Alignments n Why do Alignments Detecting Selection Evolution









- Slides: 28

Alignments n Why do Alignments?

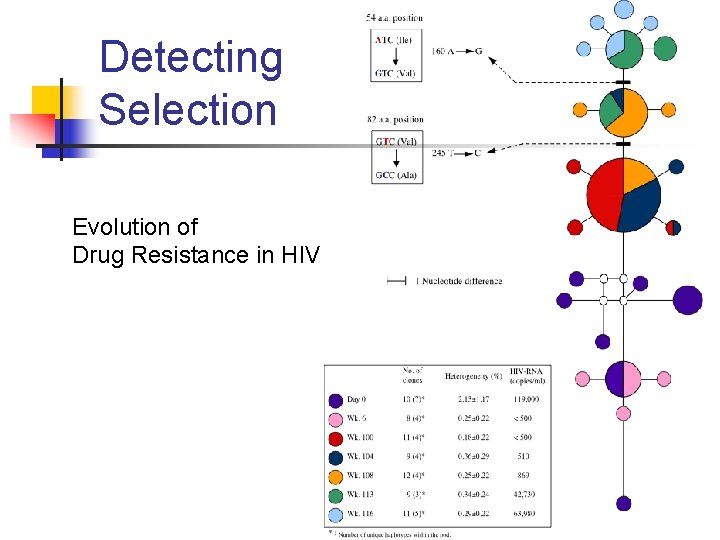
Detecting Selection Evolution of Drug Resistance in HIV

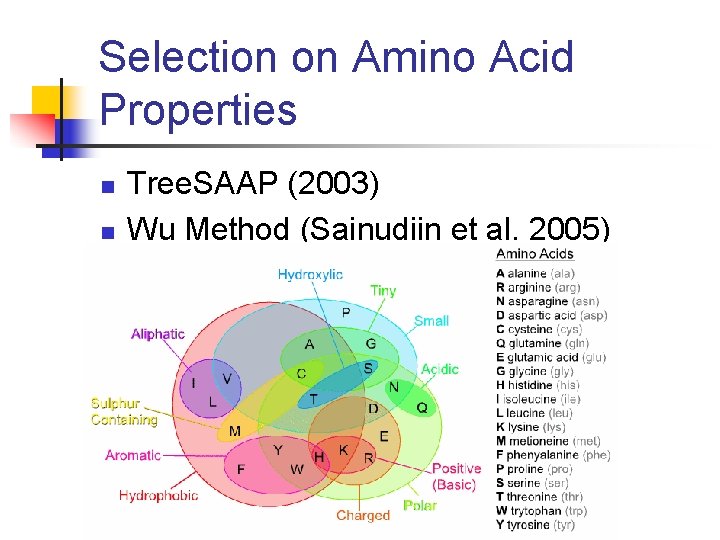
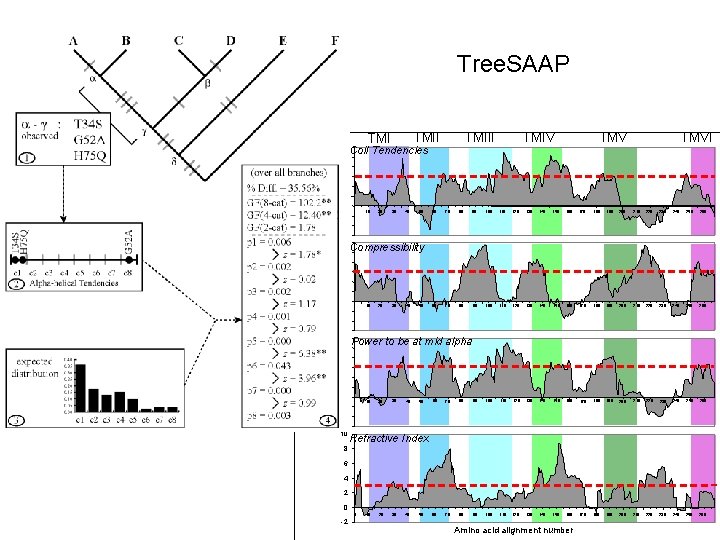
Selection on Amino Acid Properties n n Tree. SAAP (2003) Wu Method (Sainudiin et al. 2005)

