Hillary Term 04 The Human Genome 20 1




- Slides: 48

Hillary Term 04: “The Human Genome” 20. 1 The Human Genome – evolutionary issues (Hein) 27. 1 Non-Genic Selection in the Human Genome (Lunter) 3. 2 Mammalian Genes I: Conservation and slow evolution (Ponting) 10. 2 Mammalian Genes II: Functional innovation and rapid change (Ponting/Goodstadt) 17. 2 RNAs in Human Genome (Sam Griffiths-Jones) 24. 2 Population Genetics of the Human Genome (Gil Mc. Vean ) 2. 3 Association Mapping and the Human Genome (Lon Cardon) 9. 3 The Human Genome and Human Evolution (Chris Tyler-Smith)

The Human Genome – key issues The Human Genome Project Few basic facts of the human genome Grammar of Genes Basic events happening to a genome per mitosis/generation Genealogical Structures: Phylogenies, Pedigrees and the ARG Long term Dynamics of the Human Genome: The comparative aspect (Genotype Phenotype) & (Population Genetics/History) => Gene Mapping History Our interests.

History of the Human Genome Project 1956 Physical map. 24 types and total set of 46 chromosomes 1977 Sanger publishes dideoxy sequencing method 1980 Botstein proposes human genetic map using RFLPs 1987 US DOE publishes report discussing HGP 1988 HUGO is established 1990 Official start of HGP with 3 billion $ and a 15 year horizon. 1991 Genome Database GB is established 1992 Genethon publishes map based on microsatelites. 1995 Lander et al. detailed map based on sequence tagged sites. 1998 Comprehensive map based on gene markers. 1999 Sanger Centre publishes chromosome 22 2001 Draft Genome published: Celera & Public 2003 Completion (almost) of Human Genome Strachan and Read, HMG 3 p 213

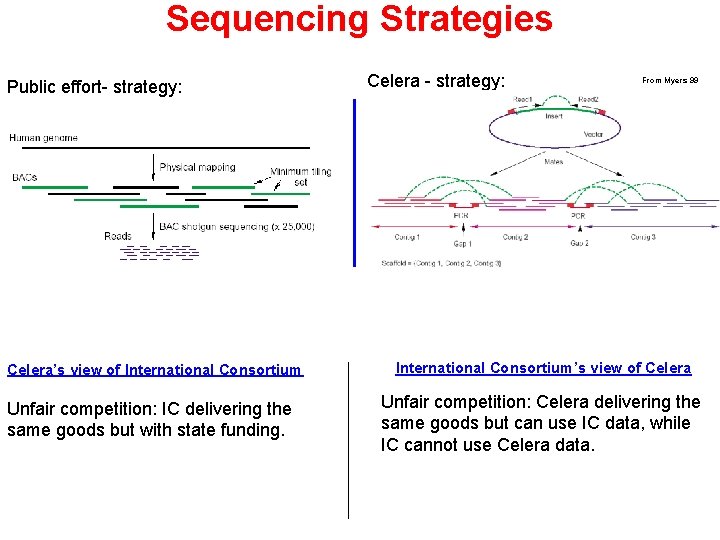
Sequencing Strategies Public effort- strategy: Celera’s view of International Consortium Unfair competition: IC delivering the same goods but with state funding. Celera - strategy: From Myers 99 International Consortium’s view of Celera Unfair competition: Celera delivering the same goods but can use IC data, while IC cannot use Celera data.

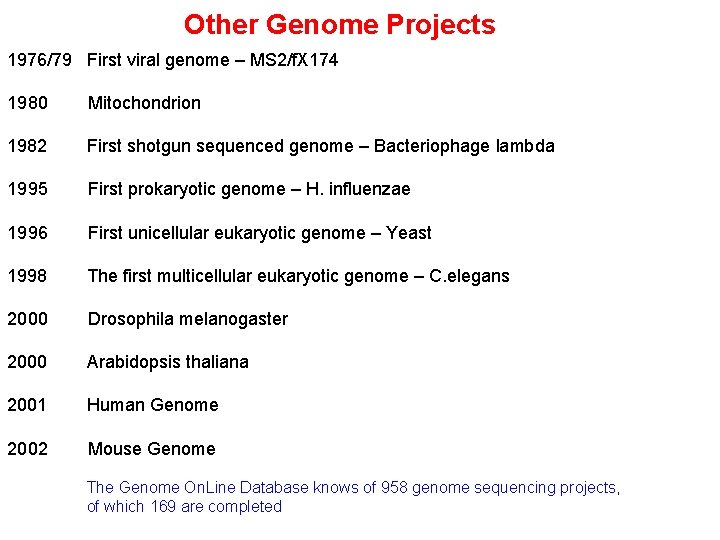
Other Genome Projects 1976/79 First viral genome – MS 2/f. X 174 1980 Mitochondrion 1982 First shotgun sequenced genome – Bacteriophage lambda 1995 First prokaryotic genome – H. influenzae 1996 First unicellular eukaryotic genome – Yeast 1998 The first multicellular eukaryotic genome – C. elegans 2000 Drosophila melanogaster 2000 Arabidopsis thaliana 2001 Human Genome 2002 Mouse Genome The Genome On. Line Database knows of 958 genome sequencing projects, of which 169 are completed

Favourite and Model Organisms Multicellular Animals Mammals Human Mouse Cow Dog Rat Chimp Pig 3. 5 3. 2 3. 0 2. 8 3. 1 3. 5 3. 0 Fish Puffer Fish Zebra Fish 0. 4 Gb 1. 9 Gb Insects Drosophila Honey Bee Yellow Fever Mosquito Malaria Mosquito Strachan and Read (2004) Chapter 8 Gb Gb Birds Chicken 1. 2 Gb Frog Xenopus Laevis 1. 7 Gb Nematodes Caenorhabdites elegans 100 Mb Caenorhabdites briggsae 80 Mb Sea Urchin Strongylocentrotus purpuratus Multicellular Plants 165 270 780 278 Mb Mb Arabidopsis thaliana Rice 125 Mb 430 Mb 800 Mb

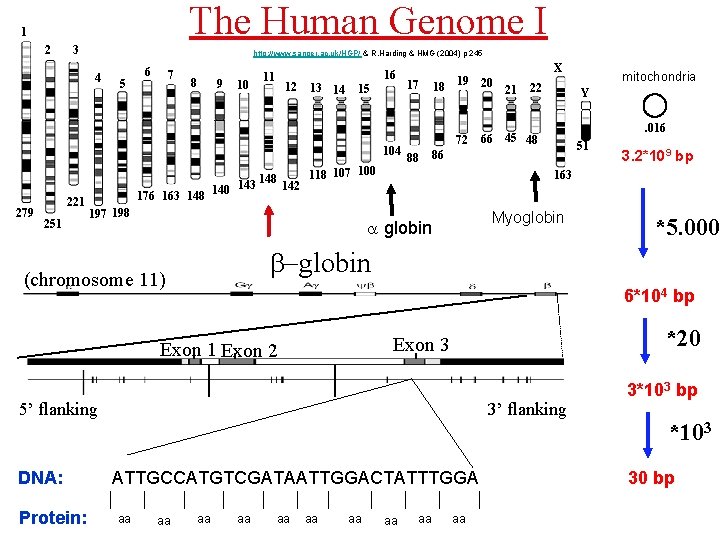
The Human Genome I 1 2 3 http: //www. sanger. ac. uk/HGP/ & R. Harding & HMG (2004) p 245 4 5 6 7 8 9 10 11 12 13 14 16 15 104 279 221 251 17 18 19 20 72 88 Y. 016 45 48 51 Myoglobin *5. 000 *20 Exon 3 3’ flanking ATTGCCATGTCGATAATTGGACTATTTGGA aa 3. 2*109 bp 6*104 bp 5’ flanking aa mitochondria b-globin Exon 1 Exon 2 Protein: 22 163 a globin (chromosome 11) DNA: 21 86 118 107 100 148 143 142 176 163 148 140 197 198 66 X aa aa 3*103 bp *103 30 bp

