Molecular Phylogenetics Dan Graur 1 2 3 4

















- Slides: 65

Molecular Phylogenetics Dan Graur 1

2

3

4

5

Molecular phylogenetic approaches: 1. distance-matrix (based on distance measures) 2. character-state (based on character states) 3. maximum likelihood (based on both character states and distances) 6

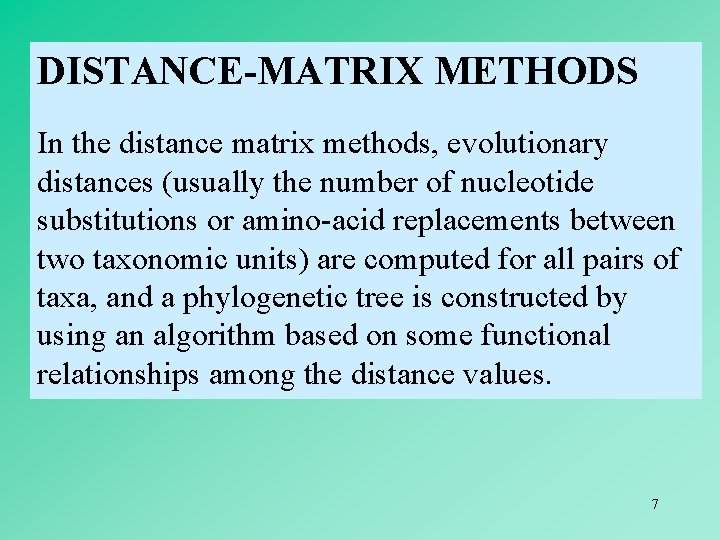
DISTANCE-MATRIX METHODS In the distance matrix methods, evolutionary distances (usually the number of nucleotide substitutions or amino-acid replacements between two taxonomic units) are computed for all pairs of taxa, and a phylogenetic tree is constructed by using an algorithm based on some functional relationships among the distance values. 7

Multiple Alignment 8

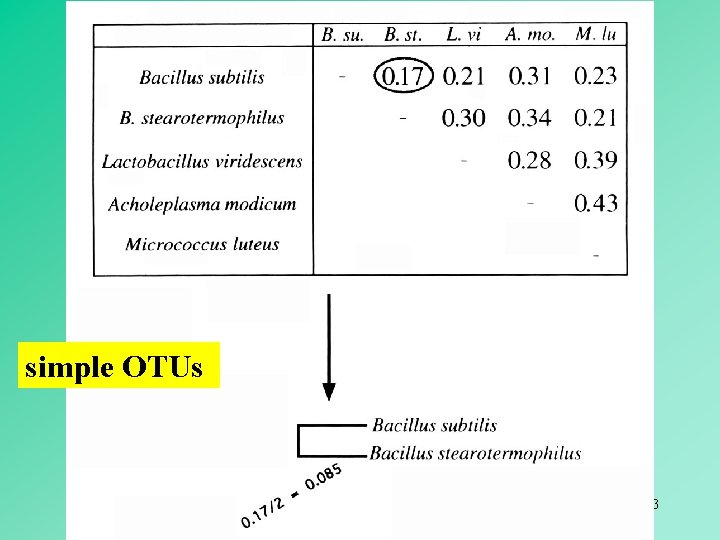
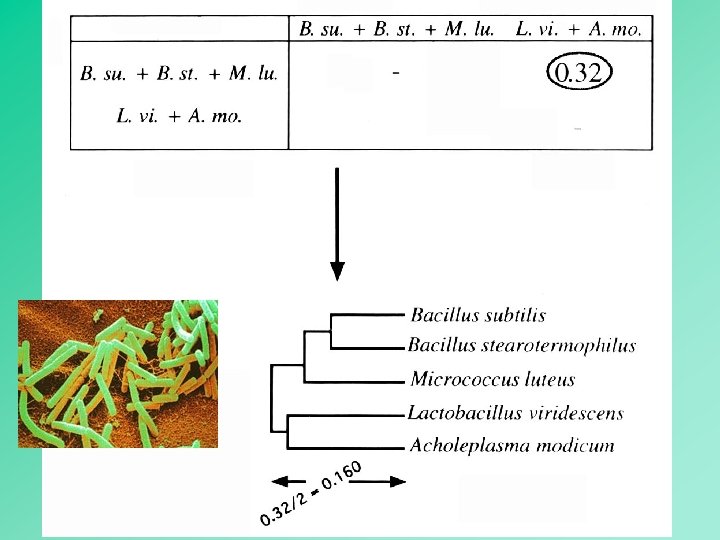
Distance Matrix* *Units: Numbers of nucleotide substitutions per 1, 000 nucleotide sites 9

Distance Methods: UPGMA Neighbor-relations Neighbor joining 10

UPGMA Unweighted pair-group method with arithmetic means 11

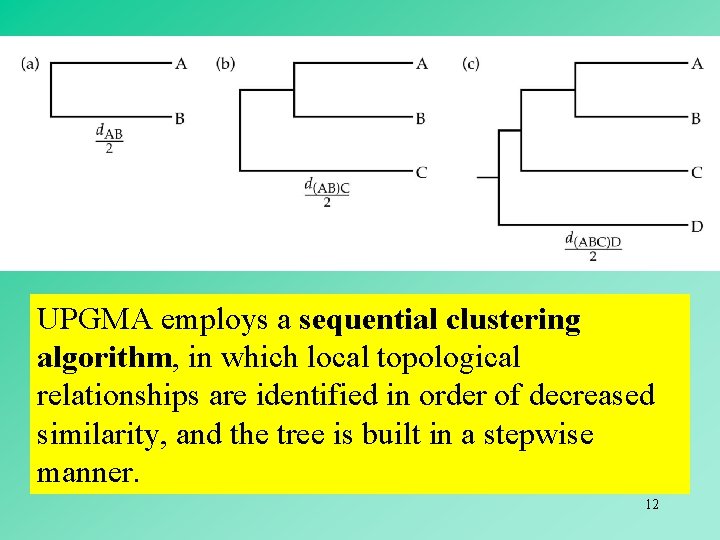
UPGMA employs a sequential clustering algorithm, in which local topological relationships are identified in order of decreased similarity, and the tree is built in a stepwise manner. 12

