Genome Projects Maps Human Genome Mapping Human Genome





- Slides: 53

Genome Projects • • • Maps Human Genome Mapping Human Genome Sequencing Landmarks in Sequencing Comparative Genomics The Tree and Our Place on It

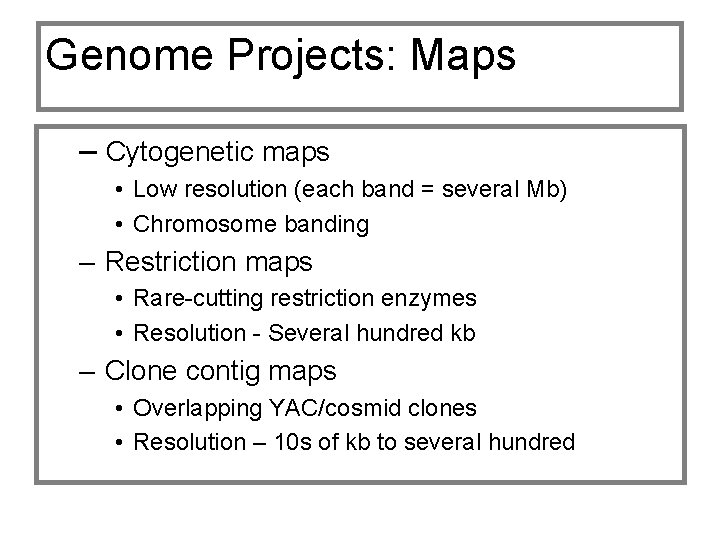
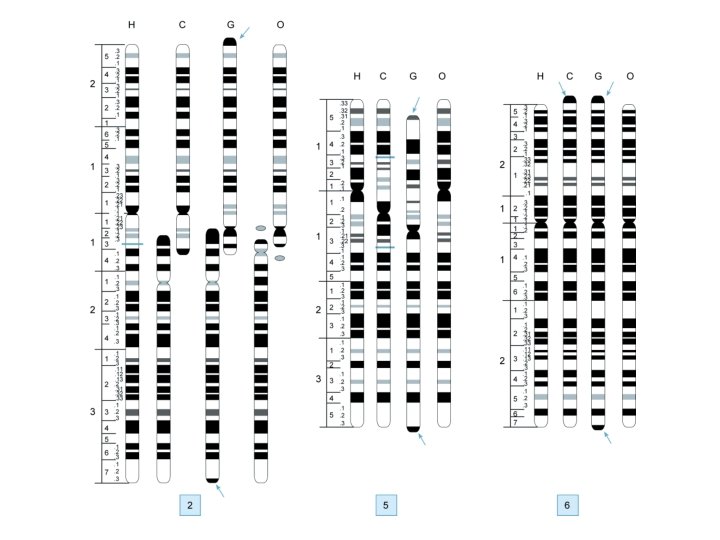
Genome Projects: Maps – Cytogenetic maps • Low resolution (each band = several Mb) • Chromosome banding – Restriction maps • Rare-cutting restriction enzymes • Resolution - Several hundred kb – Clone contig maps • Overlapping YAC/cosmid clones • Resolution – 10 s of kb to several hundred

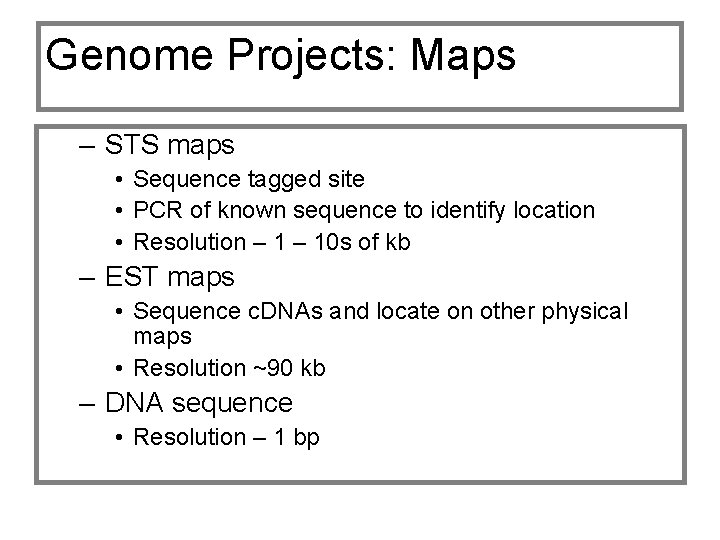
Genome Projects: Maps – STS maps • Sequence tagged site • PCR of known sequence to identify location • Resolution – 10 s of kb – EST maps • Sequence c. DNAs and locate on other physical maps • Resolution ~90 kb – DNA sequence • Resolution – 1 bp


