Genomic Sequence alignments and its application Hgchopusan ac



















![Copy Detecting System q CDS components = [K, M, P, T, A, G ] Copy Detecting System q CDS components = [K, M, P, T, A, G ]](https://slidetodoc.com/presentation_image_h/eac483fe79b99dcb9e066a3b51725058/image-44.jpg)





- Slides: 64

Genomic Sequence alignments and its application 조환규 교수 부산대학교 공과대학 정보 컴퓨터 공학부 Hgcho@pusan. ac. kr Graphics Application Lab

Biology and Informatics Mathematics : Physics = X : Biology q. X=? q Bioinformatics q Graphics Application Lab o o o Understanding Biological System with Informatics Molecular Biology Computational Biology • • • Genomics Proteomics And several –omics 2

Main features of This Talk 이미 잘 정리된 Computing methodology를 어떻 게 Bioinformatics에서 활용하는가 ? q Bioinformatics에서 잘 정리된 방법론은 CS쪽에서 어떻게 활용하는가 ? q Case Study q Graphics Application Lab • Genomic sequencing alignment와 program-copy detection과의 연관 3

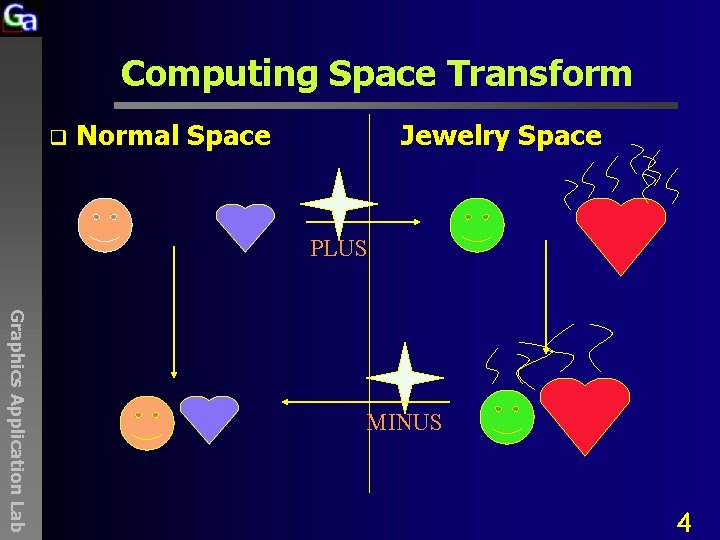
Computing Space Transform q Normal Space Jewelry Space PLUS Graphics Application Lab MINUS 4

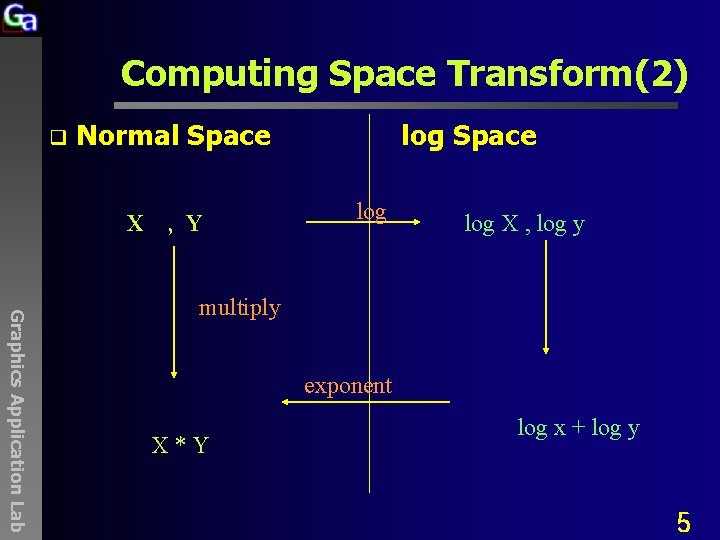
Computing Space Transform(2) q Normal Space X , Y log Space log X , log y Graphics Application Lab multiply exponent X*Y log x + log y 5

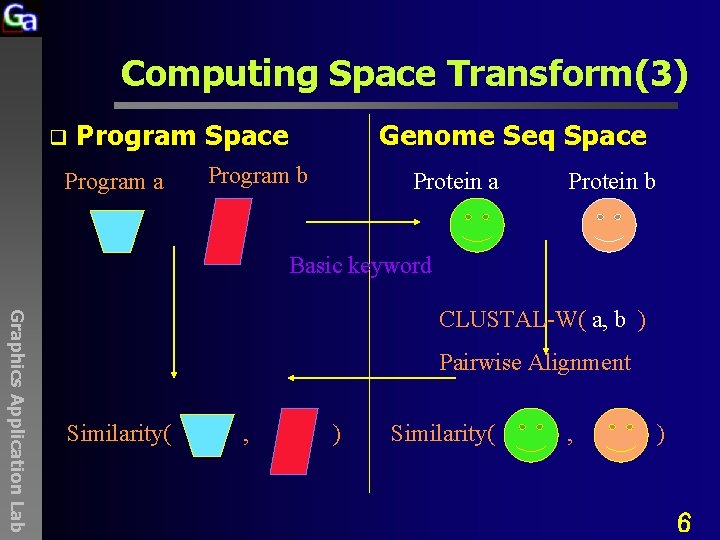
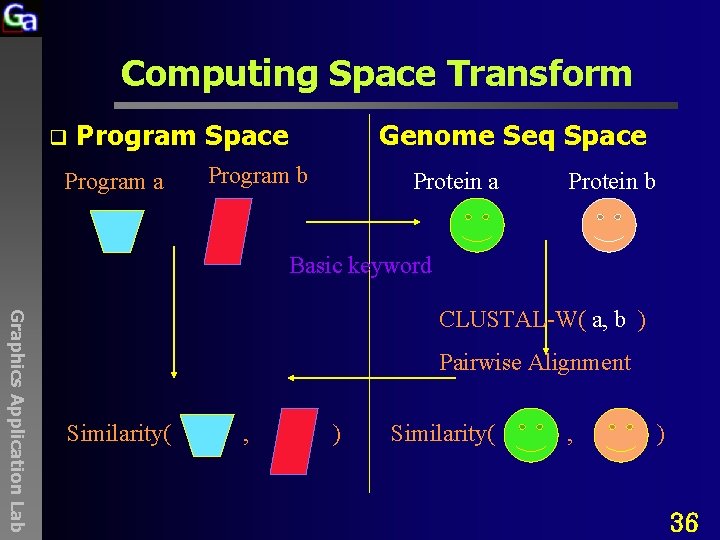
Computing Space Transform(3) q Program Space Program a Genome Seq Space Program b Protein a Protein b Basic keyword Graphics Application Lab CLUSTAL-W( a, b ) Pairwise Alignment Similarity( , ) 6


