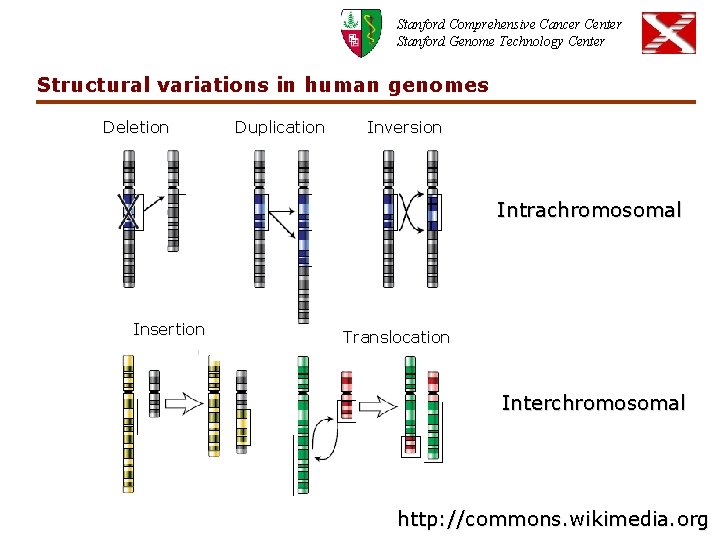
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Structural variations in human genomes Deletion Duplication Inversion Intrachromosomal Insertion Translocation Interchromosomal http: //commons. wikimedia. org

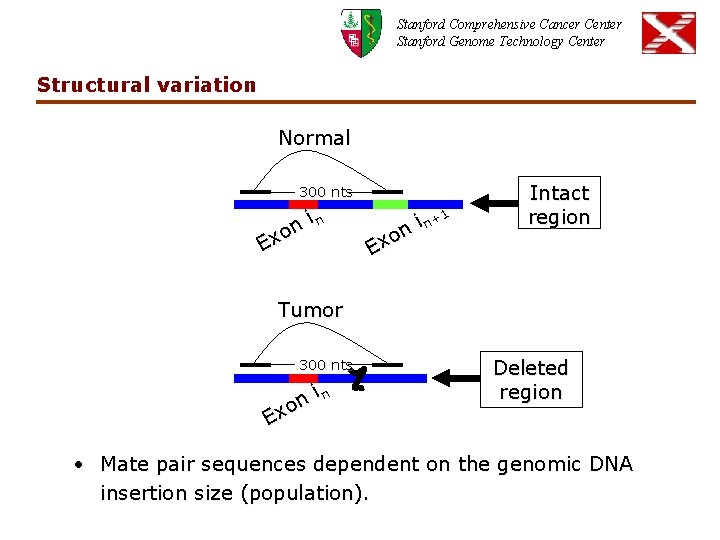
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Structural variation Normal 300 nts in n o Ex 1 i n+ on x E Intact region Tumor 300 nts in n o Ex Deleted region • Mate pair sequences dependent on the genomic DNA insertion size (population).

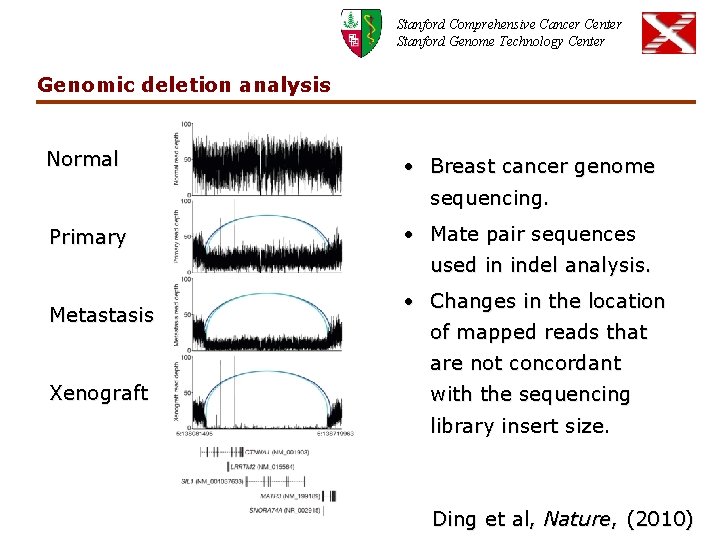
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Genomic deletion analysis Normal • Breast cancer genome sequencing. Primary • Mate pair sequences used in indel analysis. Metastasis • Changes in the location of mapped reads that are not concordant Xenograft with the sequencing library insert size. Ding et al, Nature, (2010)