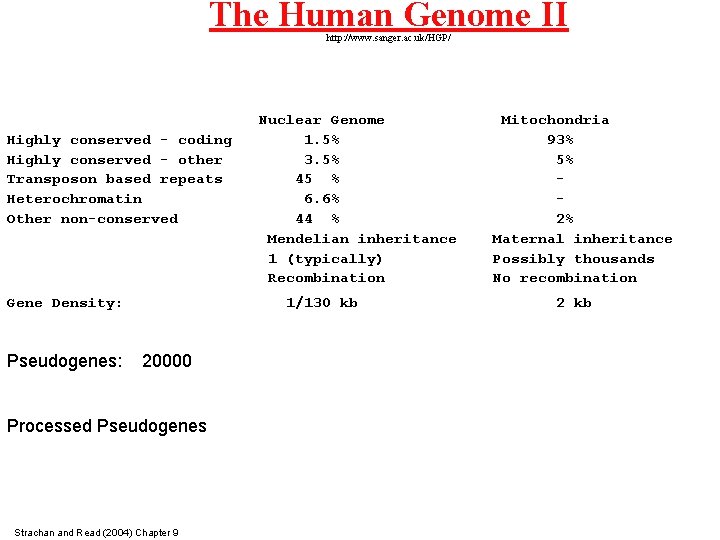
The Human Genome II http: //www. sanger. ac. uk/HGP/ Highly conserved - coding Highly conserved - other Transposon based repeats Heterochromatin Other non-conserved Gene Density: Pseudogenes: Nuclear Genome 1. 5% 3. 5% 45 % 6. 6% 44 % Mendelian inheritance 1 (typically) Recombination 1/130 kb 20000 Processed Pseudogenes Strachan and Read (2004) Chapter 9 Mitochondria 93% 5% 2% Maternal inheritance Possibly thousands No recombination 2 kb

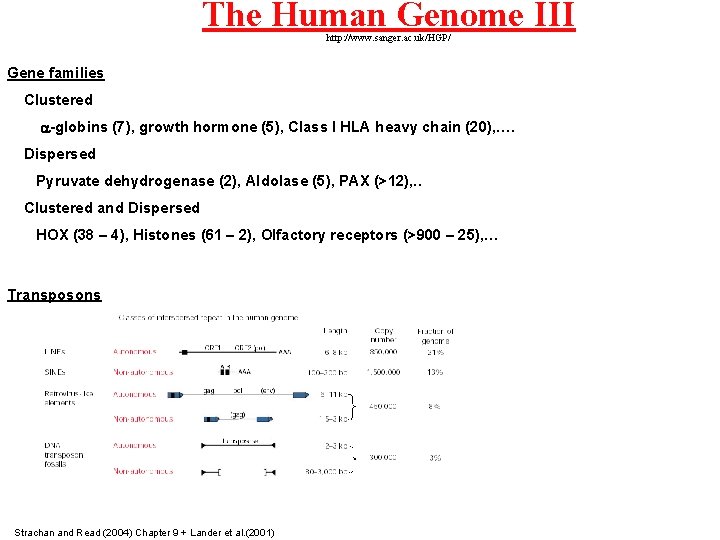
The Human Genome III http: //www. sanger. ac. uk/HGP/ Gene families Clustered a-globins (7), growth hormone (5), Class I HLA heavy chain (20), …. Dispersed Pyruvate dehydrogenase (2), Aldolase (5), PAX (>12), . . Clustered and Dispersed HOX (38 – 4), Histones (61 – 2), Olfactory receptors (>900 – 25), … Transposons Strachan and Read (2004) Chapter 9 + Lander et al. (2001)

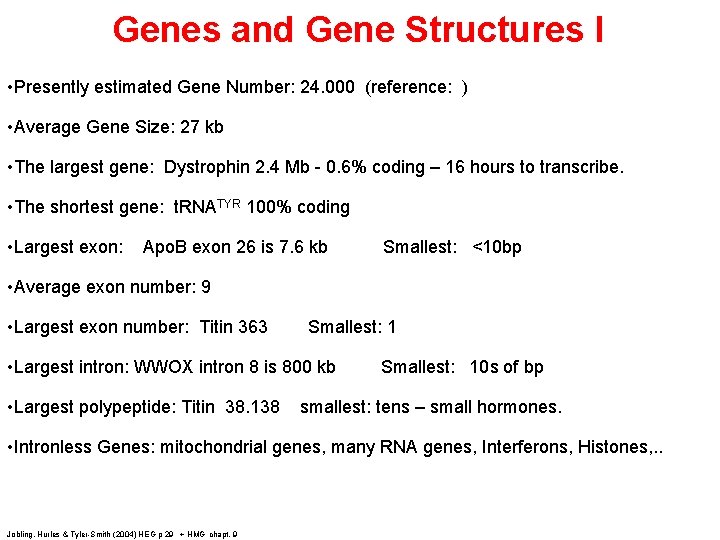
Genes and Gene Structures I • Presently estimated Gene Number: 24. 000 (reference: ) • Average Gene Size: 27 kb • The largest gene: Dystrophin 2. 4 Mb - 0. 6% coding – 16 hours to transcribe. • The shortest gene: t. RNATYR 100% coding • Largest exon: Apo. B exon 26 is 7. 6 kb Smallest: <10 bp • Average exon number: 9 • Largest exon number: Titin 363 Smallest: 1 • Largest intron: WWOX intron 8 is 800 kb • Largest polypeptide: Titin 38. 138 Smallest: 10 s of bp smallest: tens – small hormones. • Intronless Genes: mitochondrial genes, many RNA genes, Interferons, Histones, . . Jobling, Hurles & Tyler-Smith (2004) HEG p 29 + HMG chapt. 9

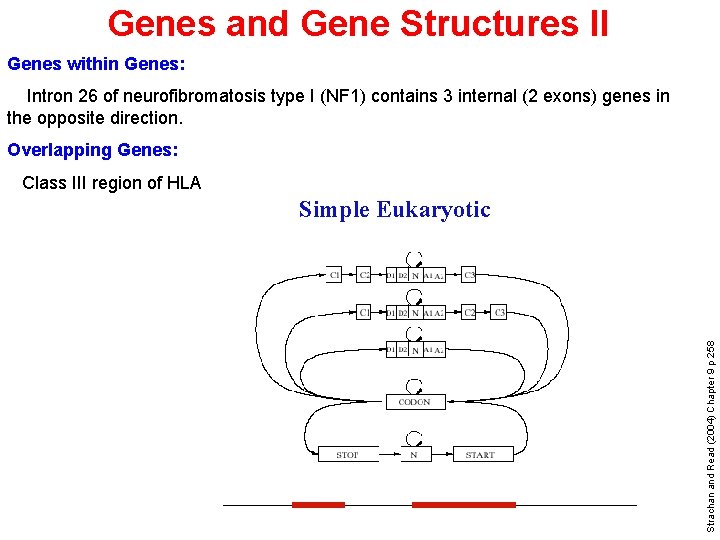
Genes and Gene Structures II Genes within Genes: Intron 26 of neurofibromatosis type I (NF 1) contains 3 internal (2 exons) genes in the opposite direction. Overlapping Genes: Class III region of HLA Strachan and Read (2004) Chapter 9 p 258 Simple Eukaryotic

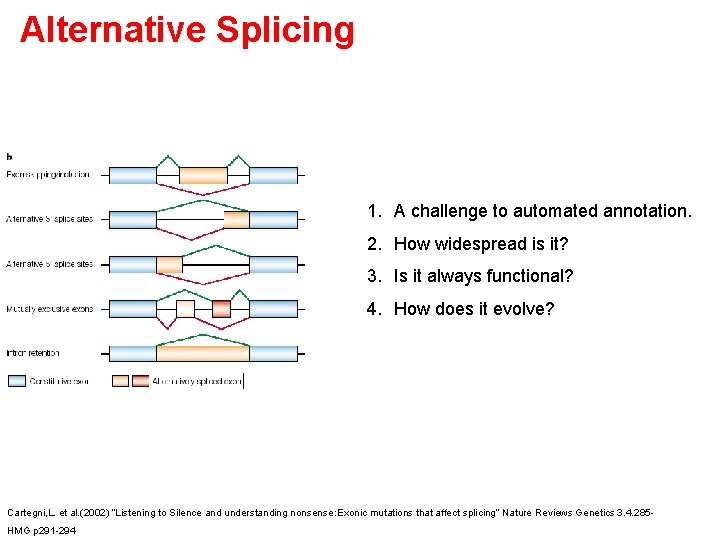
Alternative Splicing 1. A challenge to automated annotation. 2. How widespread is it? 3. Is it always functional? 4. How does it evolve? Cartegni, L. et al. (2002) “Listening to Silence and understanding nonsense: Exonic mutations that affect splicing” Nature Reviews Genetics 3. 4. 285 HMG p 291 -294

