National Human Genome Research Institute National Institutes of






















- Slides: 83

National Human Genome Research Institute National Institutes of Health U. S. Department of Health and Human Services Genome-Wide Association Studies: Hunting for Genes in the New Millennium U. S. Department of Health and Human Services National Institutes of Health National Human Genome Research Institute Teri A. Manolio, M. D. , Ph. D. Director, Office of Population Genomics Senior Advisor to the Director, NHGRI, for Population Genomics November 20, 2008

We Live in Interesting Times… “‘May he live in interesting times. ’ Like it or not we live in interesting times. ” --Robert Kennedy, June 7, 1966 May you come to the attention of those in authority. May you find what you are looking for. Wikipedia, accessed 11 Sep 07

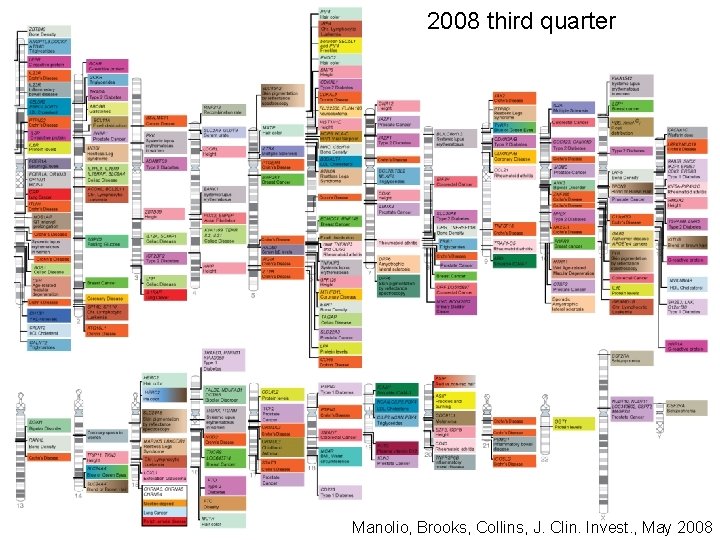
2008 third firstquarter 2008 second quarter second third quarter 2005 2007 fourth quarter 2006 2007 first quarter Manolio, Brooks, Collins, J. Clin. Invest. , May 2008

2007: The Year of GWA Studies Pennisi E, Science 2007; 318: 1842 -43.

Diseases and Traits with Published GWA Studies (n = 76, 11/17/08) • Macular Degeneration • Exfoliation Glaucoma • • • Lung Cancer Prostate Cancer Breast Cancer Colorectal Cancer Bladder Cancer Neuroblastoma Melanoma TP 53 Cancer Predispos’n Chr. Lymph. Leukemia • • Inflamm. Bowel Disease Celiac Disease Gallstones Irritable Bowel Syndrome • • • QT Prolongation Coronary Disease Coronary Spasm Atrial Fibrillation/Flutter Stroke Subarachnoid Hemorrhage Intracranial Aneurysm Hypertension Hypt. Diuretic Response Peripheral Artery Disease • Lipids and Lipoproteins • Warfarin Dosing • Ximelegatran Adv. Resp. • • • • • Parkinson Disease Amyotrophic Lat. Sclerosis Multiple Sclerosis MS Interferon-β Response Prog. Supranuclear Palsy Alzheimer’s Disease in ε 4+ Cognitive Ability Memory Hearing Restless Legs Syndrome Nicotine Dependence Methamphetamine Depend. Neuroticism Schizophrenia Sz. Iloperidone Response Bipolar Disorder Family Chaos Narcolepsy Attention Deficit Hyperactivity Personality Traits • Rheumatoid Arthritis • RA Anti-TNF Response • • • Syst. Lupus Erythematosus Sarcoidosis Pulmonary Fibrosis Psoriasis HIV Viral Setpoint Childhood Asthma • • • Type 1 Diabetes Type 2 Diabetes Diabetic Nephropathy End-St. Renal Disease Obesity, BMI, Waist, IR Height Osteoporosis Osteoarthritis Male Pattern Baldness • • • F-Cell Distribution Fetal Hgb Levels C-Reactive Protein ICAM-1 Total Ig. E Levels Uric Acid Levels, Gout Protein Levels Vitamin B 12 Levels Recombination Rate Pigmentation

“There have been few, if any, similar bursts of discovery in the history of medical research…” Hunter DJ and Kraft P, N Engl J Med 2007; 357: 436 -439.

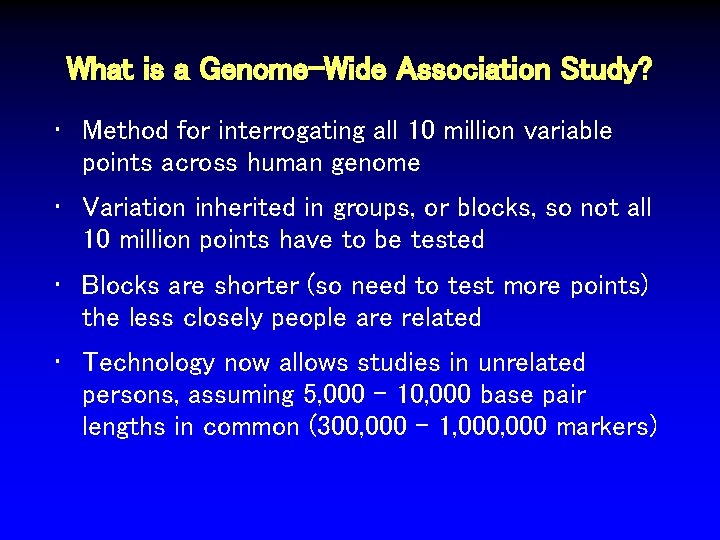
What is a Genome-Wide Association Study? • Method for interrogating all 10 million variable points across human genome • Variation inherited in groups, or blocks, so not all 10 million points have to be tested • Blocks are shorter (so need to test more points) the less closely people are related • Technology now allows studies in unrelated persons, assuming 5, 000 – 10, 000 base pair lengths in common (300, 000 – 1, 000 markers)

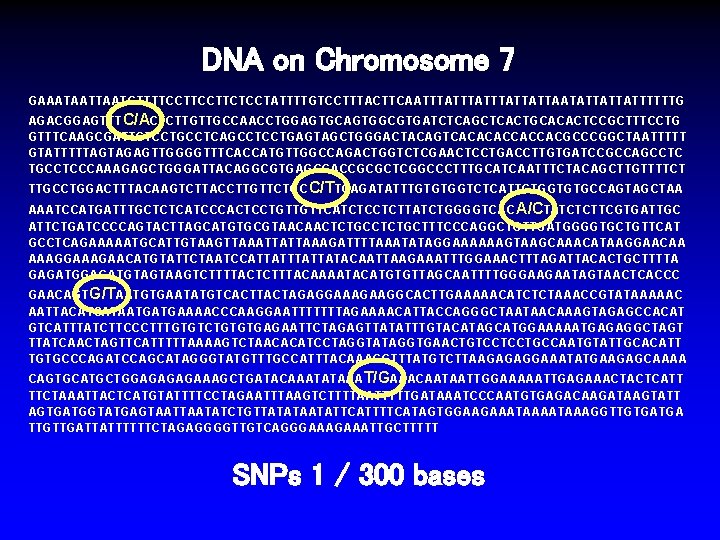
DNA on Chromosome 7 GAAATAATGTTTTCCTTCTCCTATTTTGTCCTTTACTTCAATTTATTTATTATTAATATTATTATTTTTTG AGACGGAGTTTC/ACTCTTGTTGCCAACCTGGAGTGCAGTGGCGTGATCTCAGCTCACTGCACACTCCGCTTTCCTG GTTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACTACAGTCACACACCACCACGCCCGGCTAATTTTT GTATTTTTAGTAGAGTTGGGGTTTCACCATGTTGGCCAGACTGGTCTCGAACTCCTGACCTTGTGATCCGCCAGCCTC TGCCTCCCAAAGAGCTGGGATTACAGGCGTGAGCCACCGCGCTCGGCCCTTTGCATCAATTTCTACAGCTTGTTTTCT TTGCCTGGACTTTACAAGTCTTACCTTGTTCTGC C/TTCAGATATTTGTGTGGTCTCATTCTGGTGTGCCAGTAGCTAA AAATCCATGATTTGCTCTCATCCCACTCCTGTTGTTCATCTCCTCTTATCTGGGGTCAC A/CTATCTCTTCGTGATTGC ATTCTGATCCCCAGTACTTAGCATGTGCGTAACAACTCTGCTTTCCCAGGCTGTTGATGGGGTGCTGTTCAT GCCTCAGAAAAATGCATTGTAAGTTAAATTATTAAAGATTTTAAATATAGGAAAAAAGTAAGCAAACATAAGGAACAA AAAGGAAAGAACATGTATTCTAATCCATTATTATACAATTAAGAAATTTGGAAACTTTAGATTACACTGCTTTTA GAGATGTAGTAAGTCTTTTACTCTTTACAAAATACATGTGTTAGCAATTTTGGGAAGAATAGTAACTCACCC GAACAGTG/TAATGTGAATATGTCACTTACTAGAGGAAAGAAGGCACTTGAAAAACATCTCTAAACCGTATAAAAAC AATTACATCATAATGATGAAAACCCAAGGAATTTTTTTAGAAAACATTACCAGGGCTAATAACAAAGTAGAGCCACAT GTCATTTATCTTCCCTTTGTGTCTGTGTGAGAATTCTAGAGTTATATTTGTACATAGCATGGAAAAATGAGAGGCTAGT TTATCAACTAGTTCATTTTTAAAAGTCTAACACATCCTAGGTATAGGTGAACTGTCCTCCTGCCAATGTATTGCACATT TGTGCCCAGATCCAGCATAGGGTATGTTTGCCATTTACAAACGTTTATGTCTTAAGAGAGGAAATATGAAGAGCAAAA CAGTGCATGCTGGAGAAAGCTGATACAAATATAAAT/GAAACAATAATTGGAAAAATTGAGAAACTACTCATT TTCTAAATTACTCATGTATTTTCCTAGAATTTAAGTCTTTTAATTTTTGATAAATCCCAATGTGAGACAAGATAAGTATT AGTGATGGTATGAGTAATATCTGTTATATAATATTCATTTTCATAGTGGAAGAAATAAAGGTTGTGATGA TTGTTGATTATTTTTTCTAGAGGGGTTGTCAGGGAAATTGCTTTTT SNPs 1 / 300 bases

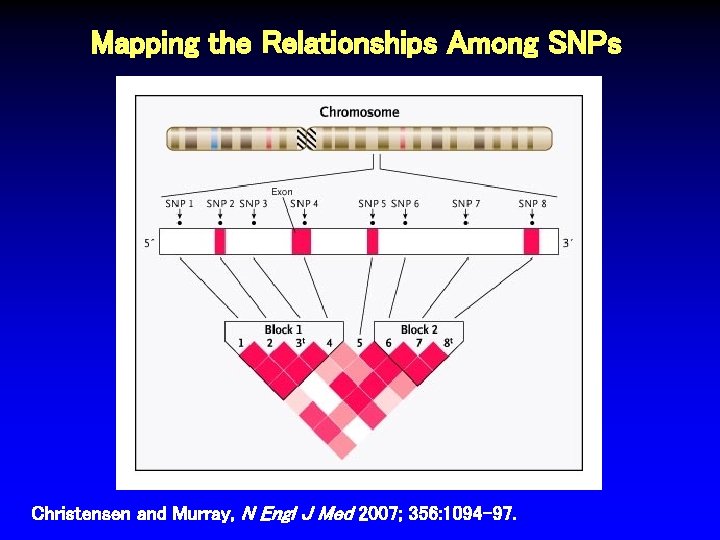
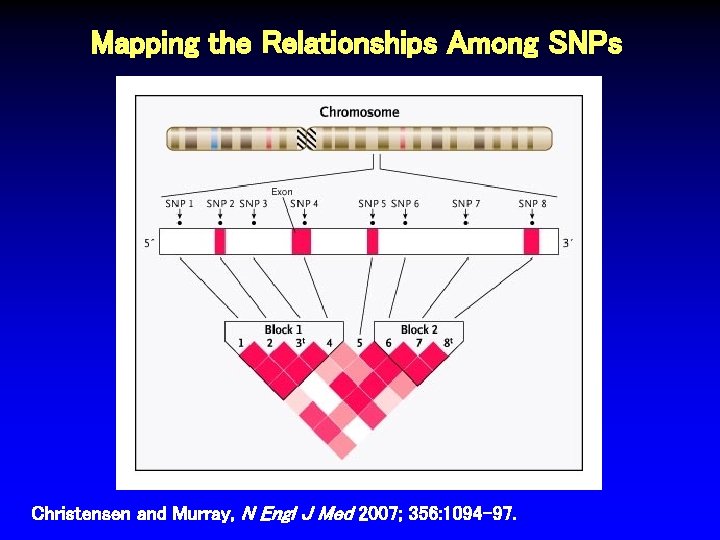
Mapping the Relationships Among SNPs Christensen and Murray, N Engl J Med 2007; 356: 1094 -97.