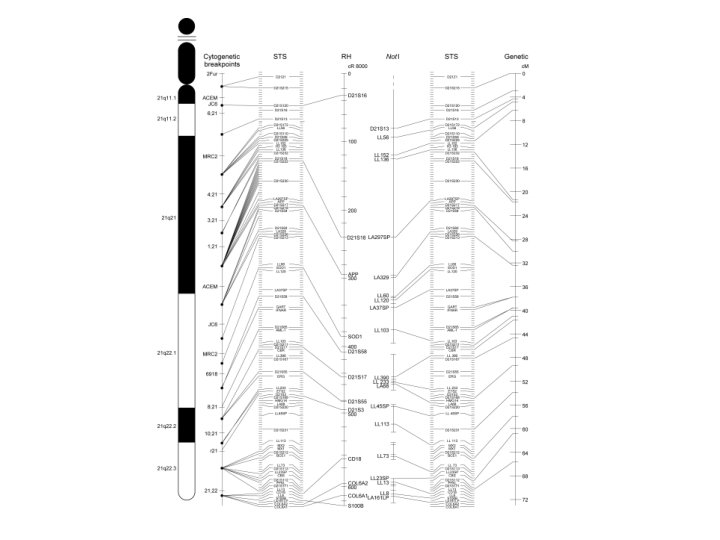
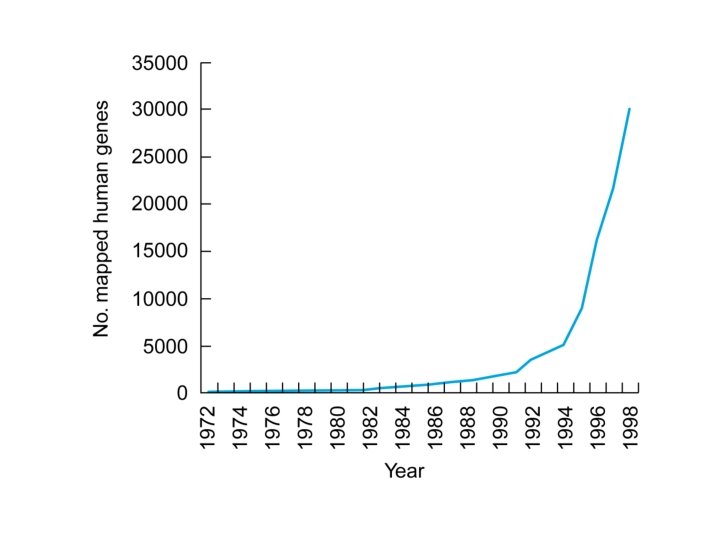
Genome Projects: Maps • Progress in human gene mapping – Slow when compared to Drosophila and mouse – Speed increased when non-coding markers began being used in conjunction with familial patterns of linkage


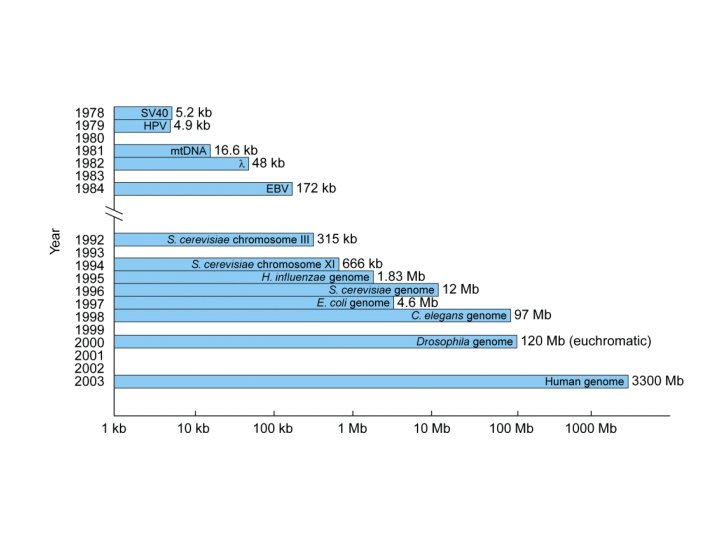
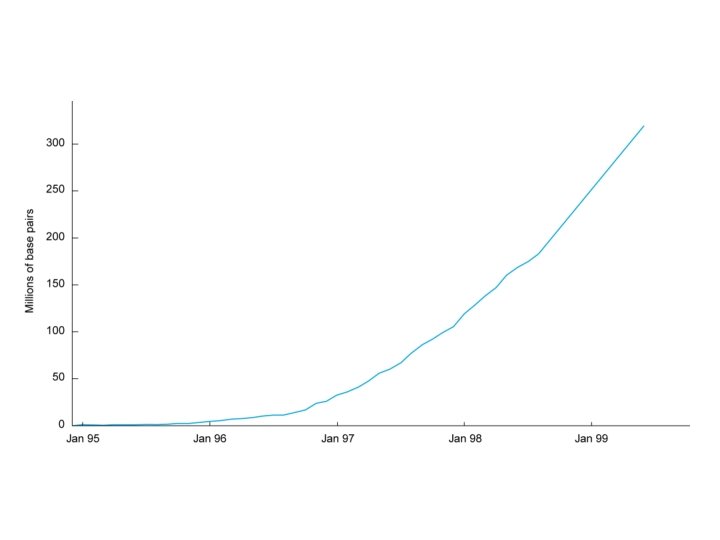
Genome Projects: Sequencing • Landmarks in genome sequencing


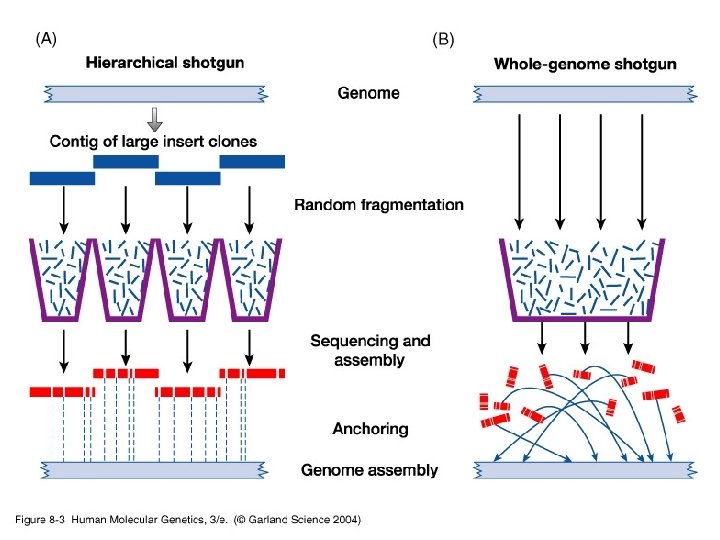
Genome Projects: Sequencing • Progress in human genome sequencing – Hierarchical vs. whole genome shotgun sequencing – Precision and accuracy – Repetitive DNA

08_03. jpg


Genome Projects: Sequencing • Progress in human genome sequencing – Hierarchical vs whole genome shotgun sequencing – Landmark papers in Nature and Science (2001) • Venter et al Science 16 February 2001; 291: 13041351 • Lander et al Nature 409 (6822): 860 -921