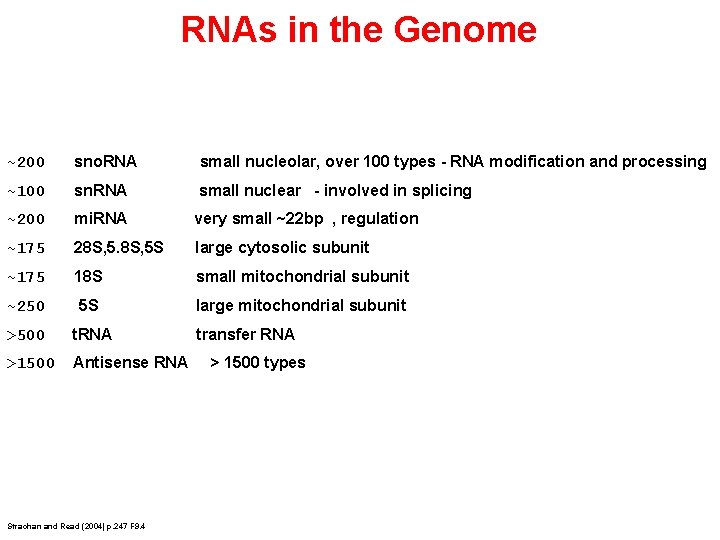
RNAs in the Genome ~200 sno. RNA small nucleolar, over 100 types - RNA modification and processing ~100 sn. RNA small nuclear - involved in splicing ~200 mi. RNA very small ~22 bp , regulation ~175 28 S, 5 S large cytosolic subunit ~175 18 S small mitochondrial subunit ~250 5 S large mitochondrial subunit >500 t. RNA >1500 Antisense RNA Strachan and Read (2004) p. 247 F 9. 4 transfer RNA > 1500 types

Genome Annotation Proteins Genomes ESTs Ensembl http: //www. ensembl. org Santa Cruz Genome Browser http: //genome. ucsc. edu/

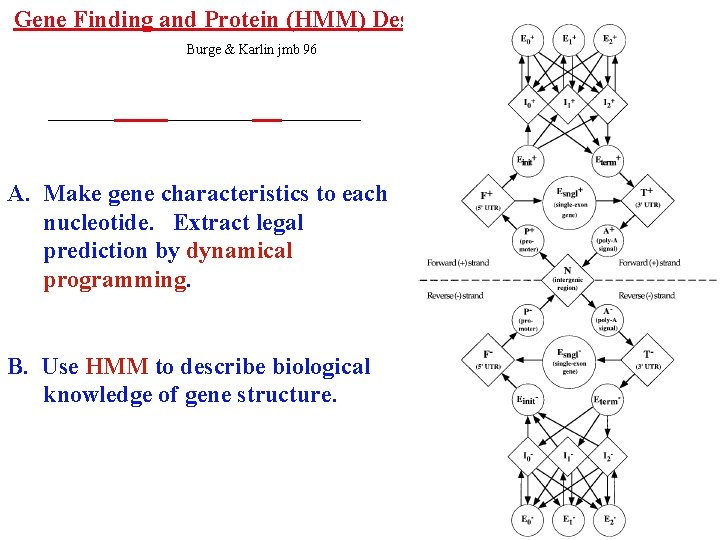
Gene Finding and Protein (HMM) Descriptors Burge & Karlin jmb 96 A. Make gene characteristics to each nucleotide. Extract legal prediction by dynamical programming. B. Use HMM to describe biological knowledge of gene structure.

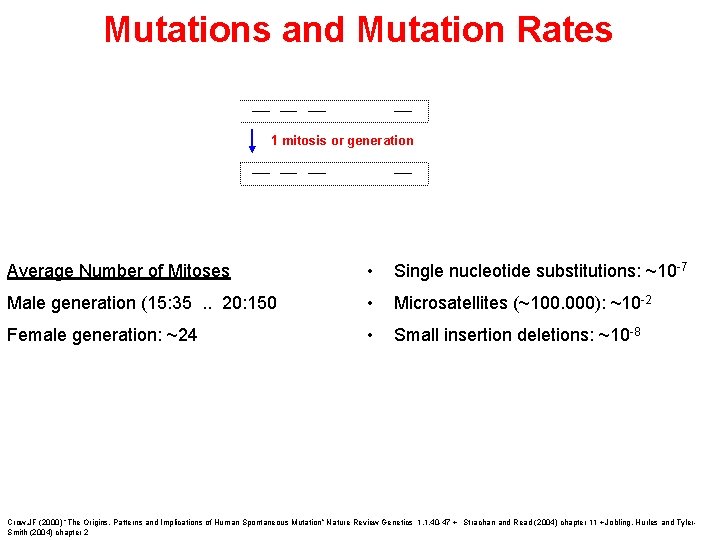
Mutations and Mutation Rates 1 mitosis or generation Average Number of Mitoses • Single nucleotide substitutions: ~10 -7 Male generation (15: 35. . 20: 150 • Microsatellites (~100. 000): ~10 -2 Female generation: ~24 • Small insertion deletions: ~10 -8 Crow, JF (2000) “The Origins, Patterns and Implications of Human Spontaneous Mutation” Nature Review Genetics 1. 1. 40 -47 + Strachan and Read (2004) chapter 11 + Jobling, Hurles and Tyler. Smith (2004) chapter 2

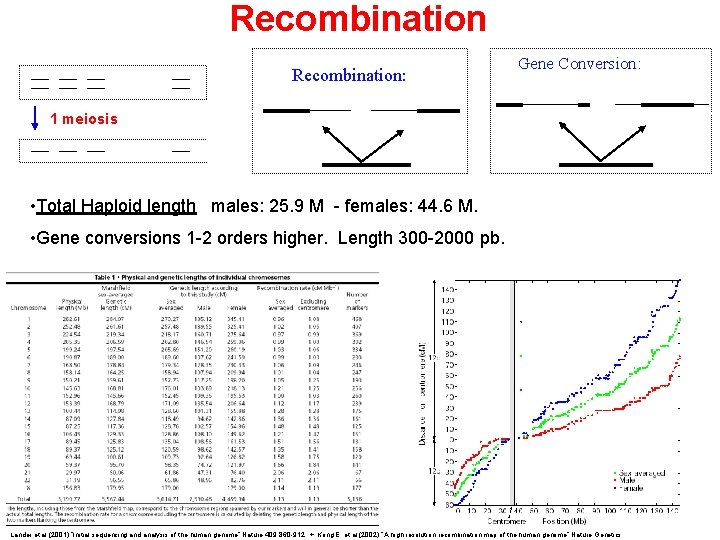
Recombination: Gene Conversion: 1 meiosis • Total Haploid length males: 25. 9 M - females: 44. 6 M. • Gene conversions 1 -2 orders higher. Length 300 -2000 pb. Lander et al. (2001) “Initial sequencing and analysis of the human genome” Nature 409. 860 -912. + Kong, E. et al. (2002) “A high resolution recombination map of the human genome” Nature Genetics

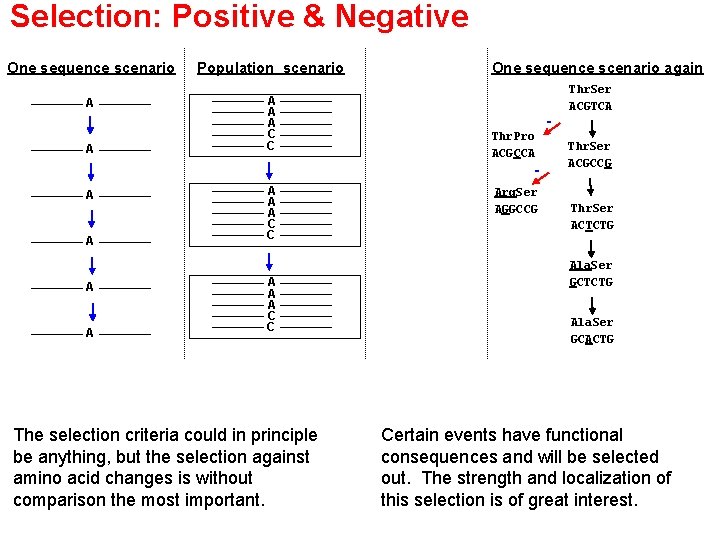
Selection: Positive & Negative One sequence scenario Population scenario A A C C A One sequence scenario again Thr. Pro ACGCCA - A A A A C C The selection criteria could in principle be anything, but the selection against amino acid changes is without comparison the most important. Arg. Ser AGGCCG Thr. Ser ACGTCA Thr. Ser ACGCCG Thr. Ser ACTCTG Ala. Ser GCACTG Certain events have functional consequences and will be selected out. The strength and localization of this selection is of great interest.