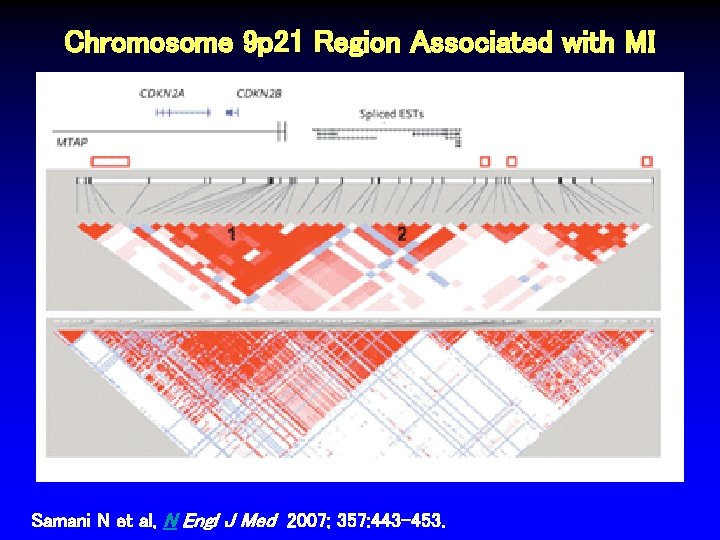
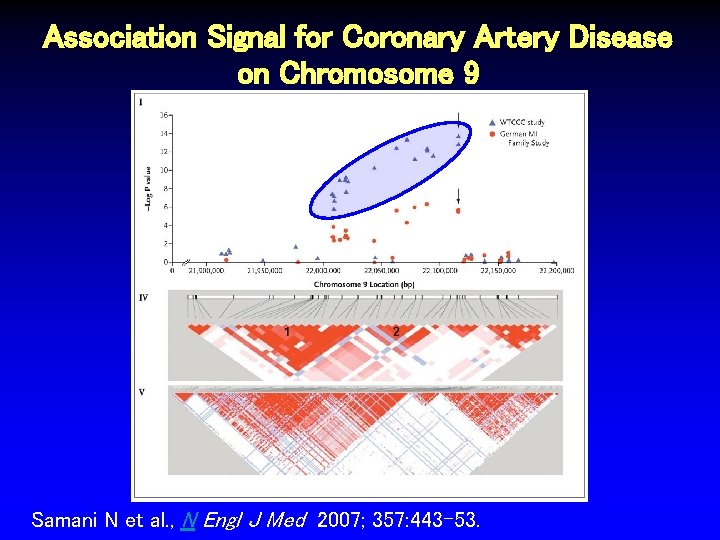
Chromosome 9 p 21 Region Associated with MI Samani N et al, N Engl J Med 2007; 357: 443 -453.

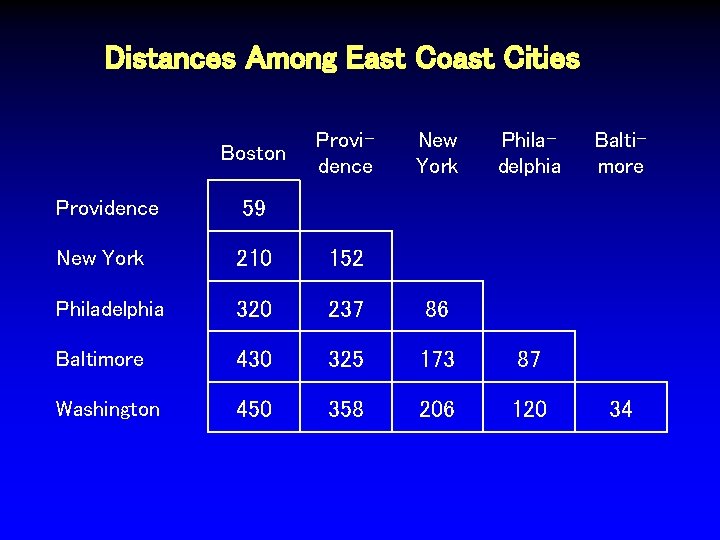
Distances Among East Coast Cities Boston Providence New York Philadelphia Providence 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 Baltimore 34

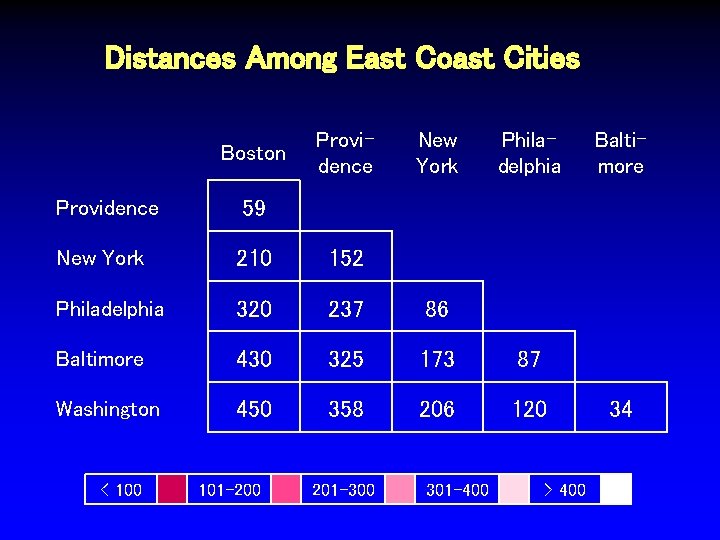
Distances Among East Coast Cities Boston Providence New York Philadelphia Providence 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 < 100 101 -200 201 -300 301 -400 > 400 Baltimore 34

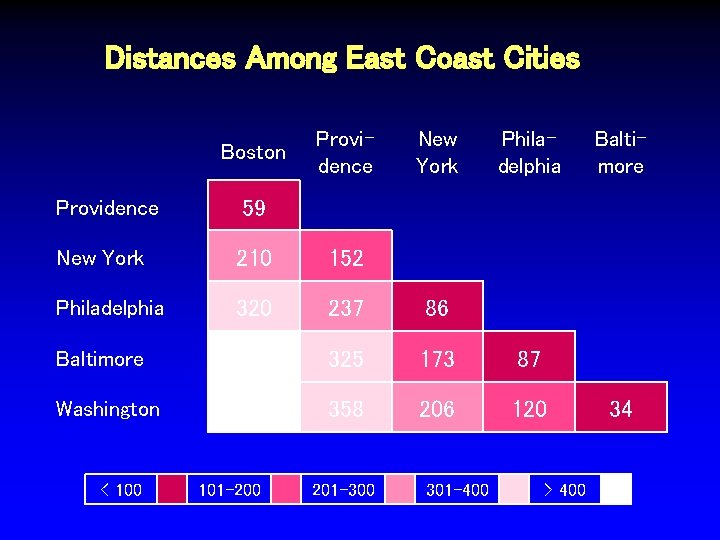
Distances Among East Coast Cities Boston Providence New York Philadelphia Providence 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 < 100 101 -200 201 -300 301 -400 > 400 Baltimore 34

Distances Among East Coast Cities n sto o B iv o Pr nce de w Ne rk Yo ila- a h P phi del ltia B re mo h s Wa ton ing

Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore Washington

Mapping the Relationships Among SNPs Christensen and Murray, N Engl J Med 2007; 356: 1094 -97.

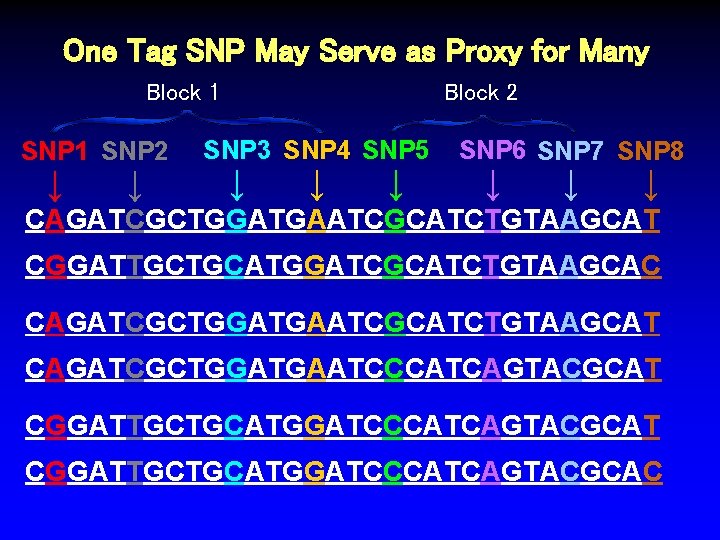
One Tag SNP May Serve as Proxy for Many Block 1 SNP 2 SNP 1 ↓ ↓ Block 2 SNP 3 SNP 4 SNP 5 ↓ ↓ ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC

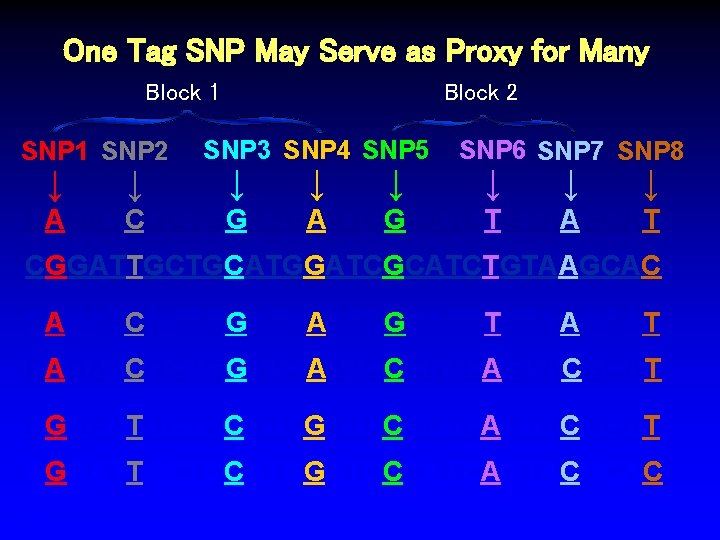
One Tag SNP May Serve as Proxy for Many Block 1 SNP 2 SNP 1 ↓ ↓ Block 2 SNP 3 SNP 4 SNP 5 ↓ ↓ ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %

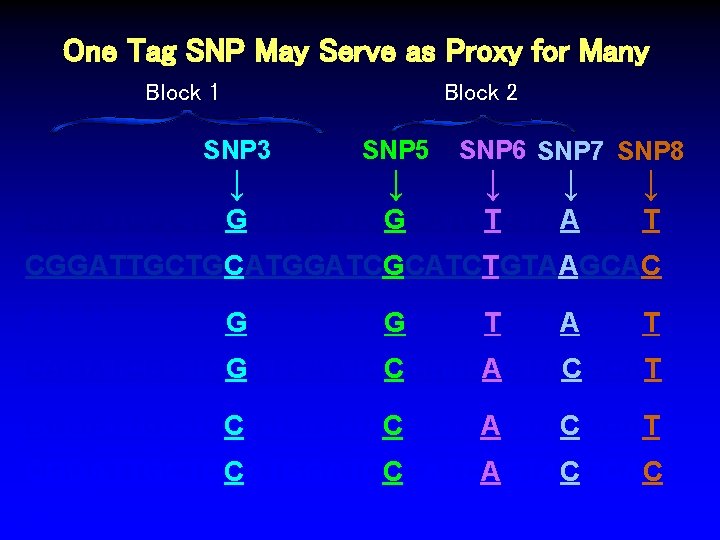
One Tag SNP May Serve as Proxy for Many Block 1 Block 2 SNP 3 ↓ SNP 5 ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %

One Tag SNP May Serve as Proxy for Many Block 1 Block 2 SNP 3 ↓ SNP 6 ↓ SNP 8 ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %

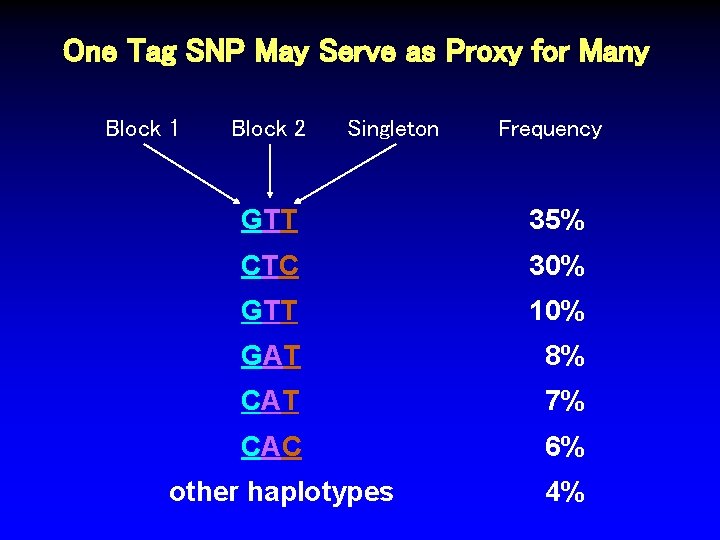
One Tag SNP May Serve as Proxy for Many Block 1 Block 2 Singleton Frequency GTT 35% CTC 30% GTT 10% GAT 8% CAT 7% CAC 6% other haplotypes 4%

www. hapmap. org Nature 2005; 437: 1299 -320. Nature 2007; 449: 851 -61.