simple OTUs 13

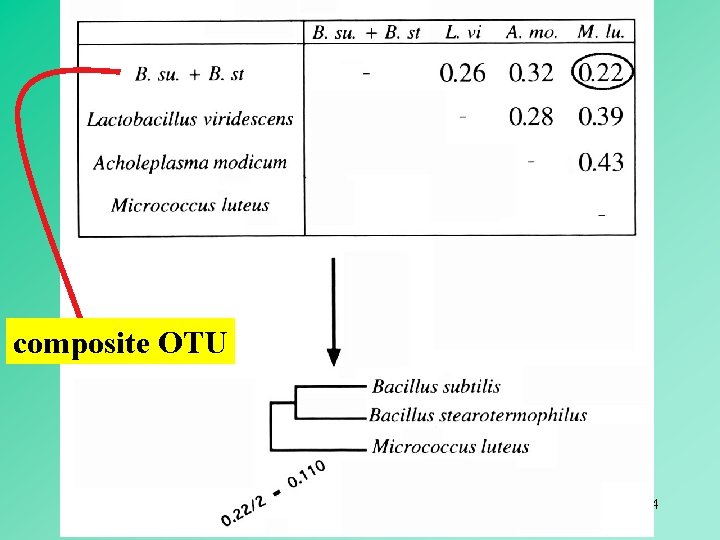
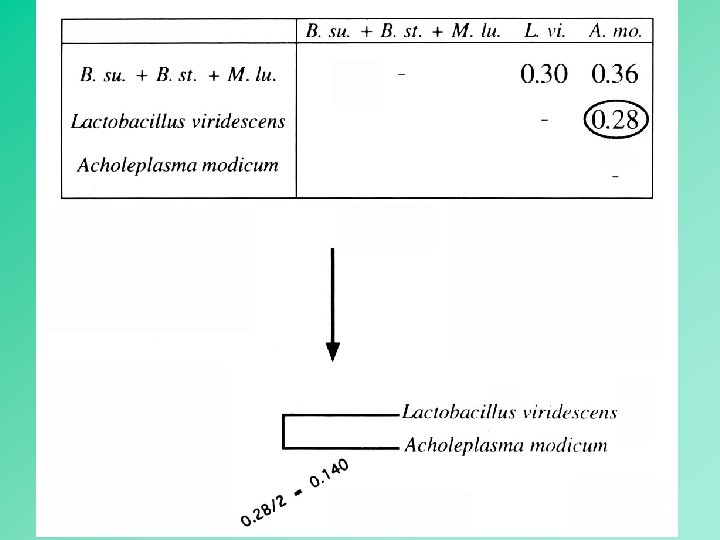
composite OTU 14

15

16

UPGMA only works if the distances are strictly ultrametric. 17

Neighborliness methods The neighbors-relation method (Sattath & Tversky) The neighbor-joining method (Saitou & Nei) 18

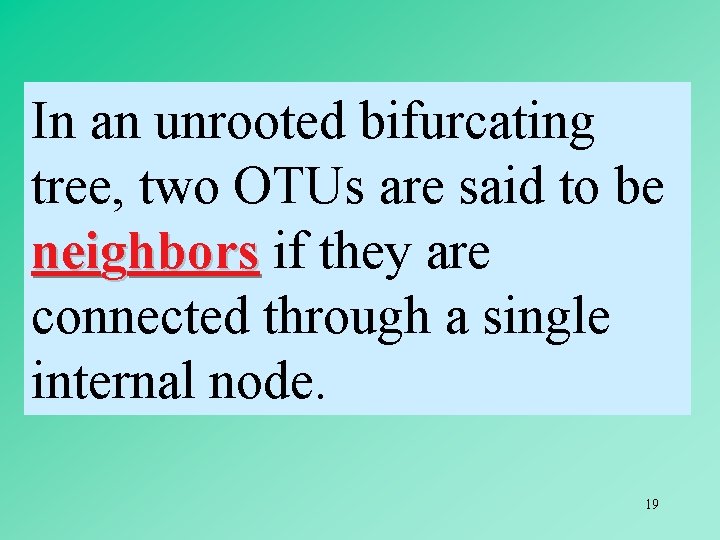
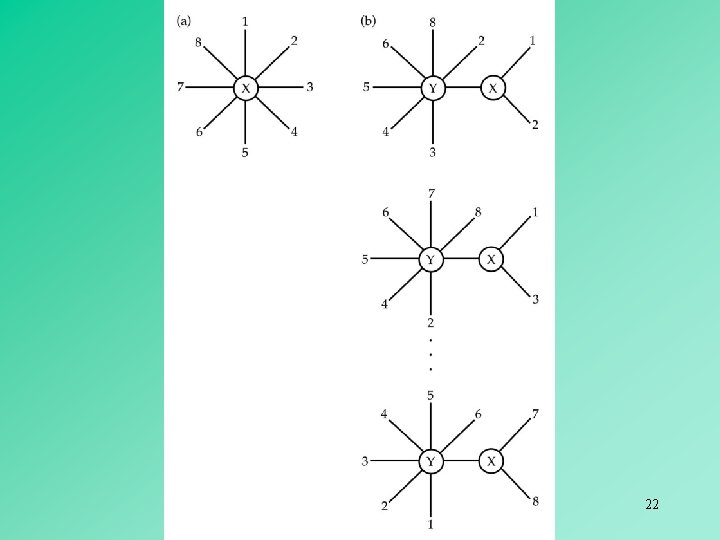
In an unrooted bifurcating tree, two OTUs are said to be neighbors if they are connected through a single internal node. 19

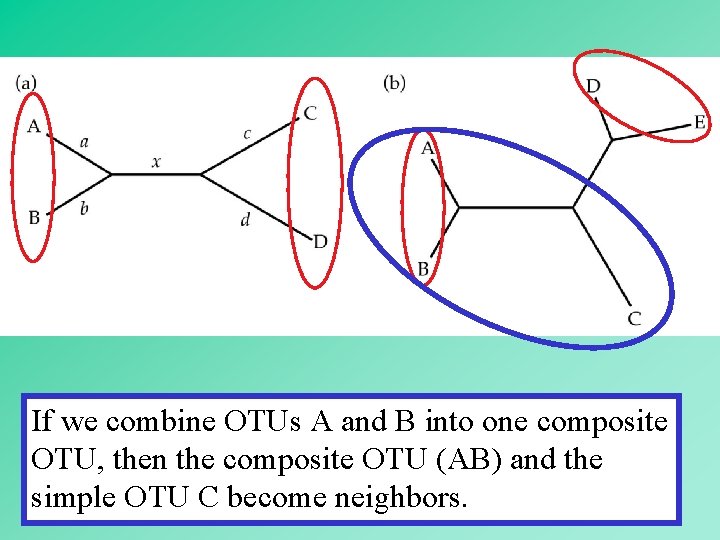
If we combine OTUs A and B into one composite OTU, then the composite OTU (AB) and the simple OTU C become neighbors. 20

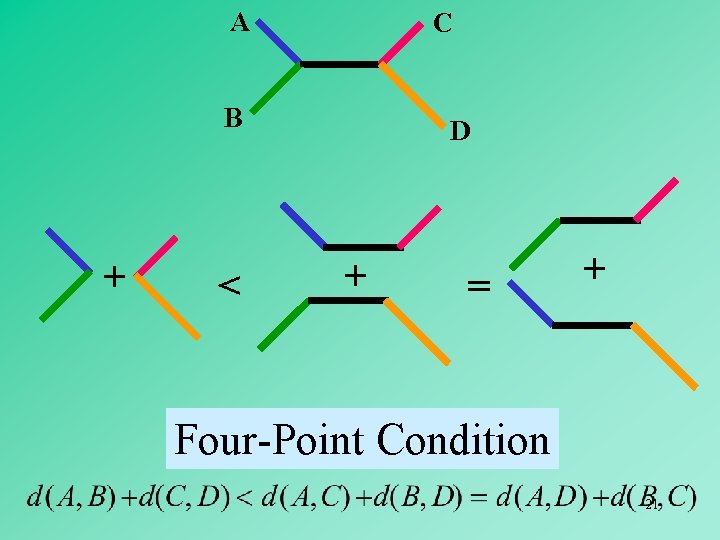
A C B + < D + = + Four-Point Condition 21