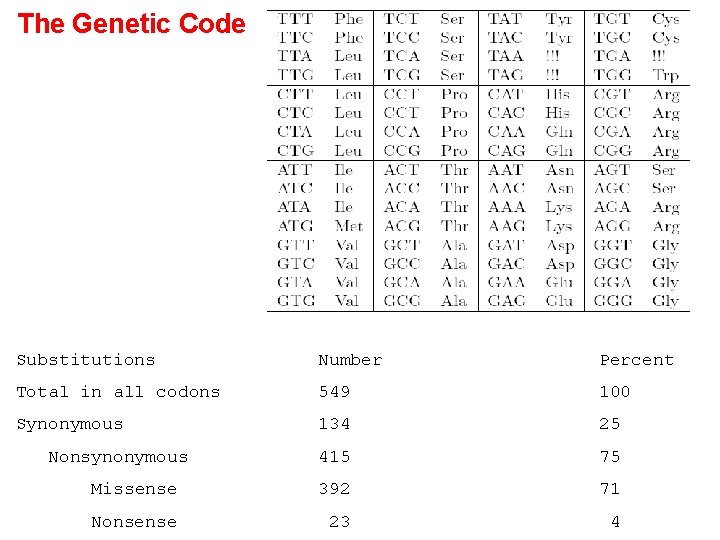
The Genetic Code Substitutions Number Percent Total in all codons 549 100 Synonymous 134 25 415 75 Missense 392 71 Nonsense 23 4 Nonsynonymous

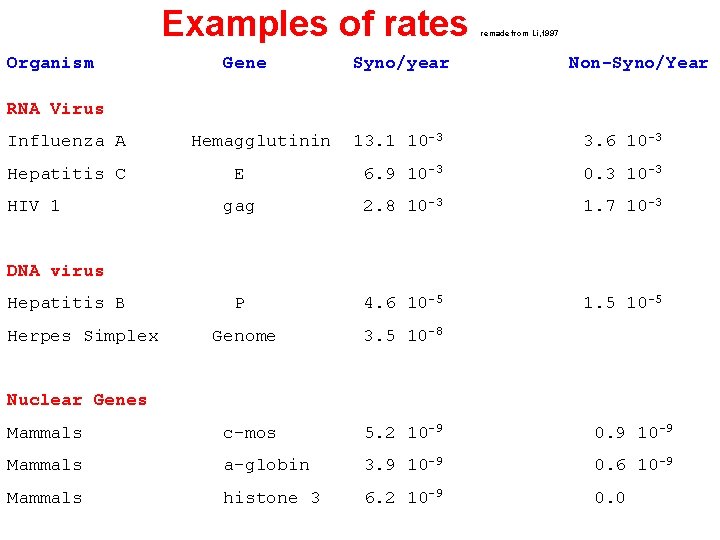
Examples of rates Organism Gene Syno/year remade from Li, 1997 Non-Syno/Year RNA Virus 13. 1 10 -3 3. 6 10 -3 E 6. 9 10 -3 0. 3 10 -3 gag 2. 8 10 -3 1. 7 10 -3 P 4. 6 10 -5 1. 5 10 -5 Genome 3. 5 10 -8 Mammals c-mos 5. 2 10 -9 0. 9 10 -9 Mammals a-globin 3. 9 10 -9 0. 6 10 -9 Mammals histone 3 6. 2 10 -9 0. 0 Influenza A Hepatitis C HIV 1 Hemagglutinin DNA virus Hepatitis B Herpes Simplex Nuclear Genes

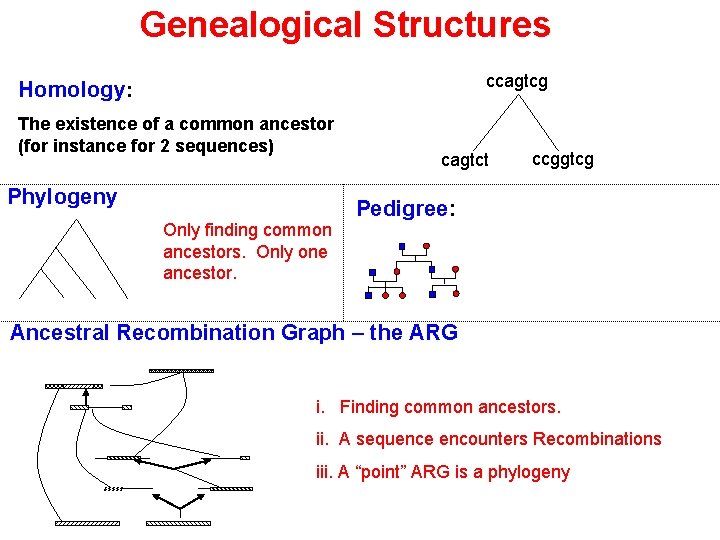
Genealogical Structures ccagtcg Homology: The existence of a common ancestor (for instance for 2 sequences) Phylogeny Only finding common ancestors. Only one ancestor. cagtct ccggtcg Pedigree: Ancestral Recombination Graph – the ARG i. Finding common ancestors. ii. A sequence encounters Recombinations iii. A “point” ARG is a phylogeny

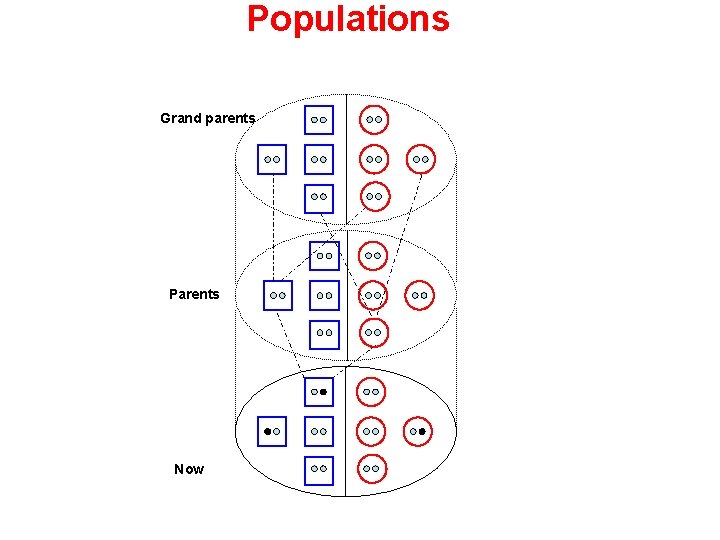
Populations Grand parents Parents Now

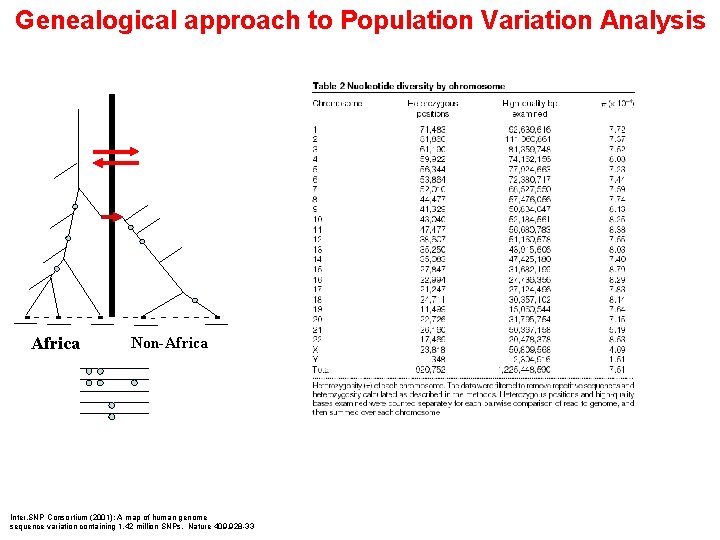
Genealogical approach to Population Variation Analysis Africa Non-Africa Inter. SNP Consortium (2001): A map of human genome sequence variation containing 1. 42 million SNPs. Nature 409. 928 -33