Genomic Sequence Alignments q Genomic Sequences, linear o o q DNA, RNA, Protein(amino acids) Why linear ? Goal of Molecular Biology or Life Science o Graphics Application Lab o o Characterizing functions of genes Understanding internal gene interactions Understanding internal & external interactions • q Drug targeting(protein interaction) Why Alignment ? 8

Human Binome BANK in 3280 Year 3280, dooms day q So many binary data files q o o Graphics Application Lab q Chips( cell-phone, cooker, TV… et al) Computer disks Figure out the contents of followings o o o 010111010001001010000101010…. ? 1000000010101111100101010010101…? 101011111110001010010100…? 9

Human-Binome-Project q They decided to establish Binary BANK. o Some are partially annotated • • q text data , Gene , garbage Junk-DNA HUMAN-Binome PROJECT…. . Starts… Graphics Application Lab o o 목적: 각종 binary sequence의 기능을 탐색 Mini-binary project ~~ E. Coli, C. Elegans • o q Object code, Protein , Cell-phone, calculator, PDA… Full sequencing of 300 Giga bytes DISK HUMAN BINOME BANK(HBB)! 10

For an Unknown Seq. X q Sequence X from a hardware o o q Find a similar pattern in HBB o Graphics Application Lab o o q Several error bits included Fragment sequencing Function ? Region ? Size ? Write a BIG paper…. o Here it comes……… 11


Dynamic Programming q A Basic Methodology o o q For all kinds of alignment Solution from all sub-partial solution 준비물 o Graphics Application Lab o Objective function Dynamic programming formula(recursion) • o o F(n) = F(n-1) + F(n-2), Base condition, F(0)=F(1)=1 Table, multi-dim. array structure 13

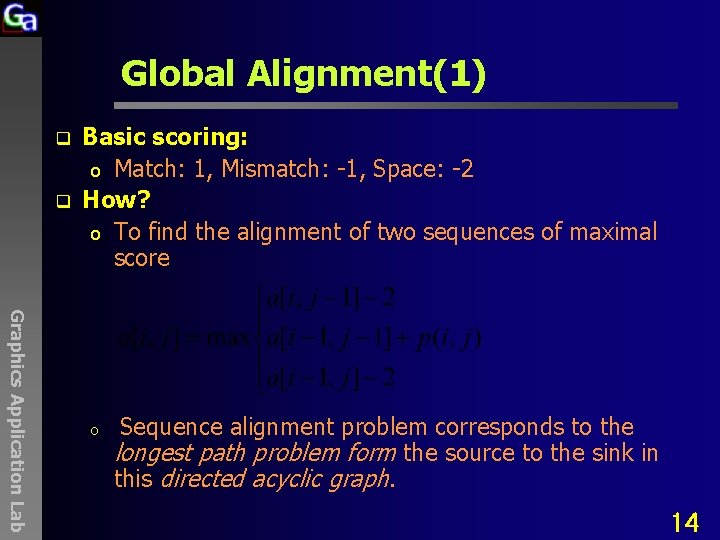
Global Alignment(1) q q Basic scoring: o Match: 1, Mismatch: -1, Space: -2 How? o To find the alignment of two sequences of maximal score Graphics Application Lab o Sequence alignment problem corresponds to the longest path problem form the source to the sink in this directed acyclic graph. 14

Global Alignment(2) q CACAGTGT 와 CAGGT Graphics Application Lab 0 C -2 A -4 C -6 A G T -8 -10 -12 -14 -16 C -2 1 -1 -3 -5 -7 -9 -11 -13 A -4 -1 2 0 -2 -4 -6 -8 -10 G -6 -3 0 1 -1 -1 -3 -5 -7 G -8 -5 -2 -1 0 0 -2 -2 -4 T -10 -7 -4 -3 -2 -1 1 -1 -1 C A G T C A - - G T 15

Local Alignment(1) q An alignment between a substring of s and a substring of t o Each entry of (I, j) will hold the highest score of an alignment between a suffix of s[1…i] and a suffix of t[1…j] Graphics Application Lab 예) AGGTATTGA - CCTATGGC 16

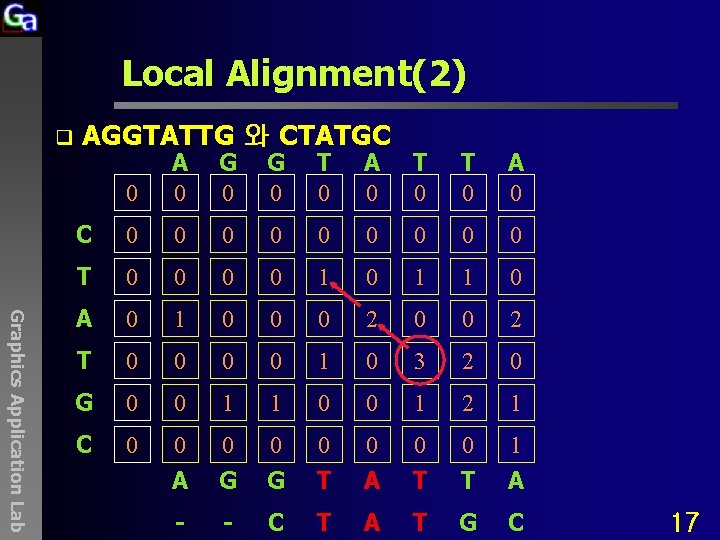
Local Alignment(2) q AGGTATTG 와 CTATGC Graphics Application Lab 0 A 0 G 0 T 0 A 0 C 0 0 0 0 0 T 0 0 1 0 1 1 0 A 0 1 0 0 0 2 T 0 0 1 0 3 2 0 G 0 0 1 1 0 0 1 2 1 C 0 0 0 0 1 A G G T A T T A - - C T A T G C 17