A Hap. Map for More Efficient Association Studies: Goals • Use just the density of SNPs needed to find associations between SNPs and diseases • Do not miss chromosomal regions with disease association • Produce a tool to assist in finding genes affecting health and disease • Use more SNPs for complete genome coverage of populations of recent African ancestry populations due to shorter LD

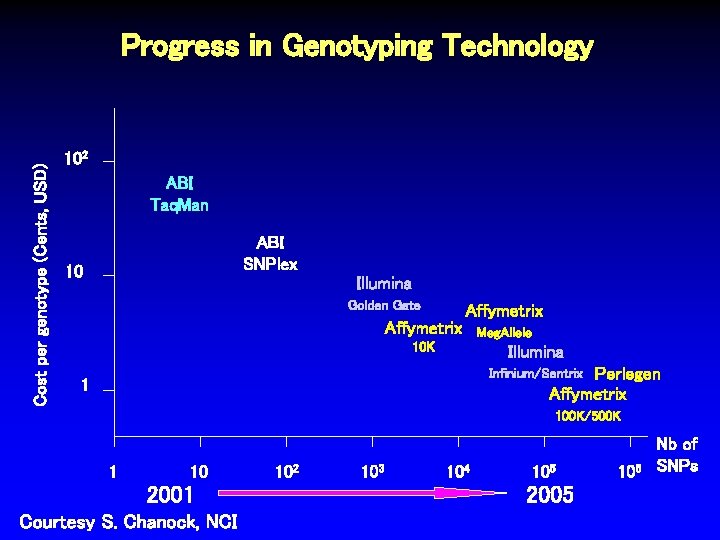
Cost per genotype (Cents, USD) Progress in Genotyping Technology 102 ABI Taq. Man ABI SNPlex 10 Illumina Golden Gate Affymetrix 10 K Meg. Allele Illumina Perlegen Affymetrix Infinium/Sentrix 1 100 K/500 K 1 10 2001 Courtesy S. Chanock, NCI 102 103 104 105 2005 106 Nb of SNPs

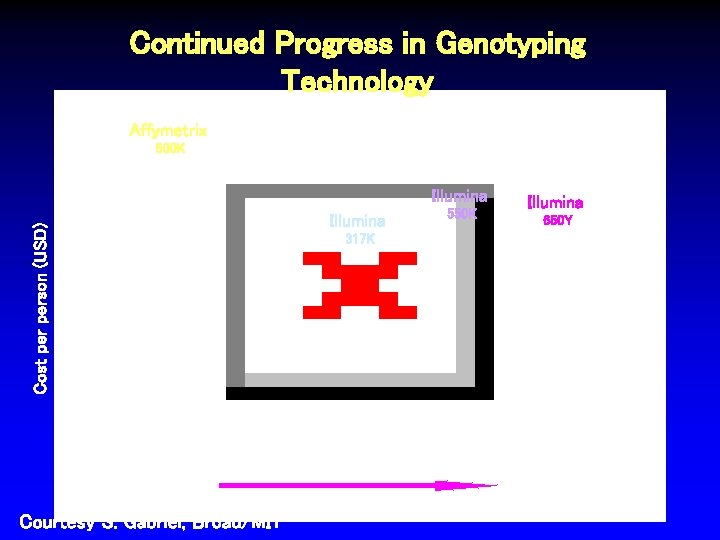
Continued Progress in Genotyping Technology Affymetrix 500 K Illumina Cost person (USD) Illumina 550 K Illumina 650 Y 317 K July 2005 Courtesy S. Gabriel, Broad/MIT Oct 2006

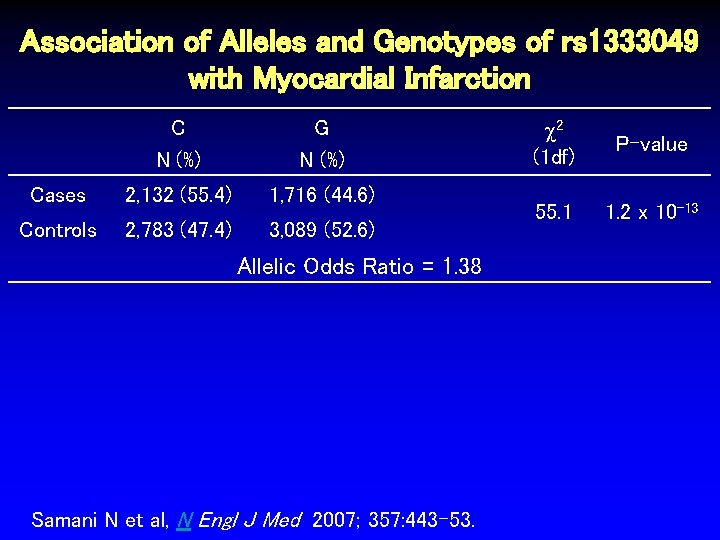
Association of Alleles and Genotypes of rs 1333049 with Myocardial Infarction C N (%) G N (%) Cases 2, 132 (55. 4) 1, 716 (44. 6) Controls 2, 783 (47. 4) 3, 089 (52. 6) Allelic Odds Ratio = 1. 38 Samani N et al, N Engl J Med 2007; 357: 443 -53. 2 (1 df) P-value 55. 1 1. 2 x 10 -13

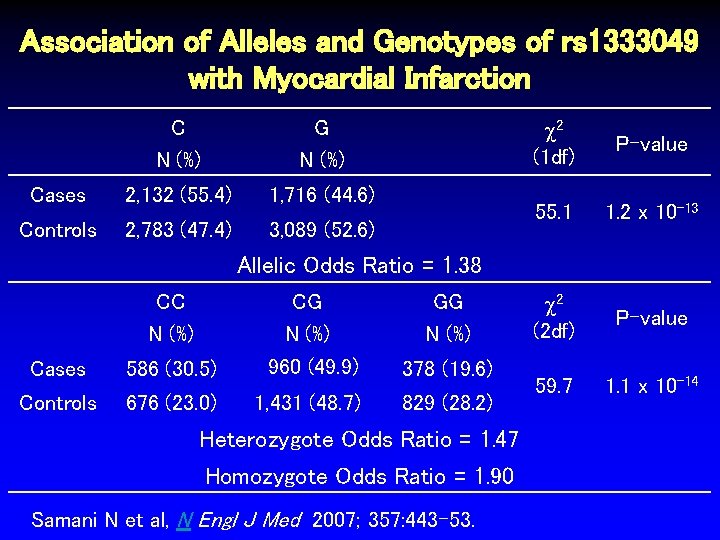
Association of Alleles and Genotypes of rs 1333049 with Myocardial Infarction C N (%) G N (%) Cases 2, 132 (55. 4) 1, 716 (44. 6) Controls 2, 783 (47. 4) 3, 089 (52. 6) 2 (1 df) P-value 55. 1 1. 2 x 10 -13 2 (2 df) P-value 59. 7 1. 1 x 10 -14 Allelic Odds Ratio = 1. 38 CC N (%) CG N (%) GG N (%) Cases 586 (30. 5) 960 (49. 9) 378 (19. 6) Controls 676 (23. 0) 1, 431 (48. 7) 829 (28. 2) Heterozygote Odds Ratio = 1. 47 Homozygote Odds Ratio = 1. 90 Samani N et al, N Engl J Med 2007; 357: 443 -53.

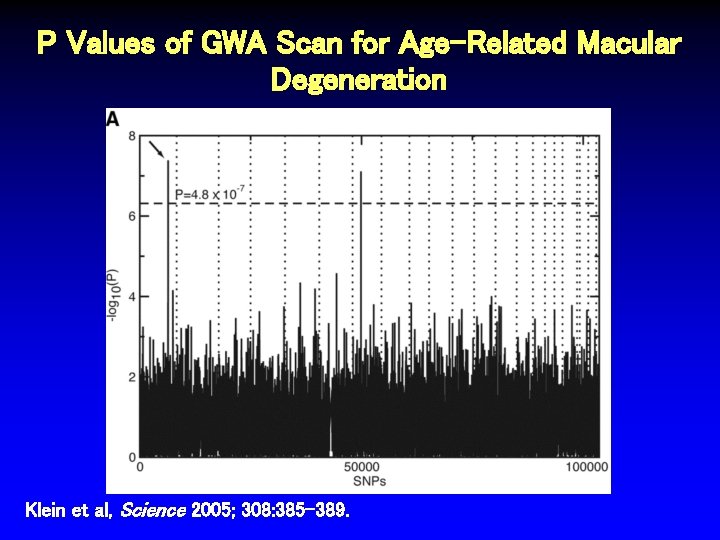
P Values of GWA Scan for Age-Related Macular Degeneration Klein et al, Science 2005; 308: 385 -389.

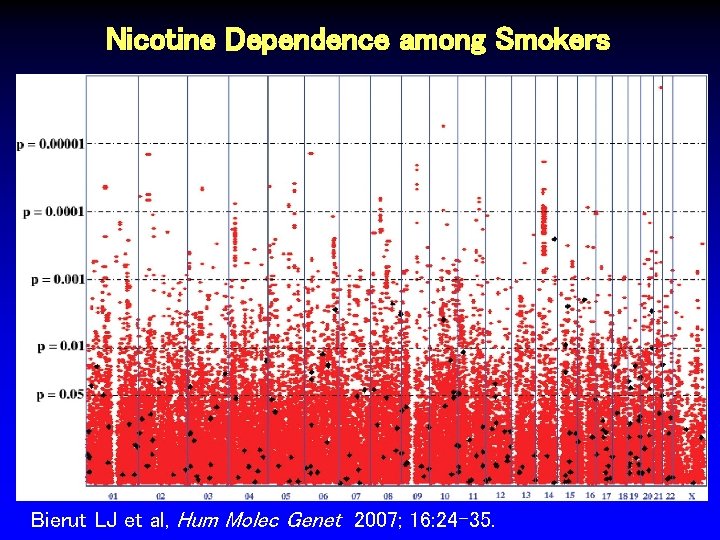
Nicotine Dependence among Smokers Bierut LJ et al, Hum Molec Genet 2007; 16: 24 -35.

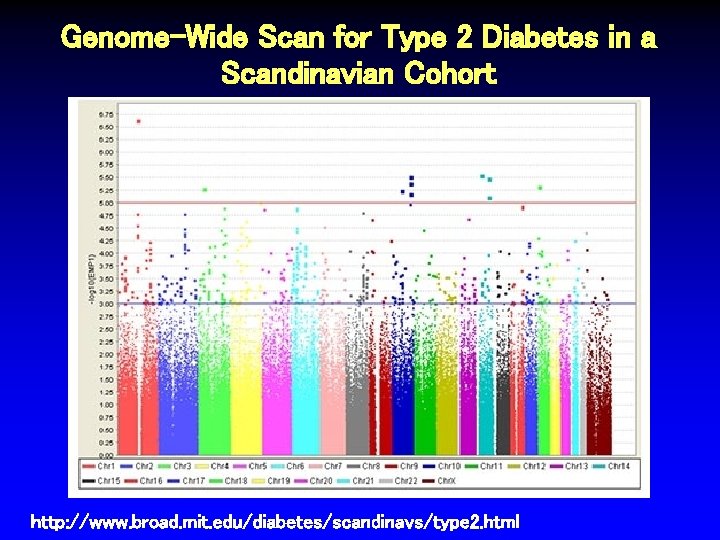
Genome-Wide Scan for Type 2 Diabetes in a Scandinavian Cohort http: //www. broad. mit. edu/diabetes/scandinavs/type 2. html

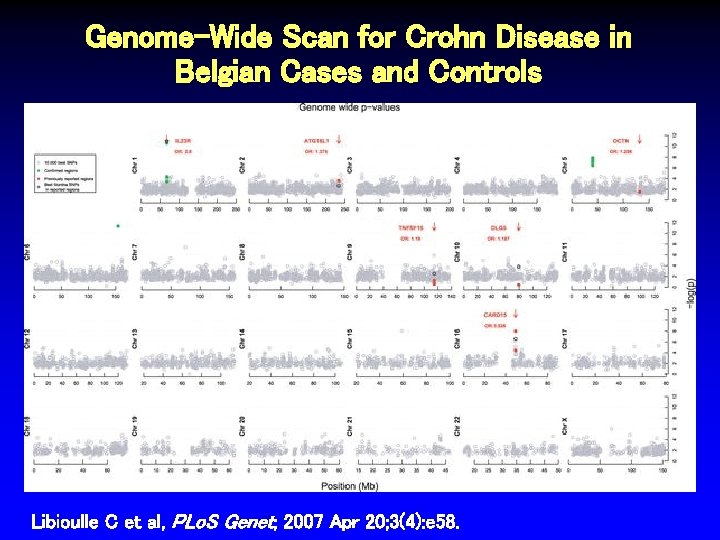
Genome-Wide Scan for Crohn Disease in Belgian Cases and Controls Libioulle C et al, PLo. S Genet; 2007 Apr 20; 3(4): e 58.

