Molecular monitoring in CML and Ph ALL Educational















- Slides: 72

Molecular monitoring in CML and Ph+ ALL Educational slide deck funded and developed by Incyte Biosciences in collaboration with a Steering Committee of European experts EU/ICLG/NP/18/0066 UK/ICLG/P/18/0034 NL/ICLG/NP/19/0011 Date of preparation: July 2018 Hosted on Think. Test. Treat. com

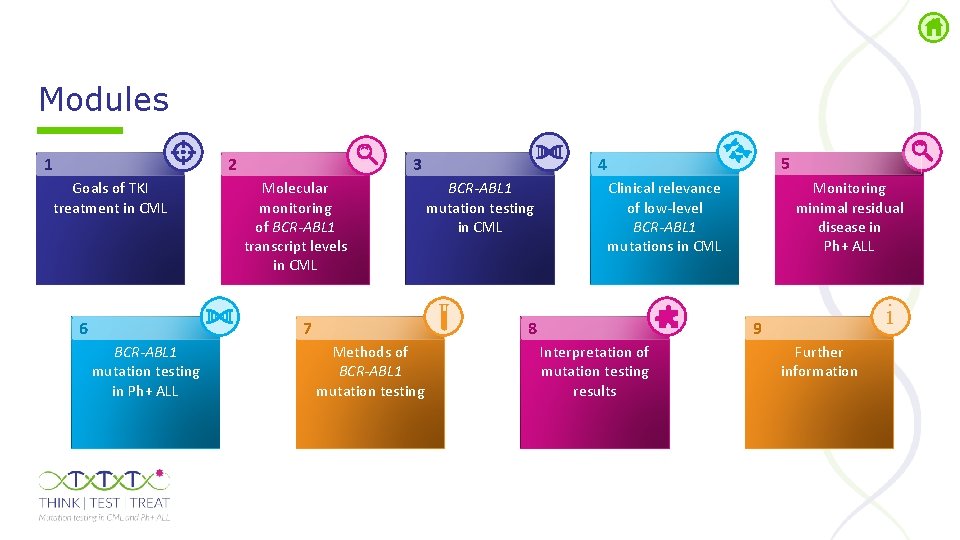
Modules 1 2 Goals of TKI treatment in CML 6 Molecular monitoring of BCR-ABL 1 transcript levels in CML 7 BCR-ABL 1 mutation testing in Ph+ ALL 5 4 3 BCR-ABL 1 mutation testing in CML 8 Methods of BCR-ABL 1 mutation testing Monitoring minimal residual disease in Ph+ ALL Clinical relevance of low-level BCR-ABL 1 mutations in CML i 9 Interpretation of mutation testing results Further information

Goals of TKI treatment in CML

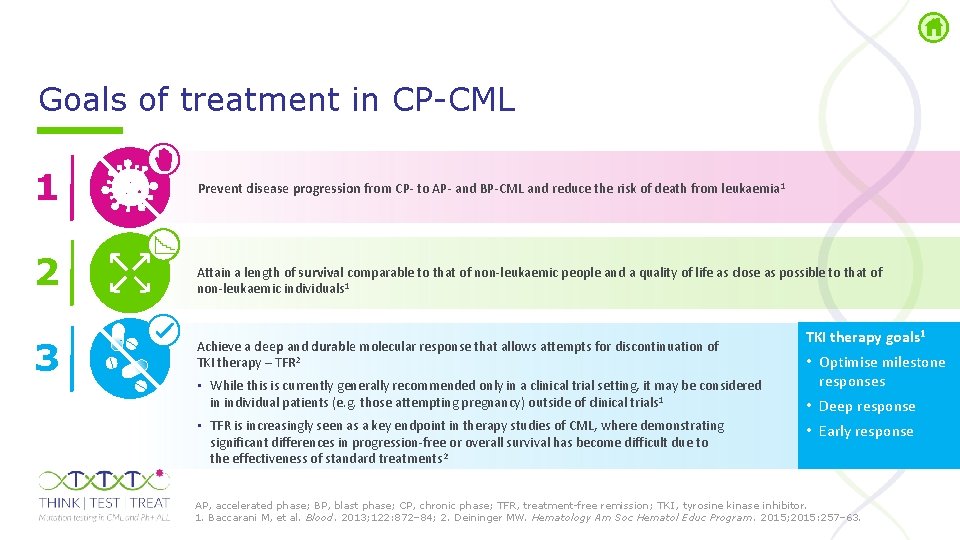
Goals of treatment in CP CML 1 2 3 Prevent disease progression from CP- to AP- and BP-CML and reduce the risk of death from leukaemia 1 Attain a length of survival comparable to that of non-leukaemic people and a quality of life as close as possible to that of non-leukaemic individuals 1 Achieve a deep and durable molecular response that allows attempts for discontinuation of TKI therapy – TFR 2 • While this is currently generally recommended only in a clinical trial setting, it may be considered in individual patients (e. g. those attempting pregnancy) outside of clinical trials 1 • TFR is increasingly seen as a key endpoint in therapy studies of CML, where demonstrating significant differences in progression-free or overall survival has become difficult due to the effectiveness of standard treatments 2 TKI therapy goals 1 • Optimise milestone responses • Deep response • Early response AP, accelerated phase; BP, blast phase; CP, chronic phase; TFR, treatment free remission; TKI , tyrosine kinase inhibitor. 1. Baccarani M, et al. Blood. 2013; 122: 872– 84; 2. Deininger MW. Hematology Am Soc Hematol Educ Program. 2015; 2015: 257– 63.

Management of CML • Monitoring response to TKI therapy is one of the key management strategies of CML 1– 3 • Long term survival outcomes cannot be used to adapt and optimise treatment – instead, early surrogate markers of response are used 1 • Effective TKI therapy has driven the need for more accurate methods to assess response, even at very low levels of disease burden 2, 3 TKI, tyrosine kinase inhibitor. 1. Baccarani M, et al. Blood. 2013; 122: 872– 84; 2. Egan D, Radich J. Am J Hematol. 2016; 91: 742– 6; 3. Morotti A, et al. Curr Hematol Malig Rep. 2015; 10: 167– 72.

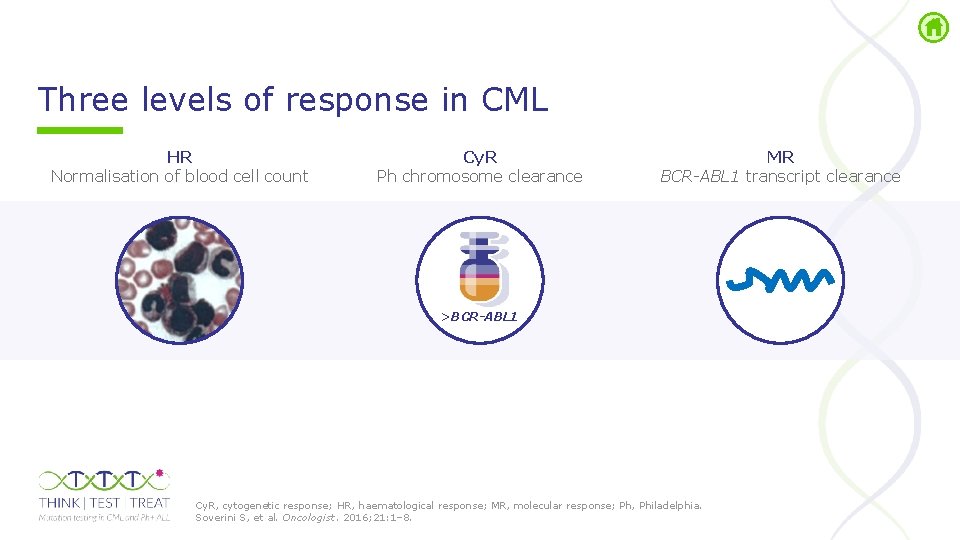
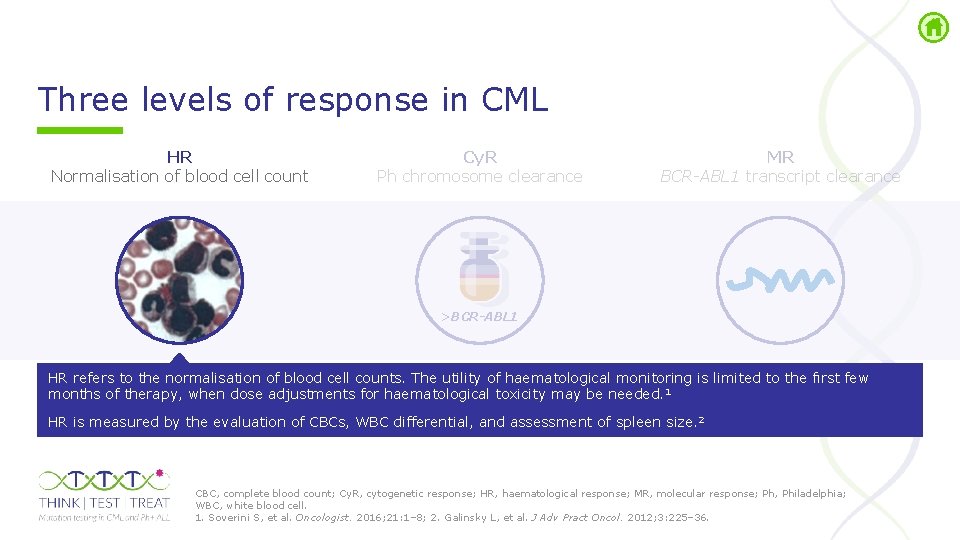
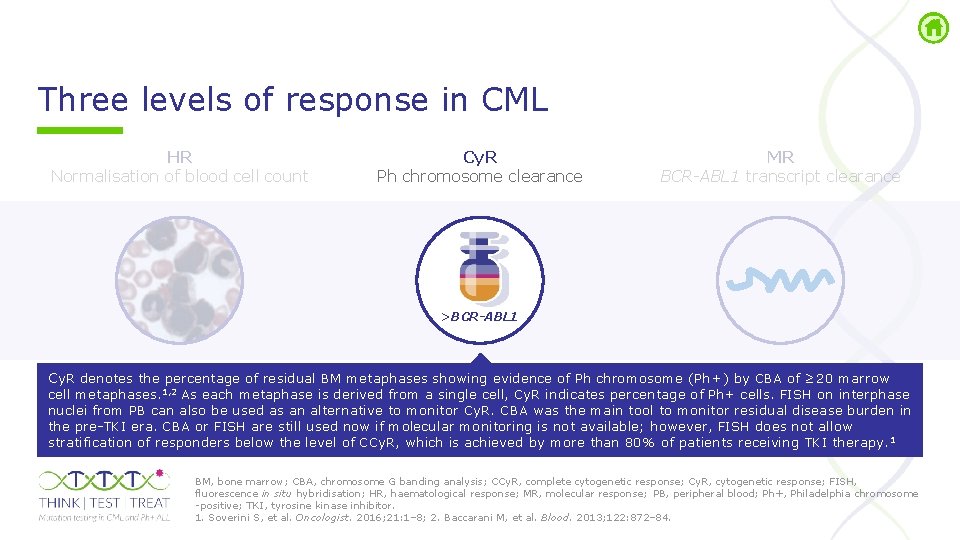
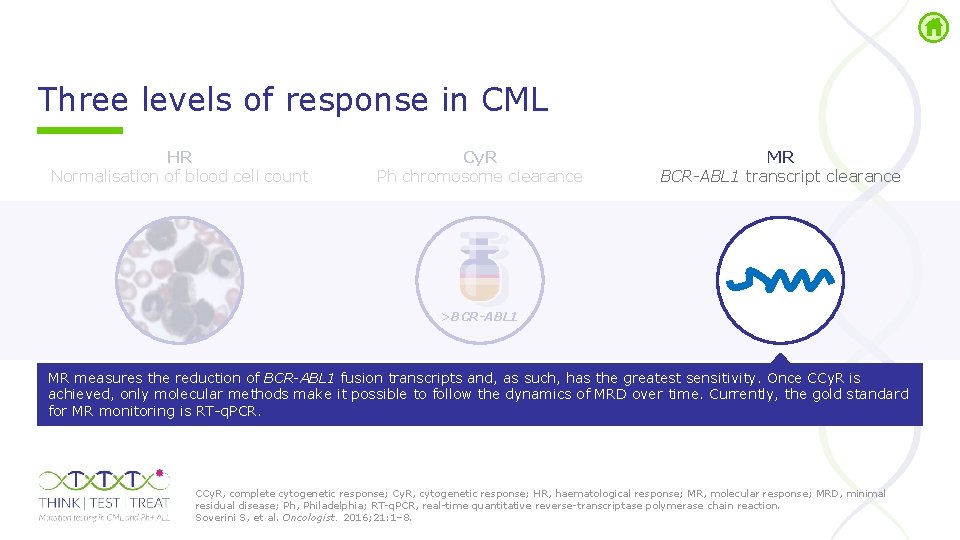
Three levels of response in CML HR Normalisation of blood cell count Cy. R Ph chromosome clearance MR BCR-ABL 1 transcript clearance >BCR-ABL 1 Cy. R, cytogenetic response; HR, haematological response; MR, molecular response; Ph, Philadelphia. Soverini S, et al. Oncologist. 2016; 21: 1– 8.