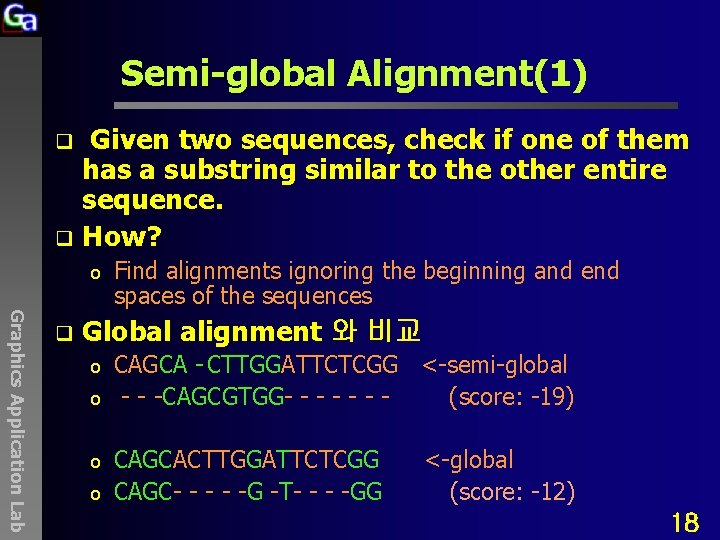
Semi-global Alignment(1) Given two sequences, check if one of them has a substring similar to the other entire sequence. q How? q o Graphics Application Lab q Find alignments ignoring the beginning and end spaces of the sequences Global alignment 와 비교 o o CAGCA - CTTGGATTCTCGG <-semi-global - - -CAGCGTGG- - - (score: -19) CAGCACTTGGATTCTCGG CAGC- - -G -T- - - -GG <-global (score: -12) 18

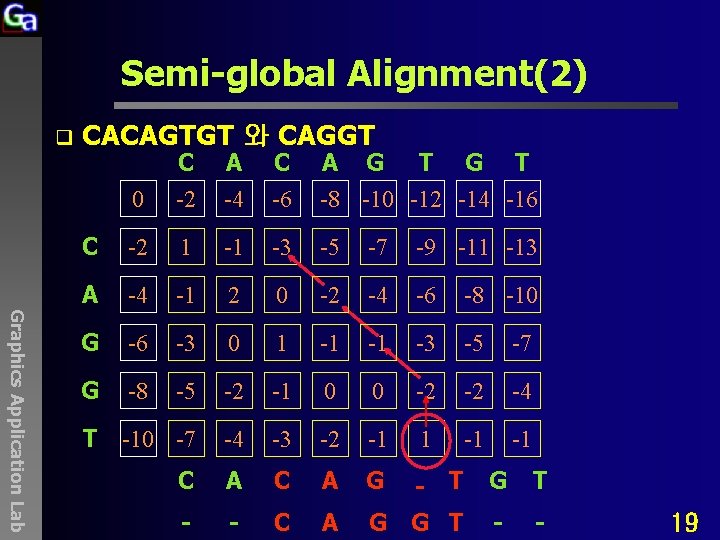
Semi-global Alignment(2) q CACAGTGT 와 CAGGT Graphics Application Lab 0 C -2 A -4 C -6 A G T -8 -10 -12 -14 -16 C -2 1 -1 -3 -5 -7 -9 -11 -13 A -4 -1 2 0 -2 -4 -6 -8 -10 G -6 -3 0 1 -1 -1 -3 -5 -7 G -8 -5 -2 -1 0 0 -2 -2 -4 T -10 -7 -4 -3 -2 -1 1 -1 -1 C A G - T G T - - C A G G T - - 19

General Gap Penalty q Definition o o Graphics Application Lab o Gap: consecutive number k > 1 of spaces When mutations are involved, the occurrence of a gap with k spaces is more probable than the occurrence of k isolated spaces w(k) : penalty associated with a gap with k spaces 20

Affine Gap Penalty Function q q Penalty for consecutive spaces <= isolated spaces Sub-additive function o q w(k 1 +k 2+…_kn) <= w(k 1) + w(k 2) +…+w(kn) Three arrays for dynamic programming o Graphics Application Lab o o a[i, j] = maximum score of an alignment between s[1…i] and t[1…i] that ends in s[i] matched with t[j] b[i, j] = maximum score of an alignment between s[1…i] and t[1…i] that ends in a space matched with t[j] c[i, j] = maximum score of an alignment between s[1…i] and t[1…i] that ends in s[i] matched with a space 21

Heuristic Alignment q Main difficulties o o o q Local search Graphics Application Lab o o q Search space, O(n^2) space or O(n^2 log n)time Optimality or Biologically-good Distance metric Multiple alignment Diagonal region searching Visualization. , e. g. , Dotlet BLAST approach for long sequence o o Small word matching And Extending from a highly matched region 22