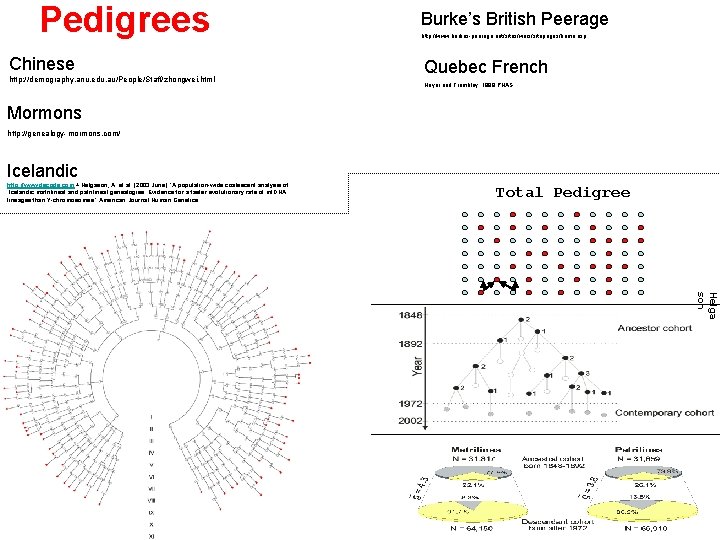
Pedigrees Chinese http: //demography. anu. edu. au/People/Staff/zhongwei. html Burke’s British Peerage http: //www. burkes-peerage. net/sites/wars/sitepages/home. asp Quebec French Heyer and Tremblay, 1998 PNAS Mormons http: //genealogy-mormons. com/ Icelandic http: //www. decode. com + Helgason, A. et al. (2003 June) “A population-wide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: Evidence for a faster evolutionary rate of mt. DNA lineages than Y-chromosomes” American Journal Human Genetics. Total Pedigree Helga son

Genealogical Questions Pedigrees Time back to first individual common ancestor to everyone ARG questions: The height of ARGs - correlation between local phylogenies Gene Phylogeny Questions Total Branch Length - Height

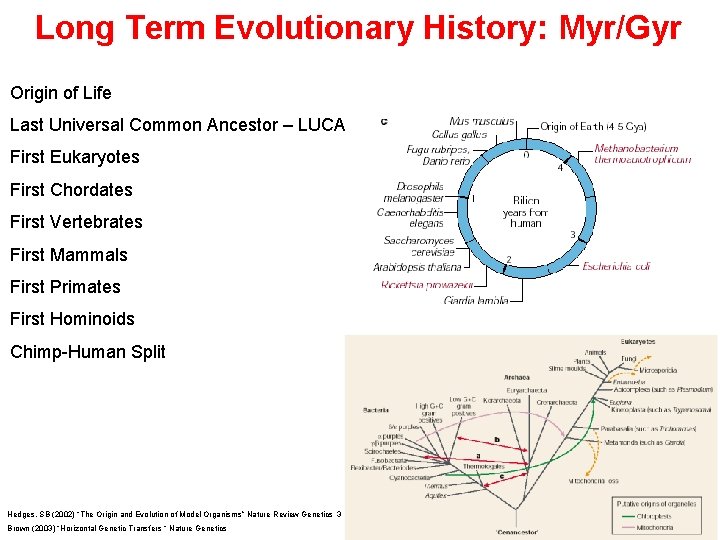
Long Term Evolutionary History: Myr/Gyr Origin of Life Last Universal Common Ancestor – LUCA First Eukaryotes First Chordates First Vertebrates First Mammals First Primates First Hominoids Chimp-Human Split Hedges, SB (2002) “The Origin and Evolution of Model Organisms” Nature Review Genetics 3. 11. 838 -848. Brown (2003) “Horizontal Genetic Transfers “ Nature Genetics

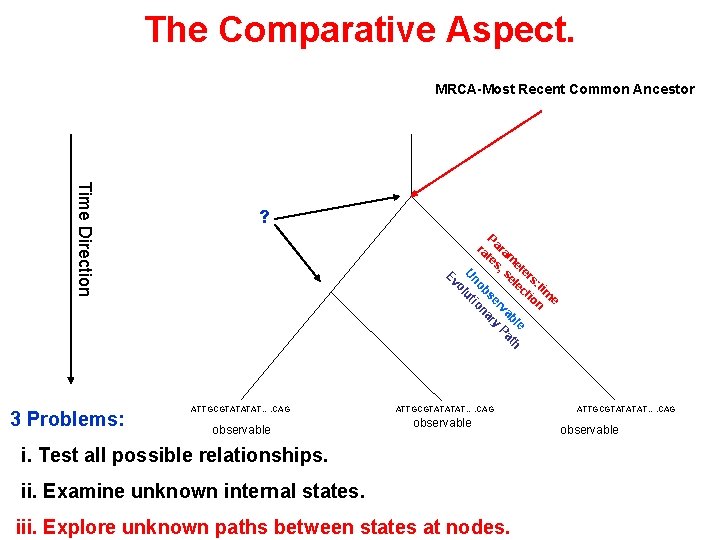
The Comparative Aspect. MRCA-Most Recent Common Ancestor P ra ara te m s, et se ers le : t ct im io e n e bl th va Pa er ry bs na no tio u ol U Ev Time Direction 3 Problems: ? ATTGCGTATATAT…. CAG observable i. Test all possible relationships. ii. Examine unknown internal states. iii. Explore unknown paths between states at nodes. ATTGCGTATATAT…. CAG observable

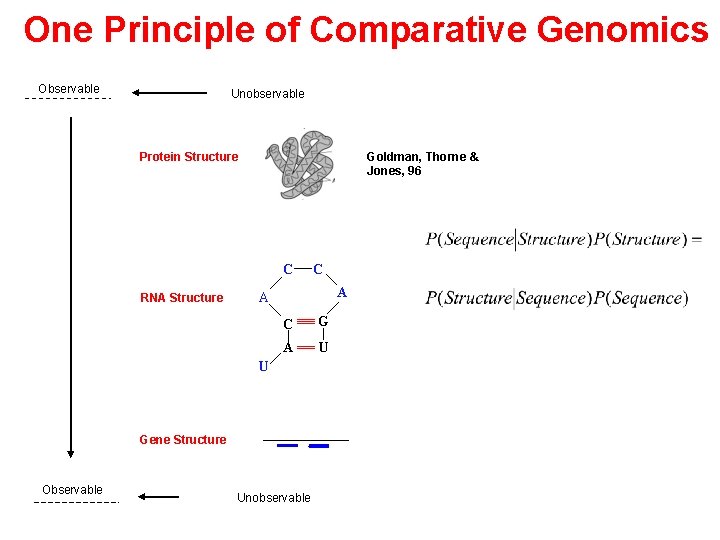
One Principle of Comparative Genomics Observable Unobservable Protein Structure Goldman, Thorne & Jones, 96 C RNA Structure A A C G A U U Gene Structure Observable C Unobservable

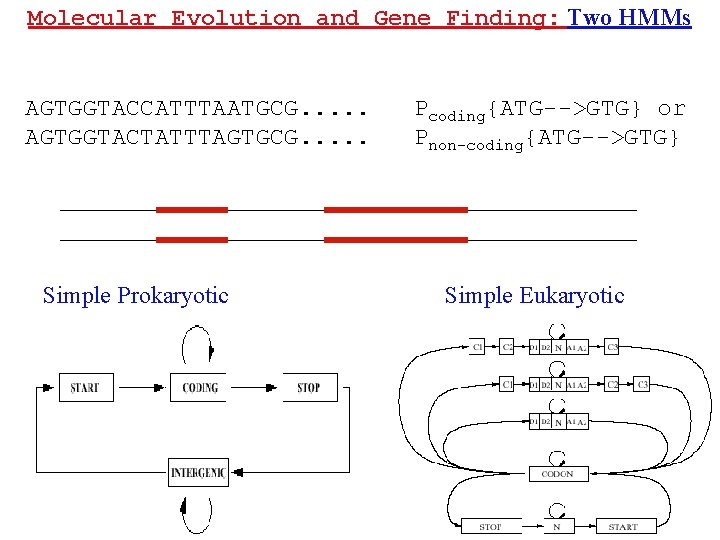
Molecular Evolution and Gene Finding: Two HMMs AGTGGTACCATTTAATGCG. . . AGTGGTACTATTTAGTGCG. . . Simple Prokaryotic Pcoding{ATG-->GTG} or Pnon-coding{ATG-->GTG} Simple Eukaryotic