Three levels of response in CML HR Normalisation of blood cell count Cy. R Ph chromosome clearance MR BCR-ABL 1 transcript clearance >BCR-ABL 1 HR refers to the normalisation of blood cell counts. The utility of haematological monitoring is limited to the fırst few months of therapy, when dose adjustments for haematological toxicity may be needed. 1 HR is measured by the evaluation of CBCs, WBC differential, and assessment of spleen size. 2 CBC, complete blood count; Cy. R, cytogenetic response; HR, haematological response; MR, molecular response; Ph, Philadelphia; WBC, white blood cell. 1. Soverini S, et al. Oncologist. 2016; 21: 1– 8; 2. Galinsky L, et al. J Adv Pract Oncol. 2012; 3: 225– 36.

Three levels of response in CML HR Normalisation of blood cell count Cy. R Ph chromosome clearance MR BCR-ABL 1 transcript clearance >BCR-ABL 1 Cy. R denotes the percentage of residual BM metaphases showing evidence of Ph chromosome (Ph+) by CBA of ≥ 20 marrow cell metaphases. 1, 2 As each metaphase is derived from a single cell, Cy. R indicates percentage of Ph+ cells. FISH on interphase nuclei from PB can also be used as an alternative to monitor Cy. R. CBA was the main tool to monitor residual disease burden in the pre TKI era. CBA or FISH are still used now if molecular monitoring is not available; however, FISH does not allow stratification of responders below the level of CCy. R, which is achieved by more than 80% of patients receiving TKI therapy. 1 BM, bone marrow; CBA, chromosome G banding analysis; CCy. R, complete cytogenetic response; Cy. R, cytogenetic response; FISH, fluorescence in situ hybridisation; HR, haematological response; MR, molecular response; PB, peripheral blood; Ph+, Philadelphia chromosome positive; TKI, tyrosine kinase inhibitor. 1. Soverini S, et al. Oncologist. 2016; 21: 1– 8; 2. Baccarani M, et al. Blood. 2013; 122: 872– 84.

Three levels of response in CML HR Normalisation of blood cell count Cy. R Ph chromosome clearance MR BCR-ABL 1 transcript clearance >BCR-ABL 1 MR measures the reduction of BCR-ABL 1 fusion transcripts and, as such, has the greatest sensitivity. Once CCy. R is achieved, only molecular methods make it possible to follow the dynamics of MRD over time. Currently, the gold standard for MR monitoring is RT q. PCR. CCy. R, complete cytogenetic response; Cy. R, cytogenetic response; HR, haematological response; MR, molecular response; MRD, minimal residual disease; Ph, Philadelphia; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. Soverini S, et al. Oncologist. 2016; 21: 1– 8.

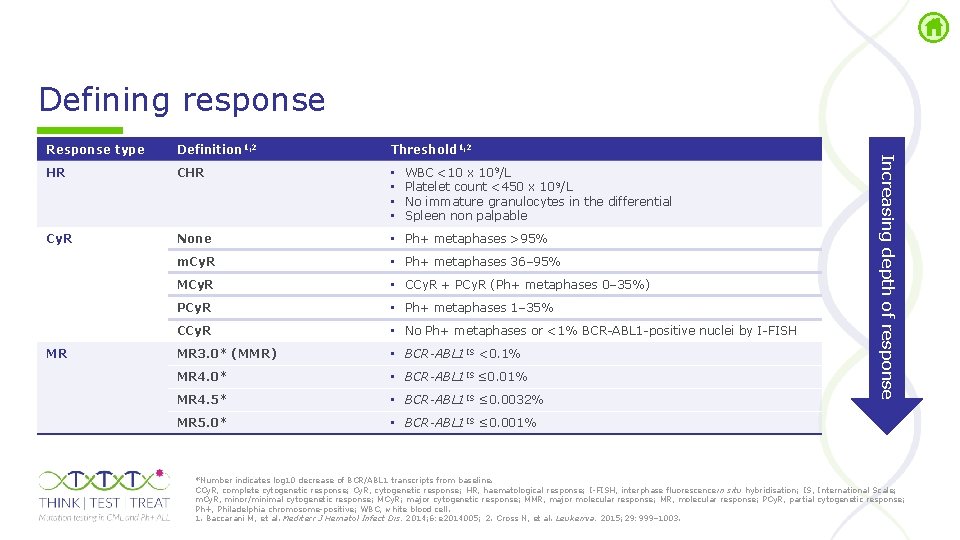
Defining response Definition 1, 2 Threshold 1, 2 HR CHR • • Cy. R None • Ph+ metaphases >95% m. Cy. R • Ph+ metaphases 36– 95% MCy. R • CCy. R + PCy. R (Ph+ metaphases 0– 35%) PCy. R • Ph+ metaphases 1– 35% CCy. R • No Ph+ metaphases or <1% BCR ABL 1 positive nuclei by I FISH MR 3. 0* (MMR) • BCR-ABL 1 IS <0. 1% MR 4. 0* • BCR-ABL 1 IS ≤ 0. 01% MR 4. 5* • BCR-ABL 1 IS ≤ 0. 0032% MR 5. 0* • BCR-ABL 1 IS ≤ 0. 001% MR WBC <10 x 109/L Platelet count <450 x 10 9/L No immature granulocytes in the differential Spleen non palpable Increasing depth of response Response type *Number indicates log 10 decrease of BCR/ABL 1 transcripts from baseline. CCy. R, complete cytogenetic response; Cy. R, cytogenetic response; HR, haematological response; I FISH, interphase fluorescence in situ hybridisation; IS, International Scale; m. Cy. R, minor/minimal cytogenetic response; MCy. R; major cytogenetic response; MMR, major molecular response; MR, molecular response; PCy. R, partial cytogenetic response; Ph+, Philadelphia chromosome positive; WBC, white blood cell. 1. Baccarani M, et al. Mediterr J Hematol Infect Dis. 2014; 6: e 2014005; 2. Cross N, et al. Leukemia. 2015; 29: 999– 1003.

Available TKIs TKI Indication 1– 5 Imatinib • • Adult and paediatric newly diagnosed Ph+ CML for whom BM transplantation is not considered for first line treatment Adult and paediatric CP CML after failure of interferon alpha therapy, or AP /BP CML Adult and paediatric newly diagnosed Ph+ ALL integrated with chemotherapy Adult relapsed or refractory Ph+ ALL as monotherapy Dasatinib • • Adult and paediatric newly diagnosed Ph+ CP CML Adult CP , AP or BP CML after resistance/intolerance to prior therapy, including imatinib Paediatric CP CML after resistance/intolerance to prior therapy, including imatinib Adult Ph+ ALL and lymphoid BP CML with resistance/intolerance to prior therapy Nilotinib • Adult and paediatric newly diagnosed Ph+ CP CML • Adults with CP /AP CML with resistance/intolerance to prior therapy including imatinib o Efficacy data in patients with CML in blast crisis are not available • Paediatric patients with Ph+ CP CML with resistance/intolerance to prior therapy including imatinib Bosutinib • Adult newly diagnosed Ph+ CP CML • Adult CP , AP or BP CML, previously treated with one or more TKI(s) and for whom imatinib, nilotinib and dasatinib are not considered appropriate treatment options Ponatinib • Adult CP , AP or BP CML resistant/intolerant to dasatinib or nilotinib and for whom subsequent treatment with imatinib is not clinically appropriate, or who have the T 315 I mutation • Adult Ph+ ALL resistant/intolerant to dasatinib and for whom subsequent treatment with imatinib is not clinically appropriate, or who have the T 315 I mutation AP, accelerated phase; BM, bone marrow; BP, blast phase; CP, chronic phase; Ph+, Philadelphia chromosome positive; TKI, tyrosine kinase inhibitor. 1. Glivec® (imatinib) – Summary of Product Characteristics. Novartis Europharm Limited 2017; 2. Sprycel® (dasatinib) – Summary of Product Characteristics. Bristol Myers Squibb Pharmaceuticals Limited 2018; 3. Tasigna® (nilotinib) – Summary of Product Characteristics. Novartis Pharmaceuticals UK Ltd 2018; 4. Bosulif® (bosutinib) – Summary of Product Characteristics. Pfizer Limited 2018; 5. Iclusig® (ponatinib) – Summary of Product Characteristics. Incyte Biosciences UK Ltd 2018. The Summary of Product Characteristics are available at http: //www. ema. europa. eu.