Genome Projects: Sequencing • A typical high-throughput genomics facility


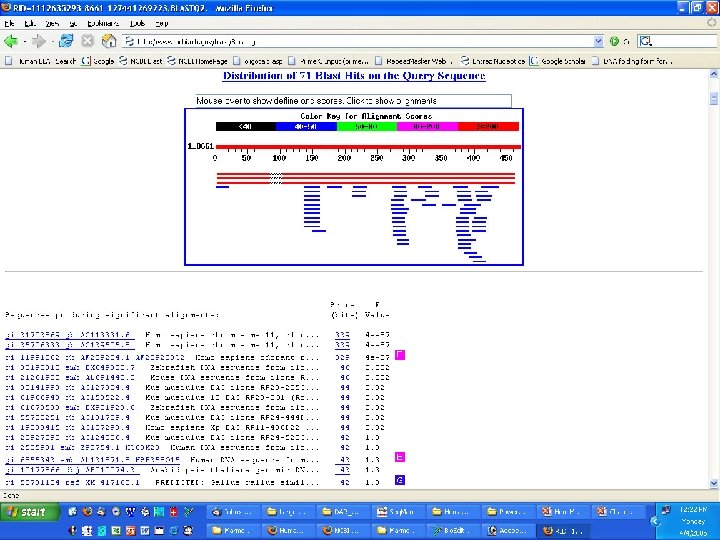
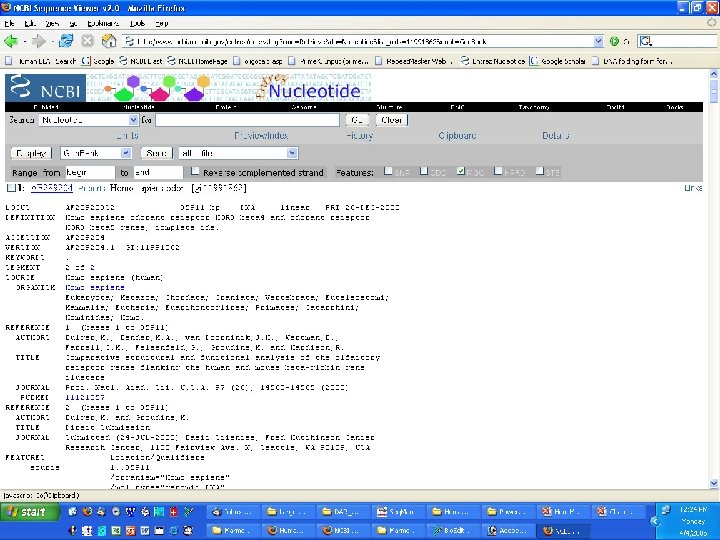
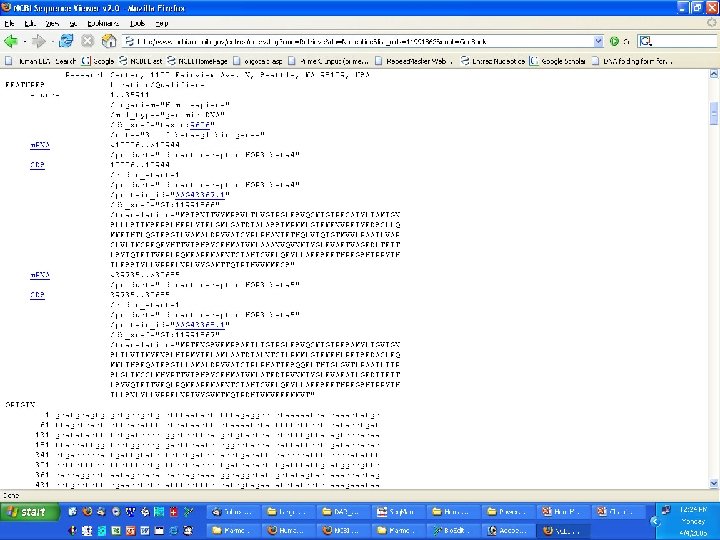
Genome Projects: Sequencing • BLAST/BLAT and other tools – BLAST - Basic local alignment search tool • Input a sequence and find matches to human or other organisms – publication information – DNA and protein sequence (if applicable)





Genome Projects: Sequencing • BLAST/BLAT and other tools – BLAT – BLAST-like alignment tool • A “genome browser” • Genomes available: – human, chimp, dog, cow, mouse, opossum, rat, chicken, Xenopus, Zebrafish, Tetraodon, Fugu, nematode (x 3), Drosophila (x 5), Apis (x 3), Saccharomyces (yeast). • Off-slide show example: chr 6: 121, 387, 504121, 720, 836

Genome Projects: BLAT • BLAT and other tools – Off-slide show example: chr 6: 121, 387, 504121, 720, 836 • • • STS and RH markers Known genes (alternative splices of genes) Conservation with other genomes Known single nucleotide polymorphisms (SNPs) Repetitive DNA

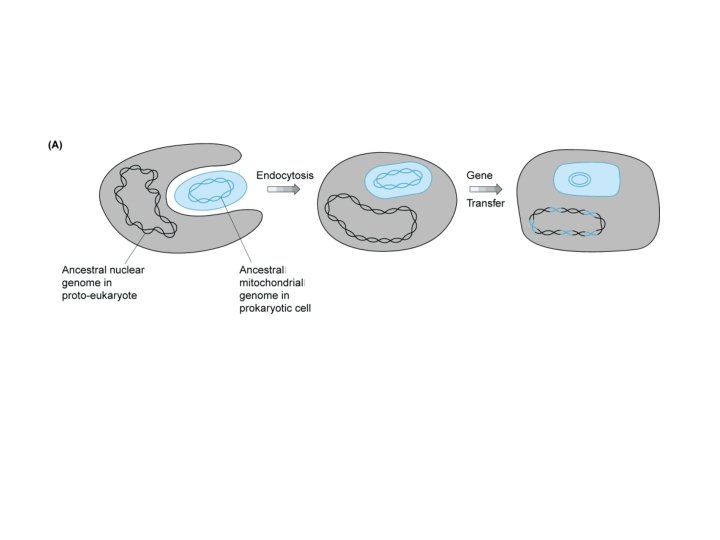
Our Place in the Tree of Life: Genome Evolution • Endosymbiont hypothesis


