Maximum parsimony with MEGA and PAUP Molecular Phylogenetics


- Slides: 5

Maximum parsimony with MEGA and PAUP* Molecular Phylogenetics – exercise Ben Stöver WS 2014/2015

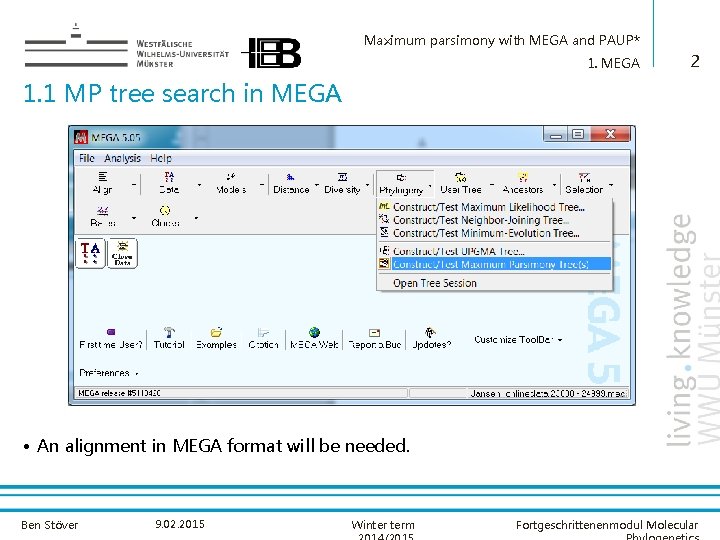
Maximum parsimony with MEGA and PAUP* 1. MEGA 2 1. 1 MP tree search in MEGA • An alignment in MEGA format will be needed. Ben Stöver 9. 02. 2015 Winter term Fortgeschrittenenmodul Molecular

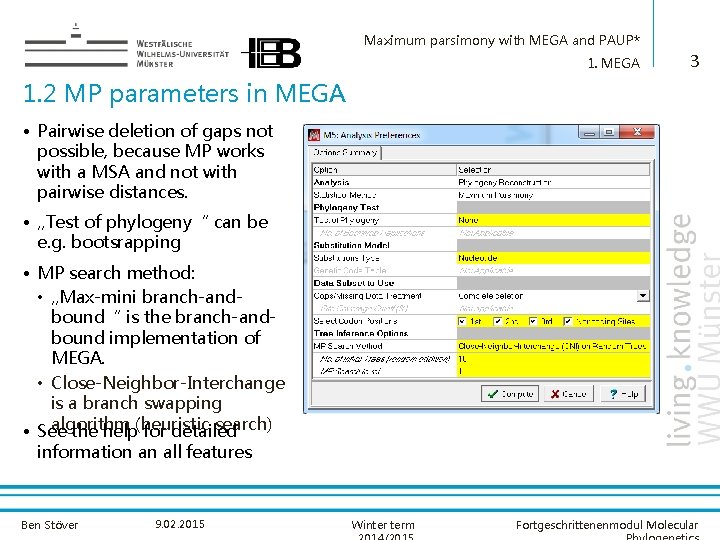
Maximum parsimony with MEGA and PAUP* 1. MEGA 3 1. 2 MP parameters in MEGA • Pairwise deletion of gaps not possible, because MP works with a MSA and not with pairwise distances. • „Test of phylogeny“ can be e. g. bootsrapping • MP search method: • „Max-mini branch-andbound“ is the branch-andbound implementation of MEGA. • Close-Neighbor-Interchange is a branch swapping algorithm search) • See the help(heuristic for detailed information an all features Ben Stöver 9. 02. 2015 Winter term Fortgeschrittenenmodul Molecular

Maximum parsimony with MEGA and PAUP* 2. PAUP* 4 2. 1 Using PAUP* • Phylogeny analysis using parsimony (* and other methods) • When starting the program you can initially open a NEXUS file with your data (e. g. your alignment). • PAUP* is used by specifying commands in the PAUP* command line in a PAUP-block of a NEXUS file. • By typing „? “ you get the list of commands. • „<command> ? “ displays the available parameters for a command. Ben Stöver 9. 02. 2015 Winter term Fortgeschrittenenmodul Molecular

Maximum parsimony with MEGA and PAUP* 2. PAUP* 5 2. 2 PAUP* commands for tree searching • All. Trees: possible). • Band. B: taxa). • HSearch: • Swap: • Start: Performs a complete tree search (not more than 12 taxa Performs a complete branch and bound search (not many Performs a heuristic search with different methods. Determines the branch-swapping algorithm. (TBR is default. ) The start tree to be used. (STEPWISE|NJ|CURRENT|tree-number) • Mul. Trees: Specifies if all optimal trees or only one shall be saved. • Set: Specifies the search criterion used by the other commands (e. g. criterion=parsimony). • Show. Trees: Shows all trees in memory on the command line. • Save. Trees: Saves the tree(s) in memory to a file • Br. Lens: Determines weather to stare branch lengths (not with • Further information can be found by typing „<command> ? “ or in the parsimony) PAUP* manual under „DownloadsinformationPAUP“ in the course folder. • File: The path to the output tree file Ben Stöver 9. 02. 2015 Winter term Fortgeschrittenenmodul Molecular