Tree. SAAP Properties n n n n Alpha-helical tendencies Average number of surrounding residues Beta-structure tendencies Bulkiness Buriedness Chromatographic Index Coil tendencies Composition Compressibility Equilibrium constant (ionization of COOH) Helical contact area Hydropathy Isoelectric point Long-range non-bonded energy Mean r. m. s. fluctuation displacement n n n n Molecular volume Molecular weight Normalized consensus hydrophobicity Partial specific volume Polar requirement Polarity Power to be at the C-terminal Power to be at the middle of alpha-helix Power to be at the N-terminal Refractive index Short and medium range non-bonded energy Solvent accessible reduction ratio Surrounding hydrophobicity Thermodynamic transfer hydrophobicity Total non-bonded energy Turn tendencies

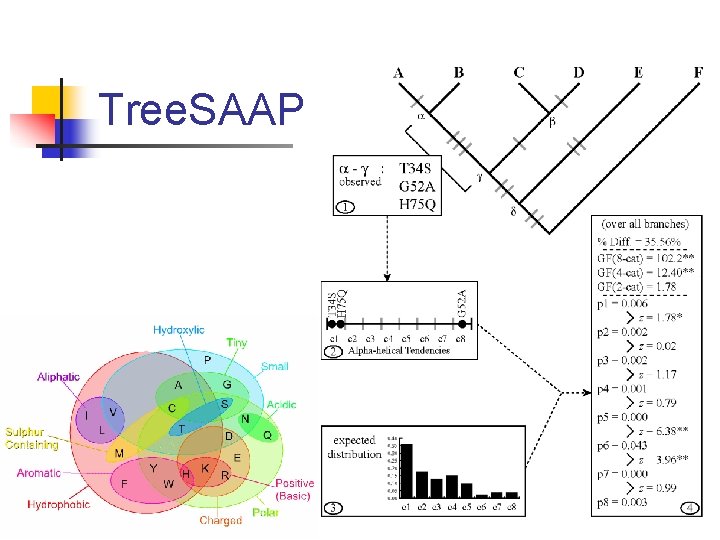
Tree. SAAP

Rhinoviruses

Selected Sites

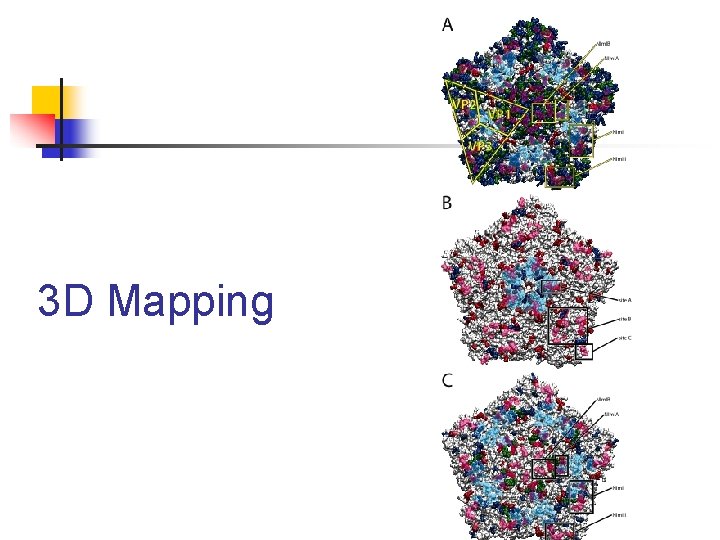
3 D Mapping

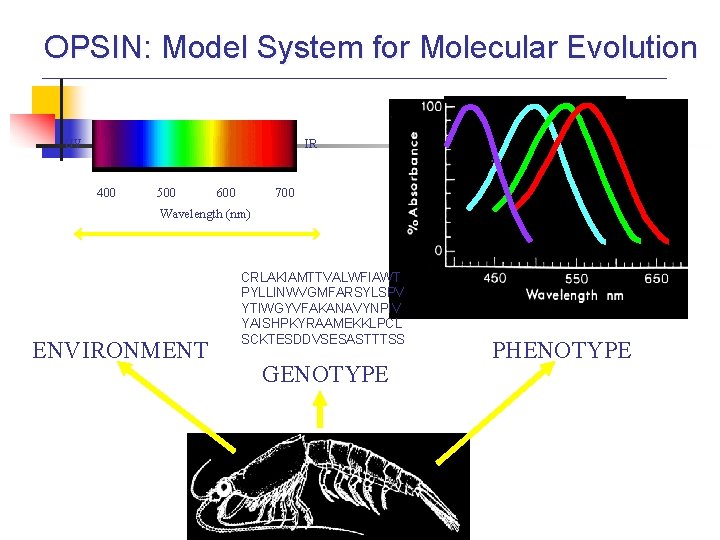
OPSIN: Model System for Molecular Evolution UV IR 400 500 600 700 Wavelength (nm) ENVIRONMENT CRLAKIAMTTVALWFIAWT PYLLINWVGMFARSYLSPV YTIWGYVFAKANAVYNPIV YAISHPKYRAAMEKKLPCL SCKTESDDVSESASTTTSS GENOTYPE PHENOTYPE