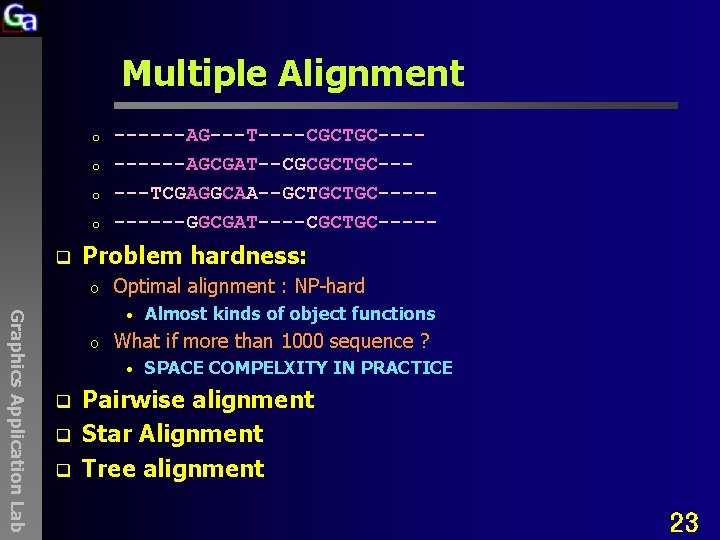
Multiple Alignment o o q ------AG---T----CGCTGC-----AGCGAT--CGCGCTGC-----TCGAGGCAA--GCTGCTGC-----GGCGAT----CGCTGC----- Problem hardness: o Optimal alignment : NP-hard Graphics Application Lab • o What if more than 1000 sequence ? • q q q Almost kinds of object functions SPACE COMPELXITY IN PRACTICE Pairwise alignment Star Alignment Tree alignment 23

Why multiple alignment ? Finding Conserved regions q Computer virus phylogeny constructing q • • • Graphics Application Lab • q tuberculosis • • q 300 sp. /year More than 10000 sp. : N Number of files in a system : M > 100000 Detecting a CV takes O( N*M) checks! 8 종, a conserved region, and polymorphic sites 김철민 교수님(부산의대) – 진단용 칩 제작 Phylogeny construct 24

Phylogeny 1 : hard Version Graphics Application Lab 25

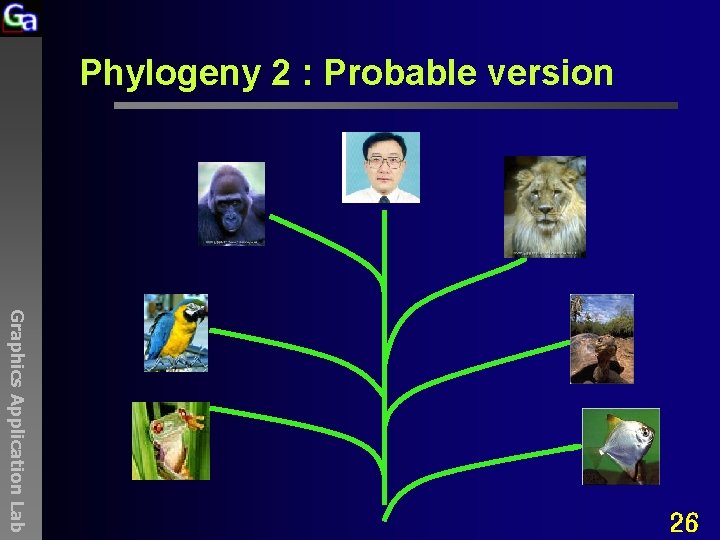
Phylogeny 2 : Probable version Graphics Application Lab 26

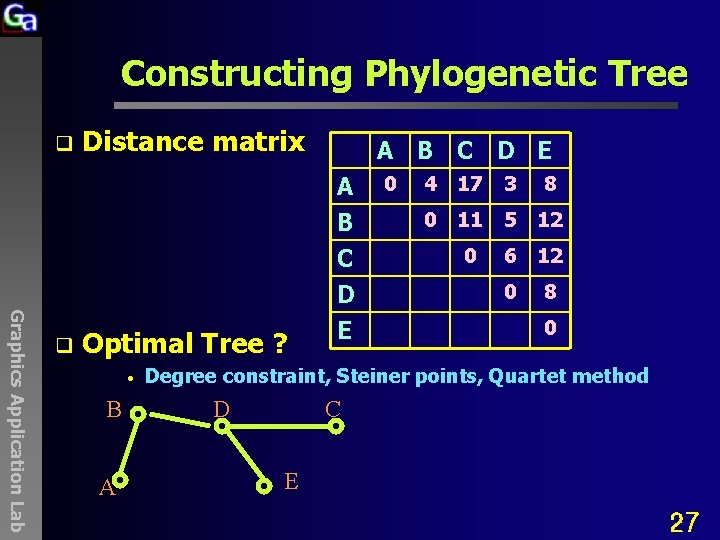
Constructing Phylogenetic Tree q Distance matrix A B C D E A B C Graphics Application Lab q Optimal Tree ? • B A D E 0 4 17 3 8 0 11 5 12 0 6 12 0 8 0 Degree constraint, Steiner points, Quartet method D C E 27

PART 2: Application Detecting Source Code Plagiarism Graphics Application Lab

Plagiarism, Plagiarism q Linear Structure o o Graphics Application Lab o Genomic sequences Plain articles Programs Human behaviors on the time-line Time-series data sets Student Reports Plagiarism q Assignment Program copying q Where is the original version of this one ? q Web searching redundancy elimination q 29



Fingerprinting Method q Keyword frequency similarity o o 특정한 단어의 사용횟수 Fingerprinting object • • Graphics Application Lab • • q Fixed size fingerprint Easy to making Database Quick searching High false positive rate Example, fingerprint vector A c x t u x g r N …. . 32

Attacking Inserting redundant words q Shuffling q Cons and Pros q o o Graphics Application Lab q Recent trends o o o q Easy to use in document application Hard to use in program file Structure-oriented similarity measure Greedy-Block-Removing methods… Is this a basic concept of local alignment ? Sample-Report-Server Building 33

Undergraduate Assignment q Programming Assignment cheating: o o q 이론적으로는 그 구별방법이 없다. Assignment cheating은 비용이 크다 • Password breaking by Mafia Assignment의 출력은 동일하다. Graphics Application Lab o Correct program들끼리만 비교 과제에 주어진 시간은 비교적 짧다(3 -4일 정도). q 수강생의 수는 적절하다(300 명 이하). q 프로그래밍 언어는 모두 동일하다. q 34