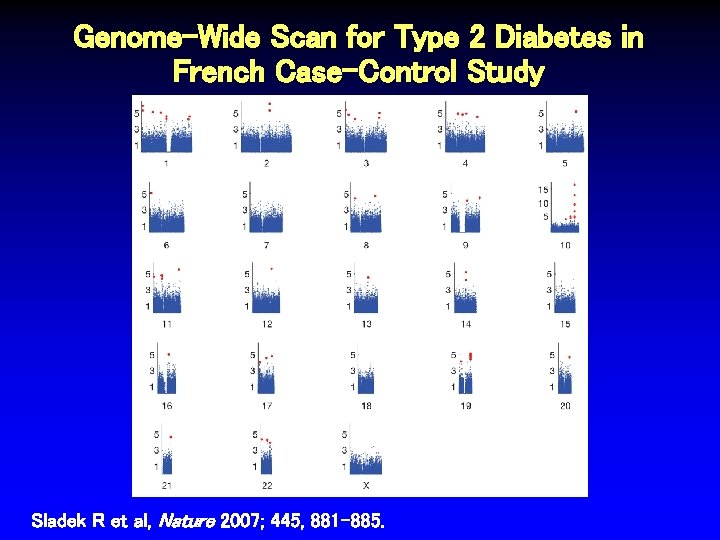
Genome-Wide Scan for Type 2 Diabetes in French Case-Control Study Sladek R et al, Nature 2007; 445, 881 -885.

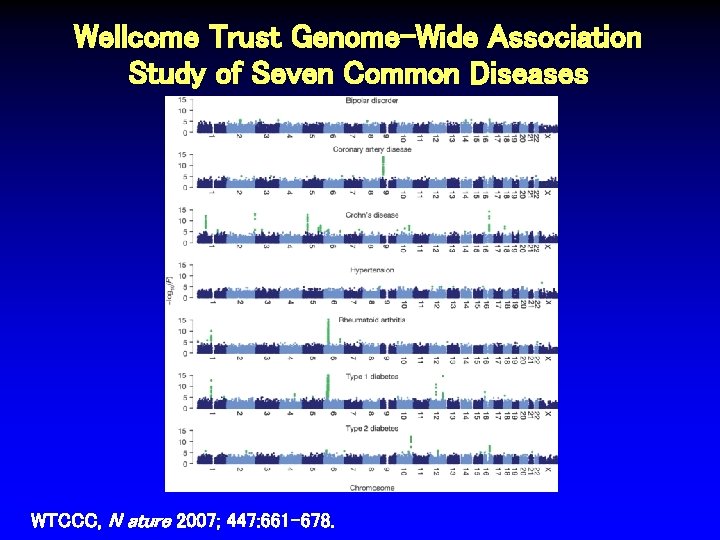
Wellcome Trust Genome-Wide Association Study of Seven Common Diseases WTCCC, N ature 2007; 447: 661 -678.

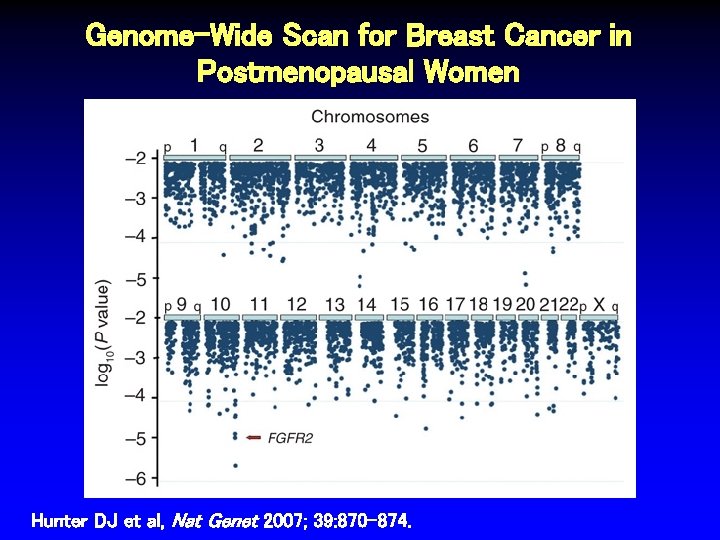
Genome-Wide Scan for Breast Cancer in Postmenopausal Women Hunter DJ et al, Nat Genet 2007; 39: 870 -874.

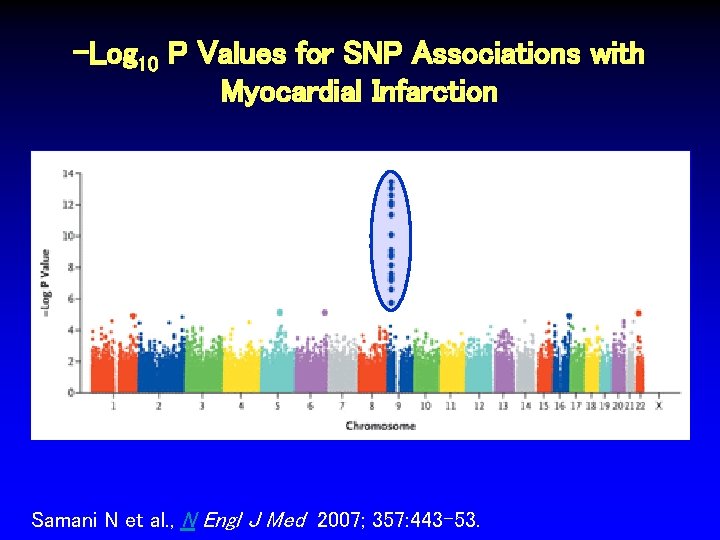
-Log 10 P Values for SNP Associations with Myocardial Infarction Samani N et al. , N Engl J Med 2007; 357: 443 -53.

Association Signal for Coronary Artery Disease on Chromosome 9 Samani N et al. , N Engl J Med 2007; 357: 443 -53.

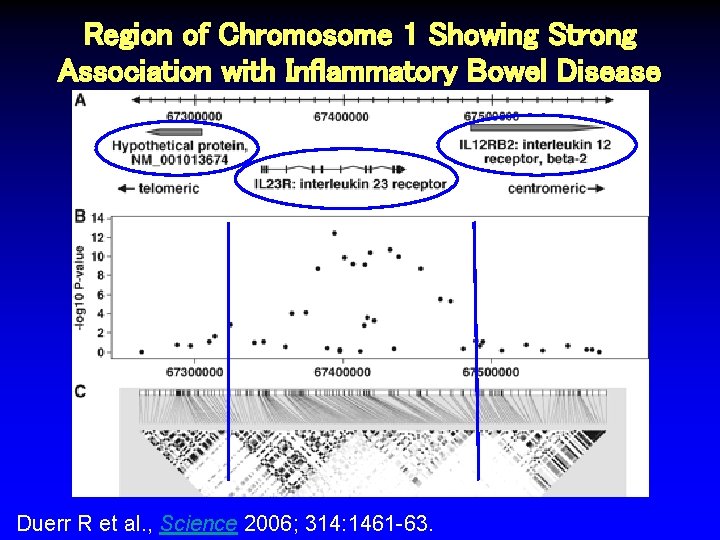
Region of Chromosome 1 Showing Strong Association with Inflammatory Bowel Disease Duerr R et al. , Science 2006; 314: 1461 -63.

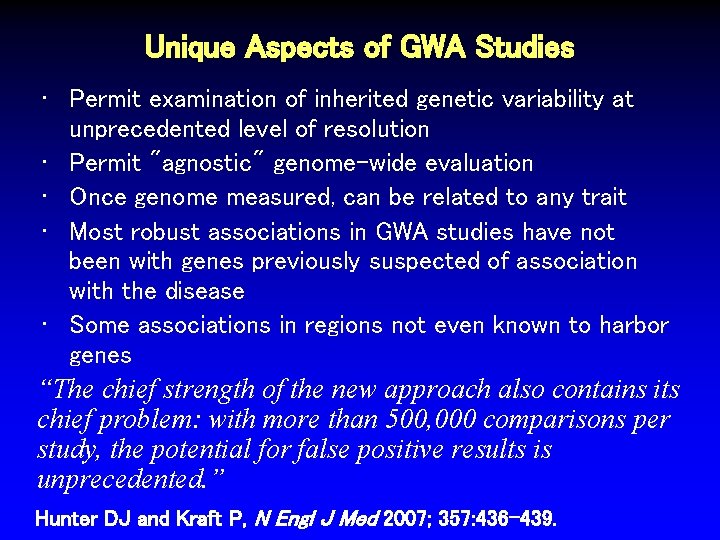
Unique Aspects of GWA Studies • Permit examination of inherited genetic variability at unprecedented level of resolution • Permit "agnostic" genome-wide evaluation • Once genome measured, can be related to any trait • Most robust associations in GWA studies have not been with genes previously suspected of association with the disease • Some associations in regions not even known to harbor genes “The chief strength of the new approach also contains its chief problem: with more than 500, 000 comparisons per study, the potential for false positive results is unprecedented. ” Hunter DJ and Kraft P, N Engl J Med 2007; 357: 436 -439.

Larson, G. The Complete Far Side. 2003.

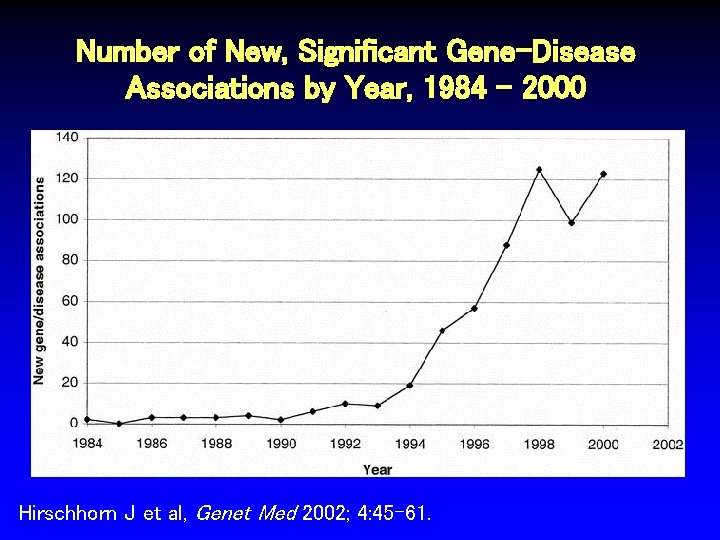
Number of New, Significant Gene-Disease Associations by Year, 1984 - 2000 Hirschhorn J et al, Genet Med 2002; 4: 45 -61.

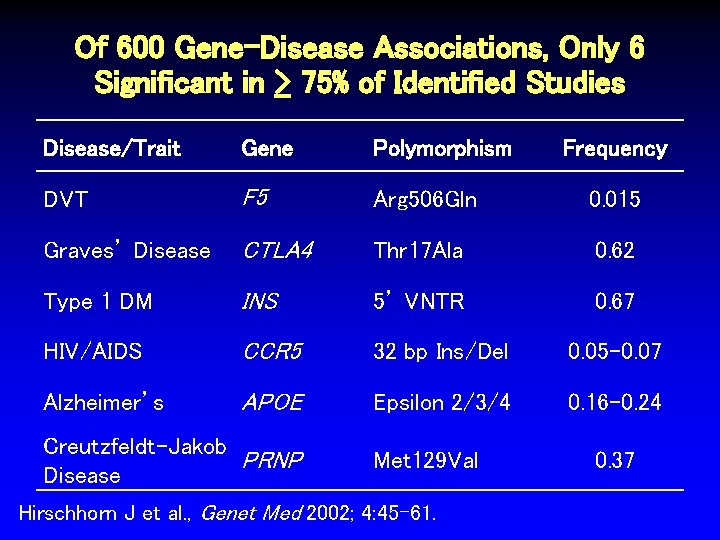
Of 600 Gene-Disease Associations, Only 6 Significant in > 75% of Identified Studies Disease/Trait Gene Polymorphism DVT F 5 Arg 506 Gln 0. 015 Graves’ Disease CTLA 4 Thr 17 Ala 0. 62 Type 1 DM INS 5’ VNTR 0. 67 HIV/AIDS CCR 5 32 bp Ins/Del 0. 05 -0. 07 Alzheimer’s APOE Epsilon 2/3/4 0. 16 -0. 24 Creutzfeldt-Jakob PRNP Disease Met 129 Val Hirschhorn J et al. , Genet Med 2002; 4: 45 -61. Frequency 0. 37

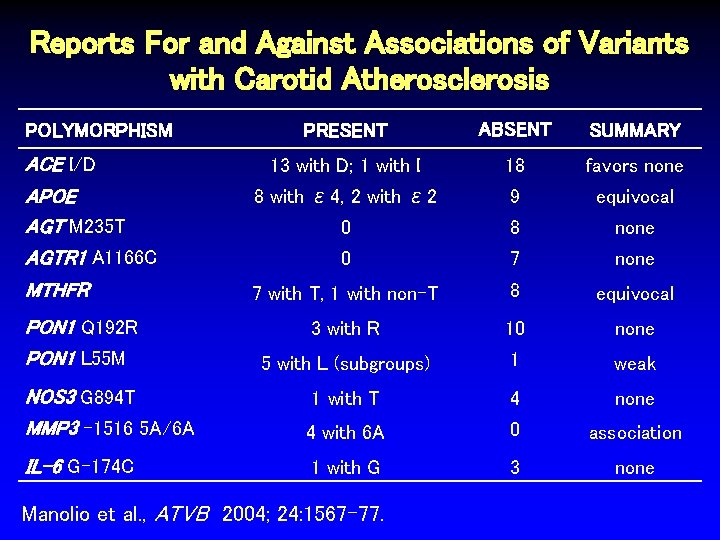
Reports For and Against Associations of Variants with Carotid Atherosclerosis PRESENT ABSENT SUMMARY 13 with D; 1 with I 18 favors none APOE AGT M 235 T AGTR 1 A 1166 C 8 with ε 4, 2 with ε 2 9 equivocal 0 8 none 0 7 none MTHFR 7 with T, 1 with non-T 8 equivocal 3 with R 10 none 5 with L (subgroups) 1 weak POLYMORPHISM ACE I/D PON 1 Q 192 R PON 1 L 55 M NOS 3 G 894 T MMP 3 -1516 5 A/6 A 1 with T 4 none 4 with 6 A 0 association IL-6 G-174 C 1 with G 3 none Manolio et al. , ATVB 2004; 24: 1567 -77.