22

23

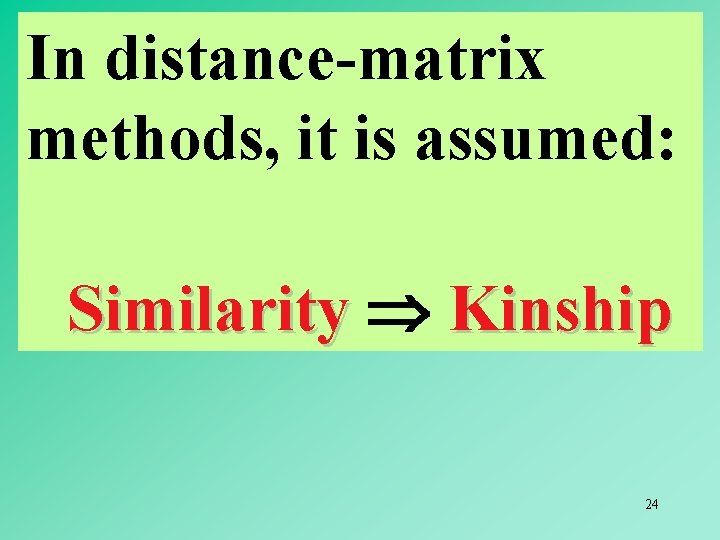
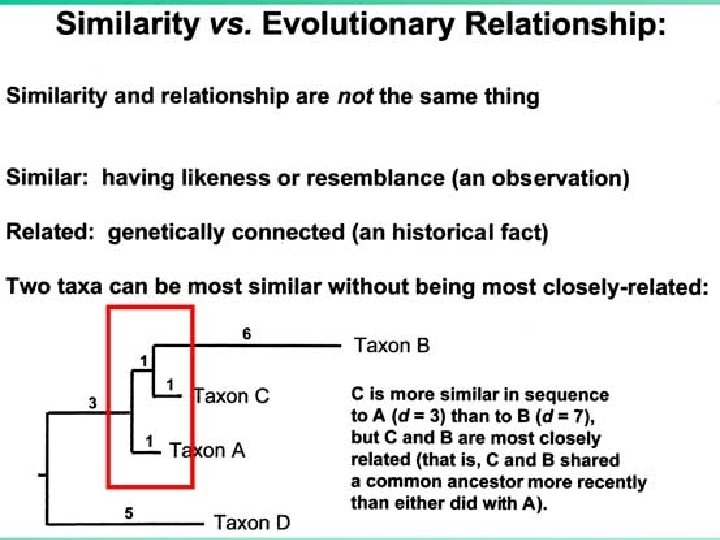
In distance-matrix methods, it is assumed: Similarity Kinship 24

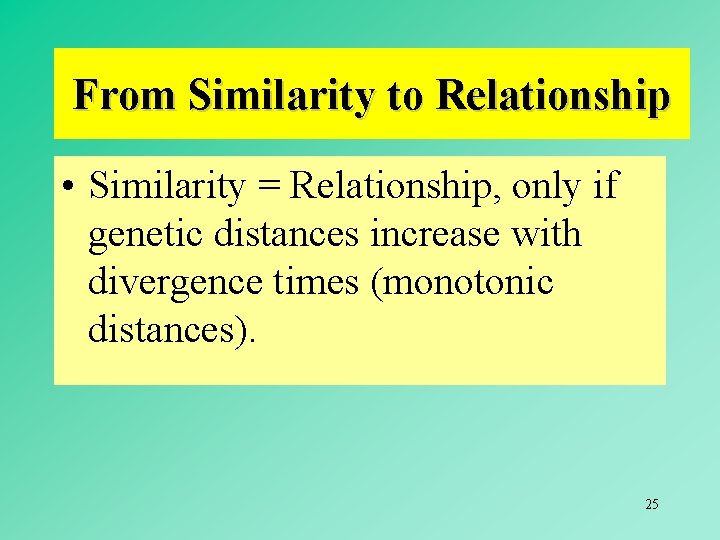
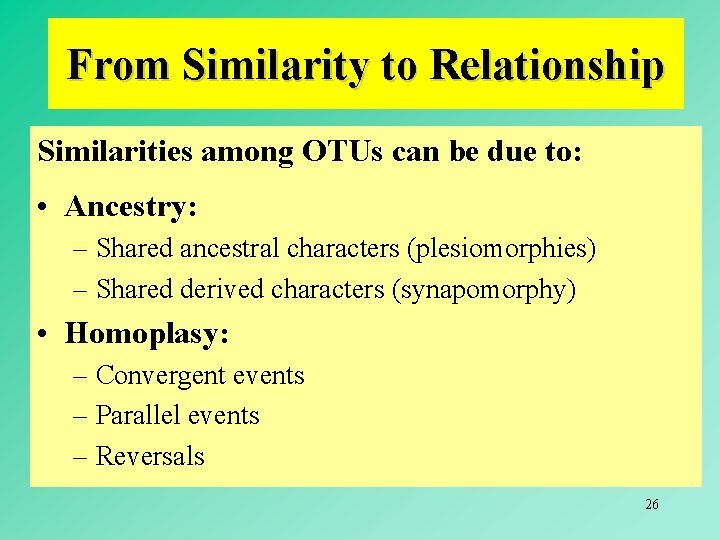
From Similarity to Relationship • Similarity = Relationship, only if genetic distances increase with divergence times (monotonic distances). 25

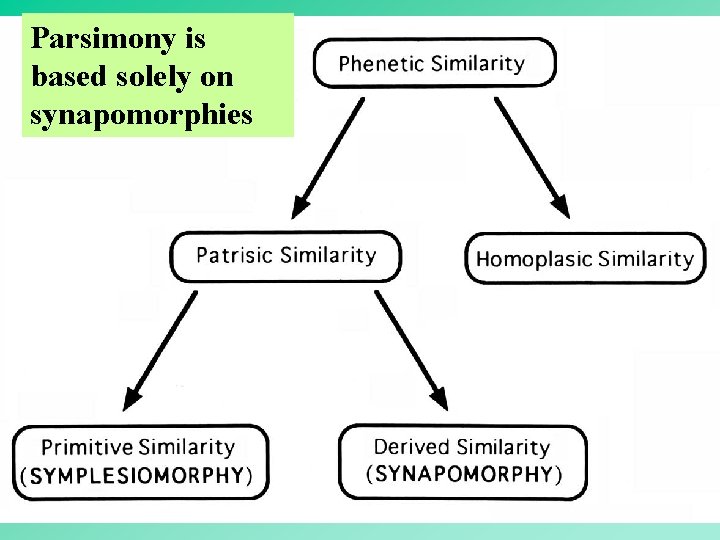
From Similarity to Relationship Similarities among OTUs can be due to: • Ancestry: – Shared ancestral characters (plesiomorphies) – Shared derived characters (synapomorphy) • Homoplasy: – Convergent events – Parallel events – Reversals 26