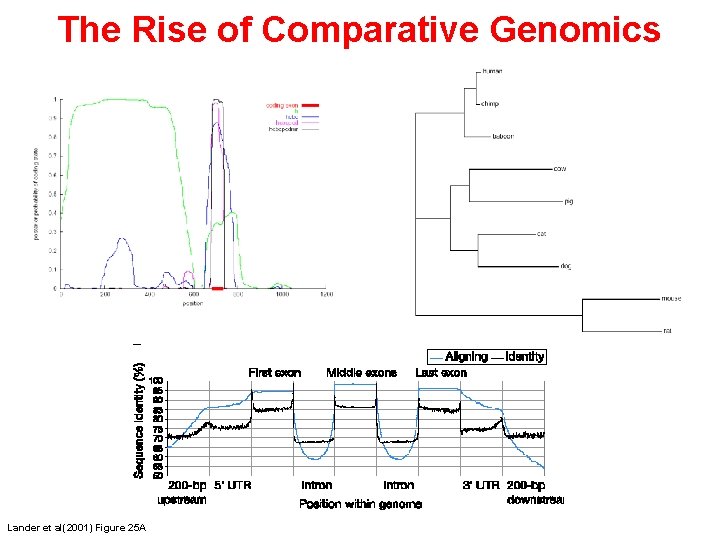
The Rise of Comparative Genomics Lander et al(2001) Figure 25 A

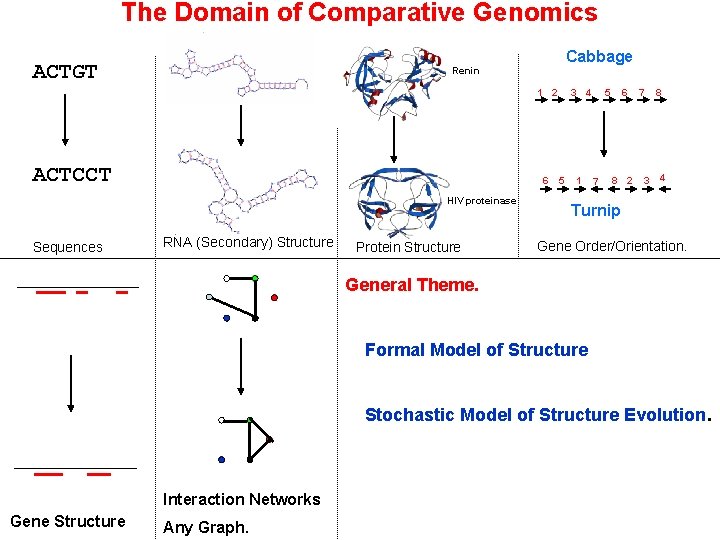
The Domain of Comparative Genomics ACTGT Cabbage Renin 1 2 ACTCCT 6 HIV proteinase Sequences RNA (Secondary) Structure Protein Structure 3 5 4 1 6 5 7 8 2 7 3 8 4 Turnip Gene Order/Orientation. General Theme. Formal Model of Structure Stochastic Model of Structure Evolution. Interaction Networks Gene Structure Any Graph.

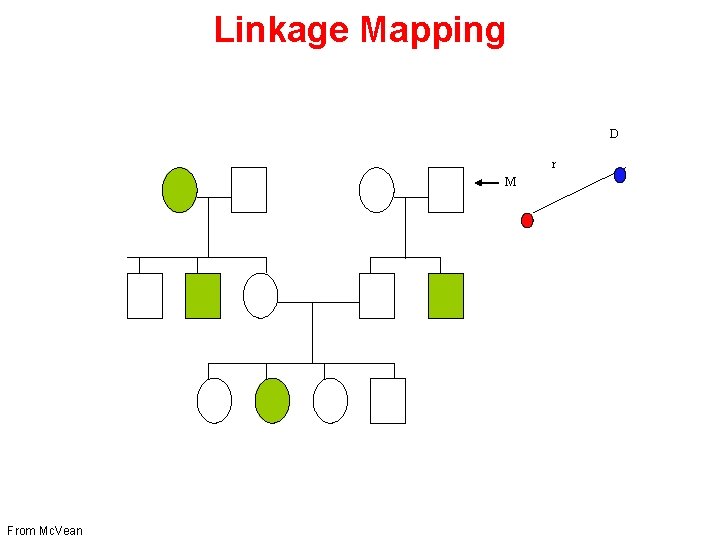
Linkage Mapping D r M From Mc. Vean

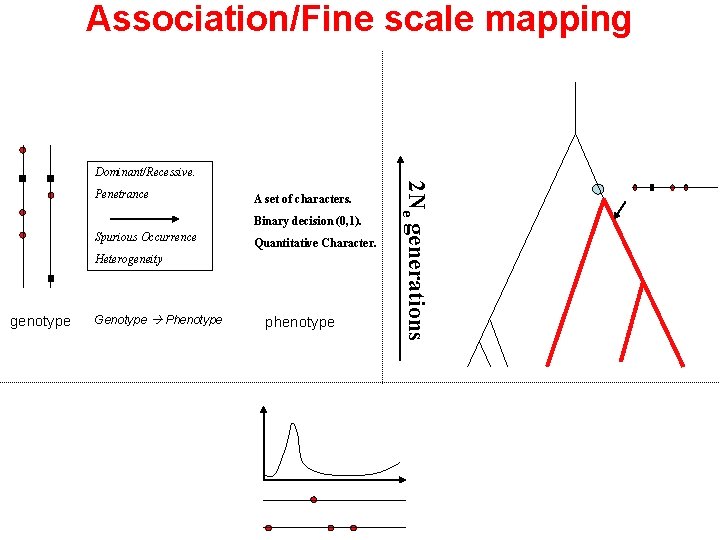
Association/Fine scale mapping Dominant/Recessive. A set of characters. Binary decision (0, 1). Spurious Occurrence Quantitative Character. Heterogeneity genotype Genotype Phenotype phenotype 2 Ne generations Penetrance

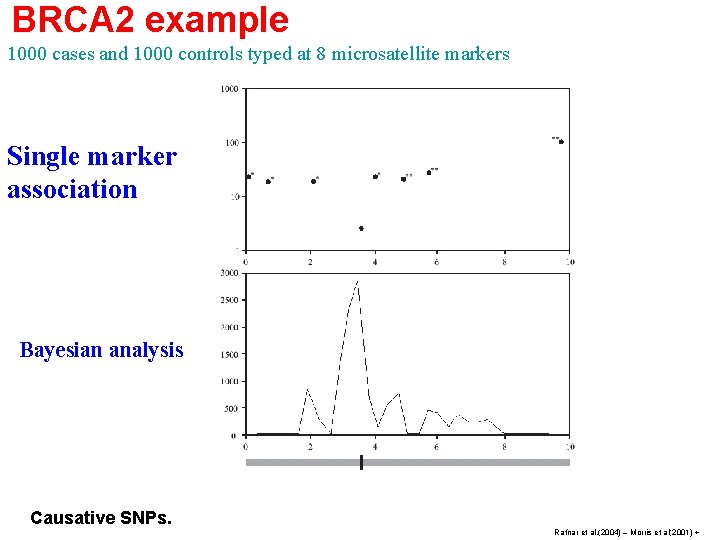
BRCA 2 example 1000 cases and 1000 controls typed at 8 microsatellite markers Single marker association Bayesian analysis Causative SNPs. Rafnar et al. (2004) – Morris et al(2001) +

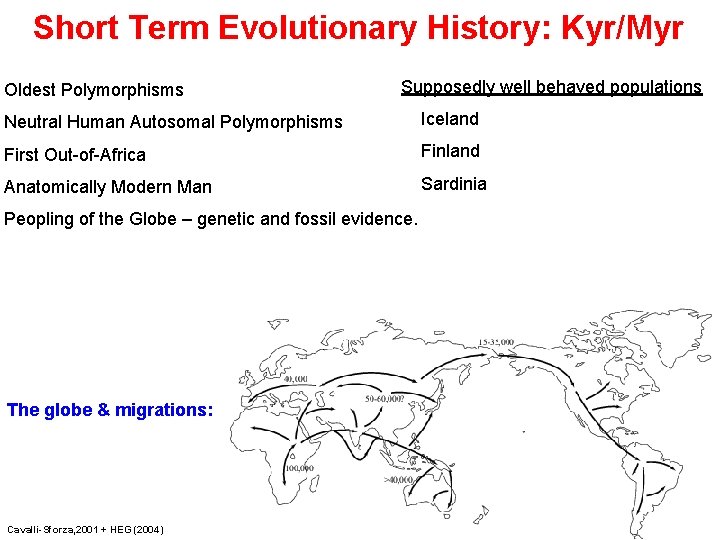
Short Term Evolutionary History: Kyr/Myr Oldest Polymorphisms Supposedly well behaved populations Neutral Human Autosomal Polymorphisms Iceland First Out-of-Africa Finland Anatomically Modern Man Sardinia Peopling of the Globe – genetic and fossil evidence. The globe & migrations: Cavalli-Sforza, 2001 + HEG (2004)