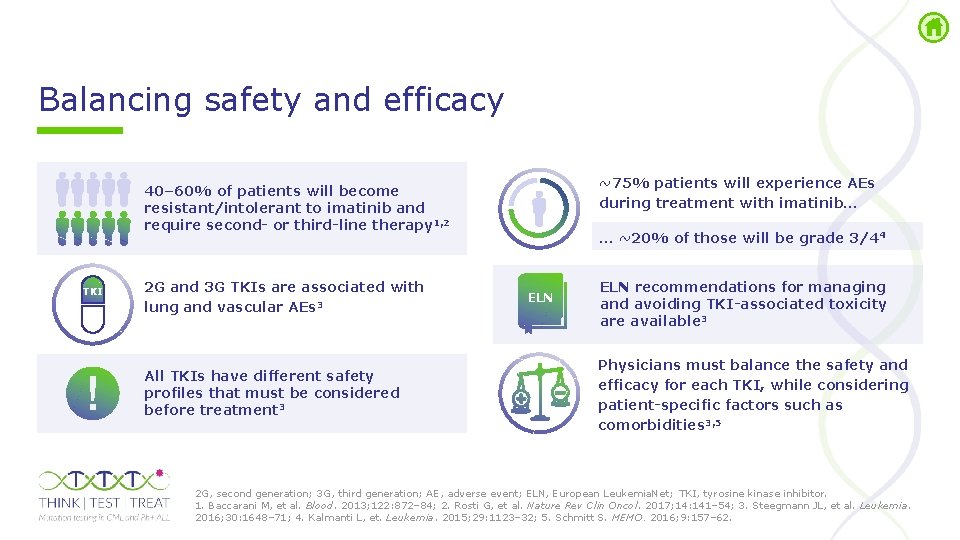
Balancing safety and efficacy ~75% patients will experience AEs during treatment with imatinib… 40– 60% of patients will become resistant/intolerant to imatinib and require second- or third-line therapy 1, 2 TKI 2 G and 3 G TKIs are associated with lung and vascular AEs 3 All TKIs have different safety profiles that must be considered before treatment 3 … ~20% of those will be grade 3/44 ELN recommendations for managing and avoiding TKI-associated toxicity are available 3 Physicians must balance the safety and efficacy for each TKI, while considering patient-specific factors such as comorbidities 3, 5 2 G, second generation; 3 G, third generation; AE, adverse event; ELN, European Leukemia. Net; TKI, tyrosine kinase inhibitor. 1. Baccarani M, et al. Blood. 2013; 122: 872– 84; 2. Rosti G, et al. Nature Rev Clin Oncol. 2017; 14: 141– 54; 3. Steegmann JL, et al. Leukemia. 2016; 30: 1648– 71; 4. Kalmanti L, et. Leukemia. 2015; 29: 1123– 32; 5. Schmitt S. MEMO. 2016; 9: 157– 62.

Balancing safety and efficacy 1 Prevent disease progression from CP- to AP- and BP-CML and reduce the risk of death from leukaemia 1 • Concerns about AEs should not compromise the main objective of CML treatment: preventing disease progression • Both safety profile and efficacy should be considered to ensure the optimum response rates for CML patients AE, adverse event; AP, accelerated phase; BP, blast phase; CP, chronic phase. 1. Steegmann JL, et al. Leukemia. 2016; 30: 1648– 71.

Molecular monitoring of BCR-ABL 1 transcript levels in CML

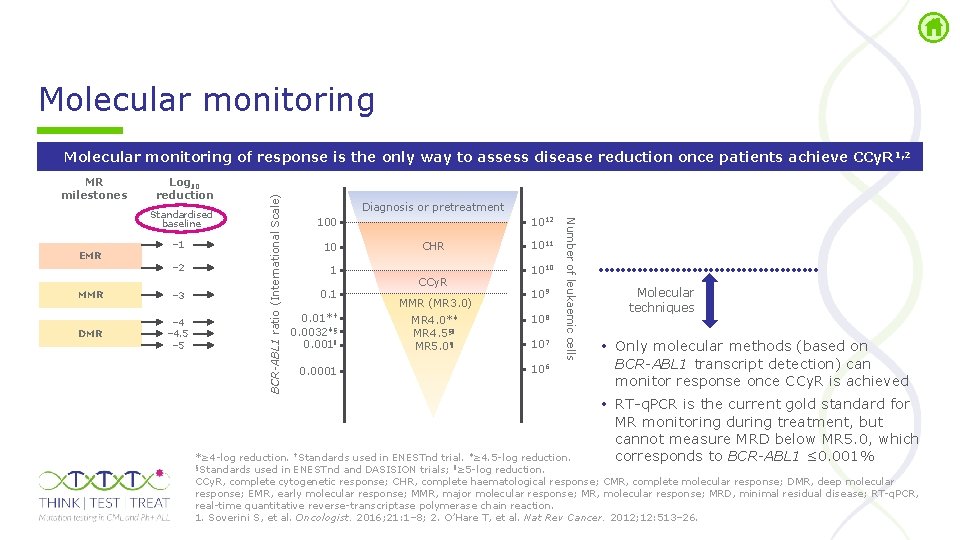
Molecular monitoring MR milestones Log 10 reduction EMR – 1 – 2 MMR – 3 DMR – 4. 5 – 5 Diagnosis or pretreatment 1012 100 10 1 0. 01*† 0. 0032‡§ 0. 001ǁ 0. 0001 CHR CCy. R MMR (MR 3. 0) MR 4. 0*‡ MR 4. 5§ǁ MR 5. 0¶ 1011 1010 109 108 107 106 Number of leukaemic cells Standardised baseline BCR-ABL 1 ratio (International Scale) Molecular monitoring of response is the only way to assess disease reduction once patients achieve CCy. R 1, 2 Molecular techniques • Only molecular methods (based on BCR-ABL 1 transcript detection) can monitor response once CCy. R is achieved • RT q. PCR is the current gold standard for MR monitoring during treatment, but cannot measure MRD below MR 5. 0, which corresponds to BCR-ABL 1 ≤ 0. 001% *≥ 4 log reduction. †Standards used in ENESTnd trial. ‡≥ 4. 5 log reduction. §Standards used in ENESTnd and DASISION trials; ǁ≥ 5 log reduction. CCy. R, complete cytogenetic response; CHR, complete haematological response; CMR, complete molecular response; DMR, deep molecular response; EMR, early molecular response; MMR, major molecular response; MR, molecular response; MRD, minimal residual disease; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. 1. Soverini S, et al. Oncologist. 2016; 21: 1– 8; 2. O’Hare T, et al. Nat Rev Cancer. 2012; 12: 513– 26.

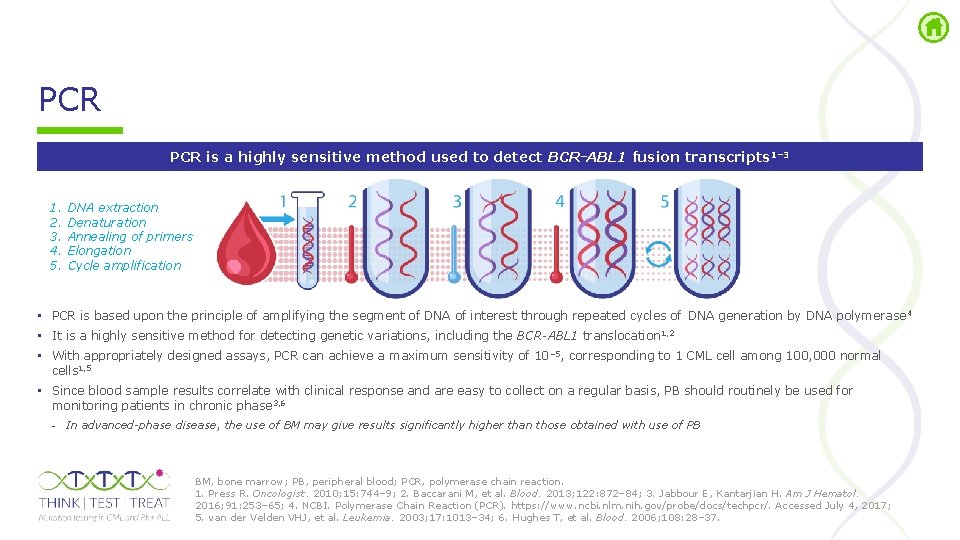
PCR is a highly sensitive method used to detect BCR-ABL 1 fusion transcripts 1– 3 1. 2. 3. 4. 5. DNA extraction Denaturation Annealing of primers Elongation Cycle amplification • PCR is based upon the principle of amplifying the segment of DNA of interest through repeated cycles of DNA generation by DNA polymerase 4 • It is a highly sensitive method for detecting genetic variations, including the BCR-ABL 1 translocation 1, 2 • With appropriately designed assays, PCR can achieve a maximum sensitivity of 10 – 5, corresponding to 1 CML cell among 100, 000 normal cells 1, 5 • Since blood sample results correlate with clinical response and are easy to collect on a regular basis, PB should routinely be used for monitoring patients in chronic phase 3, 6 - In advanced-phase disease, the use of BM may give results significantly higher than those obtained with use of PB BM, bone marrow; PB, peripheral blood; PCR, polymerase chain reaction. 1. Press R. Oncologist. 2010; 15: 744– 9; 2. Baccarani M, et al. Blood. 2013; 122: 872– 84; 3. Jabbour E, Kantarjian H. Am J Hematol. 2016; 91: 253– 65; 4. NCBI. Polymerase Chain Reaction (PCR). https: //www. ncbi. nlm. nih. gov/probe/docs/techpcr/. Accessed July 4, 2017; 5. van der Velden VHJ, et al. Leukemia. 2003; 17: 1013– 34; 6. Hughes T, et al. Blood. 2006; 108: 28– 37.

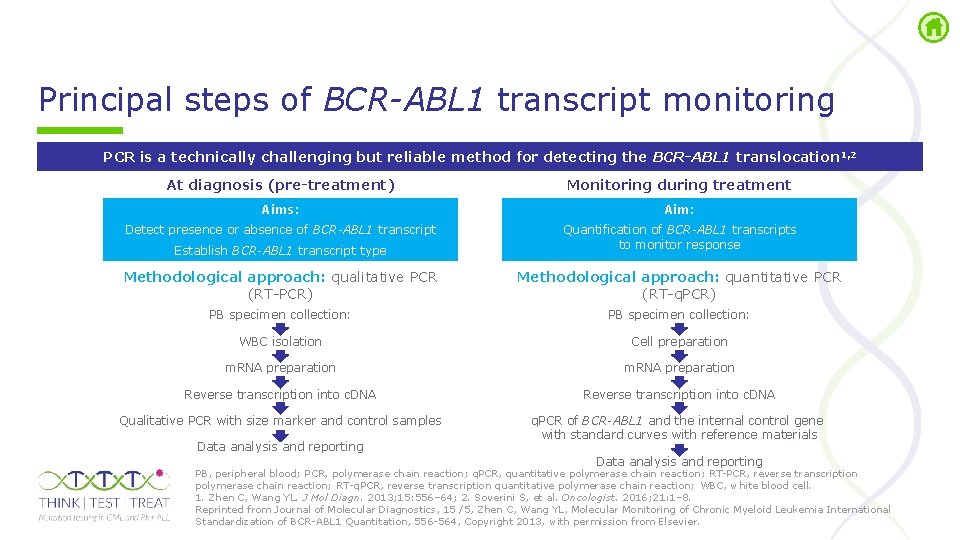
Principal steps of BCR-ABL 1 transcript monitoring PCR is a technically challenging but reliable method for detecting the BCR-ABL 1 translocation 1, 2 At diagnosis (pre-treatment) Monitoring during treatment Aims: Aim: Detect presence or absence of BCR-ABL 1 transcript Establish BCR-ABL 1 transcript type Quantification of BCR-ABL 1 transcripts to monitor response Methodological approach: qualitative PCR (RT PCR) Methodological approach: quantitative PCR (RT q. PCR) PB specimen collection: WBC isolation Cell preparation m. RNA preparation Reverse transcription into c. DNA Qualitative PCR with size marker and control samples q. PCR of BCR-ABL 1 and the internal control gene with standard curves with reference materials Data analysis and reporting PB, peripheral blood; PCR, polymerase chain reaction; q. PCR, quantitative polymerase chain reaction; RT PCR, reverse transcription polymerase chain reaction; RT q. PCR, reverse transcription quantitative polymerase chain reaction; WBC, white blood cell. 1. Zhen C, Wang YL. J Mol Diagn. 2013; 15: 556– 64; 2. Soverini S, et al. Oncologist. 2016; 21: 1– 8. Reprinted from Journal of Molecular Diagnostics, 15 /5, Zhen C, Wang YL, Molecular Monitoring of Chronic Myeloid Leukemia International Standardization of BCR ABL 1 Quantitation, 556 564, Copyright 2013, with permission from Elsevier.