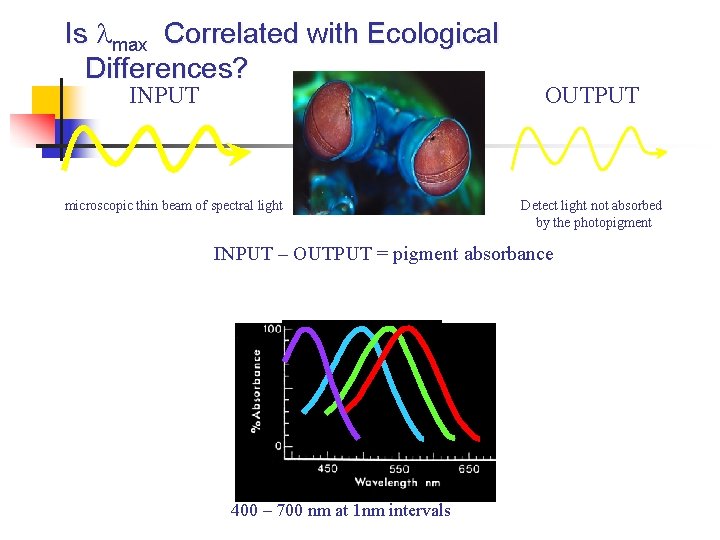
Is max Correlated with Ecological Differences? INPUT microscopic thin beam of spectral light OUTPUT Detect light not absorbed by the photopigment INPUT – OUTPUT = pigment absorbance 400 – 700 nm at 1 nm intervals