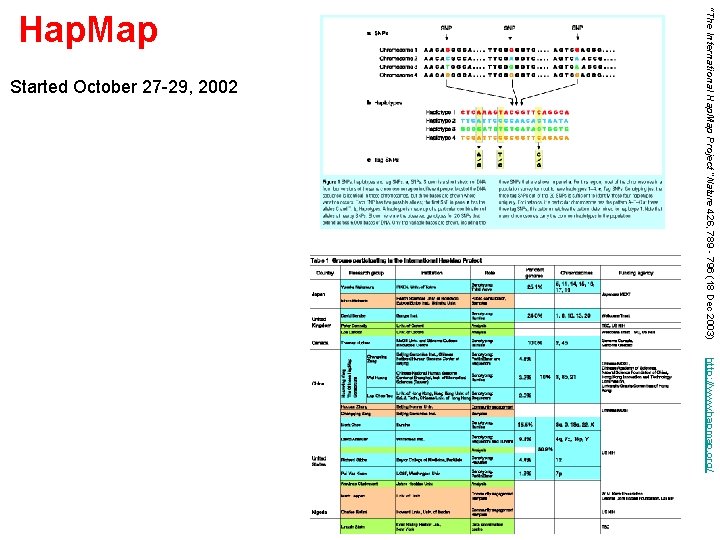
Started October 27 -29, 2002 “The International Hap. Map Project “Nature 426, 789 - 796 (18 Dec 2003) Hap. Map http: //www. hapmap. org/

Hap. Map

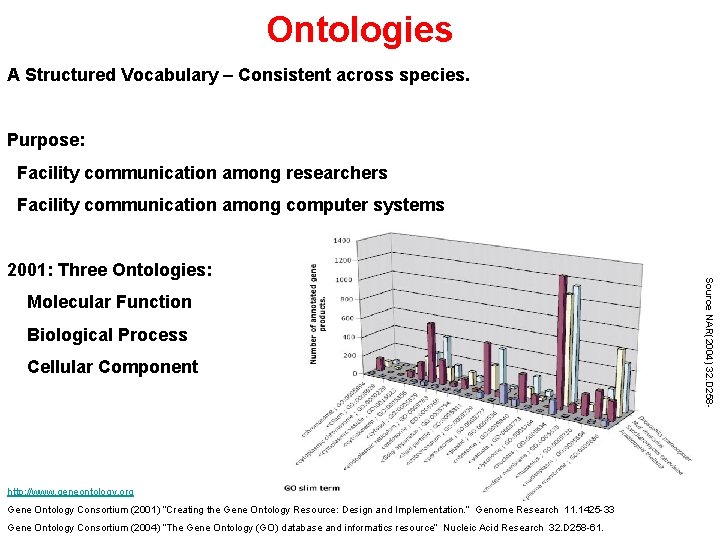
Ontologies A Structured Vocabulary – Consistent across species. Purpose: Facility communication among researchers Facility communication among computer systems Molecular Function Biological Process Cellular Component http: //www. geneontology. org Gene Ontology Consortium (2001) “Creating the Gene Ontology Resource: Design and Implementation. ” Genome Research 11. 1425 -33 Gene Ontology Consortium (2004) “The Gene Ontology (GO) database and informatics resource” Nucleic Acid Research 32. D 258 -61. Source NAR(2004) 32. D 258 - 2001: Three Ontologies:

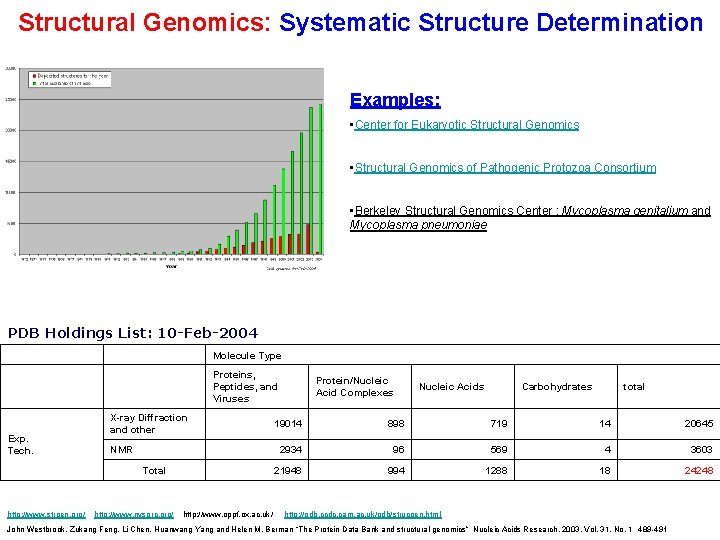
Structural Genomics: Systematic Structure Determination Examples: • Center for Eukaryotic Structural Genomics • Structural Genomics of Pathogenic Protozoa Consortium • Berkeley Structural Genomics Center : Mycoplasma genitalium and Mycoplasma pneumoniae PDB Holdings List: 10 -Feb-2004 Molecule Type Proteins, Peptides, and Viruses Exp. Tech. X-ray Diffraction and other NMR Total http: //www. strgen. org/ http: //www. nysgrc. org/ http: //www. oppf. ox. ac. uk/ Protein/Nucleic Acid Complexes Nucleic Acids Carbohydrates total 19014 898 719 14 20645 2934 96 569 4 3603 21948 994 1288 18 24248 http: //pdb. ccdc. cam. ac. uk/pdb/strucgen. html John Westbrook, Zukang Feng, Li Chen, Huanwang Yang and Helen M. Berman “The Protein Data Bank and structural genomics” Nucleic Acids Research, 2003, Vol. 31, No. 1 489 -491

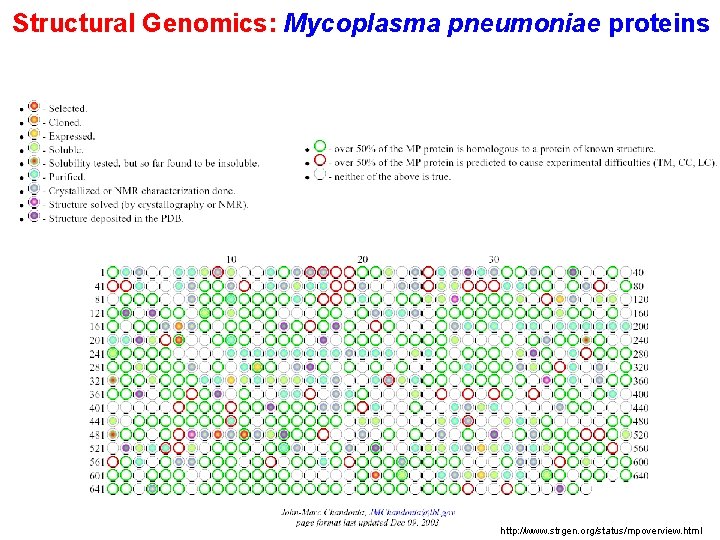
Structural Genomics: Mycoplasma pneumoniae proteins http: //www. strgen. org/status/mpoverview. html

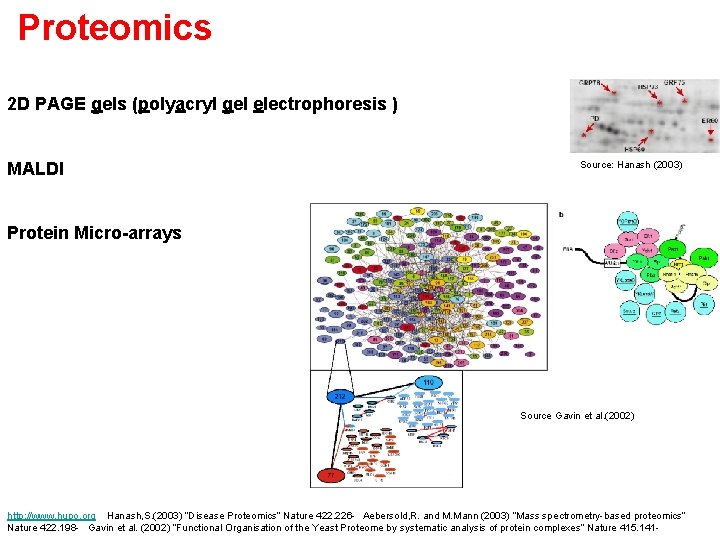
Proteomics 2 D PAGE gels (polyacryl gel electrophoresis ) MALDI Source: Hanash (2003) Protein Micro-arrays Source Gavin et al. (2002) http: //www. hupo. org Hanash, S. (2003) “Disease Proteomics” Nature 422. 226 - Aebersold, R. and M. Mann (2003) “Mass spectrometry-based proteomics” Nature 422. 198 - Gavin et al. (2002) “Functional Organisation of the Yeast Proteome by systematic analysis of protein complexes” Nature 415. 141 -