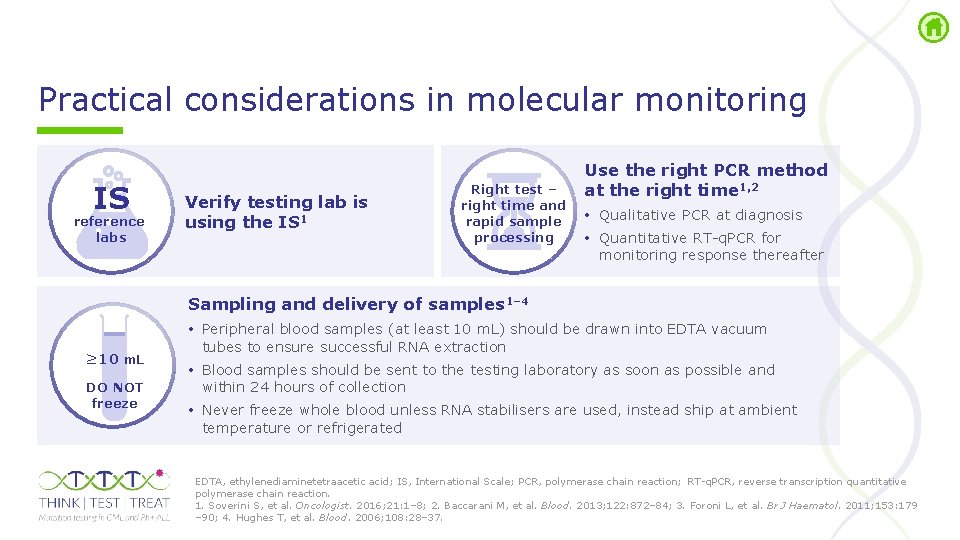
Practical considerations in molecular monitoring IS reference labs Verify testing lab is using the IS 1 Right test – right time and rapid sample processing Use the right PCR method at the right time 1, 2 • Qualitative PCR at diagnosis • Quantitative RT q. PCR for monitoring response thereafter Sampling and delivery of samples 1– 4 ≥ 10 m. L DO NOT freeze • Peripheral blood samples (at least 10 m. L) should be drawn into EDTA vacuum tubes to ensure successful RNA extraction • Blood samples should be sent to the testing laboratory as soon as possible and within 24 hours of collection • Never freeze whole blood unless RNA stabilisers are used, instead ship at ambient temperature or refrigerated EDTA, ethylenediaminetetraacetic acid; IS, International Scale; PCR, polymerase chain reaction; RT q. PCR, reverse transcription quantitative polymerase chain reaction. 1. Soverini S, et al. Oncologist. 2016; 21: 1– 8; 2. Baccarani M, et al. Blood. 2013; 122: 872– 84; 3. Foroni L, et al. Br J Haematol. 2011; 153: 179 – 90; 4. Hughes T, et al. Blood. 2006; 108: 28– 37.

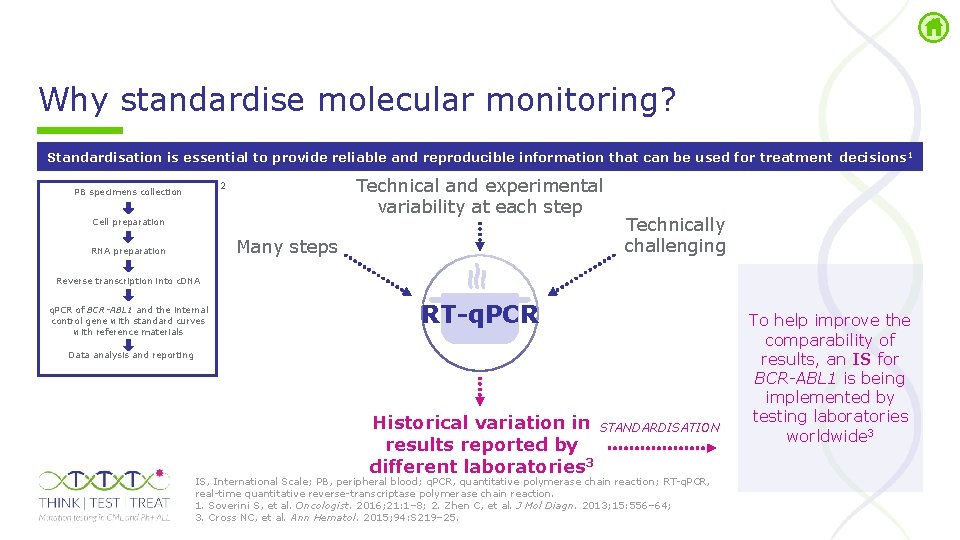
Why standardise molecular monitoring? Standardisation is essential to provide reliable and reproducible information that can be used for treatment decisions 1 Technical and experimental variability at each step 2 PB specimens collection Cell preparation Many steps RNA preparation Technically challenging Reverse transcription into c. DNA q. PCR of BCR-ABL 1 and the internal control gene with standard curves with reference materials RT-q. PCR Data analysis and reporting Historical variation in results reported by different laboratories 3 STANDARDISATION IS, International Scale; PB, peripheral blood; q. PCR, quantitative polymerase chain reaction; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. 1. Soverini S, et al. Oncologist. 2016; 21: 1– 8; 2. Zhen C, et al. J Mol Diagn. 2013; 15: 556– 64; 3. Cross NC, et al. Ann Hematol. 2015; 94: S 219– 25. To help improve the comparability of results, an IS for BCR-ABL 1 is being implemented by testing laboratories worldwide 3

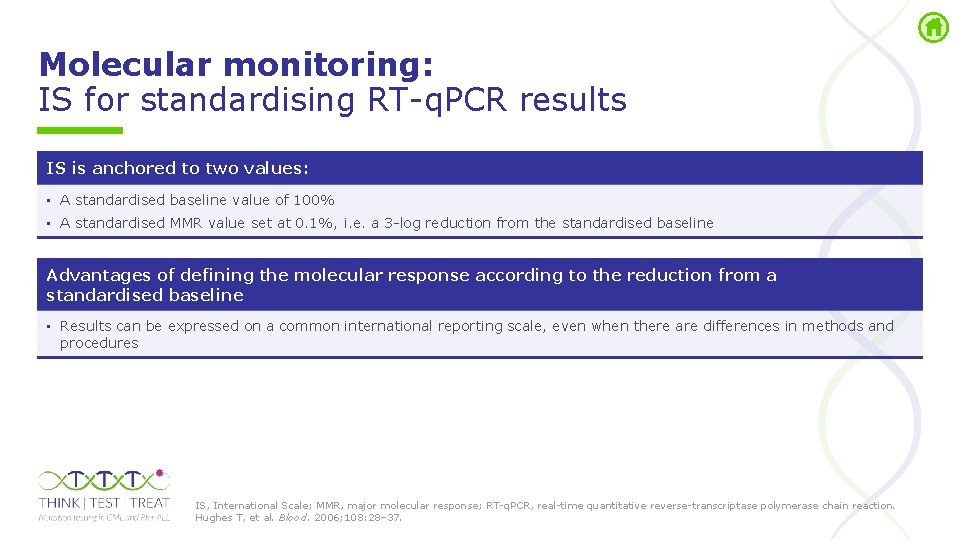
Molecular monitoring: IS for standardising RT q. PCR results IS is anchored to two values: • A standardised baseline value of 100% • A standardised MMR value set at 0. 1%, i. e. a 3 log reduction from the standardised baseline Advantages of defining the molecular response according to the reduction from a standardised baseline • Results can be expressed on a common international reporting scale, even when there are differences in methods and procedures IS, International Scale; MMR, major molecular response; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. Hughes T, et al. Blood. 2006; 108: 28– 37.

Molecular monitoring: Conversion factor Conversion to the IS is achieved by the application of laboratory-specific conversion factors: • Variation between BCR-ABL 1 testing labs can be substantially reduced by aligning data on an international reporting scale using method specific conversion factors 1 • Specific conversion factors are required for each laboratory due to differences in local instruments and protocols contributing to variation in the reported values 1 • Commercial kits and reagents have been calibrated to the World Health Organization International Genetic Reference Panel, which consists of a four level panel of e 14 a 2 positive lyophilised cell line dilutions 2 Each level has an assigned IS value, which was obtained by repeated testing of each sample IS, International Scale. 1. Brandford S, et al. Blood. 2008; 112: 3330– 8; 2. White HE, et al. Blood. 2010; 116: e 111– 7.

Optimal timing of molecular monitoring Qualitative RT-PCR Quantitative RT-q. PCR • At diagnosis to assess the transcript type • Peripheral blood adequate • Every 3 months until MMR achieved • Every 3 to 6 months afterward • Peripheral blood adequate MMR, major molecular response; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction; RT PCR, reverse transcription polymerase chain reaction. Soverini S, et al. Oncologist. 2016; 21: 1– 8.

Monitoring recommendations for patients in TFR 1 Availability of high quality internationally standardised, accurate, sensitive RT PCR laboratory 2 PCR test results available within 4 weeks 3 PCR tests every 4– 6 weeks when required 4 Regular follow up to ensure rapid intervention in response to rising BCR-ABL 1 transcript levels PCR; polymerase chain reaction; RT PCR, reverse transcription polymerase chain reaction; TFR, treatment free remission. Hochhaus A, et al. Ann Oncol. 2017; 28: iv 41– 51.

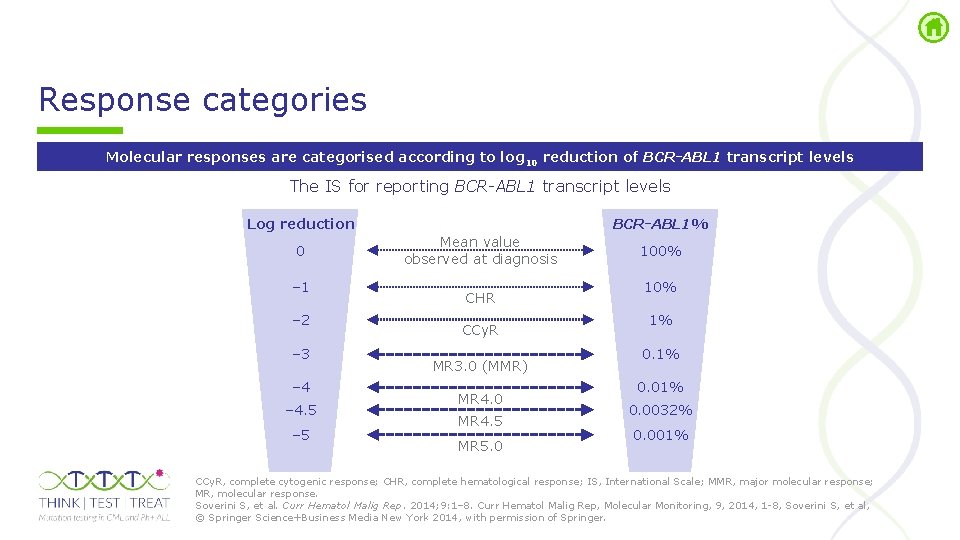
Response categories Molecular responses are categorised according to log 10 reduction of BCR-ABL 1 transcript levels The IS for reporting BCR-ABL 1 transcript levels Log reduction 0 – 1 – 2 – 3 – 4. 5 – 5 BCR-ABL 1% Mean value observed at diagnosis CHR CCy. R MR 3. 0 (MMR) MR 4. 0 MR 4. 5 MR 5. 0 100% 1% 0. 0032% 0. 001% CCy. R, complete cytogenic response; CHR, complete hematological response; IS, International Scale; MMR, major molecular response; MR, molecular response. Soverini S, et al. Curr Hematol Malig Rep. 2014; 9: 1– 8. Curr Hematol Malig Rep, Molecular Monitoring, 9, 2014, 1 8, Soverini S, et al, © Springer Science+Business Media New York 2014, with permission of Springer.