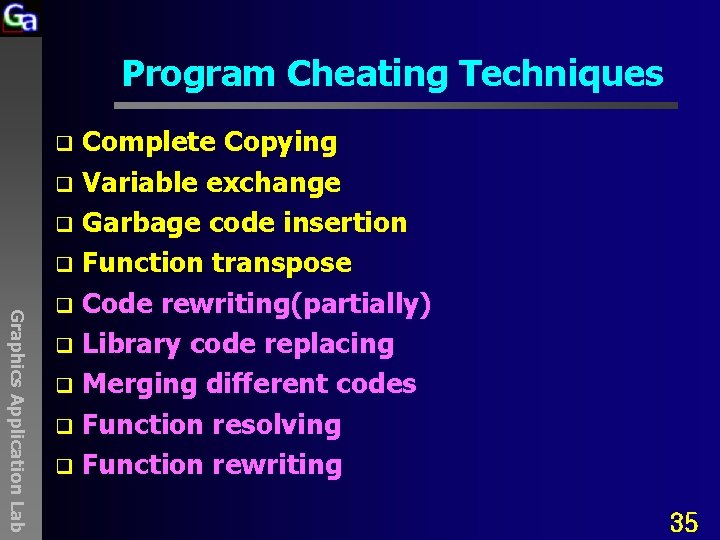
Program Cheating Techniques Complete Copying q Variable exchange q Garbage code insertion q Function transpose q Code rewriting(partially) q Library code replacing q Merging different codes q Function resolving q Function rewriting q Graphics Application Lab 35

Computing Space Transform q Program Space Program a Genome Seq Space Program b Protein a Protein b Basic keyword Graphics Application Lab CLUSTAL-W( a, b ) Pairwise Alignment Similarity( , ) 36

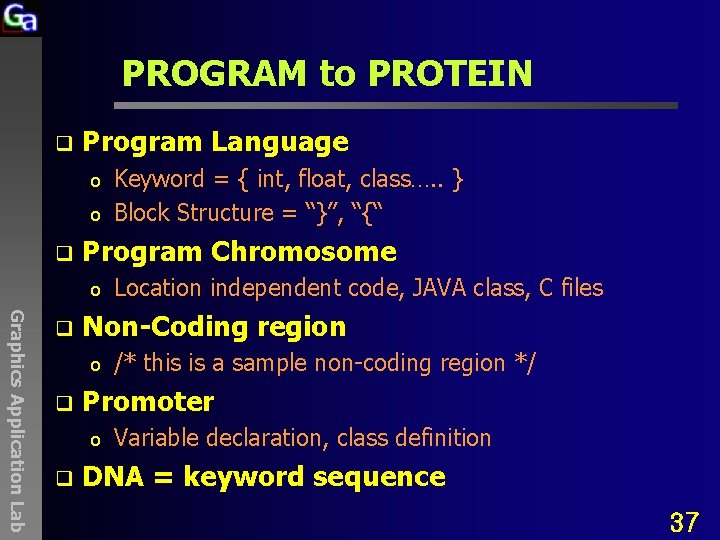
PROGRAM to PROTEIN q Program Language o o q Program Chromosome o Graphics Application Lab q /* this is a sample non-coding region */ Promoter o q Location independent code, JAVA class, C files Non-Coding region o q Keyword = { int, float, class…. . } Block Structure = “}”, “{“ Variable declaration, class definition DNA = keyword sequence 37

Extracting Program DNA Syntactic level q Semantic level q Syntactic running Graphics Application Lab syntax q Real running Program Flow-graph 38

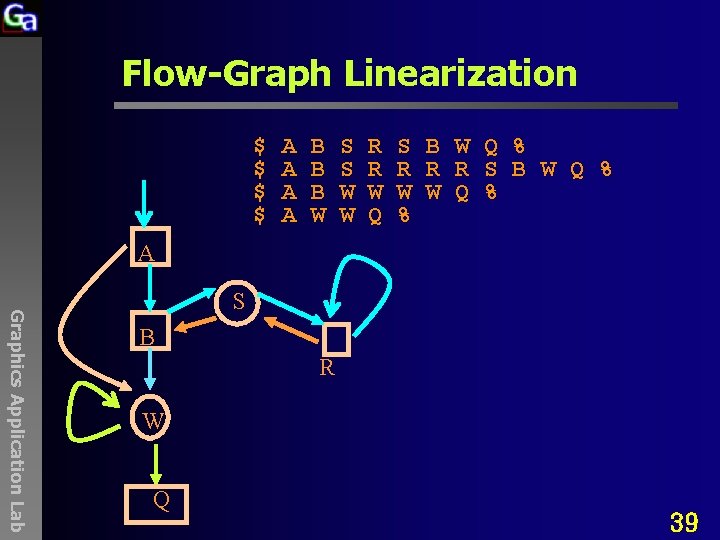
Flow-Graph Linearization $ $ A A B B B W S S W W R R W Q S B W Q % R R R S B W Q % W W Q % % A Graphics Application Lab S B R W Q 39

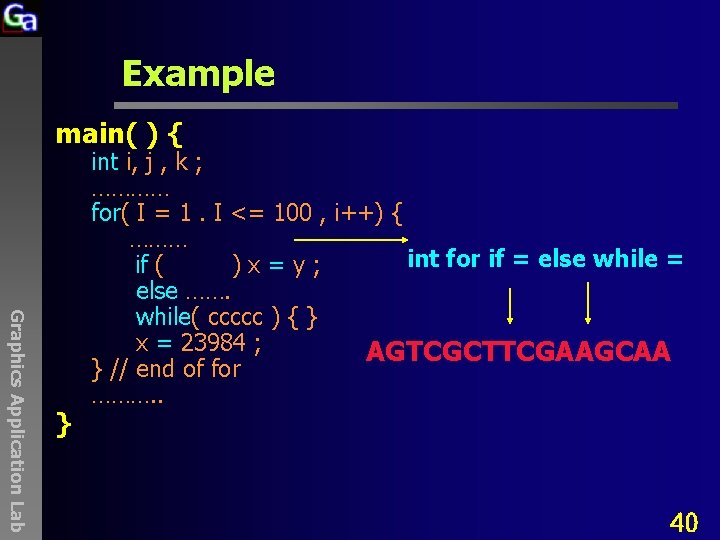
Example main( ) { Graphics Application Lab } int i, j , k ; ………… for( I = 1. I <= 100 , i++) { ……… int for if = else while = if ( )x=y; else ……. while( ccccc ) { } x = 23984 ; AGTCGCTTCGAAGCAA } // end of for ………. . 40