May 1999 J. Hirschhorn and D. Altshuler J Clin Endo Metab 2002 Am J Hum Genet July 2004 PLo. S Biol Sept 2005 Nat Genet July 2006

Chanock S, Manolio T, et al. , Nature 2007; 447: 655 -60.

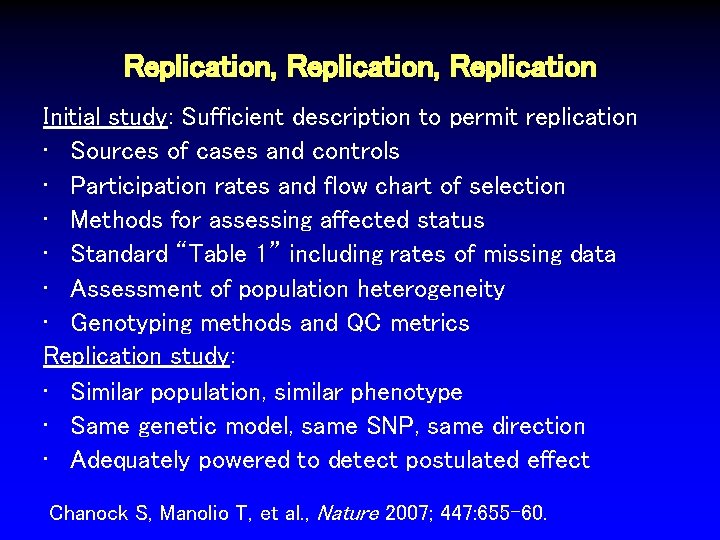
Replication, Replication Initial study: Sufficient description to permit replication • Sources of cases and controls • Participation rates and flow chart of selection • Methods for assessing affected status • Standard “Table 1” including rates of missing data • Assessment of population heterogeneity • Genotyping methods and QC metrics Replication study: • Similar population, similar phenotype • Same genetic model, same SNP, same direction • Adequately powered to detect postulated effect Chanock S, Manolio T, et al. , Nature 2007; 447: 655 -60.

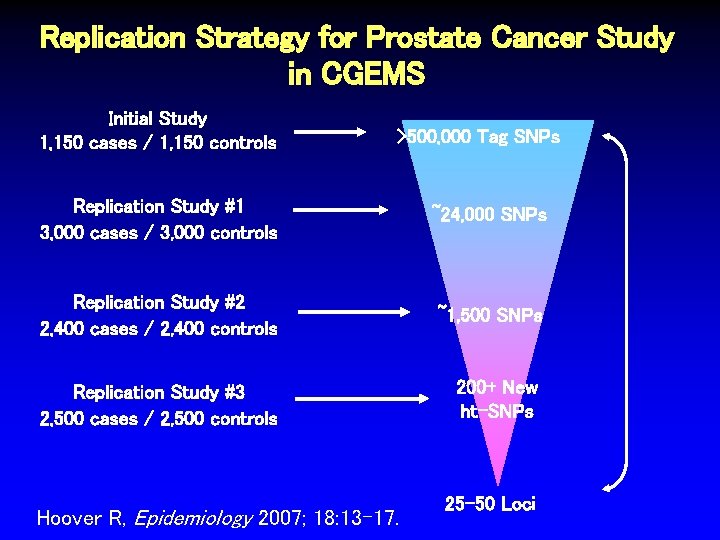
Replication Strategy for Prostate Cancer Study in CGEMS Initial Study 1, 150 cases / 1, 150 controls >500, 000 Tag SNPs Replication Study #1 3, 000 cases / 3, 000 controls ~24, 000 SNPs Replication Study #2 2, 400 cases / 2, 400 controls ~1, 500 SNPs Replication Study #3 2, 500 cases / 2, 500 controls 200+ New ht-SNPs Hoover R, Epidemiology 2007; 18: 13 -17. 25 -50 Loci

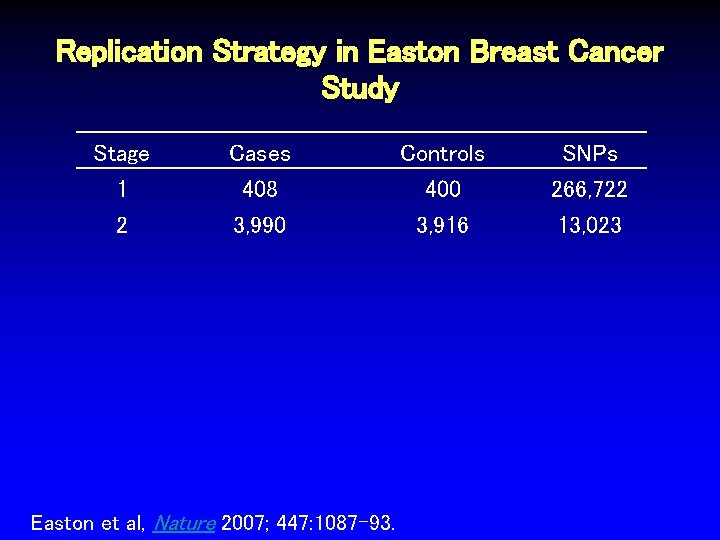
Replication Strategy in Easton Breast Cancer Study Stage 1 Cases 408 Easton et al, Nature 2007; 447: 1087 -93. Controls 400 SNPs 266, 722

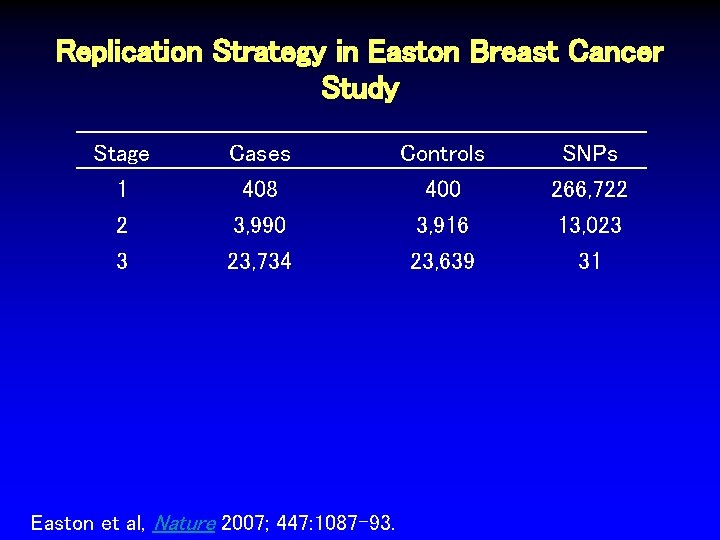
Replication Strategy in Easton Breast Cancer Study Stage 1 Cases 408 Controls 400 SNPs 266, 722 2 3, 990 3, 916 13, 023 Easton et al, Nature 2007; 447: 1087 -93.

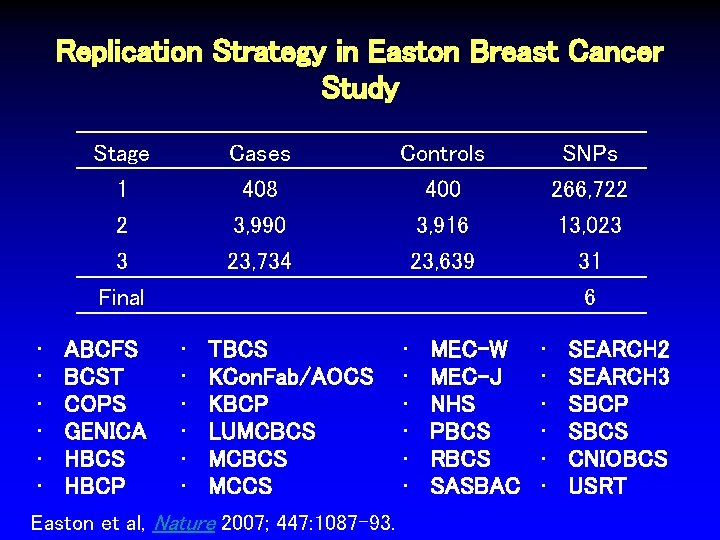
Replication Strategy in Easton Breast Cancer Study Stage 1 Cases 408 Controls 400 SNPs 266, 722 2 3 3, 990 23, 734 3, 916 23, 639 13, 023 31 Easton et al, Nature 2007; 447: 1087 -93.

Replication Strategy in Easton Breast Cancer Study Stage 1 Cases 408 Controls 400 SNPs 266, 722 2 3 3, 990 23, 734 3, 916 23, 639 13, 023 31 Final • • TBCS • KCon. Fab/AOCS • KBCP • LUMCBCS • MCCS Easton et al, Nature 2007; 447: 1087 -93. ABCFS BCST COPS GENICA HBCS HBCP 6 • • • MEC-W MEC-J NHS PBCS RBCS SASBAC • • • SEARCH 2 SEARCH 3 SBCP SBCS CNIOBCS USRT

Larson, G. The Complete Far Side. 2003.

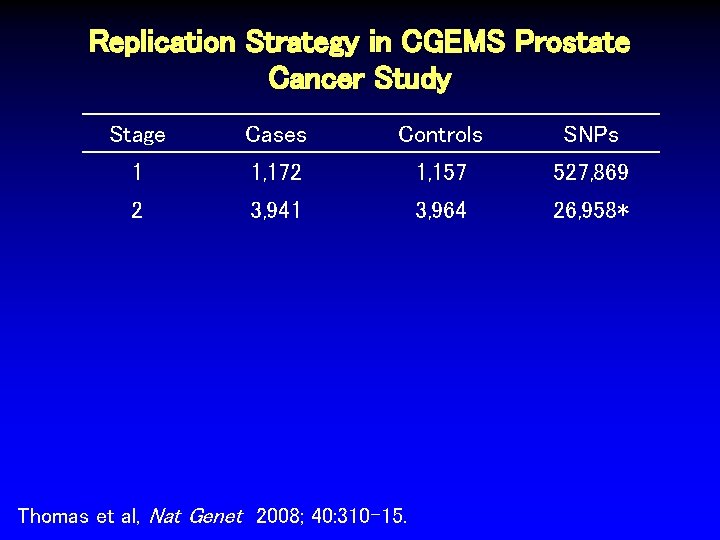
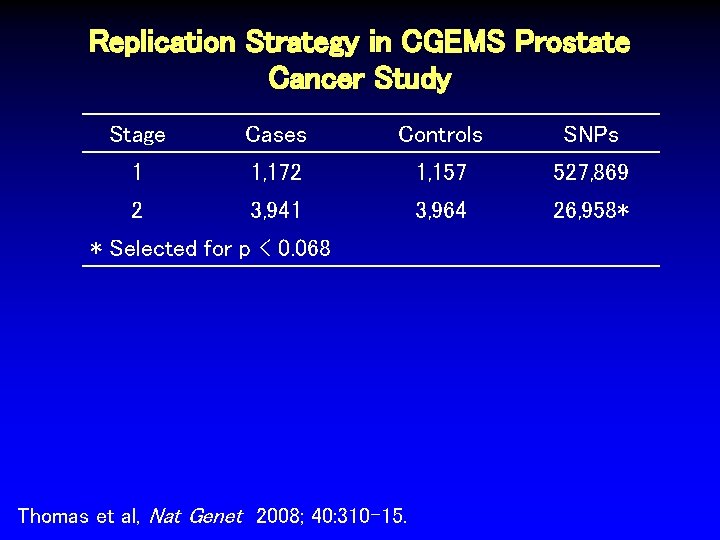
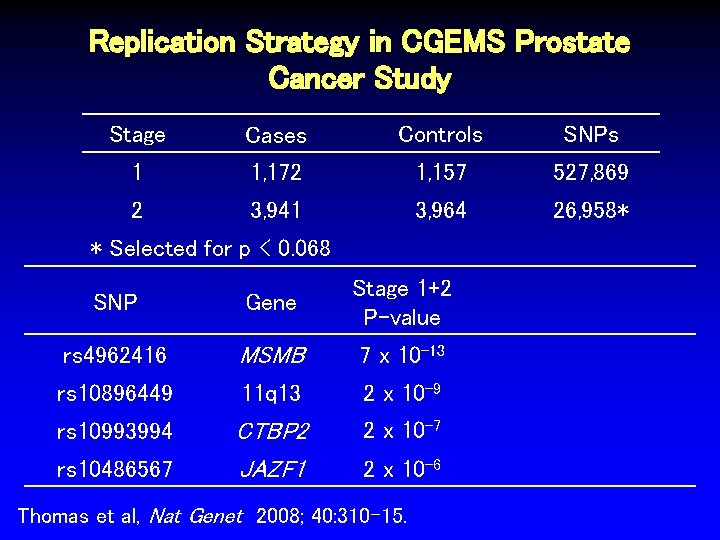
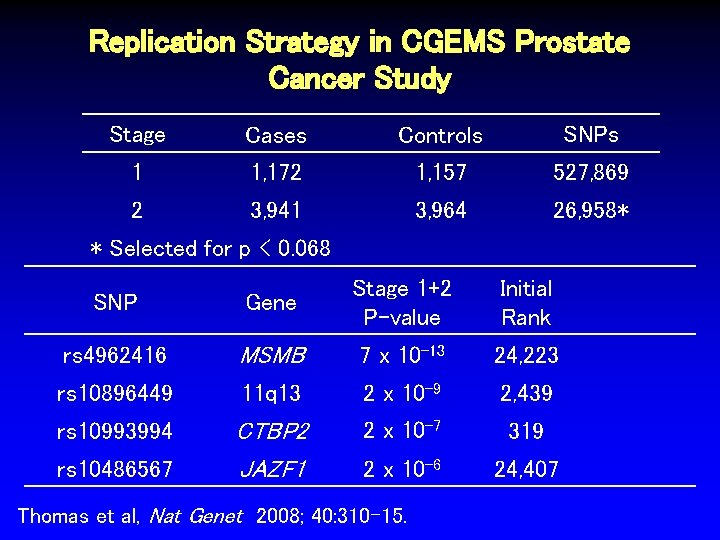
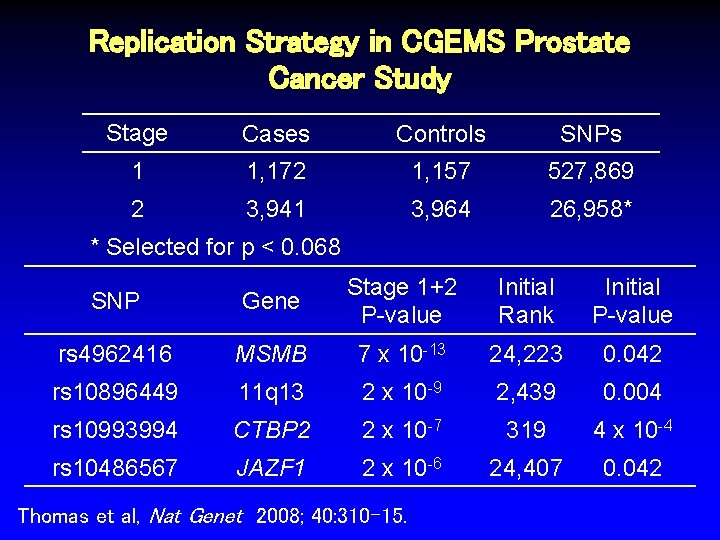
Replication Strategy in CGEMS Prostate Cancer Study Stage Cases Controls SNPs 1 1, 172 1, 157 527, 869 Thomas et al, Nat Genet 2008; 40: 310 -15.