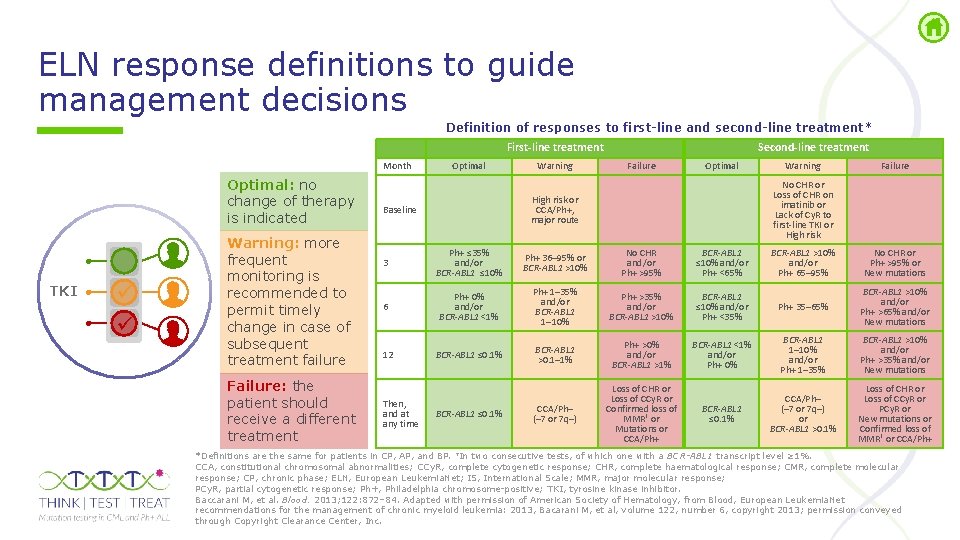
ELN response definitions to guide management decisions Definition of responses to first-line and second-line treatment* First-line treatment Month Optimal: no change of therapy is indicated TKI Warning: more frequent monitoring is recommended to permit timely change in case of subsequent treatment failure Failure: the patient should receive a different treatment Optimal Warning Second-line treatment Failure Optimal 3 Ph+ ≤ 35% and/or BCR-ABL 1 ≤ 10% Ph+ 36– 95% or BCR-ABL 1 >10% No CHR and/or Ph+ >95% BCR-ABL 1 ≤ 10% and/or Ph+ <65% 6 Ph+ 0% and/or BCR-ABL 1 <1% Ph+ 1– 35% and/or BCR-ABL 1 1– 10% Ph+ >35% and/or BCR-ABL 1 >10% BCR-ABL 1 ≤ 0. 1% BCR-ABL 1 >0. 1– 1% Ph+ >0% and/or BCR-ABL 1 >1% CCA/Ph– (– 7 or 7 q–) Loss of CHR or Loss of CCy. R or Confirmed loss of MMR† or Mutations or CCA/Ph+ 12 Then, and at any time BCR-ABL 1 ≤ 0. 1% Failure No CHR or Loss of CHR on imatinib or Lack of Cy. R to first-line TKI or High risk or CCA/Ph+, major route Baseline Warning BCR-ABL 1 >10% and/or Ph+ 65– 95% No CHR or Ph+ >95% or New mutations BCR-ABL 1 ≤ 10% and/or Ph+ <35% Ph+ 35– 65% BCR-ABL 1 >10% and/or Ph+ >65% and/or New mutations BCR-ABL 1 <1% and/or Ph+ 0% BCR-ABL 1 1– 10% and/or Ph+ 1– 35% BCR-ABL 1 >10% and/or Ph+ >35% and/or New mutations BCR-ABL 1 ≤ 0. 1% CCA/Ph– (– 7 or 7 q–) or BCR-ABL 1 >0. 1% Loss of CHR or Loss of CCy. R or PCy. R or New mutations or Confirmed loss of MMR† or CCA/Ph+ *Definitions are the same for patients in CP, AP, and BP. †In two consecutive tests, of which one with a BCR-ABL 1 transcript level ≥ 1%. CCA, constitutional chromosomal abnormalities; CCy. R, complete cytogenetic response; CHR, complete haematological response; CMR, complete molecular response; CP, chronic phase; ELN, European Leukemia. Net; IS, International Scale; MMR, major molecular response; PCy. R, partial cytogenetic response; Ph+, Philadelphia chromosome positive; TKI, tyrosine kinase inhibitor. Baccarani M, et al. Blood. 2013; 122: 872– 84. Adapted with permission of American Society of Hematology, from Blood, European Leukemia. Net recommendations for the management of chronic myeloid leukemia: 2013, Bacarani M, et al, volume 122, number 6, copyright 2013; permission conveyed through Copyright Clearance Center, Inc.

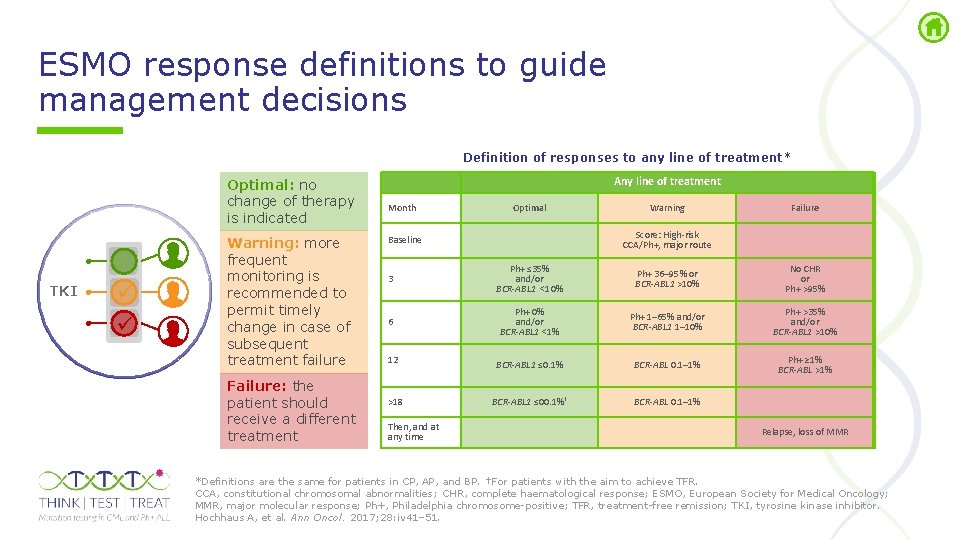
ESMO response definitions to guide management decisions Definition of responses to any line of treatment* Optimal: no change of therapy is indicated TKI Warning: more frequent monitoring is recommended to permit timely change in case of subsequent treatment failure Failure: the patient should receive a different treatment Any line of treatment Month Optimal Warning Failure Score: High-risk CCA/Ph+, major route Baseline 3 Ph+ ≤ 35% and/or BCR-ABL 1 <10% Ph+ 36– 95% or BCR-ABL 1 >10% No CHR or Ph+ >95% 6 Ph+ 0% and/or BCR-ABL 1 <1% Ph+ 1– 65% and/or BCR-ABL 1 1– 10% Ph+ >35% and/or BCR-ABL 1 >10% 12 BCR-ABL 1 ≤ 0. 1% BCR-ABL 0. 1– 1% Ph+ ≥ 1% BCR-ABL >1% >18 BCR-ABL 1 ≤ 00. 1%† BCR-ABL 0. 1– 1% Then, and at any time Relapse, loss of MMR *Definitions are the same for patients in CP, AP, and BP. †For patients with the aim to achieve TFR. CCA, constitutional chromosomal abnormalities; CHR, complete haematological response; ESMO, European Society for Medical Oncology; MMR, major molecular response; Ph+, Philadelphia chromosome positive; TFR, treatment free remission; TKI, tyrosine kinase inhibitor. Hochhaus A, et al. Ann Oncol. 2017; 28: iv 41– 51.

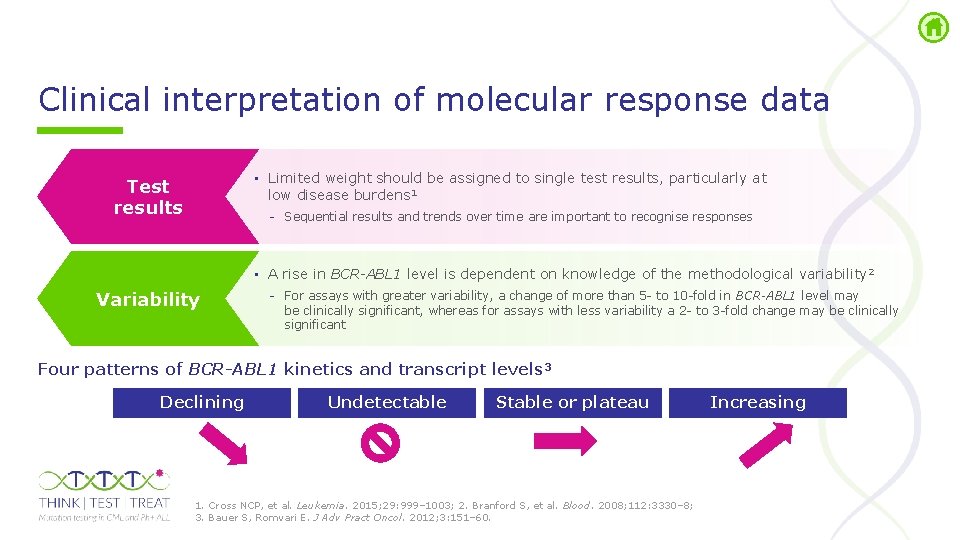
Clinical interpretation of molecular response data • Limited weight should be assigned to single test results, particularly at low disease burdens 1 Test results Sequential results and trends over time are important to recognise responses • A rise in BCR-ABL 1 level is dependent on knowledge of the methodological variability 2 Variability For assays with greater variability, a change of more than 5 to 10 fold in BCR-ABL 1 level may be clinically significant, whereas for assays with less variability a 2 to 3 fold change may be clinically significant Four patterns of BCR-ABL 1 kinetics and transcript levels 3 Declining Undetectable Stable or plateau 1. Cross NCP, et al. Leukemia. 2015; 29: 999– 1003; 2. Branford S, et al. Blood. 2008; 112: 3330– 8; 3. Bauer S, Romvari E. J Adv Pract Oncol. 2012; 3: 151– 60. Increasing

Possible reasons for increasing BCR-ABL 1 levels Reasons Quality control • Resistance, non adherence, drug–drug or drug–food interactions, assay error, different laboratory or different laboratory procedure, or impending relapse 1 • RT q. PCR assays generate variable BCR-ABL 1 values (inherent variability of molecular assays), so robust quality control process to assess the degree of variability and to recognise failed technical procedures is an essential component of these molecular techniques 2 If a trend of rising BCR-ABL 1 transcripts emerges, signalling loss of MMR, a change in treatment strategy may be warranted 1 MMR, major molecular response; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. 1. Bauer S, Romvari E. J Adv Pract Oncol. 2012; 3: 151– 60; 2. Branford S. Hematology Am Soc Hematol Educ Program. 2016; 2016: 156– 63.