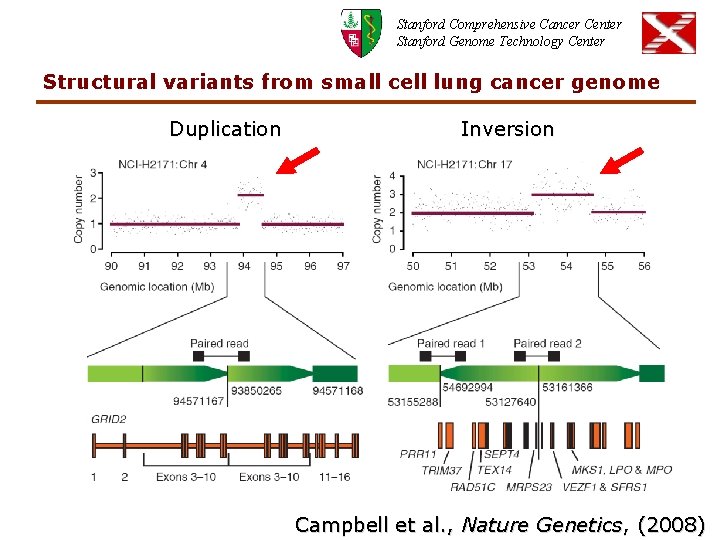
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Structural variants from small cell lung cancer genome Duplication Inversion Campbell et al. , Nature Genetics, (2008)

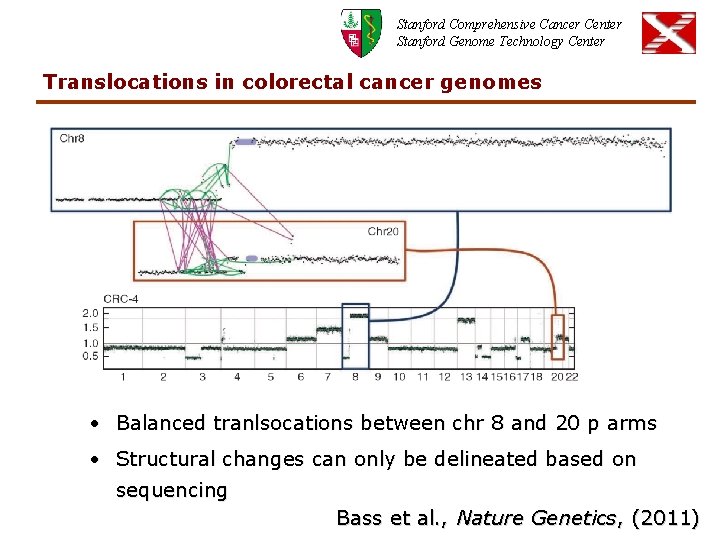
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Translocations in colorectal cancer genomes • Balanced tranlsocations between chr 8 and 20 p arms • Structural changes can only be delineated based on sequencing Bass et al. , Nature Genetics, (2011)

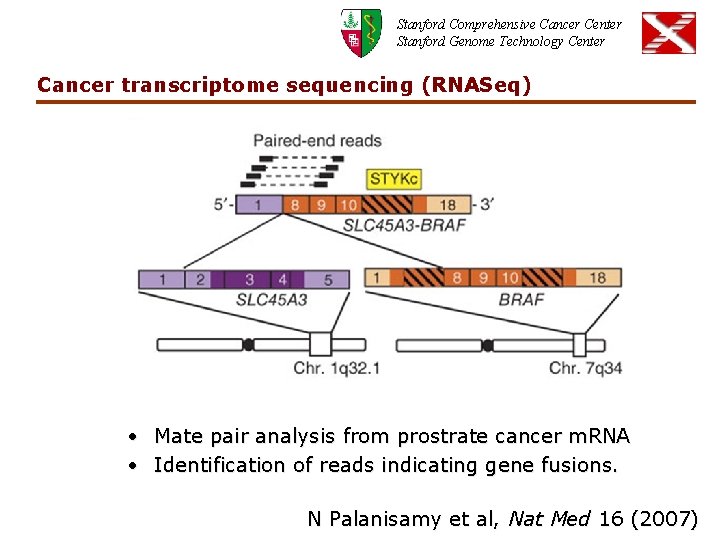
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Cancer transcriptome sequencing (RNASeq) • Mate pair analysis from prostrate cancer m. RNA • Identification of reads indicating gene fusions. N Palanisamy et al, Nat Med 16 (2007)