Why Protein mapping ? q DNA sequence overlap o o if = AA, then = AG, * = GA, return = GG AAGGA = AG + GA or AA + GG + A • q Ambiguity resolving 20 Amino acid bases Graphics Application Lab o o About 20 keywords 2 -3 groups • • polar, non-polar hydrophobic, hydrophilic Charged, uncharged Small, large 41

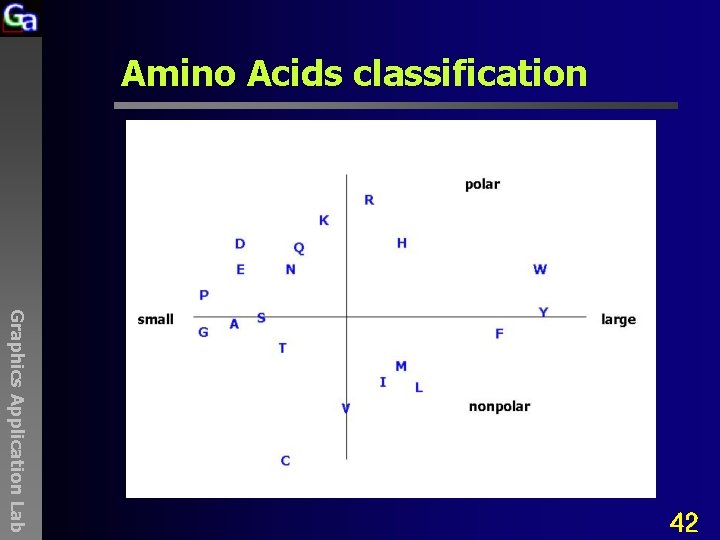
Amino Acids classification Graphics Application Lab 42

Keyword Mapping Strategy q Convertibility = { for , while } Easy q Convertibility = { for, then } Hard Convertibility = { if, ‘=‘ } Impossible? q Procedure q Graphics Application Lab o o Preprocessing Chromosome arrangement Keyword selection Protein mapping 43
![Copy Detecting System q CDS components K M P T A G Copy Detecting System q CDS components = [K, M, P, T, A, G ]](https://slidetodoc.com/presentation_image_h/eac483fe79b99dcb9e066a3b51725058/image-44.jpg)
Copy Detecting System q CDS components = [K, M, P, T, A, G ] o o o Graphics Application Lab o o o Keyword table Matching Score matrix Affine Gap Penalty Threshold length Alignment Set Scoring ma. Ximum Gap allowing 20 keyword borrow from Protein(PAM) 44

Experiment Overview Sample programs “data structure” q Students , 60 q Programming assignments, 12 set q 1 semester q On-line evaluation system = ESPA q Graphics Application Lab o o q Java-based on-line evaluation system Due, 1 week We do not monitor all programs 45

Clustal-W (1)(www 2. ebi. ac. uk/clusterw) q Input Fasta file Graphics Application Lab 46

Clustal-W(2) (www 2. ebi. ac. uk/clusterw) q Output Graphics Application Lab 47

Phylo. Draw (cho et al, Bioinformatics 2001. ) Graphics Application Lab 48

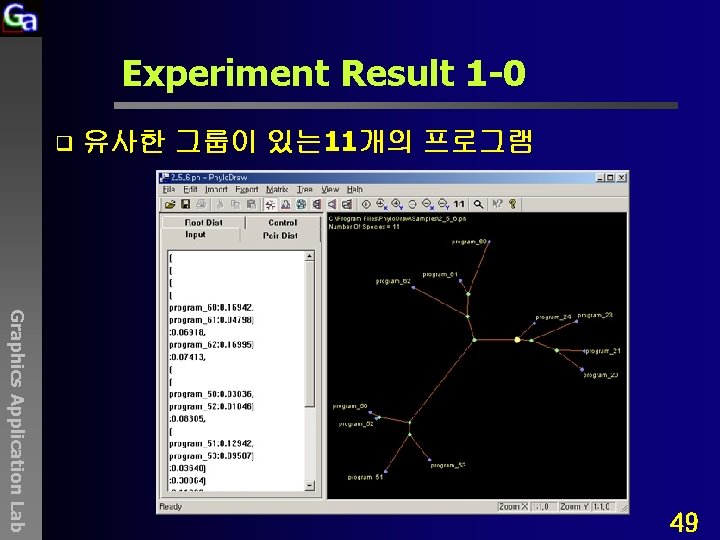
Experiment Result 1 -0 q 유사한 그룹이 있는 11개의 프로그램 Graphics Application Lab 49

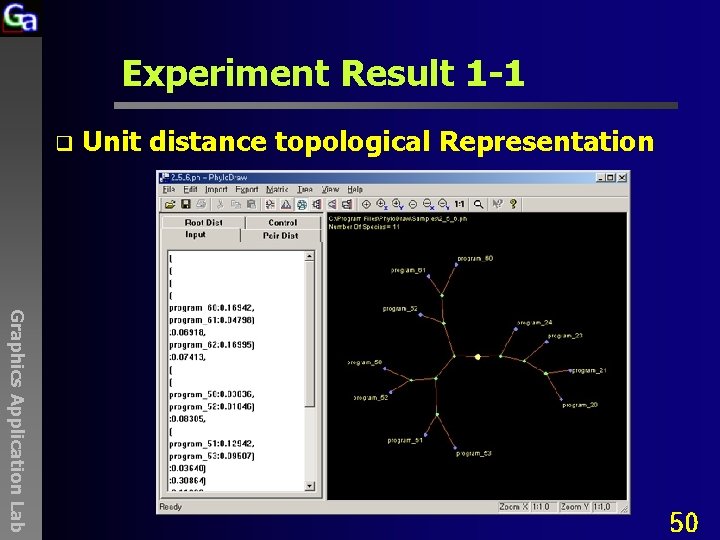
Experiment Result 1 -1 q Unit distance topological Representation Graphics Application Lab 50