Replication Strategy in CGEMS Prostate Cancer Study Stage Cases Controls SNPs 1 1, 172 1, 157 527, 869 2 3, 941 3, 964 26, 958* Thomas et al, Nat Genet 2008; 40: 310 -15.

Replication Strategy in CGEMS Prostate Cancer Study Stage Cases Controls SNPs 1 1, 172 1, 157 527, 869 2 3, 941 3, 964 26, 958* * Selected for p < 0. 068 Thomas et al, Nat Genet 2008; 40: 310 -15.

Replication Strategy in CGEMS Prostate Cancer Study Stage Cases Controls SNPs 1 1, 172 1, 157 527, 869 2 3, 941 3, 964 26, 958* * Selected for p < 0. 068 SNP Gene Stage 1+2 P-value rs 4962416 MSMB 7 x 10 -13 rs 10896449 11 q 13 2 x 10 -9 rs 10993994 CTBP 2 JAZF 1 2 x 10 -7 rs 10486567 2 x 10 -6 Thomas et al, Nat Genet 2008; 40: 310 -15.

Replication Strategy in CGEMS Prostate Cancer Study Stage Cases Controls SNPs 1 1, 172 1, 157 527, 869 2 3, 941 3, 964 26, 958* * Selected for p < 0. 068 SNP Gene Stage 1+2 P-value rs 4962416 MSMB 7 x 10 -13 24, 223 rs 10896449 11 q 13 2 x 10 -9 2, 439 rs 10993994 CTBP 2 JAZF 1 2 x 10 -7 319 2 x 10 -6 24, 407 rs 10486567 Thomas et al, Nat Genet 2008; 40: 310 -15. Initial Rank

Replication Strategy in CGEMS Prostate Cancer Study Stage Cases Controls SNPs 1 1, 172 1, 157 527, 869 2 3, 941 3, 964 26, 958* * Selected for p < 0. 068 SNP Gene Stage 1+2 P-value rs 4962416 MSMB 7 x 10 -13 24, 223 0. 042 rs 10896449 11 q 13 2 x 10 -9 2, 439 0. 004 rs 10993994 CTBP 2 2 x 10 -7 319 4 x 10 -4 rs 10486567 JAZF 1 2 x 10 -6 24, 407 0. 042 Thomas et al, Nat Genet 2008; 40: 310 -15. Initial Rank Initial P-value

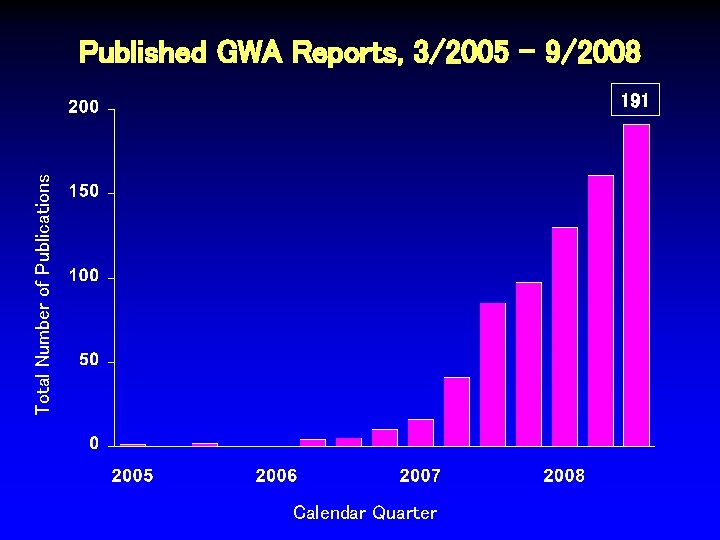
Published GWA Reports, 3/2005 - 9/2008 Total Number of Publications 191 Calendar Quarter

NHGRI Catalog of GWA Studies: http: //www. genome. gov/gwastudies/

NHGRI GWA Catalog - Objectives • Identify and track all GWA publications attempting to assay > 100, 000 SNPs • Extract key information regarding associations • Provide widely as scientific resource, including downloadable datafile • Seek commonalities across associations genomewide rather than disease by disease • Describe approach clearly so others can replicate or expand upon it • Maintain consistency in approach • Adapt to evolving technologies: CNVs?

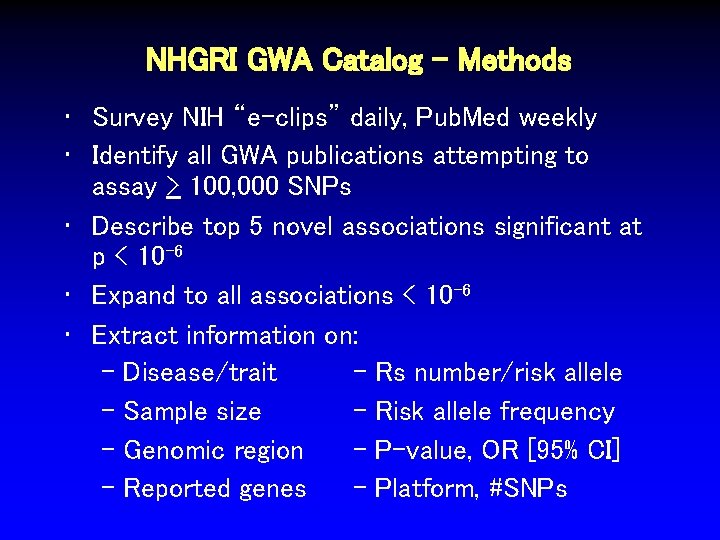
NHGRI GWA Catalog - Methods • Survey NIH “e-clips” daily, Pub. Med weekly • Identify all GWA publications attempting to assay > 100, 000 SNPs • Describe top 5 novel associations significant at p < 10 -6 • Expand to all associations < 10 -6 • Extract information on: - Disease/trait - Rs number/risk allele - Sample size - Risk allele frequency - Genomic region - P-value, OR [95% CI] - Reported genes - Platform, #SNPs

Reports Included in this Analysis • 180 published papers through 9/18/2008 • 34 did not report SNP • 1 reported haplotypes without specific SNPs • 145 reports – 782 unique (“index”) SNPs – 3, 841 unique perfect LD SNPs (“linked”) – 4, 623 index + linked • 83 index SNPs reported 2 -7 times • 10 multiple reports in “unrelated” traits

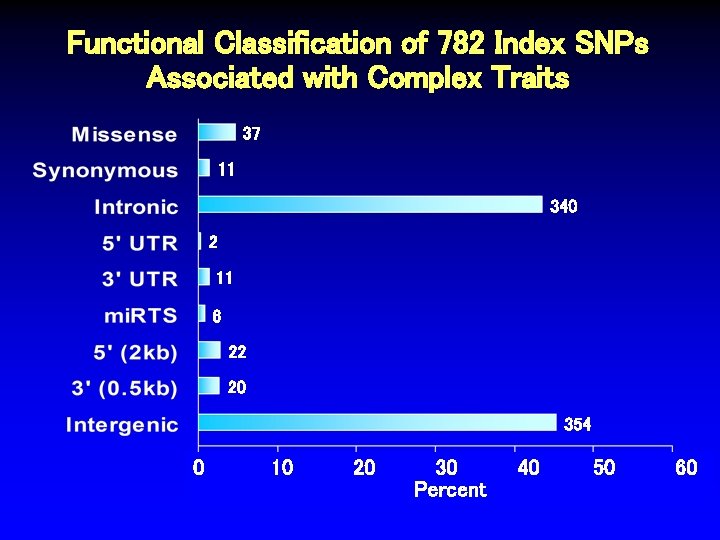
Functional Classification of 782 Index SNPs Associated with Complex Traits 37 11 340 2 11 6 22 20 354 0 10 20 30 Percent 40 50 60

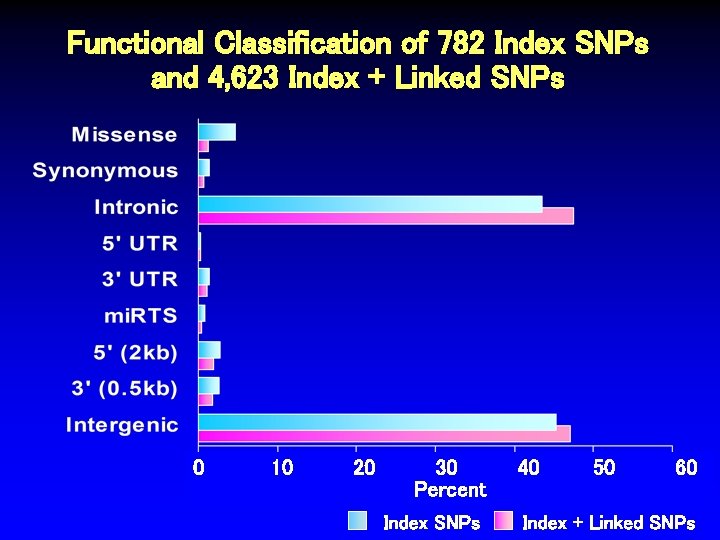
Functional Classification of 782 Index SNPs and 4, 623 Index + Linked SNPs 0 10 20 30 Percent Index SNPs 40 50 60 Index + Linked SNPs

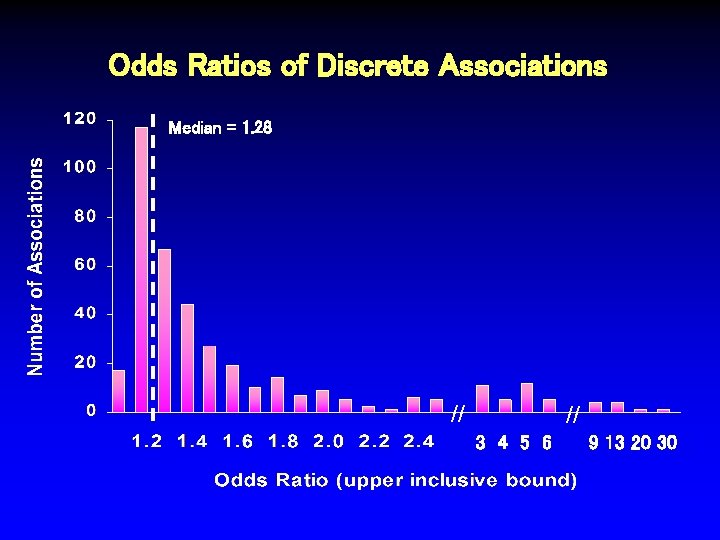
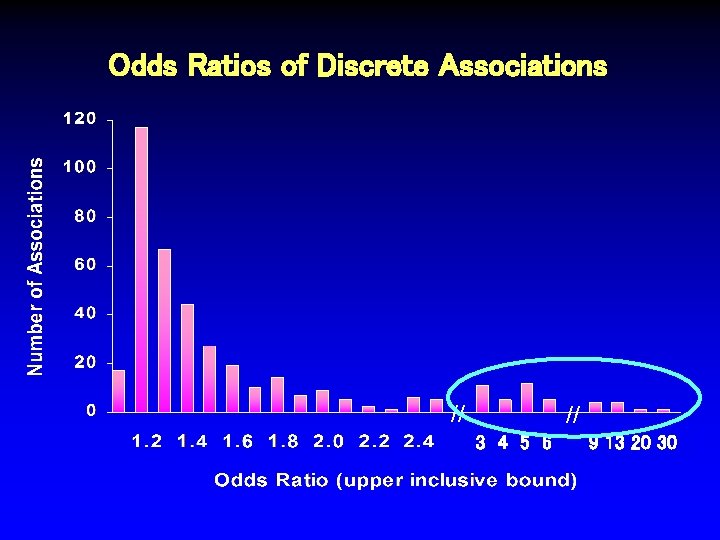
Odds Ratios of Discrete Associations Median = 1. 28 // // 3 4 5 6 9 13 20 30