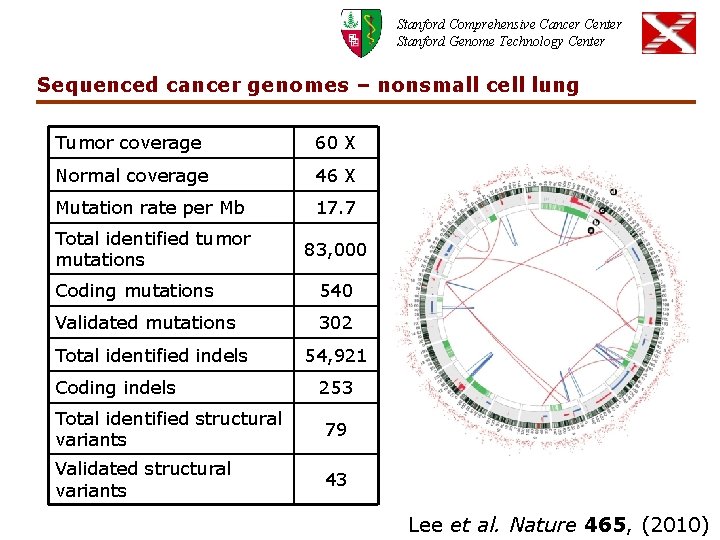
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequenced cancer genomes – nonsmall cell lung Tumor coverage 60 X Normal coverage 46 X Mutation rate per Mb 17. 7 Total identified tumor mutations 83, 000 Coding mutations 540 Validated mutations 302 Total identified indels Coding indels 54, 921 253 Total identified structural variants 79 Validated structural variants 43 Lee et al. Nature 465, (2010)

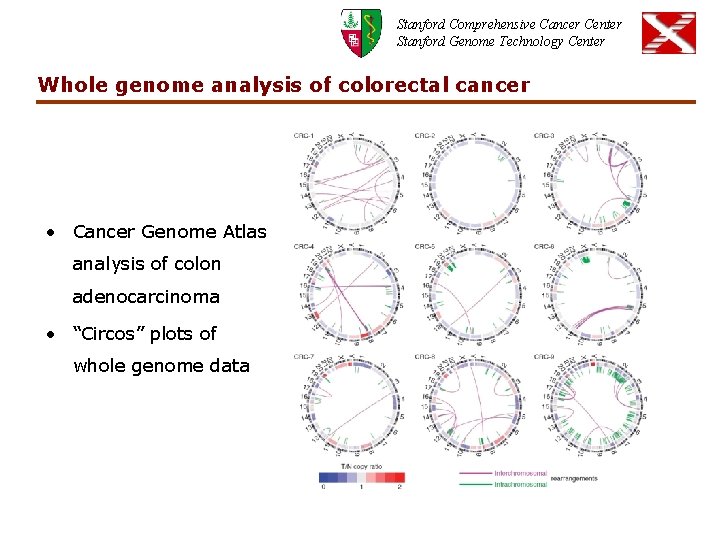
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Whole genome analysis of colorectal cancer • Cancer Genome Atlas analysis of colon adenocarcinoma • “Circos” plots of whole genome data