Communicating with patients 1 Patients should be evaluated for, and educated as to the possible reasons for, a rising BCR-ABL 1 level such as: non adherence, drug–drug or drug–food interactions, assay error, different laboratory or different laboratory procedure, or impending relapse 2 Molecular monitoring results can be used to educate patients, similar to the use of monitoring blood pressure in patients with hypertension, or glucose readings in patients with diabetes 3 BCR-ABL 1 results reflect response to therapy and may signal when a change in treatment is needed 4 Patients should know that an increase in the BCR-ABL 1 transcript number is not necessarily an immediate cause for concern Bauer S, Romvari E. J Adv Pract Oncol. 2012; 3: 151– 60. 5

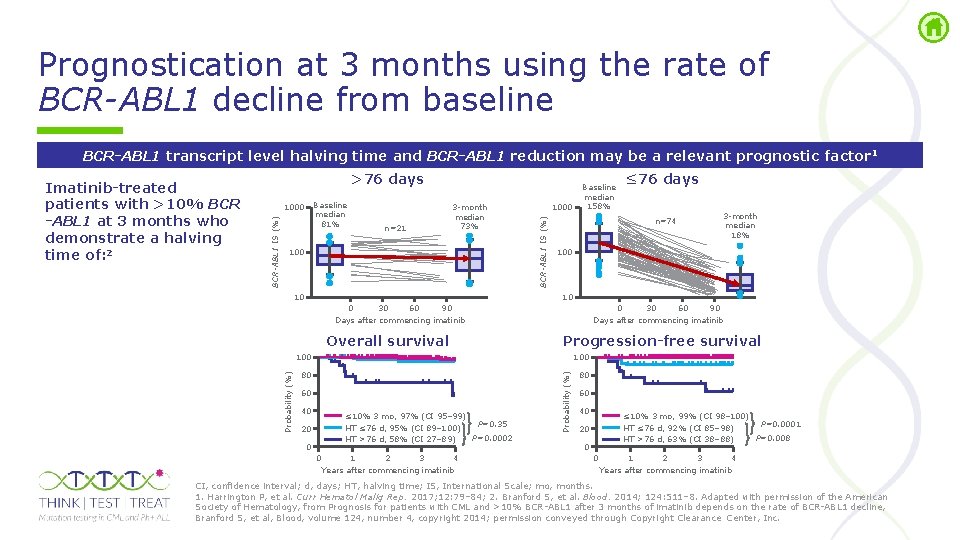
Prognostication at 3 months using the rate of BCR-ABL 1 decline from baseline BCR-ABL 1 transcript level halving time and BCR-ABL 1 reduction may be a relevant prognostic factor 1 >76 days n=21 3 month median 73% 100 10 1000 BCR-ABL 1 IS (%) Baseline median 81% 1000 BCR-ABL 1 IS (%) Imatinib-treated patients with >10% BCR -ABL 1 at 3 months who demonstrate a halving time of: 2 Baseline median 158% n=74 3 month median 18% 100 10 0 30 60 90 Days after commencing imatinib Overall survival 0 30 60 90 Days after commencing imatinib Progression-free survival 100 80 80 60 40 ≤ 10% 3 mo, 97% (CI 95– 99) P=0. 35 HT ≤ 76 d, 95% (CI 89– 100) P=0. 0002 HT >76 d, 58% (CI 27– 89) 20 0 0 1 2 3 4 Years after commencing imatinib Probability (%) ≤ 76 days 60 40 ≤ 10% 3 mo, 99% (CI 98– 100) P=0. 0001 HT ≤ 76 d, 92% (CI 85– 98) P=0. 008 HT >76 d, 63% (CI 38– 88) 20 0 0 1 2 3 4 Years after commencing imatinib CI, confidence interval; d, days; HT, halving time; IS, International Scale; mo, months. 1. Harrington P, et al. Curr Hematol Malig Rep. 2017; 12: 79– 84; 2. Branford S, et al. Blood. 2014; 124: 511– 8. Adapted with permission of the American Society of Hematology, from Prognosis for patients with CML and >10% BCR ABL 1 after 3 months of imatinib depends on the rate of BCR ABL 1 decline, Branford S, et al, Blood, volume 124, number 4, copyright 2014; permission conveyed through Copyright Clearance Center, Inc.

BCR-ABL 1 mutation testing in CML

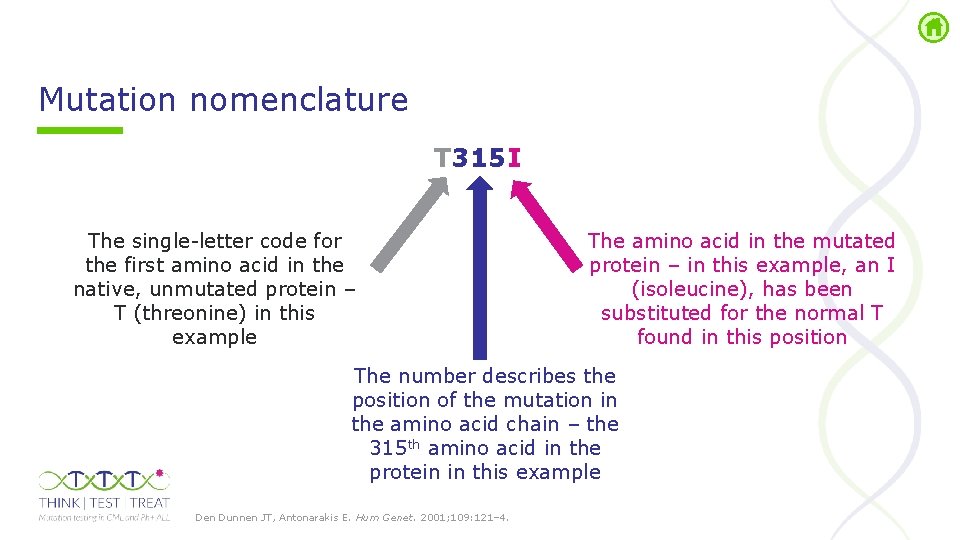
Mutation nomenclature T 315 I The single letter code for the first amino acid in the native, unmutated protein – T (threonine) in this example The amino acid in the mutated protein – in this example, an I (isoleucine), has been substituted for the normal T found in this position The number describes the position of the mutation in the amino acid chain – the 315 th amino acid in the protein in this example Den Dunnen JT, Antonarakis E. Hum Genet. 2001; 109: 121– 4.

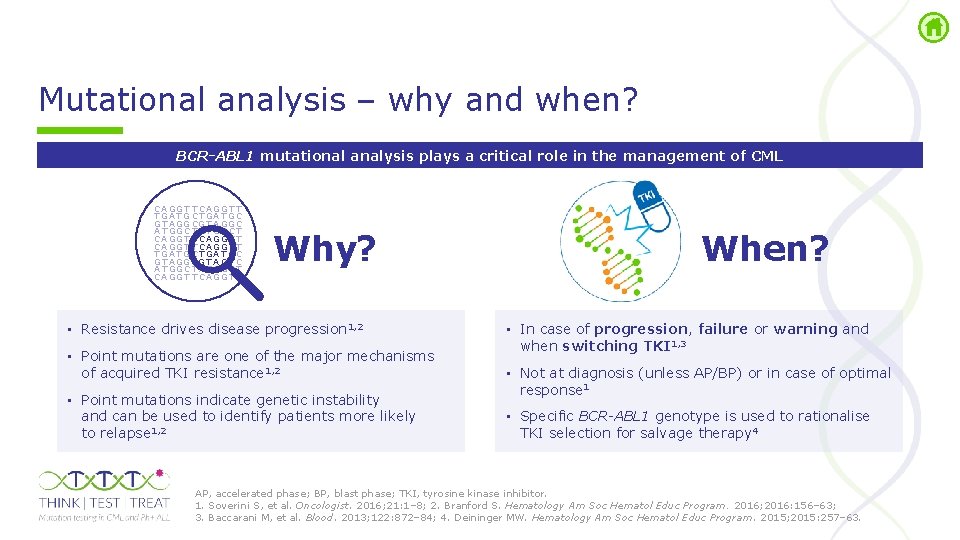
Mutational analysis – why and when? BCR-ABL 1 mutational analysis plays a critical role in the management of CML C A GG T T C A G G T T T GA T G C T GA T GC G T A GG C A T GG C T A T G G C T C A GG T T C A G G T T T GA T G C T GA T GC G T A GG CG T A G T C A T GG C T A T G G C T C A GG T T C A G G T T Why? • Resistance drives disease progression 1, 2 • Point mutations are one of the major mechanisms of acquired TKI resistance 1, 2 • Point mutations indicate genetic instability and can be used to identify patients more likely to relapse 1, 2 When? • In case of progression, failure or warning and when switching TKI 1, 3 • Not at diagnosis (unless AP/BP) or in case of optimal response 1 • Specific BCR-ABL 1 genotype is used to rationalise TKI selection for salvage therapy 4 AP, accelerated phase; BP, blast phase; TKI, tyrosine kinase inhibitor. 1. Soverini S, et al. Oncologist. 2016; 21: 1– 8; 2. Branford S. Hematology Am Soc Hematol Educ Program. 2016; 2016: 156– 63; 3. Baccarani M, et al. Blood. 2013; 122: 872– 84; 4. Deininger MW. Hematology Am Soc Hematol Educ Program. 2015; 2015: 257– 63.