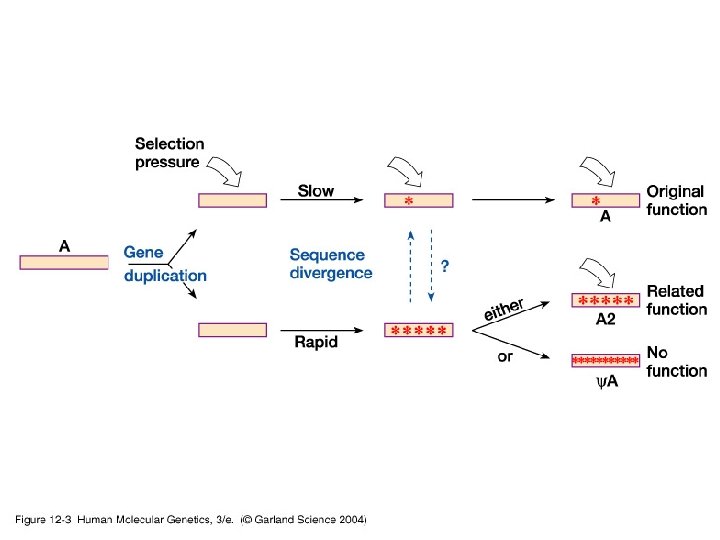
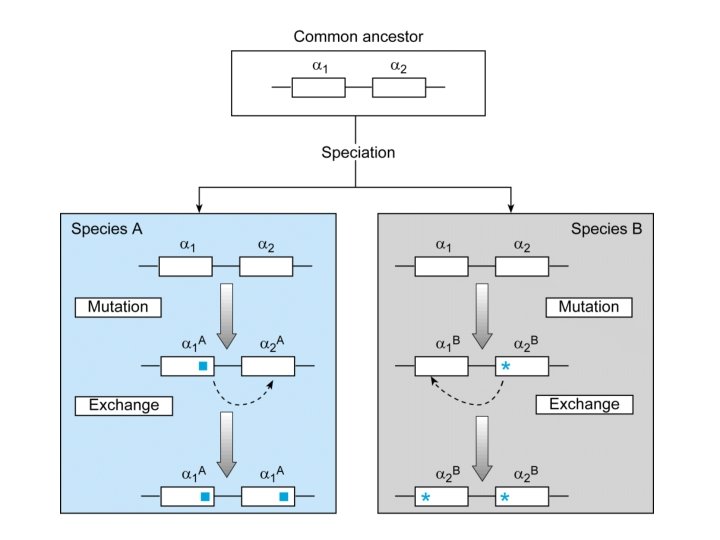
Our Place in the Tree of Life: Genome Evolution • Gene duplication – Acquisition of novel functions


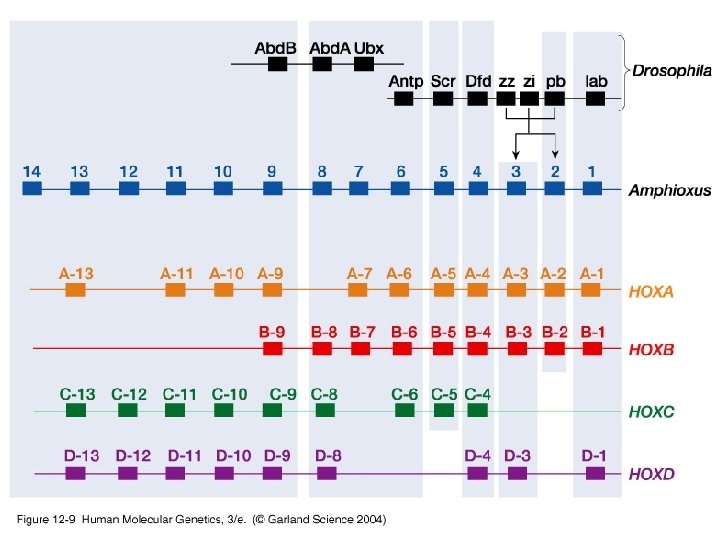
Our Place in the Tree of Life: Genome Evolution • Gene duplication – Paralogous chromosome segments – Ancient genome duplications – Hox gene clusters


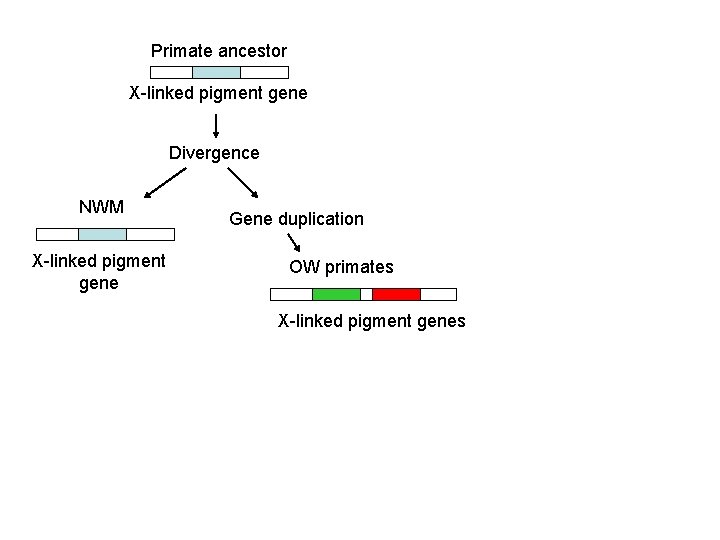
Our Place in the Tree of Life: Genome Evolution • Concerted evolution – – members of a sequence of families in a species evolve together so that they maintain high similarities among themselves while diverging from members of other species – via gene conversion – example primate photoreceptors