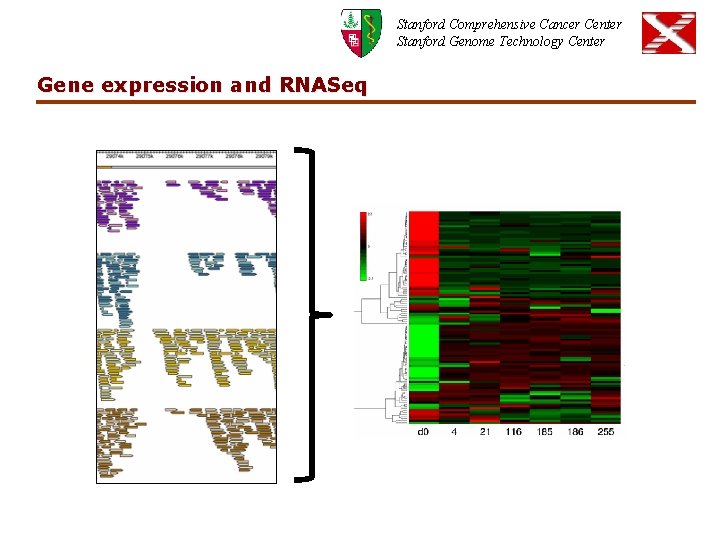
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Gene expression and RNASeq

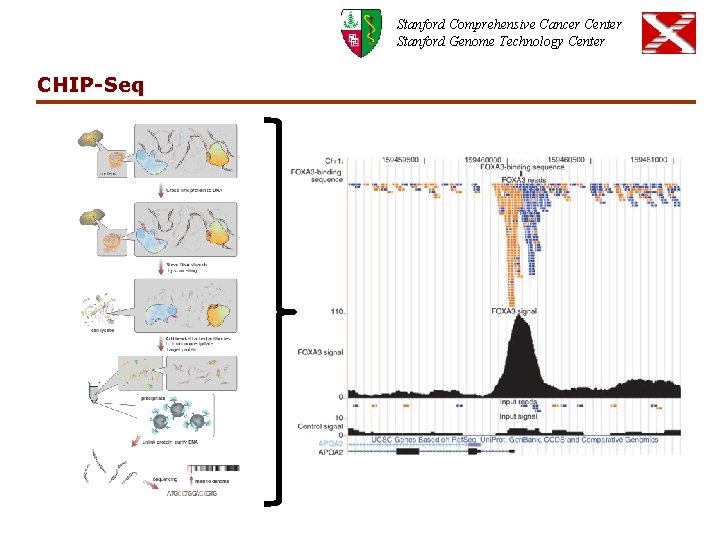
Stanford Comprehensive Cancer Center Stanford Genome Technology Center CHIP-Seq

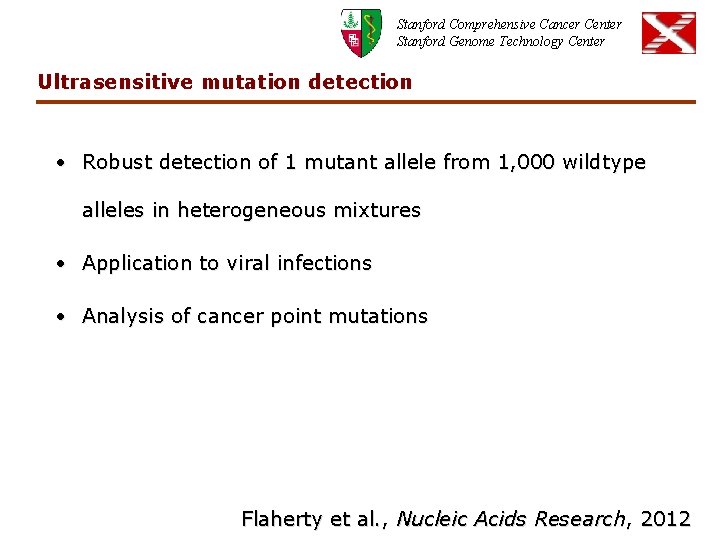
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Ultrasensitive mutation detection • Robust detection of 1 mutant allele from 1, 000 wildtype alleles in heterogeneous mixtures • Application to viral infections • Analysis of cancer point mutations Flaherty et al. , Nucleic Acids Research, 2012