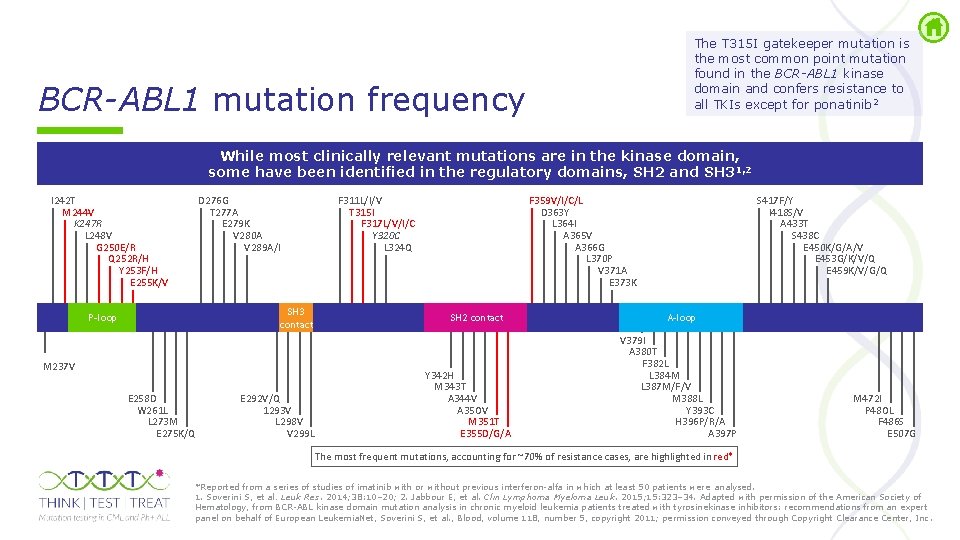
The T 315 I gatekeeper mutation is the most common point mutation found in the BCR-ABL 1 kinase domain and confers resistance to all TKIs except for ponatinib 2 BCR-ABL 1 mutation frequency While most clinically relevant mutations are in the kinase domain, some have been identified in the regulatory domains, SH 2 and SH 31, 2 I 242 T M 244 V K 247 R L 248 V G 250 E/R Q 252 R/H Y 253 F/H E 255 K/V D 276 G T 277 A E 279 K V 280 A V 289 A/I SH 3 contact P-loop M 237 V E 258 D W 261 L L 273 M E 275 K/Q E 292 V/Q 1293 V L 298 V V 299 L F 311 L/I/V T 315 I F 317 L/V/I/C Y 320 C L 324 Q F 359 V/I/C/L D 363 Y L 364 I A 365 V A 366 G L 370 P V 371 A E 373 K SH 2 contact Y 342 H M 343 T A 344 V A 35 OV M 351 T E 355 D/G/A S 417 F/Y I 418 S/V A 433 T S 438 C E 450 K/G/A/V E 453 G/K/V/Q E 459 K/V/G/Q A-loop V 379 I A 380 T F 382 L L 384 M L 387 M/F/V M 388 L Y 393 C H 396 P/R/A A 397 P M 472 I P 48 OL F 486 S E 507 G The most frequent mutations, accounting for ~70% of resistance cases, are highlighted in red* *Reported from a series of studies of imatinib with or without previous interferon alfa in which at least 50 patients were analysed. 1. Soverini S, et al. Leuk Res. 2014; 38: 10– 20; 2. Jabbour E, et al. Clin Lymphoma Myeloma Leuk. 2015; 15: 323– 34. Adapted with permission of the American Society of Hematology, from BCR ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosinekinase inhibitors: recommendations from an expert panel on behalf of European Leukemia. Net, Soverini S, et al. , Blood, volume 118, number 5, copyright 2011; permission conveyed through Copyright Clearance Center, Inc.

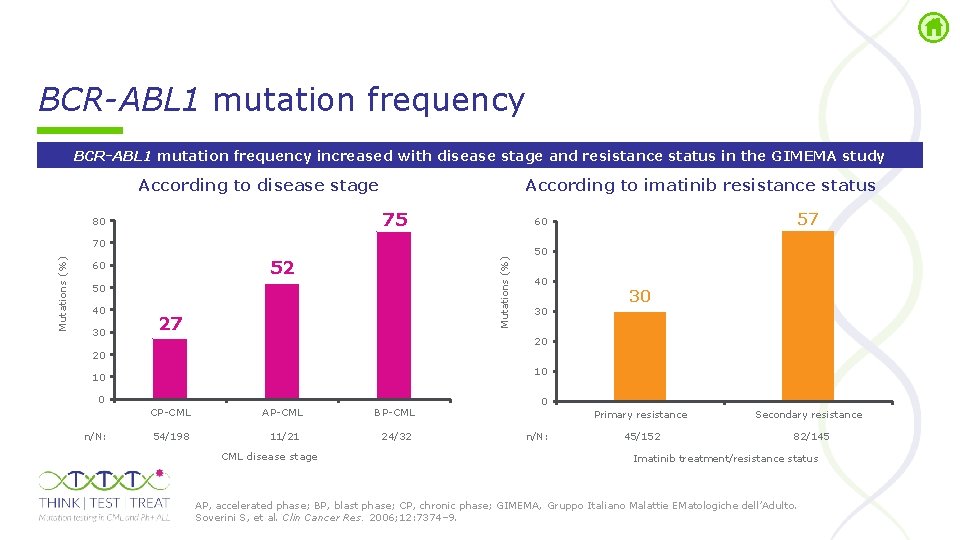
BCR-ABL 1 mutation frequency increased with disease stage and resistance status in the GIMEMA study According to disease stage According to imatinib resistance status 75 80 52 Mutations (%) 70 60 50 40 30 27 57 60 50 40 30 30 20 20 10 10 0 n/N: CP CML AP CML BP CML 54/198 11/21 24/32 CML disease stage 0 Primary resistance n/N: 45/152 Secondary resistance 82/145 Imatinib treatment/resistance status AP, accelerated phase; BP, blast phase; CP, chronic phase; GIMEMA, Gruppo Italiano Malattie EMatologiche dell’Adulto. Soverini S, et al. Clin Cancer Res. 2006; 12: 7374– 9.

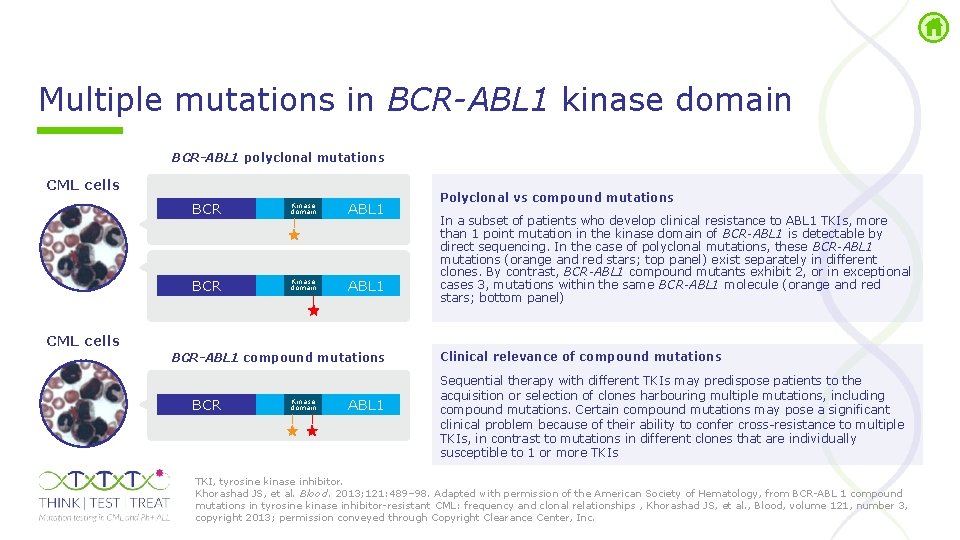
Multiple mutations in BCR-ABL 1 kinase domain BCR-ABL 1 polyclonal mutations CML cells BCR Kinase domain ABL 1 CML cells BCR-ABL 1 compound mutations BCR Kinase domain ABL 1 Polyclonal vs compound mutations In a subset of patients who develop clinical resistance to ABL 1 TKIs, more than 1 point mutation in the kinase domain of BCR-ABL 1 is detectable by direct sequencing. In the case of polyclonal mutations, these BCR-ABL 1 mutations (orange and red stars; top panel) exist separately in different clones. By contrast, BCR-ABL 1 compound mutants exhibit 2, or in exceptional cases 3, mutations within the same BCR-ABL 1 molecule (orange and red stars; bottom panel) Clinical relevance of compound mutations Sequential therapy with different TKIs may predispose patients to the acquisition or selection of clones harbouring multiple mutations, including compound mutations. Certain compound mutations may pose a significant clinical problem because of their ability to confer cross resistance to multiple TKIs, in contrast to mutations in different clones that are individually susceptible to 1 or more TKIs TKI, tyrosine kinase inhibitor. Khorashad JS, et al. Blood. 2013; 121: 489– 98. Adapted with permission of the American Society of Hematology, from BCR ABL 1 compound mutations in tyrosine kinase inhibitor resistant CML: frequency and clonal relationships , Khorashad JS, et al. , Blood, volume 121, number 3, copyright 2013; permission conveyed through Copyright Clearance Center, Inc.

Compound mutations in BCR-ABL 1 kinase domain • Compound mutations are ≥ 2 mutations on the same BCR-ABL 1 allele 1, 2 - Can confer resistance to multiple TKIs 2 • Most compound mutations that include the T 315 I mutation are resistant to ALL TKIs 1 • Sanger (direct) sequencing is unable to definitively identify compound mutations 1 • NGS identifies compound mutations as long as the average read length exceeds the distance between the two single nucleotide variations 1 In an analysis of the frequency of multiple mutations among 1, 700 CML patients on TKI therapy, 11. 4% were shown to have ≥ 2 mutations • 70% of double mutations in the BCR ABL 1 kinase domain detected by direct sequencing are compound mutations 3 Using a more sensitive mutation assessment technique – NGS – no single or compound mutation was identified that consistently conferred primary and/or secondary resistance to ponatinib in CP-CML patients 1 NGS, next generation sequencing; TKI, tyrosine kinase inhibitor. 1. Deininger MW, et al. Blood. 2016; 127: 703– 12; 2. Zabriskie MS, et al. Cancer Cell. 2014; 26: 428– 42; 3. Khorashad JS, et al. Blood. 2013; 121: 489– 98.

Clinical relevance of low-level BCR-ABL 1 mutations in CML

Why test for low level mutations? Earlier detection and clinical relevance • Sensitive methods can detect mutations earlier than Sanger sequencing 1 • Although the clinical relevance of low level mutations has been debated, 2 there is increasing evidence that they have prognostic significance 3 May identify patients failing to achieve early and/or optimal response milestones Expansion of resistant subclones • Several studies have shown that low level mutations can expand cause resistance when treated with the wrong TKI 4– 9 • Monitoring kinetics of low level mutations may be more revealing than single time points and may inform interventions that aim to prevent resistant subclones from becoming dominant 7, 10 TKI, tyrosine kinase inhibitor. 1. Baer C, et al. Haematologica. 2016; 101: 830– 8; 2. Ai J, Tiu RV. Ther Adv Hematol. 2014; 5107– 20; 3. Kizilors A, et al. Lancet Haematol. 2019; 6: e 276– 84; 4. Machova PK, et al. J Cancer Res Clin Oncol. 2015; 141: 887– 99; 5. Parker WT, et al. J Clin Oncol. 2011; 29: 4250– 9; 6. Parker WT, et al. Br J Cancer. 2013; 109: 1593– 8; 7. Soverini S, et al. Blood. 2013; 122: 1634– 48; 8. Soverini S, et al. Oncotarget. 2016; 7: 21982– 90; 9. Soverini S, et al. BMC Cancer. 2016; 16: 572; 10. Preuner S, et al. Int J Mol Sci. 2016; 17: 642.

Definition of low level mutations • Low level mutations are below the detection limit of Sanger (direct) sequencing and are detected only with more sensitive methods 1 The detection limit of direct sequencing is ~20% Therefore, any mutation present at <20% BCR ABL 1 may be considered a low-level mutation • How low can we go? 2 Challenge for NGS is finding a true signal when mutation incidence is very low • Detection limit depends on technology used • PCR artefacts can be an issue and must be considered: error rates of 1– 3% limit the sensitivity of low-level mutations detectable by NGS, next generation sequencing; PCR, polymerase chain reaction. 1. de Lavallade H, Kizilors A. EMJ Oncol. 2016; 4: 86– 95; 2. Flaherty P, et al. Nucleic Acids Res. 2012; 40: e 2.