Primate ancestor X-linked pigment gene Divergence NWM X-linked pigment gene Gene duplication OW primates X-linked pigment genes

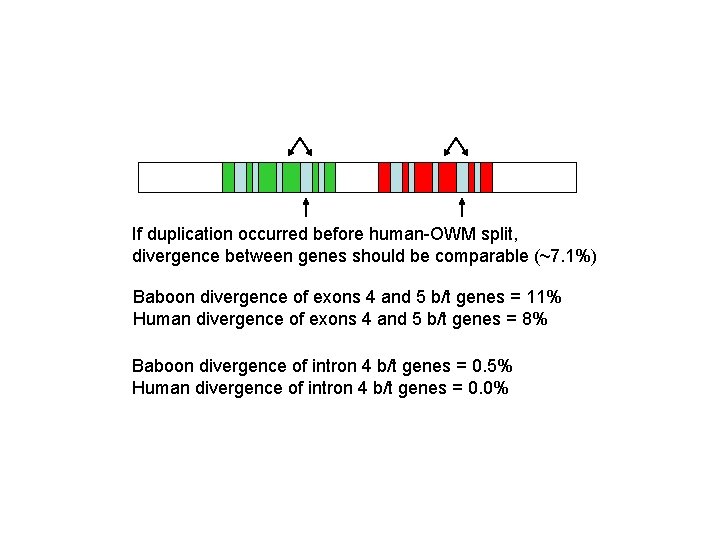
If duplication occurred before human-OWM split, divergence between genes should be comparable (~7. 1%) Baboon divergence of exons 4 and 5 b/t genes = 11% Human divergence of exons 4 and 5 b/t genes = 8% Baboon divergence of intron 4 b/t genes = 0. 5% Human divergence of intron 4 b/t genes = 0. 0%

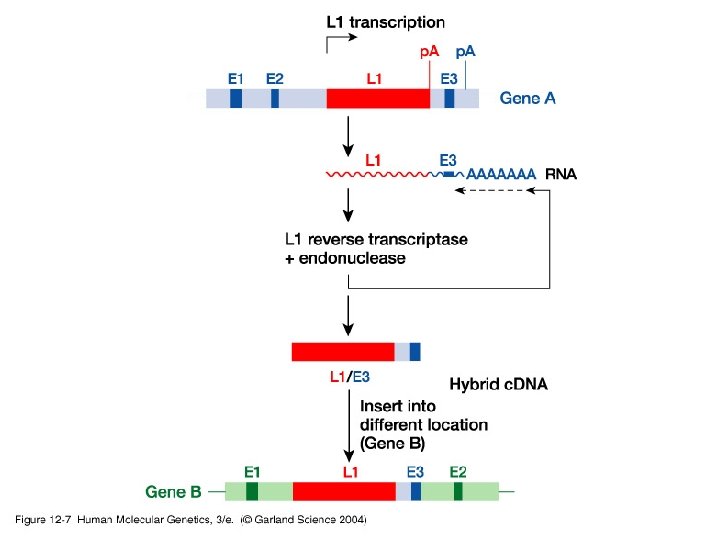
Our place in the tree of life • Exon duplication and shuffling – Examples – May be mediated by mobile elements


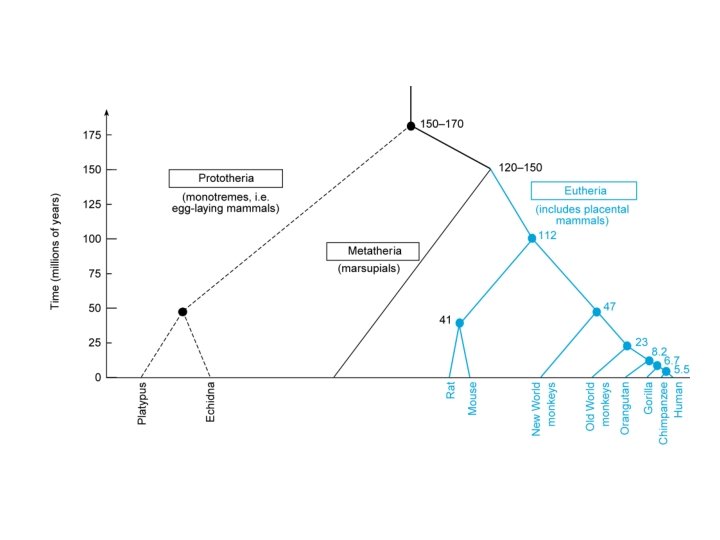
Our Place in the Tree of Life: Genome Evolution • Mammalian phylogeny