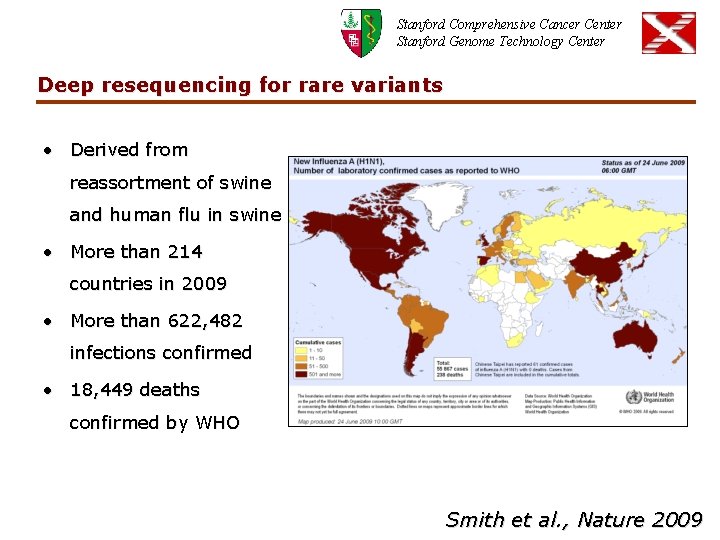
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Deep resequencing for rare variants • Derived from reassortment of swine and human flu in swine • More than 214 countries in 2009 • More than 622, 482 infections confirmed • 18, 449 deaths confirmed by WHO Smith et al. , Nature 2009

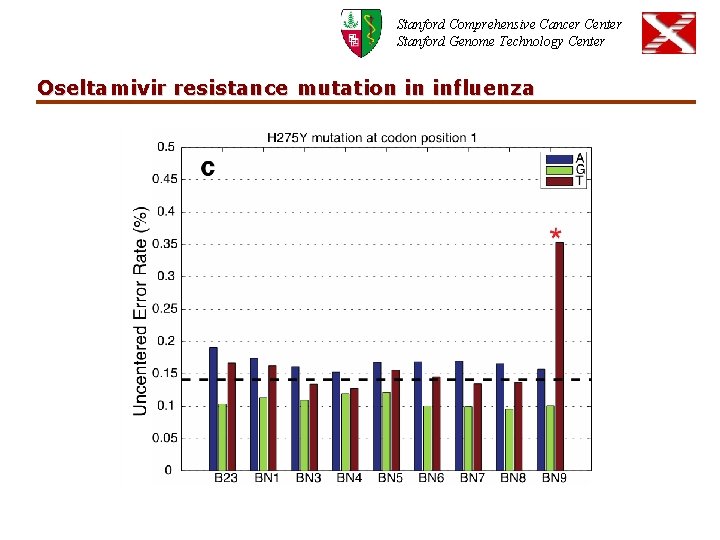
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Oseltamivir resistance mutation in influenza

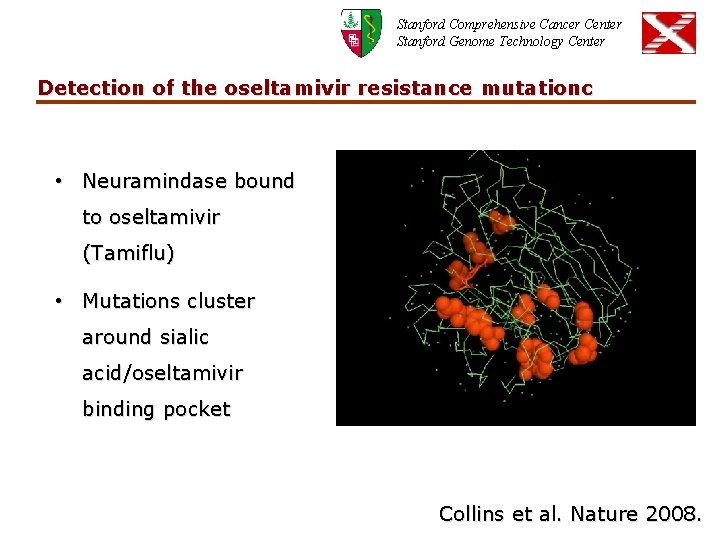
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Detection of the oseltamivir resistance mutationc • Neuramindase bound to oseltamivir (Tamiflu) • Mutations cluster around sialic acid/oseltamivir binding pocket Collins et al. Nature 2008.