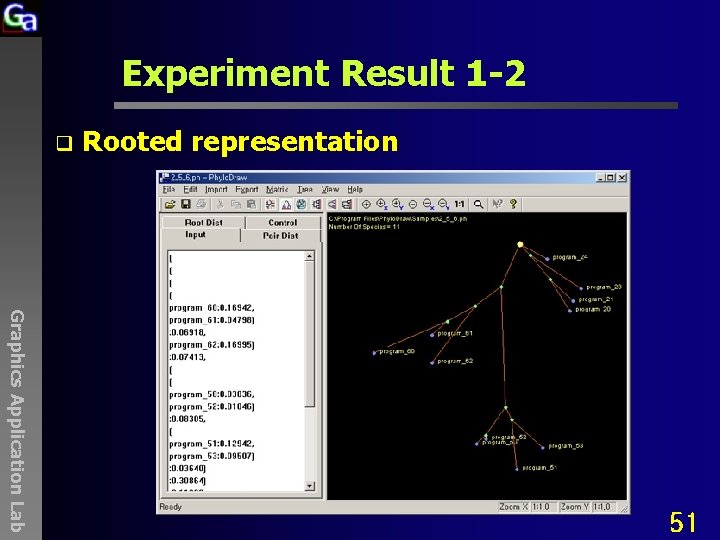
Experiment Result 1 -2 q Rooted representation Graphics Application Lab 51

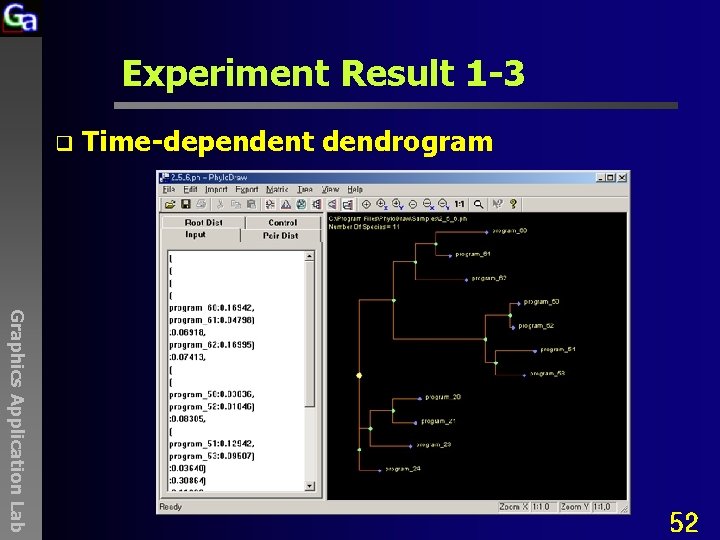
Experiment Result 1 -3 q Time-dependent dendrogram Graphics Application Lab 52

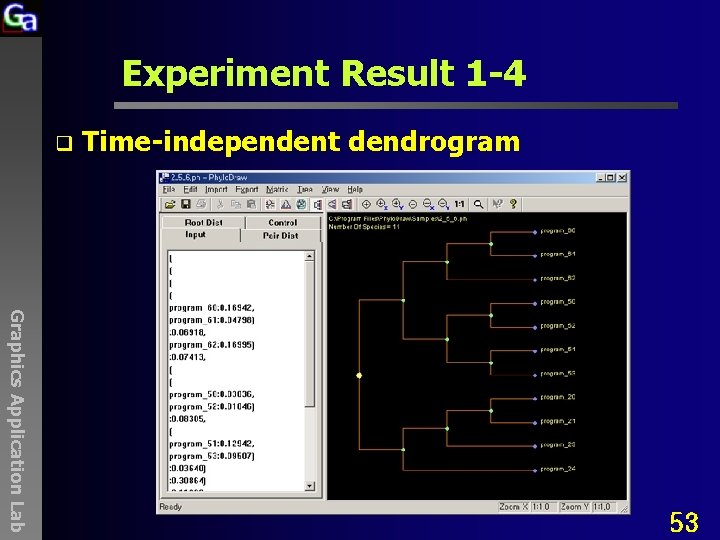
Experiment Result 1 -4 q Time-independent dendrogram Graphics Application Lab 53

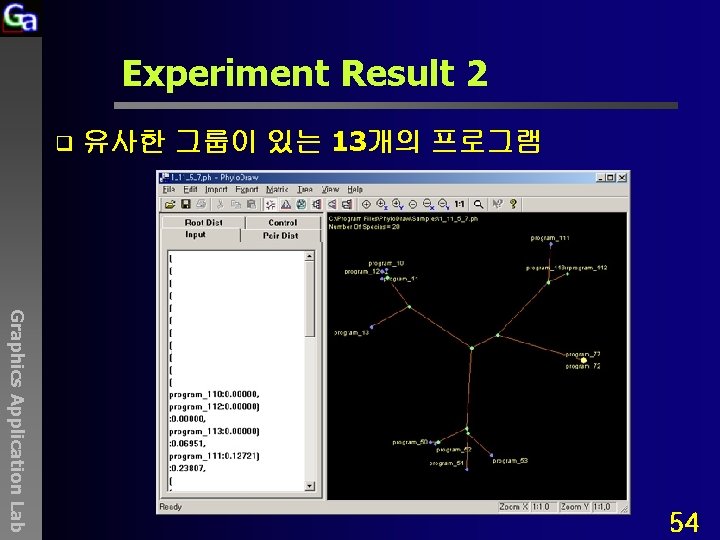
Experiment Result 2 q 유사한 그룹이 있는 13개의 프로그램 Graphics Application Lab 54