27

Parsimony Methods: Willi Hennig 1913 -1976 28

“Pluralitas non est ponenda sine neccesitate. ” (Plurality should not be posited without necessity. ) Occam’s razor William of Occam or Ockham (ca. 1285 -1349) English philosopher & Franciscan monk Excommunicated by Pope John XXII in 1328. Officially rehabilitated by Pope Innocent VI in 1359. 29

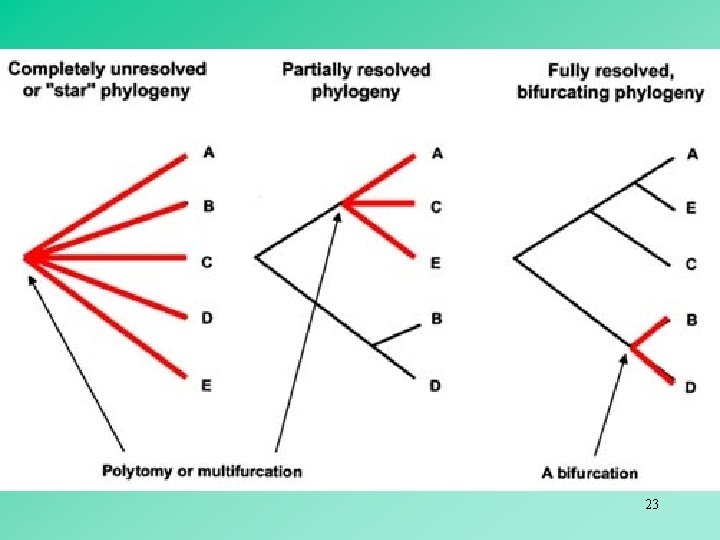
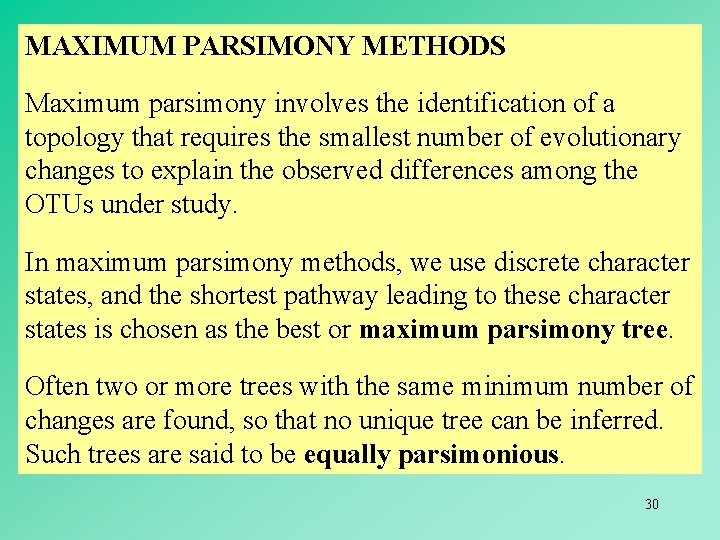
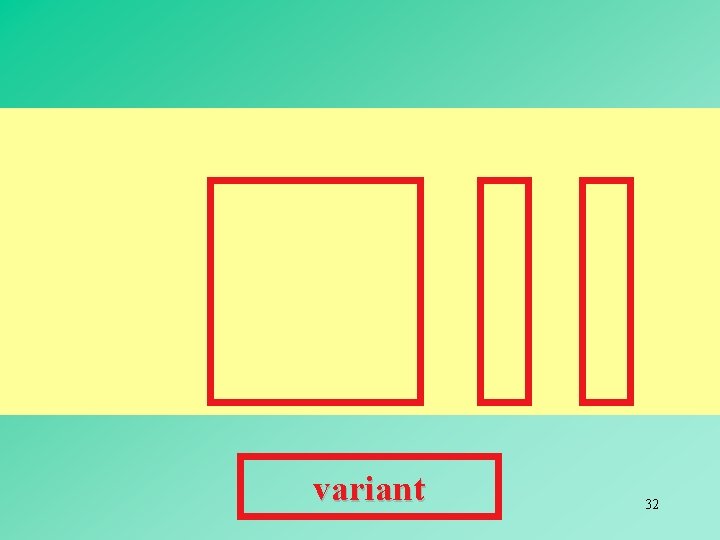
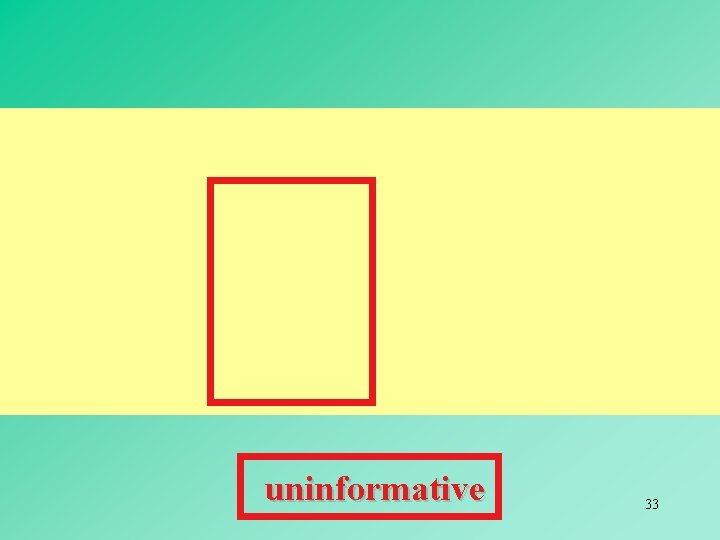
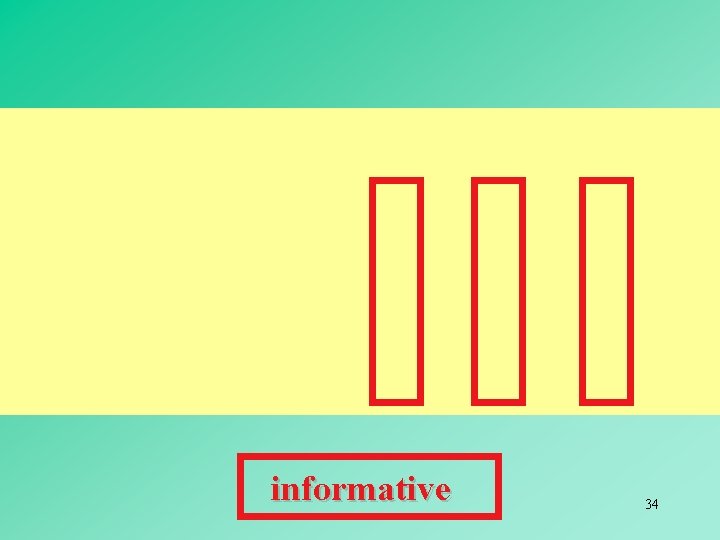
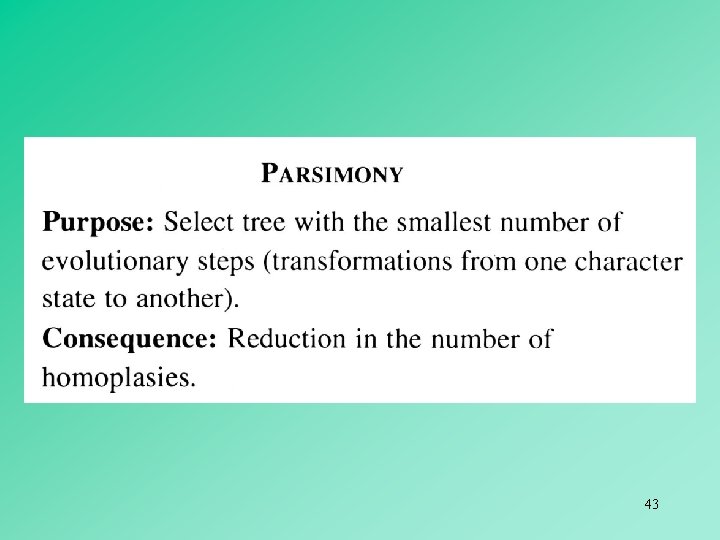
MAXIMUM PARSIMONY METHODS Maximum parsimony involves the identification of a topology that requires the smallest number of evolutionary changes to explain the observed differences among the OTUs under study. In maximum parsimony methods, we use discrete character states, and the shortest pathway leading to these character states is chosen as the best or maximum parsimony tree. Often two or more trees with the same minimum number of changes are found, so that no unique tree can be inferred. Such trees are said to be equally parsimonious. 30