Summary The Genomes: Variation and long term evolution. Genealogical Structures: Phylogenies, Pedigrees and the ARG Long term Dynamics of the Human Genome: The comparative aspect (Genotype Phenotype) & (Population Genetics/History) => Gene Mapping

Our Genomically Motivated Projects 1. Comparative gene annotation (Meyer, Skou Pedersen) 2. Superimposed selective constraints (Forsberg, Meyer, Skou Pedersen) * 3. Haplotype Blocks (Song) * 4. Genome transformations (Miklos) 5. Ancestral Blocks* 6. Statistical Sequence Comparison (Drummond, Lunter, Miklos) 7. Substitutions and insertion-deletions at the Genome Level (Lunter) Next week

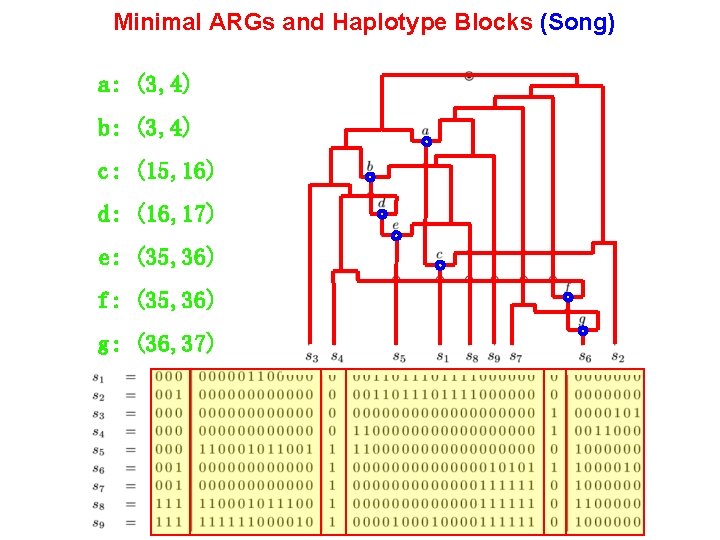
Minimal ARGs and Haplotype Blocks (Song) a: (3, 4) b: (3, 4) c: (15, 16) d: (16, 17) e: (35, 36) f: (35, 36) g: (36, 37)

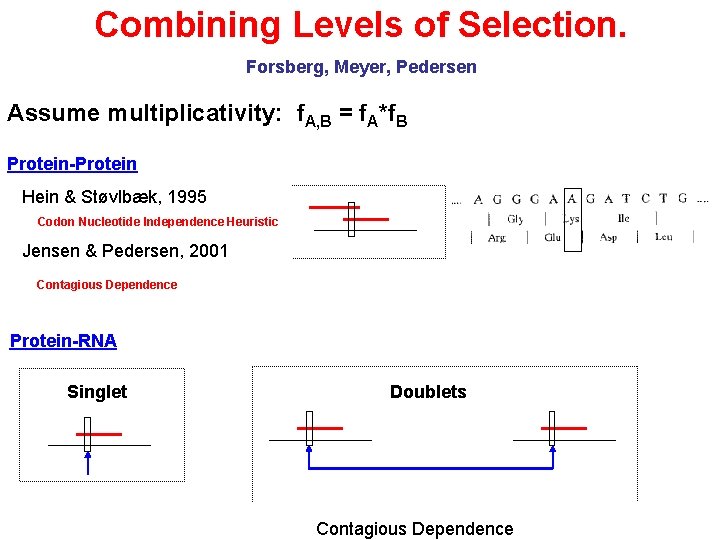
Combining Levels of Selection. Forsberg, Meyer, Pedersen Assume multiplicativity: f. A, B = f. A*f. B Protein-Protein Hein & Støvlbæk, 1995 Codon Nucleotide Independence Heuristic Jensen & Pedersen, 2001 Contagious Dependence Protein-RNA Singlet Doublets Contagious Dependence

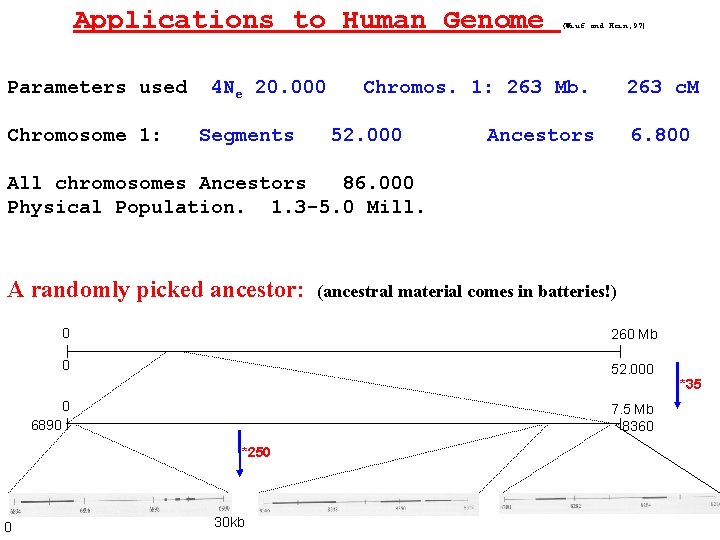
Applications to Human Genome Parameters used Chromosome 1: 4 Ne 20. 000 Segments (Wiuf and Hein, 97) Chromos. 1: 263 Mb. 52. 000 263 c. M Ancestors 6. 800 All chromosomes Ancestors 86. 000 Physical Population. 1. 3 -5. 0 Mill. A randomly picked ancestor: 0 260 Mb 0 52. 000 0 7. 5 Mb 8360 6890 *250 0 (ancestral material comes in batteries!) 30 kb *35

References: Books & www-pages. Books: Strachan and Read (2004) “Human Molecular Genetics” (3 rd Ed. ) Bioscience Jobling, Hurles and Tyler-Smith (2004) “Human Evolutionary Genetics” Bioscience Sulston, J. (2002) “Our Common Thread” Corgi Books Ridley, Matt (2001) “Genome” “Encyclopedia of the Human Genome” (2003) Nature Publishing Group Cavalli-Sforza, L. (2001) “Genes, People and Language” Penguin Key articles: Lander et al. (2001) “Initial Sequencing and Analysis of the Human Genome” Nature Venter et al. (2001)”The Sequence of the Human Genome” Science 291. 1304 -1351

References: www-pages. Major sequencing centers: Baylor College of Medicine Genome Sequencing Center hgsc. bcm. tcm. edu/ Celera www. celera. com Do. E Joint Genome Institute www. jgi. doe. gov Genoscope www. genoscope. cns. fr TIGR www. tigr. org Washington University Genome Sequencing Center www. genome. wustl. edu Wellcome Trust Sanger Institute www. sanger. ac. uk Whitehead Institute/MIT Center for Genome Research www. -genome. wi. mit. edu Ensembl genome annotator European Bionformatics Institute NCBI - www. ensembl. org www. ebi. ac. uk www. ncbi. nlm. nih. gov Nature Genome Gateway http: //www. nature. com/genomics/human/ Integrated Genomics http: //wit. integratedgenomics. com/GOLD/ Ebi genome databases http: //www 2. ebi. ac. uk/genomes/ Primate Sequencing Projects http: //sayer. lab. nig. jp/~silver/index. html European Bioinformatics Institute Proteomics http: //www. ebi. ac. uk/proteome/ National Center for Biotechnology Information http: //www. ncbi. nlm. nih. gov/ Hap. Map Project Homepage http: //www. hapmap. org/ Online Inheritance in Man http: //www. ncbi. nlm. nih. gov/omim/