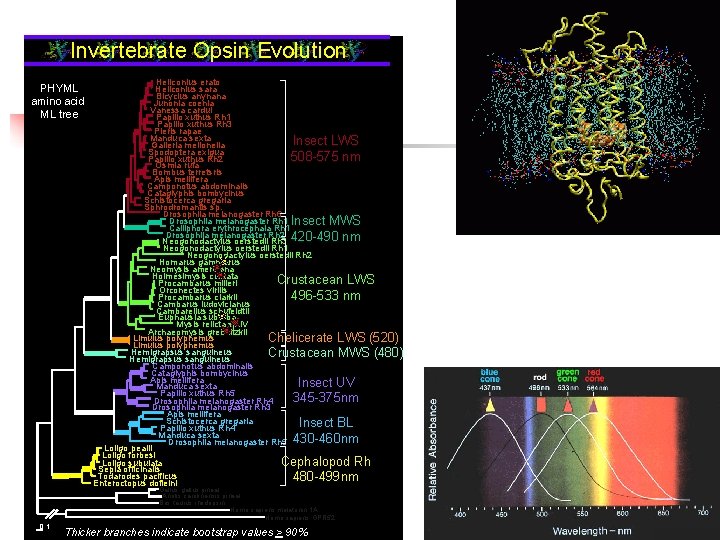
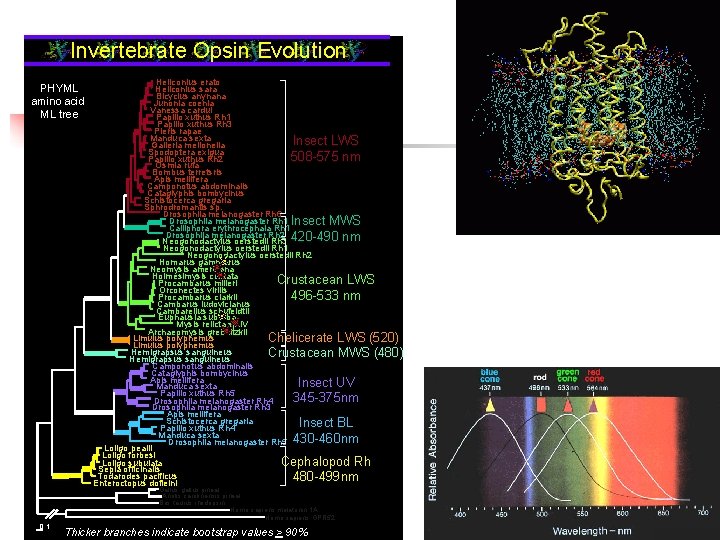
Invertebrate Opsin Evolution PHYML amino acid ML tree Heliconius erato Heliconius sara Bicyclus anynana Junonia coenia Vanessa cardui Papilio xuthus Rh 1 Papilio xuthus Rh 3 Pieris rapae Manduca sexta Insect LWS Galleria mellonella Spodoptera exigua 508 -575 nm Papilio xuthus Rh 2 Osmia rufa Bombus terretsris Apis mellifera Camponotus abdominalis Cataglyphis bombycinus Schistocerca gregaria Sphrodromantis sp. Drosophila melanogaster Rh 6 Drosophila melanogaster Rh 1 Insect MWS Calliphora erythrocephala Rh 1 Drosophila melanogaster Rh 2 Neogonodactylus oerstedii Rh 3 420 -490 nm Neogonodactylus oerstedii Rh 1 Neogonodactylus oerstedii Rh 2 Homarus gammarus Neomysis americana Holmesimysis costata Crustacean LWS Procambarus milleri Orconectes virilis 496 -533 nm Procambarus clarkii Cambarus ludovicianus Cambarellus schufeldtii Euphausia suberba Mysis relicta sp. IV Archaeomysis grebnitzkii Limulus polyphemus Chelicerate LWS (520) Limulus polyphemus Hemigrapsus sanguineus Crustacean MWS (480) Hemigrapsus sanguineus Camponotus abdominalis Cataglyphis bombycinus Apis mellifera Insect UV Manduca sexta Papilio xuthus Rh 5 345 -375 nm Drosophila melanogaster Rh 4 Drosophila melanogaster Rh 3 Apis mellifera Schistocerca gregaria Insect BL Papilio xuthus Rh 4 Manduca sexta Drosophila melanogaster Rh 5 430 -460 nm Loligo pealii Loligo forbesi Loligo subulata Cephalopod Rh Sepia officinalis Todarodes pacificus 480 -499 nm Enteroctopus dofleini Gallus gallus pineal Anolis carolinensis pineal Bos taurus rhodopsin Homo sapiens melatonin 1 A Homo sapiens GPR 52 0. 1 Thicker Thickbranchesindicatebootstrapvalues> >90%

Coil Tendencies, Compressibility, Alpha-Helix

Tree. SAAP 6 TMIII Coil Tendencies TMIV TMVI 4 2 0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210 220 230 240 250 260 -2 6 Compressibility 4 2 Z-score 0 10 20 30 40 50 -2 6 Power to be at mid alpha 4 2 0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210 220 230 240 250 260 -2 10 Refractive Index 8 6 4 2 0 0 -2 10 20 30 40 50 Amino acid alignment number

Invertebrate Opsin Evolution PHYML amino acid ML tree Heliconius erato Heliconius sara Bicyclus anynana Junonia coenia Vanessa cardui Papilio xuthus Rh 1 Papilio xuthus Rh 3 Pieris rapae Manduca sexta Insect LWS Galleria mellonella Spodoptera exigua 508 -575 nm Papilio xuthus Rh 2 Osmia rufa Bombus terretsris Apis mellifera Camponotus abdominalis Cataglyphis bombycinus Schistocerca gregaria Sphrodromantis sp. Drosophila melanogaster Rh 6 Drosophila melanogaster Rh 1 Insect MWS Calliphora erythrocephala Rh 1 Drosophila melanogaster Rh 2 Neogonodactylus oerstedii Rh 3 420 -490 nm Neogonodactylus oerstedii Rh 1 Neogonodactylus oerstedii Rh 2 Homarus gammarus Neomysis americana Holmesimysis costata Crustacean LWS Procambarus milleri Orconectes virilis 496 -533 nm Procambarus clarkii Cambarus ludovicianus Cambarellus schufeldtii Euphausia suberba Mysis relicta sp. IV Archaeomysis grebnitzkii Limulus polyphemus Chelicerate LWS (520) Limulus polyphemus Hemigrapsus sanguineus Crustacean MWS (480) Hemigrapsus sanguineus Camponotus abdominalis Cataglyphis bombycinus Apis mellifera Insect UV Manduca sexta Papilio xuthus Rh 5 345 -375 nm Drosophila melanogaster Rh 4 Drosophila melanogaster Rh 3 Apis mellifera Schistocerca gregaria Insect BL Papilio xuthus Rh 4 Manduca sexta Drosophila melanogaster Rh 5 430 -460 nm Loligo pealii Loligo forbesi Loligo subulata Cephalopod Rh Sepia officinalis Todarodes pacificus 480 -499 nm Enteroctopus dofleini Gallus gallus pineal Anolis carolinensis pineal Bos taurus rhodopsin Homo sapiens melatonin 1 A Homo sapiens GPR 52 0. 1 Thicker Thickbranchesindicatebootstrapvalues> >90%