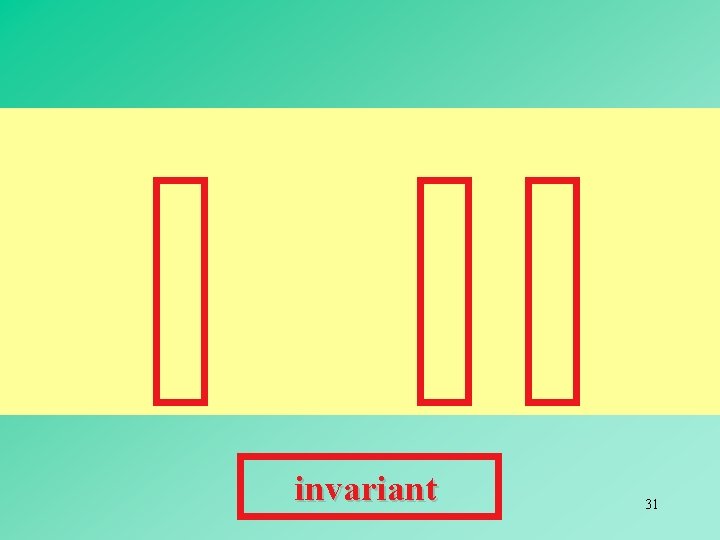
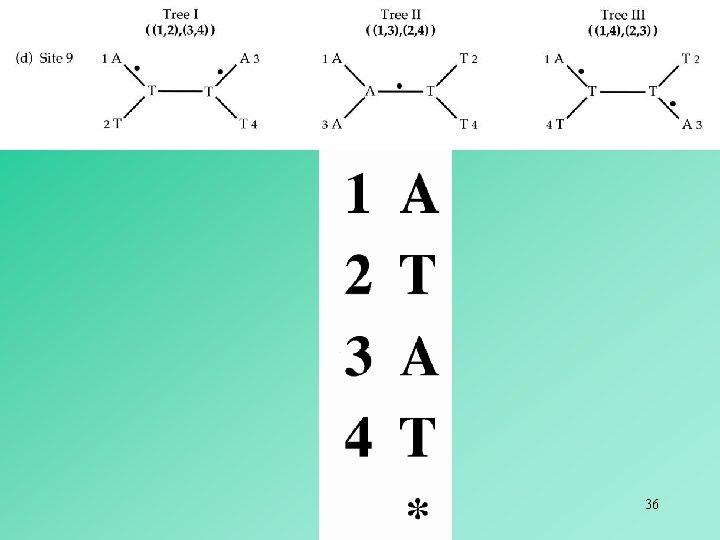
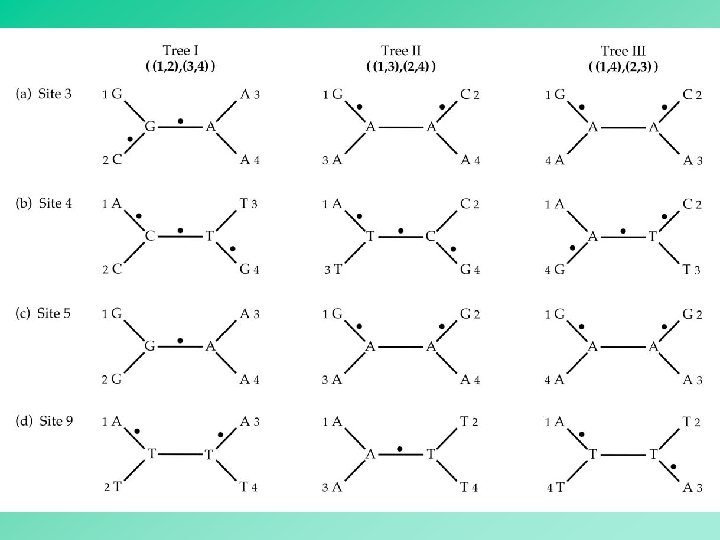
invariant 31