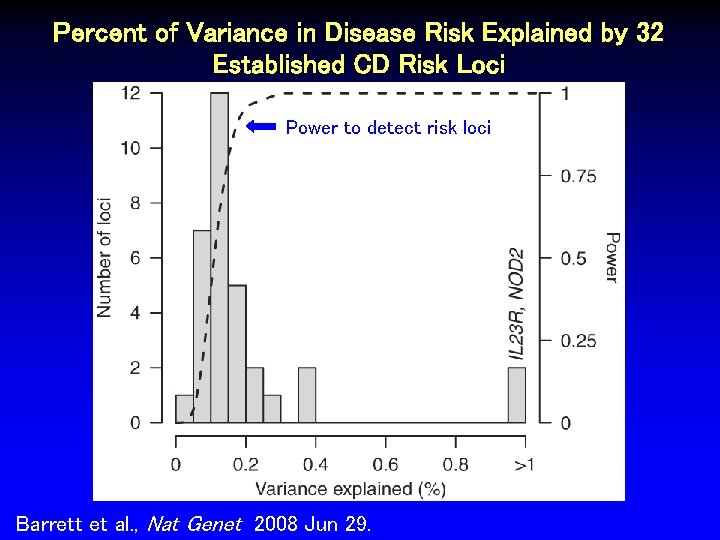
Percent of Variance in Disease Risk Explained by 32 Established CD Risk Loci Power to detect risk loci Barrett et al. , Nat Genet 2008 Jun 29.

Odds Ratios of Discrete Associations // // 3 4 5 6 9 13 20 30

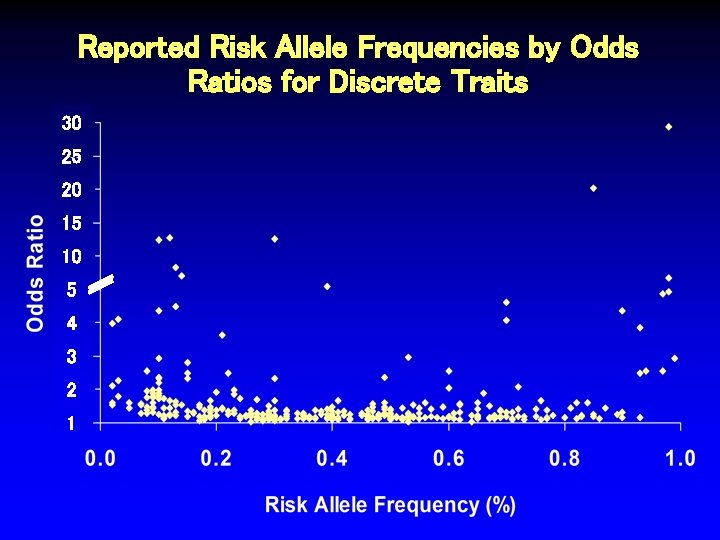
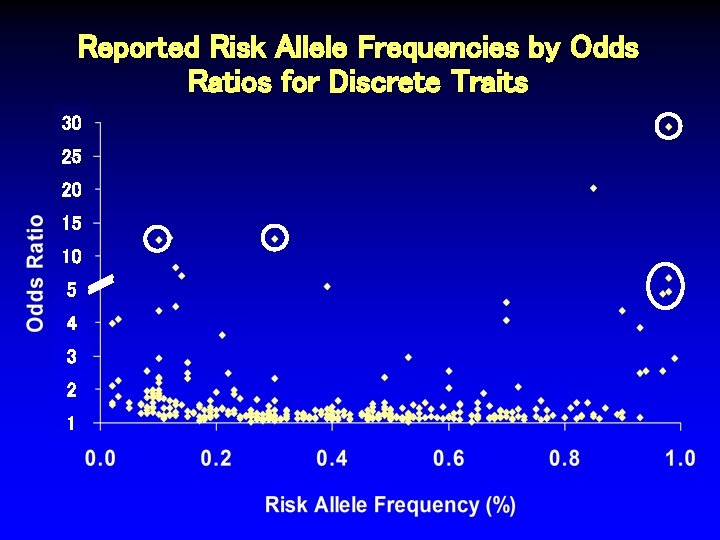
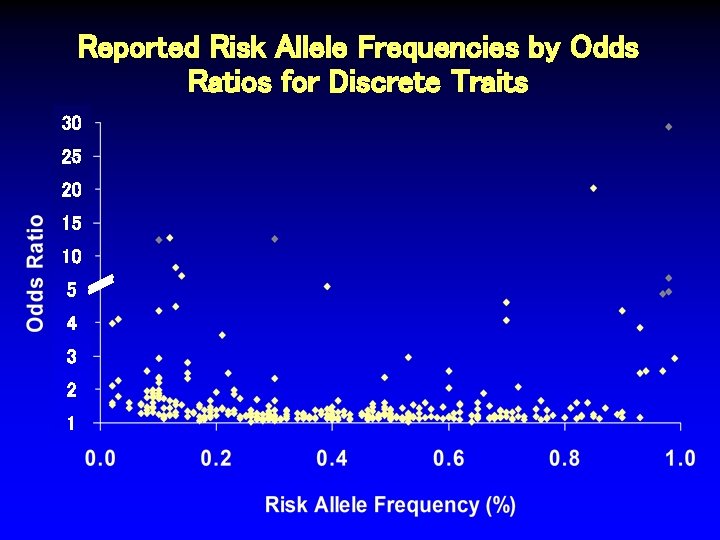
Reported Risk Allele Frequencies by Odds Ratios for Discrete Traits 30 25 20 15 10 5 4 3 2 1

Reported Risk Allele Frequencies by Odds Ratios for Discrete Traits 30 25 20 15 10 5 4 3 2 1

Reported Risk Allele Frequencies by Odds Ratios for Discrete Traits 30 25 20 15 10 5 4 3 2 1

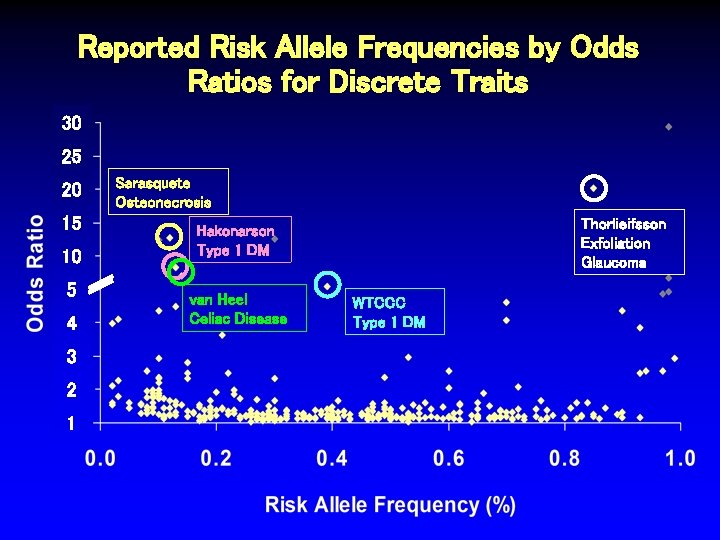
Reported Risk Allele Frequencies by Odds Ratios for Discrete Traits 30 25 20 15 10 5 4 3 2 1 Sarasquete Osteonecrosis Thorlieifsson Exfoliation Glaucoma Hakonarson Type 1 DM van Heel Celiac Disease WTCCC Type 1 DM

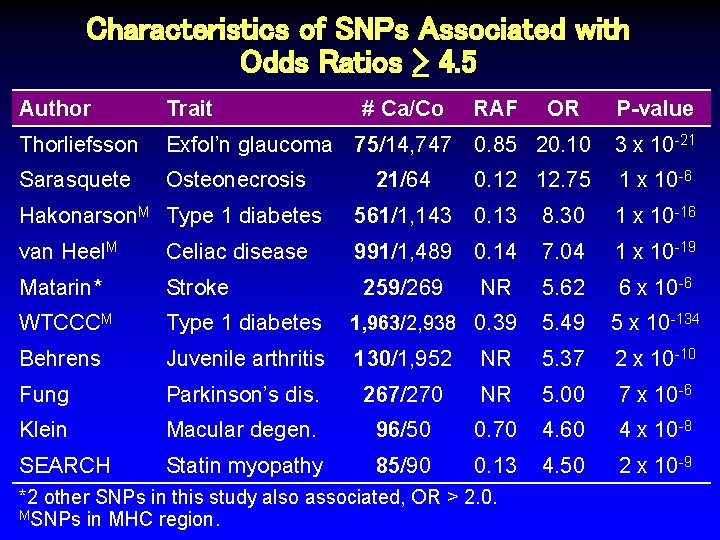
Characteristics of SNPs Associated with Odds Ratios > 4. 5 Author Trait # Ca/Co Thorliefsson Exfol’n glaucoma 75/14, 747 0. 85 20. 10 3 x 10 -21 Sarasquete Osteonecrosis 0. 12 12. 75 1 x 10 -6 21/64 RAF OR P-value Hakonarson. M Type 1 diabetes 561/1, 143 0. 13 8. 30 1 x 10 -16 van Heel. M Celiac disease 991/1, 489 0. 14 7. 04 1 x 10 -19 Matarin* Stroke 259/269 NR 5. 62 6 x 10 -6 WTCCCM Type 1 diabetes 1, 963/2, 938 0. 39 5. 49 5 x 10 -134 Behrens Juvenile arthritis 130/1, 952 NR 5. 37 2 x 10 -10 Fung Parkinson’s dis. 267/270 NR 5. 00 7 x 10 -6 Klein Macular degen. 96/50 0. 70 4. 60 4 x 10 -8 SEARCH Statin myopathy 85/90 0. 13 4. 50 2 x 10 -9 *2 other SNPs in this study also associated, OR > 2. 0. MSNPs in MHC region.

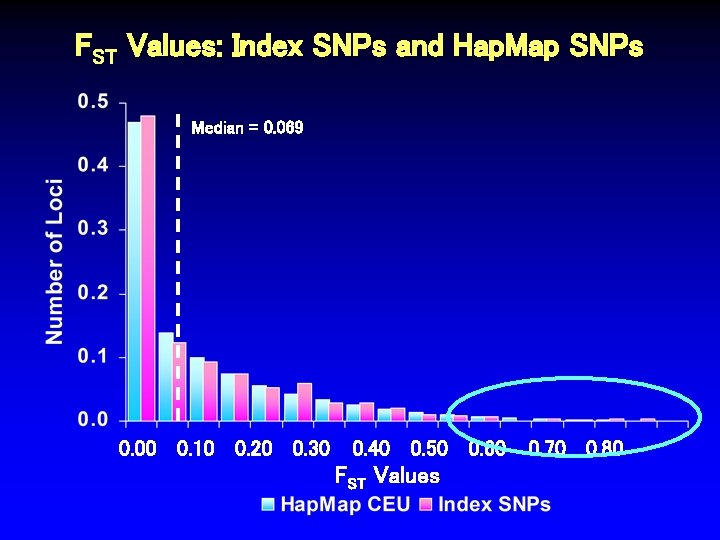
FST Values: Index SNPs and Hap. Map SNPs Median = 0. 069 0. 00 0. 10 0. 20 0. 30 0. 40 0. 50 0. 60 FST Values 0. 70 0. 80

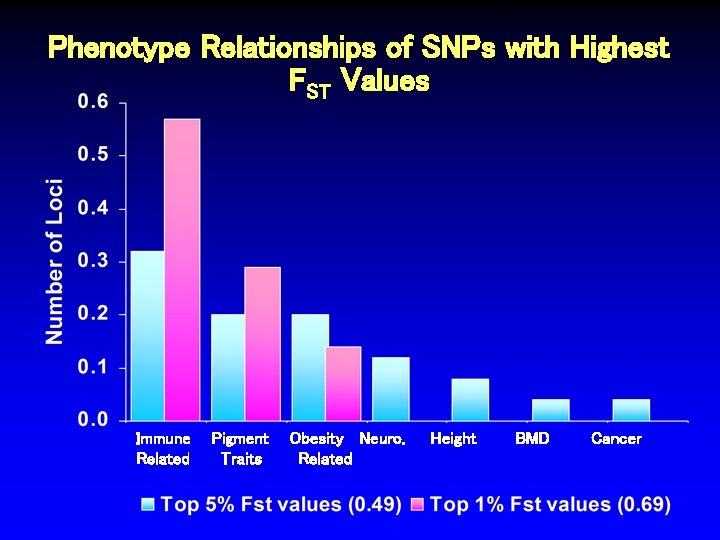
Phenotype Relationships of SNPs with Highest FST Values Immune Related Pigment Traits Obesity Neuro. Related Height BMD Cancer