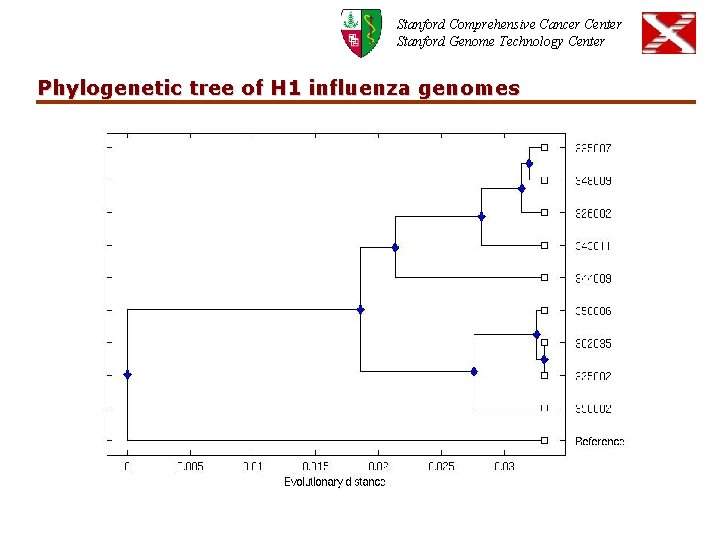
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Phylogenetic tree of H 1 influenza genomes

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Conclusion • Multiple approaches available for analysis of genomes • Scale of sequence data requires extensive computational, bioinformatic and statistical data analysis • Methods, technologies and analysis continue to improve and become simpler.

Stanford Comprehensive Cancer Center Stanford Genome Technology Center

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Genetics via large scale sequencing • mutations and other genomic DNA aberrations contribute to neoplastic development • Specific genetic variants and other indicate clinical phenotype • Utility as diagnostics

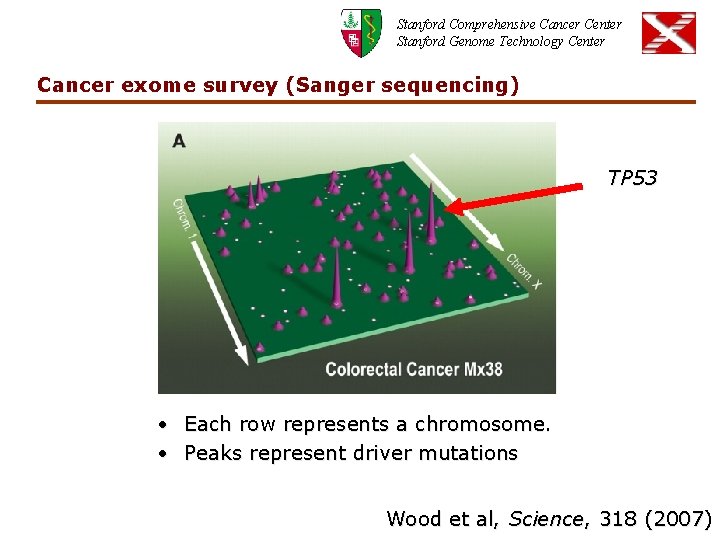
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Cancer exome survey (Sanger sequencing) TP 53 • Each row represents a chromosome. • Peaks represent driver mutations Wood et al, Science, 318 (2007)

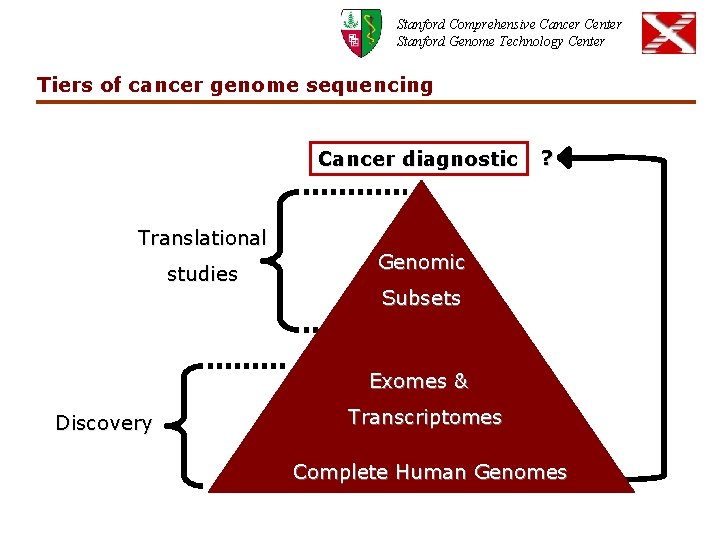
Stanford Comprehensive Cancer Center Stanford Genome Technology Center Tiers of cancer genome sequencing Cancer diagnostic Translational studies ? Genomic Subsets Exomes & Discovery Transcriptomes Complete Human Genomes