variant 32

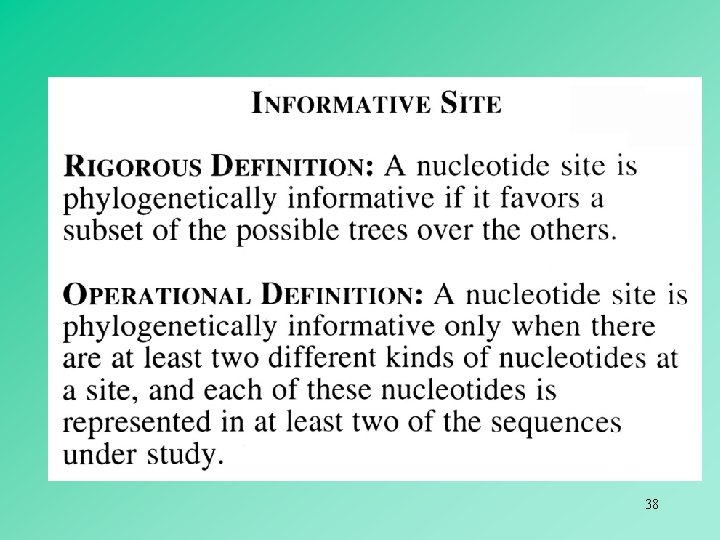
uninformative 33

informative 34

35

36

37

38

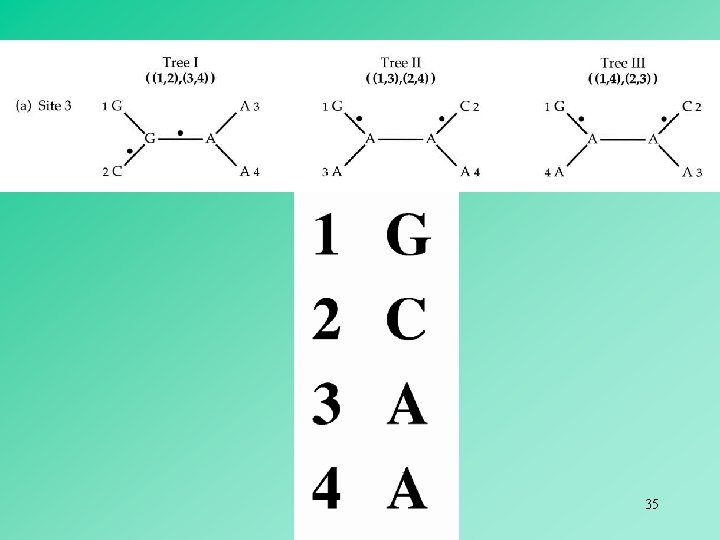
Inferring the maximum parsimony tree: 1. Identify all the informative sites. 2. For each possible tree, calculate the minimum number of substitutions at each informative site. 3. Sum up the number of changes over all the informative sites for each possible tree. 4. Choose the tree associated with the smallest number of changes as the maximum parsimony tree. 39

In the case of four OTUs, an informative site can only favor one of the three possible alternative trees. Thus, the tree supported by the largest number of informative sites is the most parsimonious tree. 40

With more than 4 OTUs, an informative site may favor more than one tree, and the maximum parsimony tree may not necessarily be the one supported by the largest number of informative sites. 41

The informative sites that support the internal branches in the inferred tree are deemed to be synapomorphies. All other informative sites are deemed to be homoplasies. 42

43

Parsimony is based solely on synapomorphies 44

45

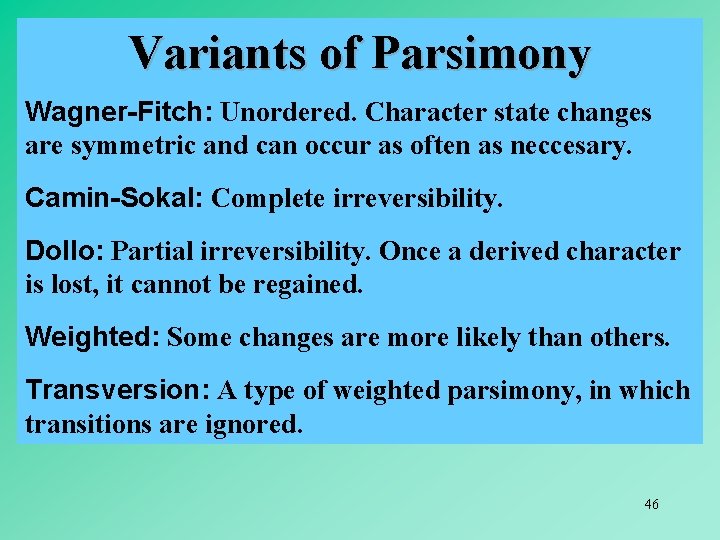
Variants of Parsimony Wagner-Fitch: Unordered. Character state changes are symmetric and can occur as often as neccesary. Camin-Sokal: Complete irreversibility. Dollo: Partial irreversibility. Once a derived character is lost, it cannot be regained. Weighted: Some changes are more likely than others. Transversion: A type of weighted parsimony, in which transitions are ignored. 46

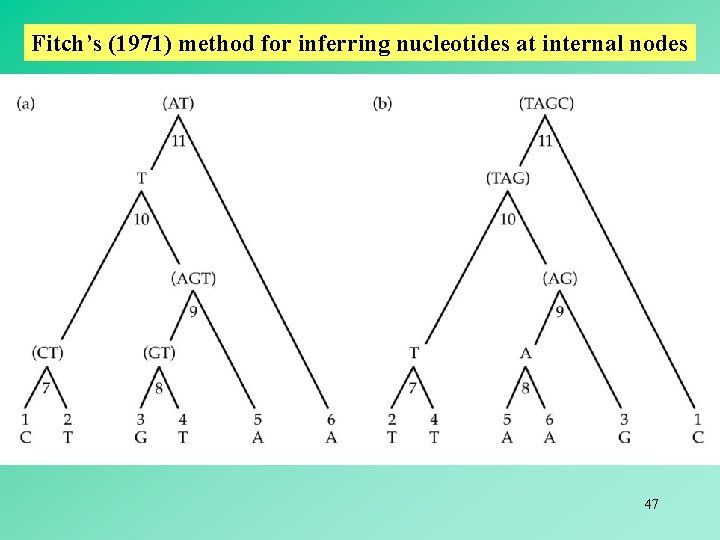
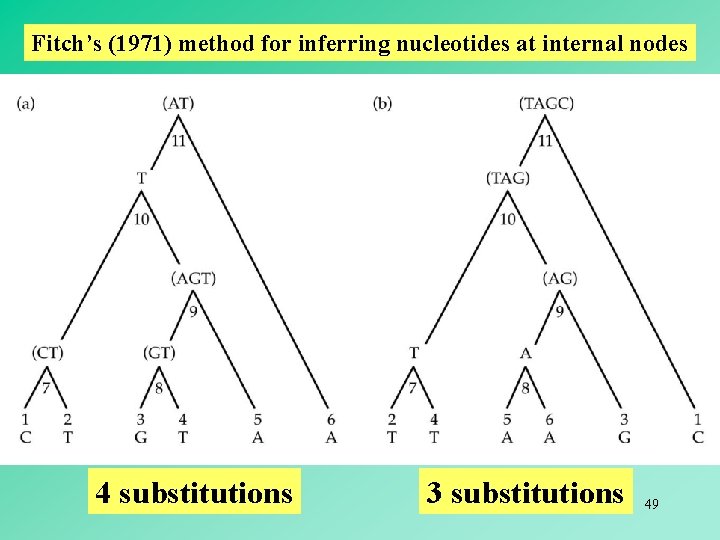
Fitch’s (1971) method for inferring nucleotides at internal nodes 47