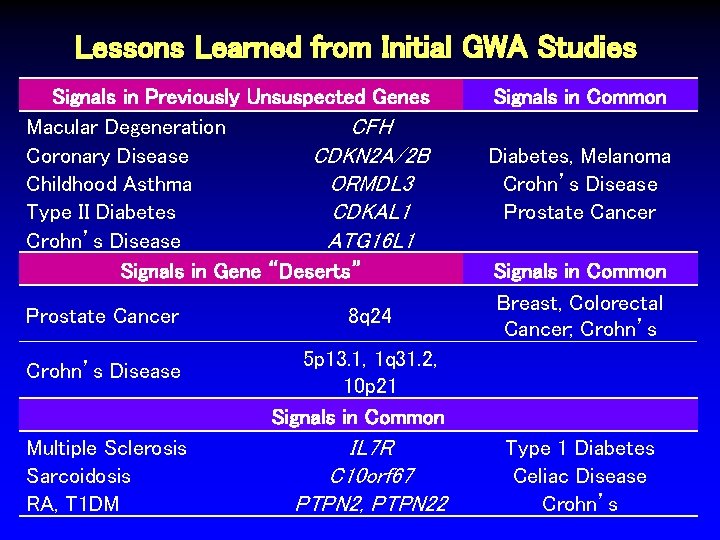
Lessons Learned from Initial GWA Studies Signals in Previously Unsuspected Genes Macular Degeneration CFH Coronary Disease CDKN 2 A/2 B Childhood Asthma ORMDL 3 Type II Diabetes CDKAL 1 Crohn’s Disease ATG 16 L 1

Lessons Learned from Initial GWA Studies Signals in Previously Unsuspected Genes Macular Degeneration CFH Coronary Disease CDKN 2 A/2 B Childhood Asthma ORMDL 3 Type II Diabetes CDKAL 1 Crohn’s Disease ATG 16 L 1 Signals in Gene “Deserts” Prostate Cancer 8 q 24 Crohn’s Disease 5 p 13. 1, 1 q 31. 2, 10 p 21

Lessons Learned from Initial GWA Studies Signals in Previously Unsuspected Genes Macular Degeneration CFH Coronary Disease CDKN 2 A/2 B Childhood Asthma ORMDL 3 Type II Diabetes CDKAL 1 Crohn’s Disease ATG 16 L 1 Signals in Gene “Deserts” Prostate Cancer 8 q 24 Crohn’s Disease 5 p 13. 1, 1 q 31. 2, 10 p 21 Signals in Common Diabetes, Melanoma Crohn’s Disease Prostate Cancer

Lessons Learned from Initial GWA Studies Signals in Previously Unsuspected Genes Macular Degeneration CFH Coronary Disease CDKN 2 A/2 B Childhood Asthma ORMDL 3 Type II Diabetes CDKAL 1 Crohn’s Disease ATG 16 L 1 Signals in Gene “Deserts” Prostate Cancer 8 q 24 Crohn’s Disease 5 p 13. 1, 1 q 31. 2, 10 p 21 Signals in Common Diabetes, Melanoma Crohn’s Disease Prostate Cancer Signals in Common Breast, Colorectal Cancer; Crohn’s

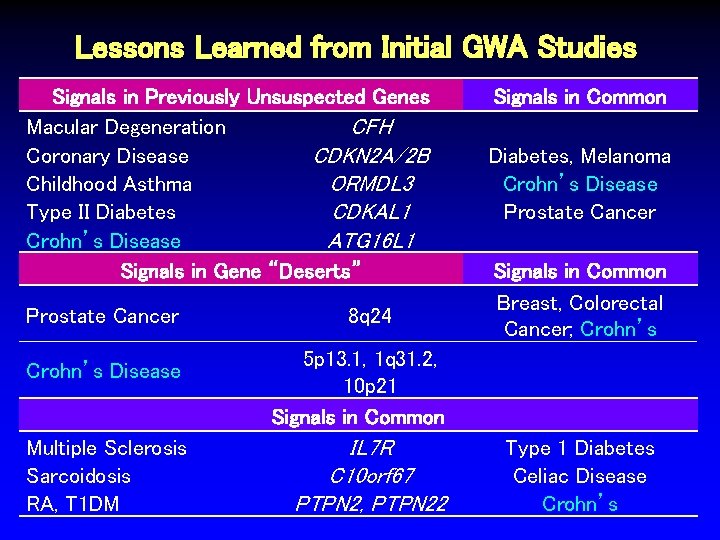
Lessons Learned from Initial GWA Studies Signals in Previously Unsuspected Genes Macular Degeneration CFH Coronary Disease CDKN 2 A/2 B Childhood Asthma ORMDL 3 Type II Diabetes CDKAL 1 Crohn’s Disease ATG 16 L 1 Signals in Gene “Deserts” Prostate Cancer Crohn’s Disease Multiple Sclerosis Sarcoidosis RA, T 1 DM 8 q 24 Signals in Common Diabetes, Melanoma Crohn’s Disease Prostate Cancer Signals in Common Breast, Colorectal Cancer; Crohn’s 5 p 13. 1, 1 q 31. 2, 10 p 21 Signals in Common IL 7 R C 10 orf 67 PTPN 2, PTPN 22 Type 1 Diabetes Celiac Disease Crohn’s

Lessons Learned from Initial GWA Studies Signals in Previously Unsuspected Genes Macular Degeneration CFH Coronary Disease CDKN 2 A/2 B Childhood Asthma ORMDL 3 Type II Diabetes CDKAL 1 Crohn’s Disease ATG 16 L 1 Signals in Gene “Deserts” Prostate Cancer Crohn’s Disease Multiple Sclerosis Sarcoidosis RA, T 1 DM 8 q 24 Signals in Common Diabetes, Melanoma Crohn’s Disease Prostate Cancer Signals in Common Breast, Colorectal Cancer; Crohn’s 5 p 13. 1, 1 q 31. 2, 10 p 21 Signals in Common IL 7 R C 10 orf 67 PTPN 2, PTPN 22 Type 1 Diabetes Celiac Disease Crohn’s

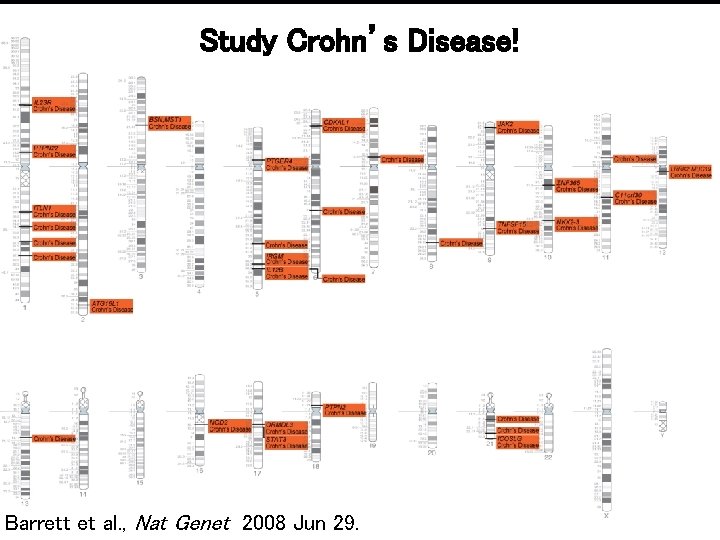
Study Crohn’s Disease! Barrett et al. , Nat Genet 2008 Jun 29.

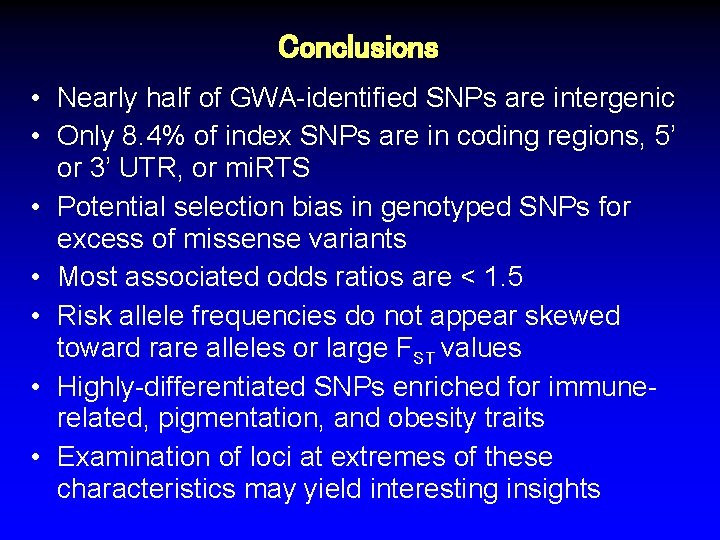
Conclusions • Nearly half of GWA-identified SNPs are intergenic • Only 8. 4% of index SNPs are in coding regions, 5’ or 3’ UTR, or mi. RTS • Potential selection bias in genotyped SNPs for excess of missense variants • Most associated odds ratios are < 1. 5 • Risk allele frequencies do not appear skewed toward rare alleles or large FST values • Highly-differentiated SNPs enriched for immunerelated, pigmentation, and obesity traits • Examination of loci at extremes of these characteristics may yield interesting insights

“The more we find, the more we see, the more we come to learn. The more that we explore, the more we shall return. ” Sir Tim Rice, Aida, 2000
 National human genome research institute
National human genome research institute National human genome research institute
National human genome research institute Translation
Translation Encode
Encode Semi-global alignment
Semi-global alignment Go foods
Go foods National institutes for health
National institutes for health The human genome consists of
The human genome consists of Human genome size
Human genome size Human genome size
Human genome size Bacterial artificial chromosome slideshare
Bacterial artificial chromosome slideshare Genome organization
Genome organization Human genome structure
Human genome structure Human genome project source code
Human genome project source code Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key Sequencing human genome
Sequencing human genome History of human genome project
History of human genome project Chapter 15 the human genome answer key
Chapter 15 the human genome answer key Chapter 14 the human genome
Chapter 14 the human genome Human genome project
Human genome project Section 14-3 human molecular genetics answer key
Section 14-3 human molecular genetics answer key Slidetodoc
Slidetodoc Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Plant genome research program
Plant genome research program Genome research limited
Genome research limited Genome research limited
Genome research limited National assistive technology research institute
National assistive technology research institute National research institute of brewing
National research institute of brewing Swedish national road and transport research institute
Swedish national road and transport research institute National research center kurchatov institute
National research center kurchatov institute National research center kurchatov institute
National research center kurchatov institute List of nih institutes
List of nih institutes Coaching institutes for iit jee in agra
Coaching institutes for iit jee in agra Wisconsin institutes of discovery
Wisconsin institutes of discovery European network of forensic science institutes
European network of forensic science institutes Bank po coaching in hyderabad
Bank po coaching in hyderabad What is a genome
What is a genome Stanford genome technology center
Stanford genome technology center Mash distance
Mash distance Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Dna
Dna Patric bioinformatics
Patric bioinformatics Genome modification ustaz auni
Genome modification ustaz auni Genome klick
Genome klick Genome assembly and annotation ppt
Genome assembly and annotation ppt Genome.ucsc.edu tutorial
Genome.ucsc.edu tutorial Genome
Genome Genome identification
Genome identification Genome sequencing
Genome sequencing Savant genome browser
Savant genome browser 1000 genome project
1000 genome project Alternate splicing
Alternate splicing Open reading frame
Open reading frame Genome.gov
Genome.gov Innovation genome project
Innovation genome project Genome mapping
Genome mapping Igv genome browser
Igv genome browser Integrated microbial genome
Integrated microbial genome Artemis genome
Artemis genome Genome project
Genome project Scalable annotation pipeline
Scalable annotation pipeline Genome definition
Genome definition Tomato genome annotation
Tomato genome annotation Genome adalah
Genome adalah Integrative genomics viewer
Integrative genomics viewer Savant genome browser
Savant genome browser Duke human vaccine institute
Duke human vaccine institute Australian primary health care research institute
Australian primary health care research institute Telethon institute for child health research
Telethon institute for child health research Croatian forest research institute
Croatian forest research institute Where is happiness
Where is happiness Bill harris centerpointe
Bill harris centerpointe Primary control vs secondary control
Primary control vs secondary control Samsung biomedical research institute
Samsung biomedical research institute Sagar institute of research and technology
Sagar institute of research and technology Cardiovascular research institute basel
Cardiovascular research institute basel Kavi kenya
Kavi kenya Environmental systems research institute
Environmental systems research institute Electronics and telecommunications research institute
Electronics and telecommunications research institute Digital enterprise research institute
Digital enterprise research institute Heilbronn institute for mathematical research
Heilbronn institute for mathematical research Target institute of medical education and research
Target institute of medical education and research