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Whole cancer genome sequencing • Pros – Most comprehensive coverage of the genome. – Least bias – most objective analysis – Highest resolution at base pair level – Identification of complex structural variants – Experimentally straightforward… • Cons – Cost (rapidly dropping!) – Rapidly evolution of technologies – Challenging data management and analysis

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sample and genetic complexity of cancer • Sample variability – Normal stroma contamination – Mixtures of variable lineages – Degradation of DNA • Intratumoral genetic heterogeneity – Clonal subpopulations carrying different mutations • Background random mutations (e. g. passengers) • Complex genomic structure

Stanford Comprehensive Cancer Center Stanford Genome Technology Center Sequencing cancer genomes – clinical samples • Type of samples – Cancer cell lines – Xenografts – Primary tumors – Purified cancer cells • Requirement for matched samples – Normal diploid genome

Stanford Comprehensive Cancer Center Stanford Genome Technology Center False positive mutation rates in genome sequencing • Mutation false positive rate requires high accuracy. – 1 base / 10, 000 error = 300, 000 false mutations – 1 base / 100, 000 error = 30, 000 false mutations • Ideal false positive rate – 1 base / 1, 000 error – ~50% of candidate mutations are correct!
 Stanford genome technology center
Stanford genome technology center Genome-to-genome distance calculator
Genome-to-genome distance calculator Wagholi cancer research center
Wagholi cancer research center Howard university cancer center
Howard university cancer center Genomics
Genomics Plant genome research program
Plant genome research program Euphenics
Euphenics Human genome size
Human genome size Mash genome
Mash genome Human genome size
Human genome size Future of human genome project
Future of human genome project Vntrs vs strs
Vntrs vs strs Perpartes
Perpartes Human genome structure
Human genome structure Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Shotgun sequencing
Shotgun sequencing Genome sequencing
Genome sequencing Human genome project source code
Human genome project source code Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key Patric bioinformatics
Patric bioinformatics National human genome research institute
National human genome research institute Genome modification ustaz auni
Genome modification ustaz auni National human genome research institute
National human genome research institute Human genome project
Human genome project Genome klick
Genome klick History of human genome project
History of human genome project Chapter 15 the human genome answer key
Chapter 15 the human genome answer key Chapter 14 the human genome
Chapter 14 the human genome Human genome project
Human genome project Translation
Translation Sequence assembly ppt
Sequence assembly ppt Encode
Encode Ucsc genome browser tutorial
Ucsc genome browser tutorial Genome
Genome Genome identification
Genome identification Genome sequencing
Genome sequencing Savant genome browser
Savant genome browser 1000 genome project
1000 genome project Ribosomes structures
Ribosomes structures Alternate splicing
Alternate splicing Consensus sequence in prokaryotes
Consensus sequence in prokaryotes Genome.gov
Genome.gov Genome research limited
Genome research limited Innovation genome project
Innovation genome project Genome mapping
Genome mapping Igv genome browser
Igv genome browser Integrated microbial genome
Integrated microbial genome Chrl3
Chrl3 Genome project
Genome project Scalable annotation pipeline
Scalable annotation pipeline 14-3 human molecular genetics answer key
14-3 human molecular genetics answer key Genome definition
Genome definition What is a human genome
What is a human genome Tomato genome annotation
Tomato genome annotation Genome adalah
Genome adalah Igv broad institute
Igv broad institute Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Savant genome browser
Savant genome browser Solar center stanford
Solar center stanford Comprehensive report writing
Comprehensive report writing Comprehensive science villanova
Comprehensive science villanova Comprehensive care plan
Comprehensive care plan Washington comprehensive assessment program
Washington comprehensive assessment program