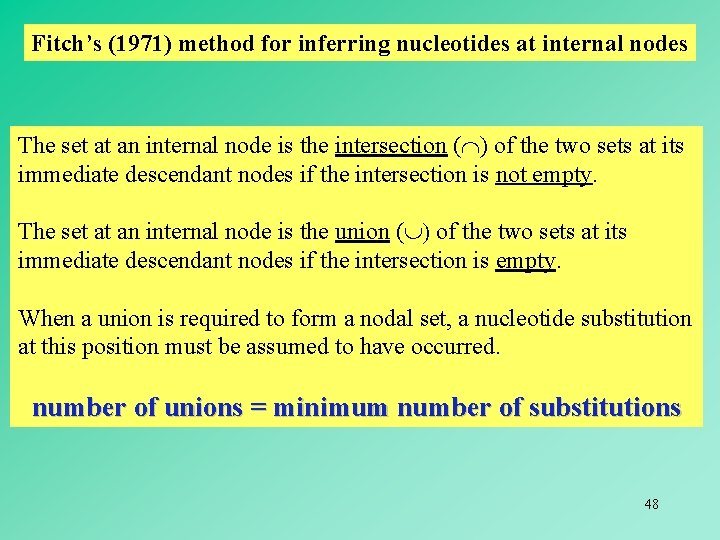
Fitch’s (1971) method for inferring nucleotides at internal nodes The set at an internal node is the intersection ( ) of the two sets at its immediate descendant nodes if the intersection is not empty. The set at an internal node is the union ( ) of the two sets at its immediate descendant nodes if the intersection is empty. When a union is required to form a nodal set, a nucleotide substitution at this position must be assumed to have occurred. number of unions = minimum number of substitutions 48

Fitch’s (1971) method for inferring nucleotides at internal nodes 4 substitutions 3 substitutions 49

50

total number of substitutions in a tree = tree length 51

Searching for the maximum-parsimony tree 52

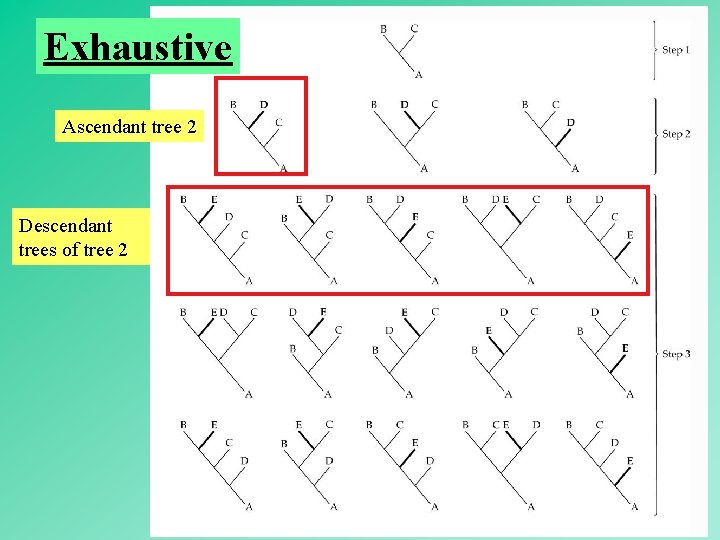
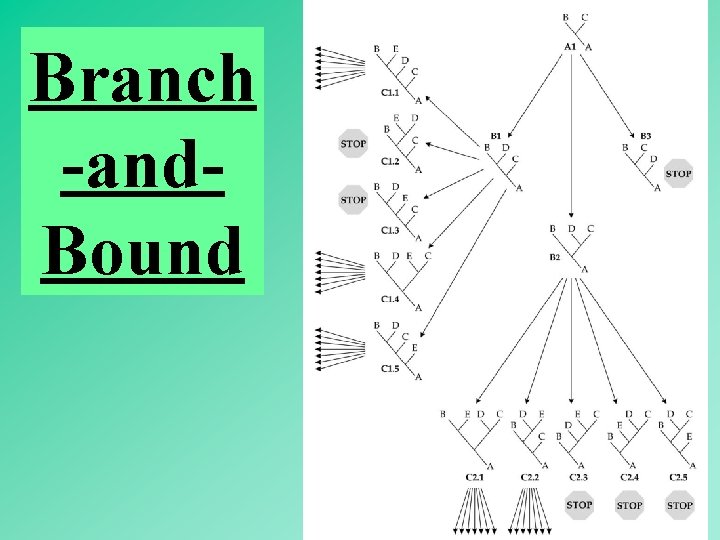
Exhaustive = Examine all trees, get the best tree (guaranteed). Branch-and-Bound = Examine some trees, get the best tree (guaranteed). Heuristic = Examine some trees, get a tree that may or may not be the best tree. 53

Exhaustive Ascendant tree 2 Descendant trees of tree 2 54

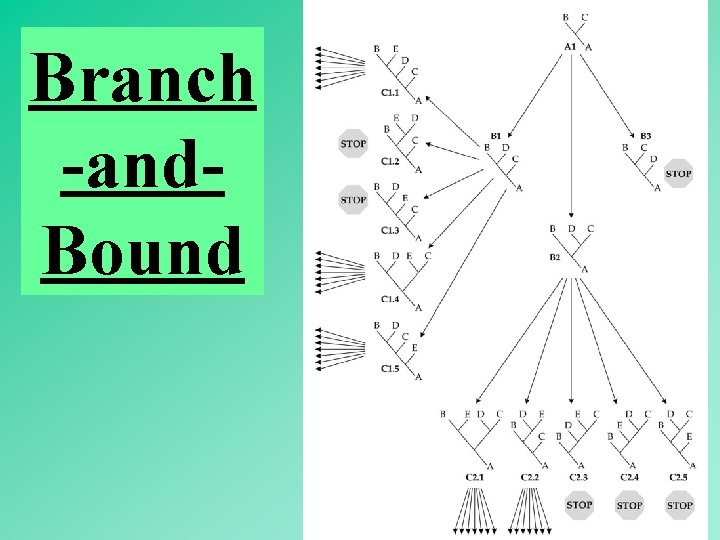
Branch -and. Bound 55

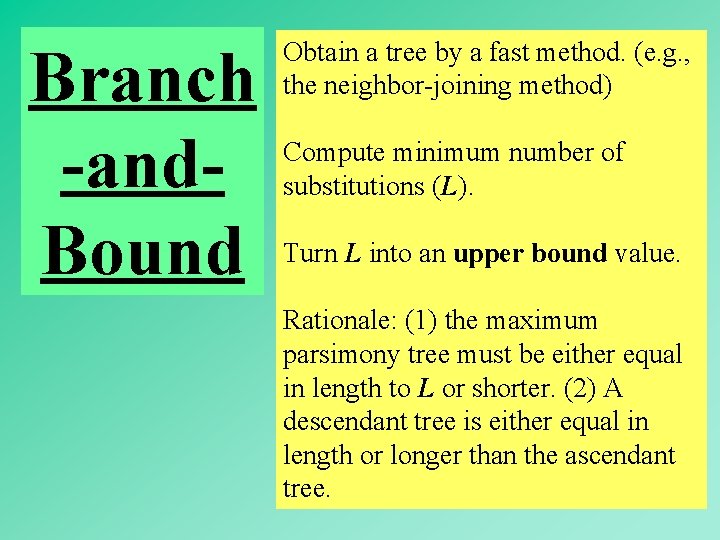
Branch -and. Bound Obtain a tree by a fast method. (e. g. , the neighbor-joining method) Compute minimum number of substitutions (L). Turn L into an upper bound value. Rationale: (1) the maximum parsimony tree must be either equal in length to L or shorter. (2) A descendant tree is either equal in length or longer than the ascendant tree. 56