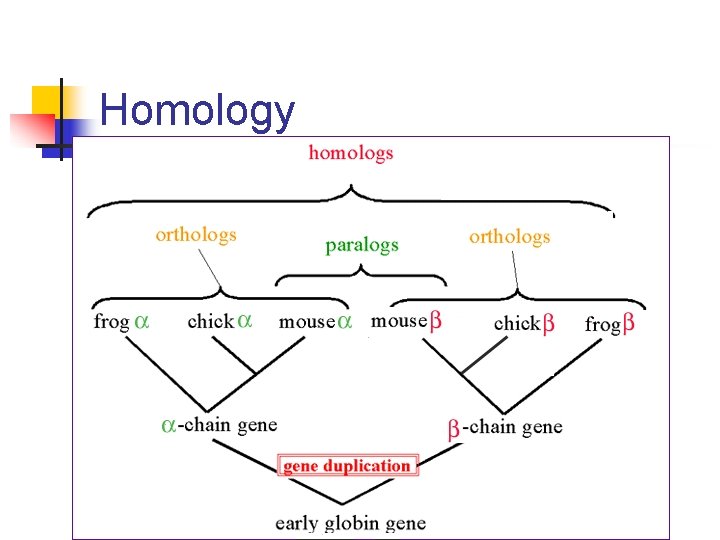
Homology

Homology definitions n n Homology is an evolutionary relationship that either exists or does not. It cannot be partial. An ortholog is a homolog that arose through a speciation event A paralog is a homolog that arose through a gene duplication event. Paralogs often have divergent function. Similarity is a measure of the quality of alignment between two sequences. High similarity is evidence for homology. Similar sequences may be orthologs or paralogs.

One More Homology type n n Xenology – similarity due to horizontal gene transfer (HGT) How do you discover this?

Alignment Problem n n (Optimal) pairwise alignment consists of considering all possible alignments of two sequences and choosing the optimal one. Sub-optimal (heuristic) alignment algorithms are also very important: eg BLAST

Key Issues Types of alignments (local vs. global) n The scoring system n The alignment algorithm n Measuring alignment significance n

Types of Alignment n n Global—sequences aligned from end-to -end. Local—alignments may start in the middle of either sequence Ungapped—no insertions or deletions are allowed Other types: overlap alignments, repeated match alignments

Local vs. Global Pairwise Alignments n A global alignment includes all elements of the sequences and includes gaps. n n n A global alignment may or may not include "end gap" penalties. Global alignments are better indicators of homology and take longer to compute. A local alignment includes only subsequences, and sometimes is computed without gaps. n Local alignments can find shared domains in divergent proteins and are fast to compute

How do you compare alignments? n Scoring scheme n What events do we score? n n n Matches Mismatches Gaps What scores will you give these events? What assumptions are you making? Score your alignment

Scoring Matrices n n n How do you determine scores? What is out there already for your use? DNA versus Amino Acids? n n TTACGGAGCTTC CTGAGATCC

Multiple Sequence Alignment n Global versus Local Alignments Progressive alignment Estimate guide tree n Do pairwise alignment on subtrees Clustal. X n

Improvements n Consistency-based Algorithms n T-Coffee - consistency-based objective function to minimize potential errors n n n Generates pair-wise global (Clustal) Local (Lalign) Then combine, reweight, progressive alignment

Iterative Algorithms n n Estimate draft progressive alignment (uncorrected distances) Improved progressive (reestimate guide tree using Kimura 2 -parameter) Refinement - divide into 2 subtrees, estimate two profiles, then re-align 2 profiles Continue refinement until convergence

Software n n Clustal T-Coffee MUSCLE (limited models) MAFFT (wide variety of models)

Comparisons n Speed n n Accuracy n n Muscle>MAFFT>CLUSTALW>T-COFFEE MAFFT>Muscle>T-COFFEE>CLUSTALW Lots more work to do here!