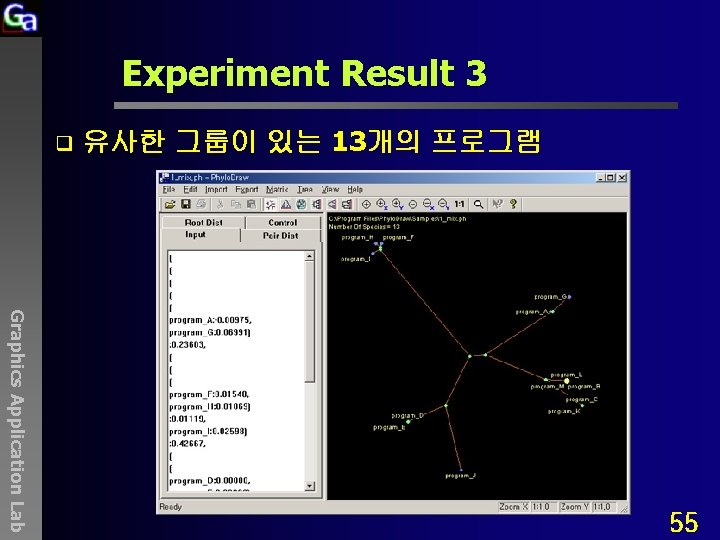
Experiment Result 3 q 유사한 그룹이 있는 13개의 프로그램 Graphics Application Lab 55

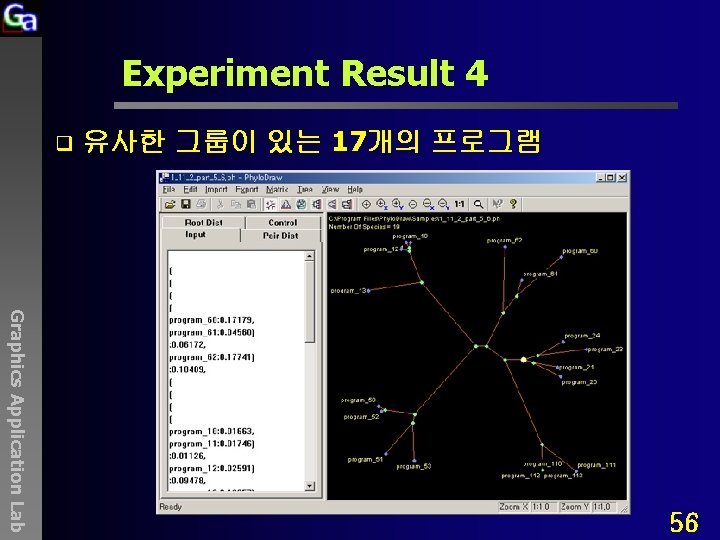
Experiment Result 4 q 유사한 그룹이 있는 17개의 프로그램 Graphics Application Lab 56

Experiment Result 5 q 유사한 그룹이 있는 21개의 프로그램 Graphics Application Lab 57

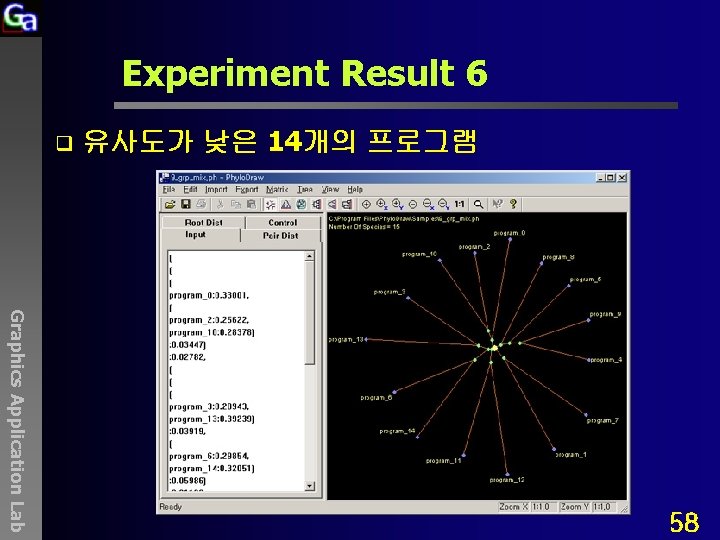
Experiment Result 6 q 유사도가 낮은 14개의 프로그램 Graphics Application Lab 58

Another Application q Music Score Plagiarism o o Tempo, melody line…. C major, A minor , key-transformation Credit Card Bankruptcy Alert q Drinking Alert q Graphics Application Lab annotated time-line 59

Application q Web searching Engine o o Eliminate redundant documents Eg. ) Query = “썬베드“ in EMPASS search engine • o Graphics Application Lab q 탐색된 상위 10 개의 관련 문서 중에서 8개는 동일한 문서 신문기사 검색에서도 유사한 경우 Original Paper 60

Further Work q q q Program DNA-Bank server Copying Phylogenetics Building Parametric Method o o o Graphics Application Lab q q University Report Oracle Music Plagiarism o q Phylogeny tree for Old classical music(Palestria to Brahms) How to linearize a procedure call ? o o q Fixed-size fingerprinting = program protein PAM for program copying behavior Real Practice Parameter tuning Procedure call is a sort of directed graph Fast and moderate size Program DNA 61

Conclusion q Bioinformatics o Bioinformatics • Bioinformatics § Bioinformatics ü ü Graphics Application Lab q Linear Structure Similarity o o o q Bioinformatics A brave new world…… Local Alignment Gap penalty Structure-based similarity Good Application 62

PUSAN BIOINFORMATICS JIHAD Graphics Application Lab 63

Realtime Home-Bioinformatics Graphics Application Lab 64