Branch -and. Bound 57

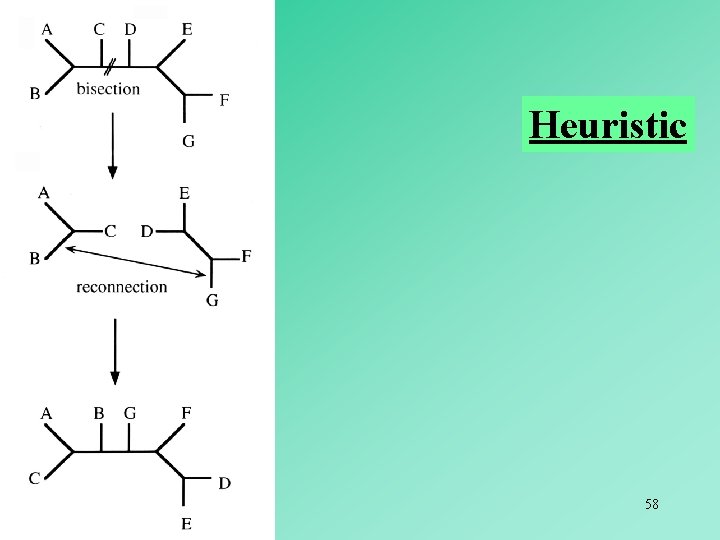
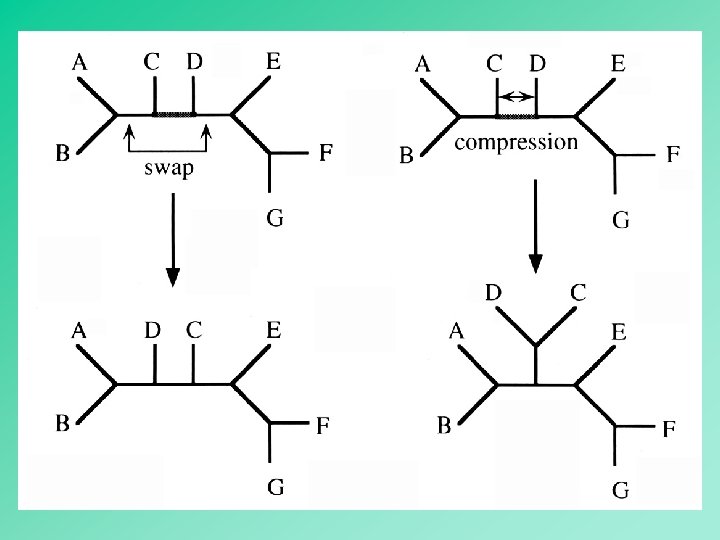
Heuristic 58

59

60

Likelihood • Example: • Data: • Hypothesis: Coin tossing Outcome of 10 tosses: 6 heads + 4 tails Binomial distribution 61

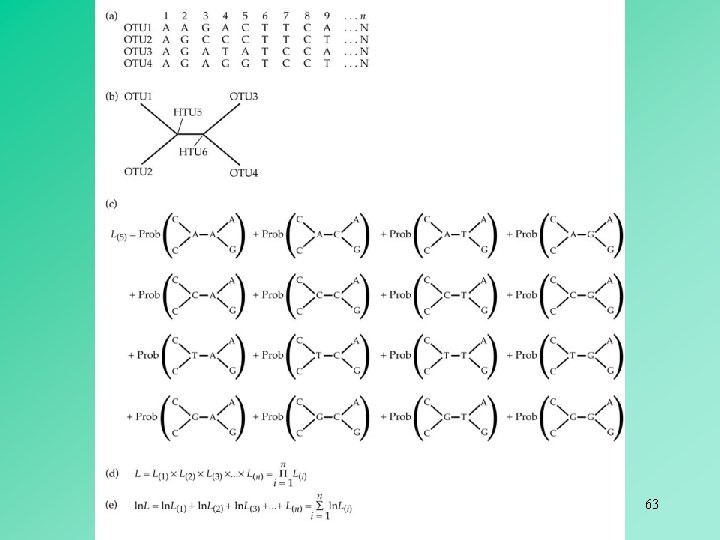
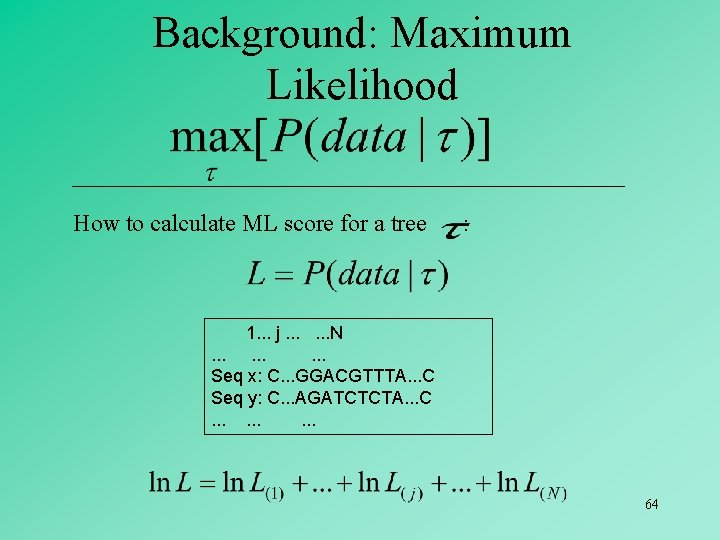
LIKELIHOOD IN MOLECULAR PHYLOGENETICS • The data are the aligned sequences • The model is the probability of change from one character state to another (e. g. , Jukes & Cantor 1 -P model). • The parameters to be estimated are: Topology & Branch Lengths 62

63

Background: Maximum Likelihood How to calculate ML score for a tree : 1. . . j. . . N. . Seq x: C. . . GGACGTTTA. . . C Seq y: C. . . AGATCTCTA. . . C. . 64

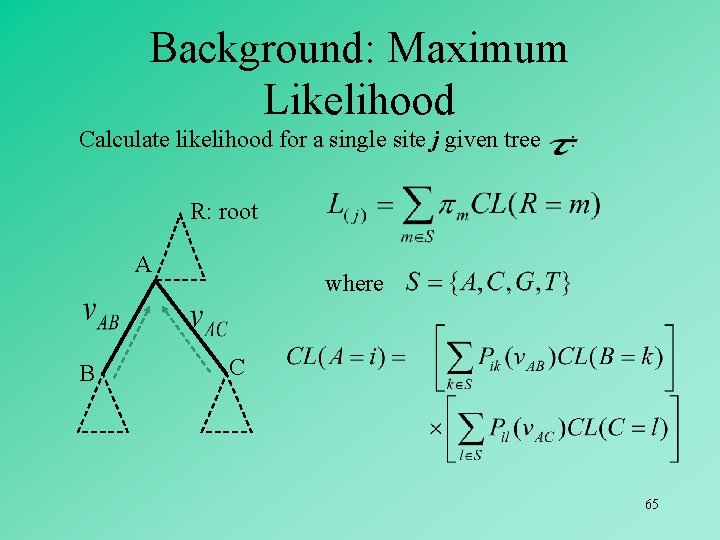
Background: Maximum Likelihood Calculate likelihood for a single site j given tree : R: root A B where C 65