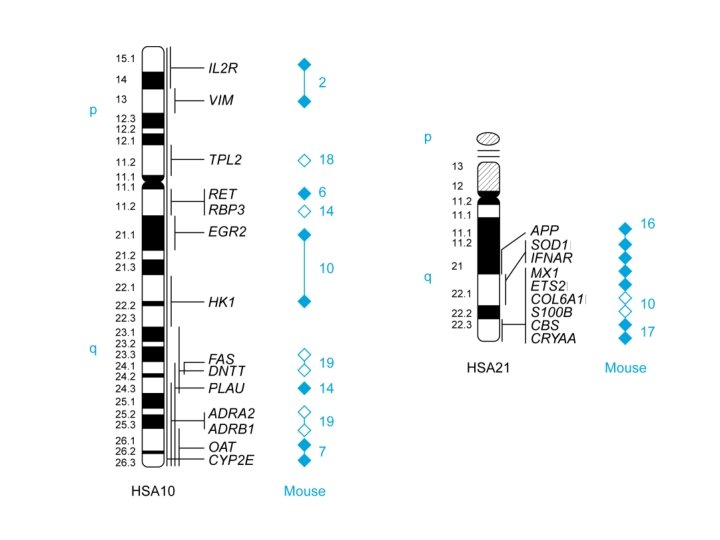
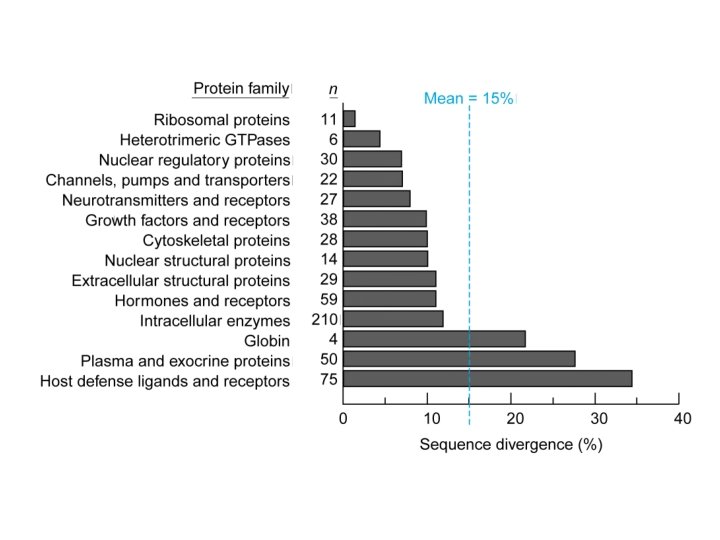
Our Place in the Tree of Life: Genome Evolution • Conservation between mouse and man – Limited to subchromosomal regions – Variable rates of amino acid sequence divergence



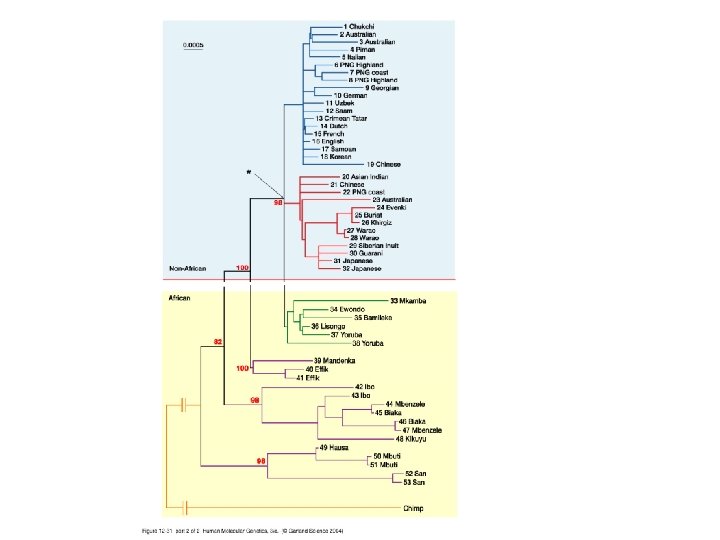
Our Place in the Tree of Life: Genome Evolution • Human evolution – Determining our place in the tree • Cytogenetic analysis • Gene content analysis • Sequence analysis – Intron and exon sequences, mt. DNA sequences • SINE analysis


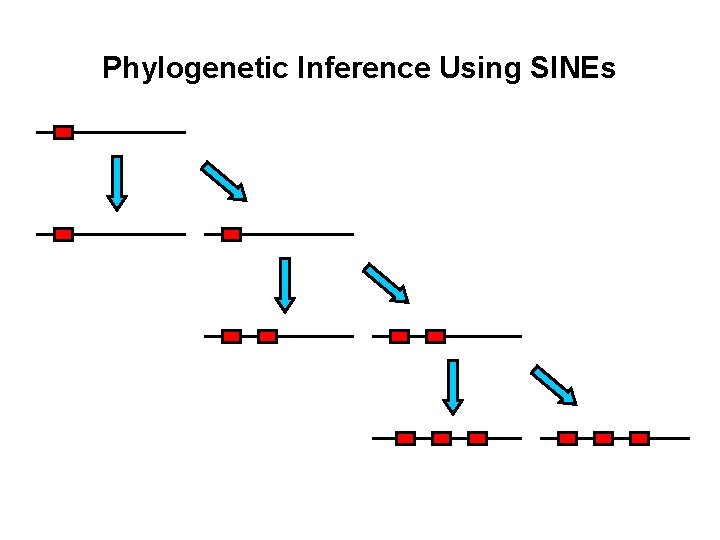
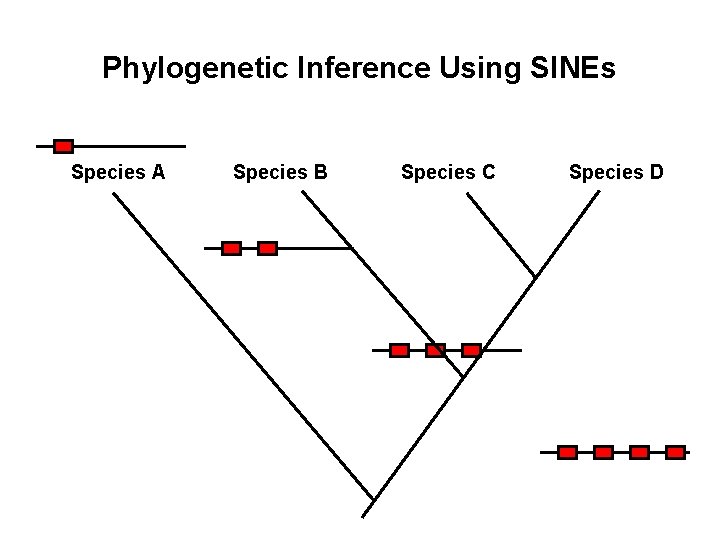
Phylogenetic Inference Using SINEs

Phylogenetic Inference Using SINEs Species A Species B Species C Species D

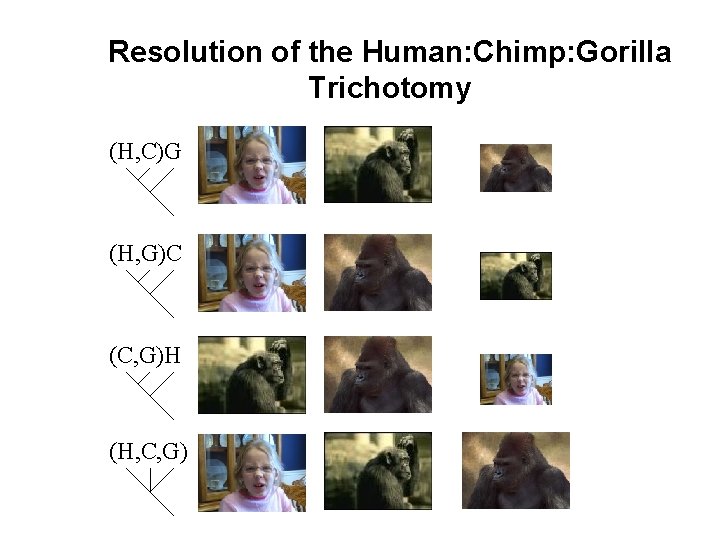
Resolution of the Human: Chimp: Gorilla Trichotomy (H, C)G (H, G)C (C, G)H (H, C, G)

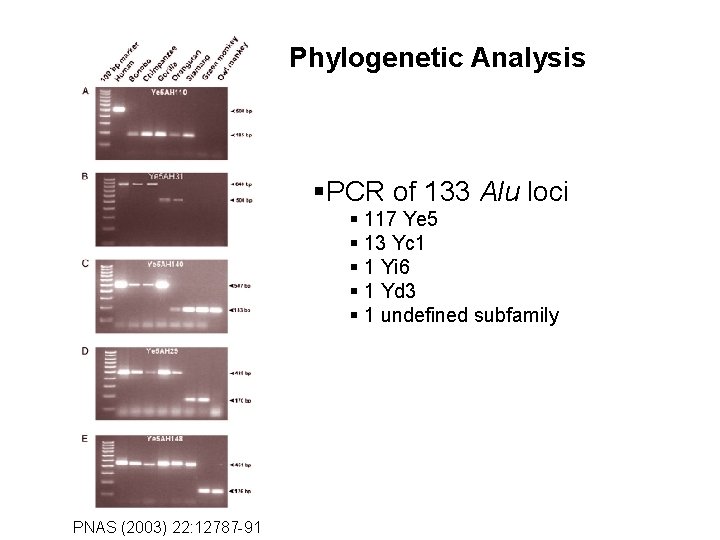
Phylogenetic Analysis §PCR of 133 Alu loci § 117 Ye 5 § 13 Yc 1 § 1 Yi 6 § 1 Yd 3 § 1 undefined subfamily PNAS (2003) 22: 12787 -91

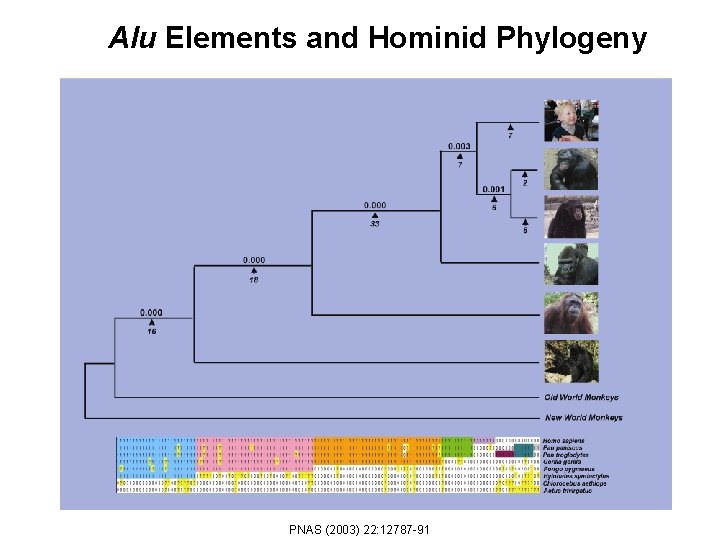
Alu Elements and Hominid Phylogeny PNAS (2003) 22: 12787 -91

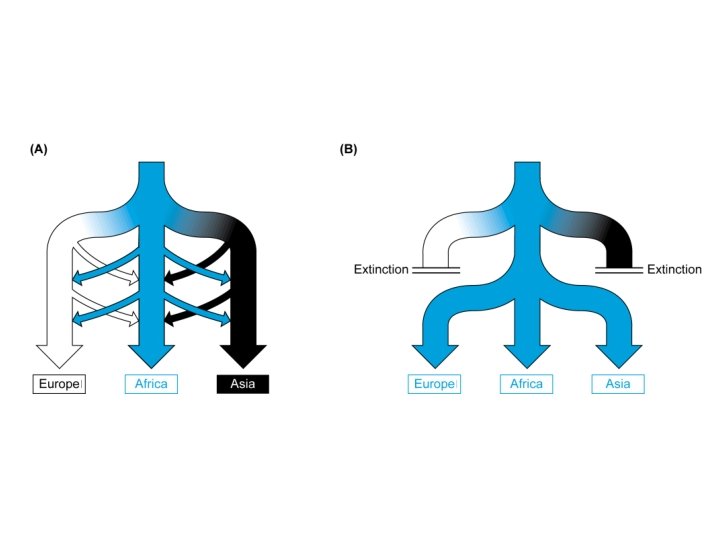
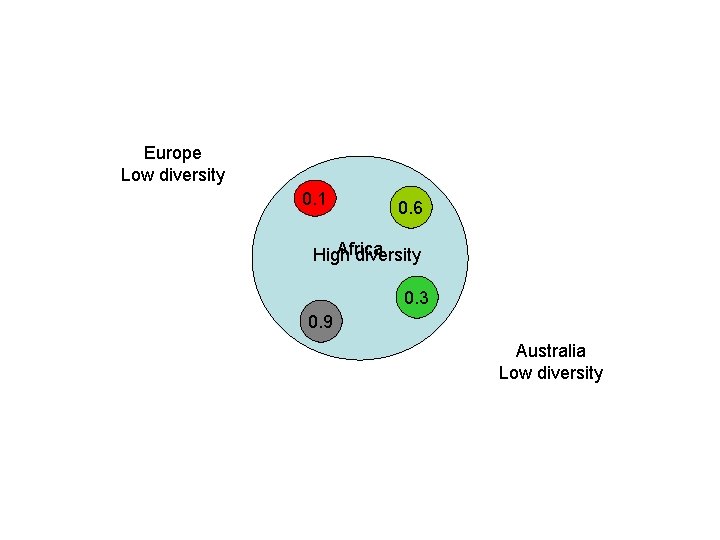
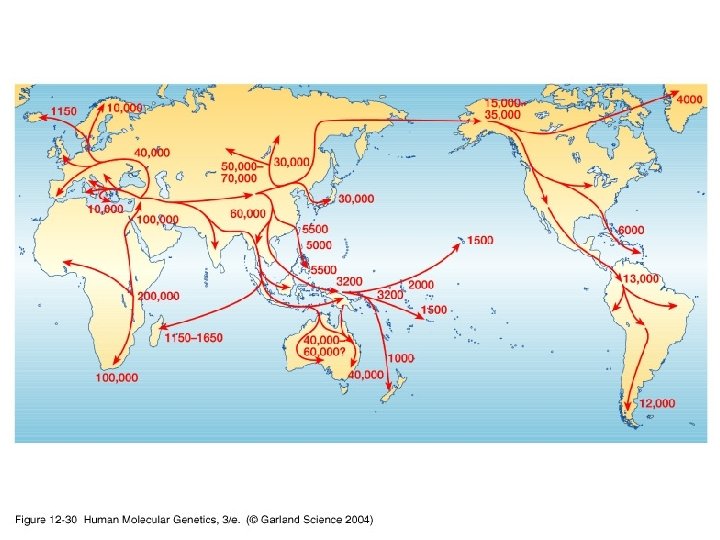
Our Place in the Tree of Life: Genome Evolution • Human evolution – Coalescence analyses (mt. DNA and Y chromosome) – Mutiregional vs. Out of Africa • Predictions of the Multiregional Hypothesis – Equal diversity in human subpopulations – No obvious root to the human tree • Predictions of the Out of Africa Hypothesis – Higher diversity in African subpopulations – Root of the human tree in Africa



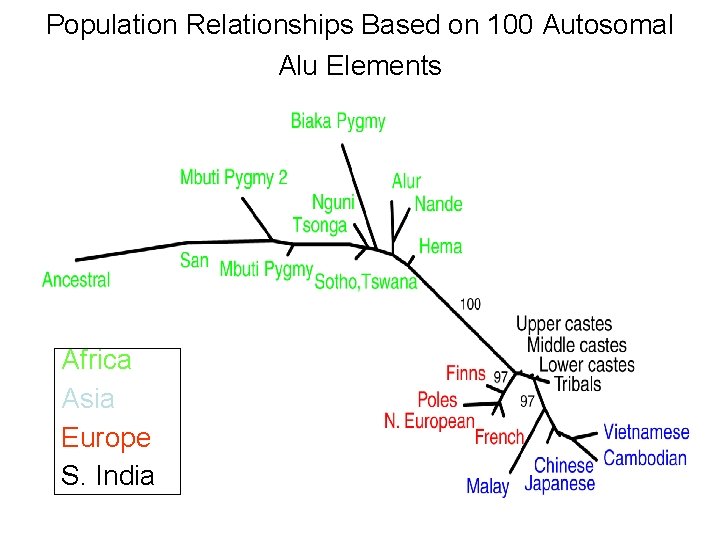
Population Relationships Based on 100 Autosomal Alu Elements Africa Asia Europe S. India

Europe Low diversity 0. 1 0. 6 Africa High diversity 0. 3 0. 9 Australia Low diversity

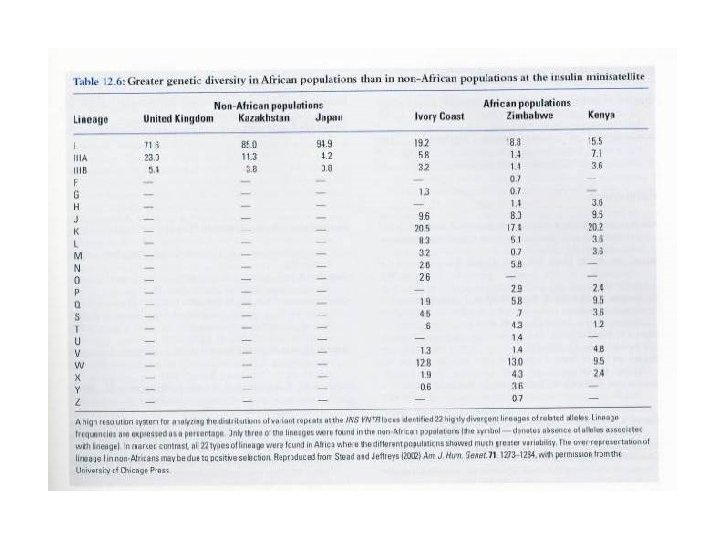
Our Place in the Tree of Life: Genome Evolution • Human evolution – Higher diversity in African subpopulations • Insulin minisatellite Table 12. 6 in text • 22 divergent lineages exist in the human population • All are found in Africa. Only 3 are found outside of Africa.