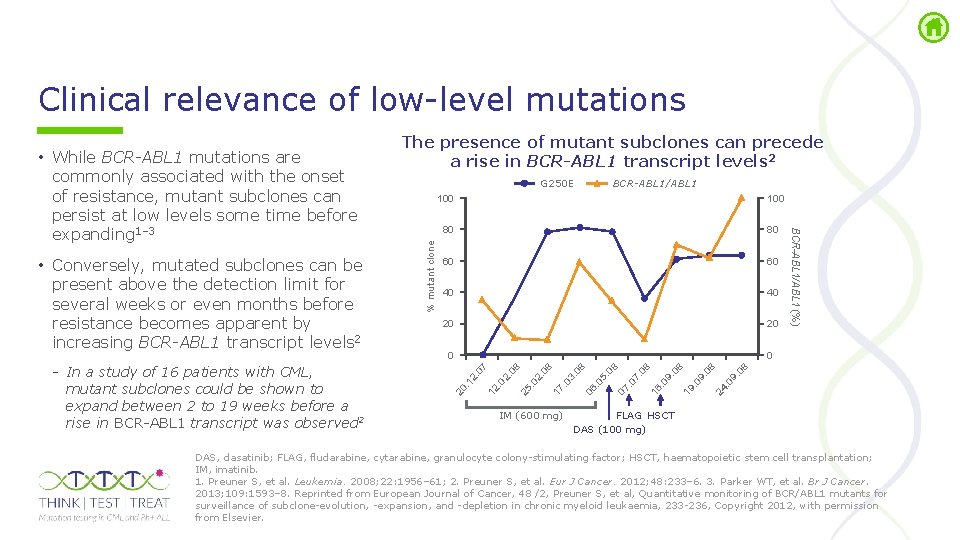
Clinical relevance of low level mutations BCR-ABL 1/ABL 1 40 40 20 20 0 0 9. . 0 08 24 9. . 0 19 9. . 0 16 7. . 0 07 5. . 0 06 . 0 3. 08 08 17 2. 25 . 0 2. 12 2. . 1 . 0 IM (600 mg) 08 60 08 80 08 100 07 100 20 In a study of 16 patients with CML, mutant subclones could be shown to expand between 2 to 19 weeks before a rise in BCR ABL 1 transcript was observed 2 % mutant clone • Conversely, mutated subclones can be present above the detection limit for several weeks or even months before resistance becomes apparent by increasing BCR-ABL 1 transcript levels 2 G 250 E BCR-ABL 1/ABL 1 (%) • While BCR-ABL 1 mutations are commonly associated with the onset of resistance, mutant subclones can persist at low levels some time before expanding 1– 3 The presence of mutant subclones can precede a rise in BCR-ABL 1 transcript levels 2 FLAG HSCT DAS (100 mg) DAS, dasatinib; FLAG, fludarabine, cytarabine, granulocyte colony stimulating factor; HSCT, haematopoietic stem cell transplantation; IM, imatinib. 1. Preuner S, et al. Leukemia. 2008; 22: 1956– 61; 2. Preuner S, et al. Eur J Cancer. 2012; 48: 233– 6. 3. Parker WT, et al. Br J Cancer. 2013; 109: 1593– 8. Reprinted from European Journal of Cancer, 48 /2, Preuner S, et al, Quantitative monitoring of BCR/ABL 1 mutants for surveillance of subclone evolution, expansion, and depletion in chronic myeloid leukaemia, 233 236, Copyright 2012, with permission from Elsevier.

Monitoring minimal residual disease in Ph+ ALL

MRD is an extraordinarily important prognosticator of outcome in ALL, regardless of cytogenetic subset Used to monitor residual leukaemic cells Increasing evidence for role of MRD status in determining: • Which patients should undergo allogeneic transplantation • Which patients should be considered for post-transplant therapies (especially in Ph+ ALL) MRD positivity remains a strong predictor of haematological relapse after allogeneic transplantation, even in the TKI era Ph+, Philadelphia chromosome positive; MRD, minimal residual disease; TKI, tyrosine kinase inhibitor. Leonard JT, Stock W. Biol Blood Marrow Transplant. 2016; 22: 1913– 4.

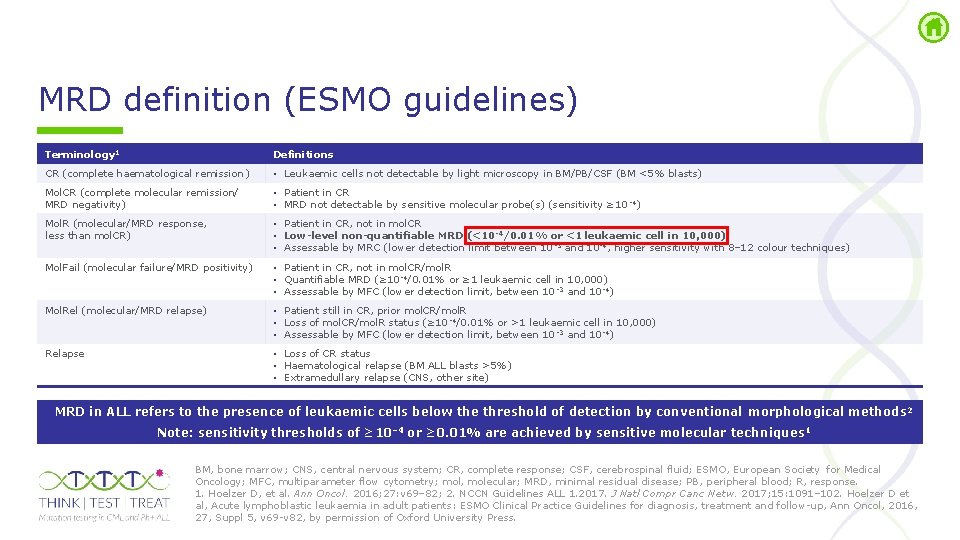
MRD definition (ESMO guidelines) Terminology 1 Definitions CR (complete haematological remission ) • Leukaemic cells not detectable by light microscopy in BM/PB/CSF (BM <5% blasts) Mol. CR (complete molecular remission/ MRD negativity) • Patient in CR • MRD not detectable by sensitive molecular probe(s) (sensitivity ≥ 10 4) Mol. R (molecular/MRD response, less than mol. CR) • Patient in CR, not in mol. CR • Low-level non-quantifiable MRD (<10 -4/0. 01% or <1 leukaemic cell in 10, 000) • Assessable by MRC (lower detection limit between 10 3 and 10 4, higher sensitivity with 8– 12 colour techniques) Mol. Fail (molecular failure/MRD positivity) • Patient in CR, not in mol. CR/mol. R • Quantifiable MRD (≥ 10 4/0. 01% or ≥ 1 leukaemic cell in 10, 000) • Assessable by MFC (lower detection limit, between 10 3 and 10 4) Mol. Rel (molecular/MRD relapse) • Patient still in CR, prior mol. CR/mol. R • Loss of mol. CR/mol. R status (≥ 10 4/0. 01% or >1 leukaemic cell in 10, 000) • Assessable by MFC (lower detection limit, between 10 3 and 10 4) Relapse • Loss of CR status • Haematological relapse (BM ALL blasts >5%) • Extramedullary relapse (CNS, other site) MRD in ALL refers to the presence of leukaemic cells below the threshold of detection by conventional morphological methods 2 Note: sensitivity thresholds of ≥ 10– 4 or ≥ 0. 01% are achieved by sensitive molecular techniques 1 BM, bone marrow; CNS, central nervous system; CR, complete response; CSF, cerebrospinal fluid; ESMO, European Society for Medical Oncology; MFC, multiparameter flow cytometry; mol, molecular; MRD, minimal residual disease; PB, peripheral blood; R, response. 1. Hoelzer D, et al. Ann Oncol. 2016; 27: v 69– 82; 2. NCCN Guidelines ALL 1. 2017. J Natl Compr Canc Netw. 2017; 15: 1091– 102. Hoelzer D et al, Acute lymphoblastic leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow up, Ann Oncol, 2016, 27, Suppl 5, v 69 v 82, by permission of Oxford University Press.

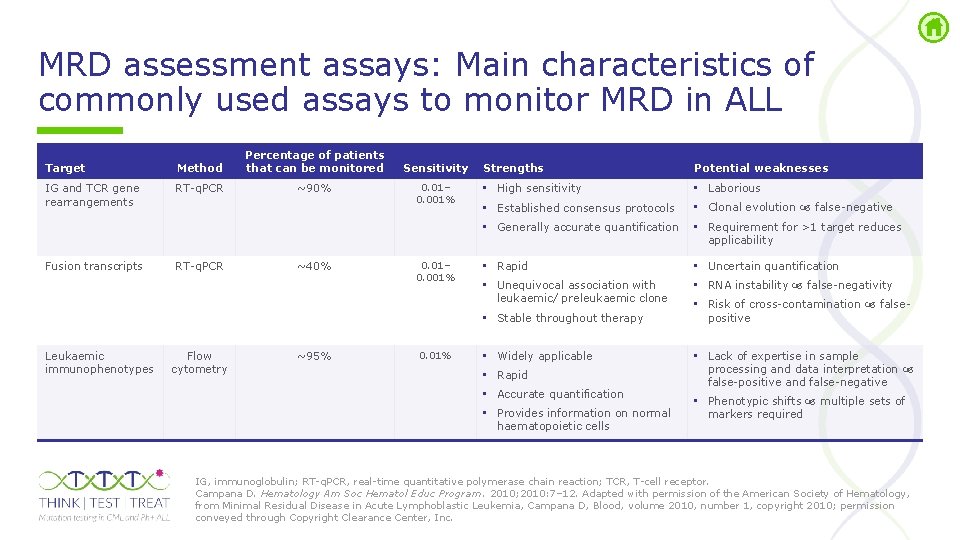
MRD assessment assays: Main characteristics of commonly used assays to monitor MRD in ALL Target Method Percentage of patients that can be monitored IG and TCR gene rearrangements RT q. PCR ~90% Sensitivity 0. 01– 0. 001% Strengths Potential weaknesses • High sensitivity • Laborious • Established consensus protocols • Clonal evolution false negative • Generally accurate quantification • Requirement for >1 target reduces applicability Fusion transcripts RT q. PCR ~40% 0. 01– 0. 001% • Rapid • Uncertain quantification • Unequivocal association with leukaemic/ preleukaemic clone • RNA instability false negativity • Stable throughout therapy Leukaemic immunophenotypes Flow cytometry ~95% 0. 01% • Widely applicable • Rapid • Accurate quantification • Provides information on normal haematopoietic cells • Risk of cross contamination false positive • Lack of expertise in sample processing and data interpretation false positive and false negative • Phenotypic shifts multiple sets of markers required IG, immunoglobulin; RT q. PCR, real time quantitative polymerase chain reaction; TCR, T cell receptor. Campana D. Hematology Am Soc Hematol Educ Program. 2010; 2010: 7– 12. Adapted with permission of the American Society of Hematology, from Minimal Residual Disease in Acute Lymphoblastic Leukemia, Campana D, Blood, volume 2010, number 1, copyright 2010; permission conveyed through Copyright Clearance Center, Inc.

MRD monitoring in practice • Monitoring of treatment response by BCR-ABL 1 transcript level is important in Ph+ ALL • BCR-ABL 1 rearrangement is found in ~20– 30% of adult ALL • RT q. PCR is the best approach for MRD in Ph+ ALL monitoring BUT - No harmonisation of methodology - No established definition for MR • In addition, some centres also use clonal DNA markers for detection of MRD MR, molecular response; MRD, minimal residual disease; Ph+, Philadelphia chromosome positive; RT q. PCR, real time quantitative polymerase chain reaction. Chiaretti S, Foà R. Hematology Am Soc Hematol Educ Program. 2015; 2015: 406– 13.

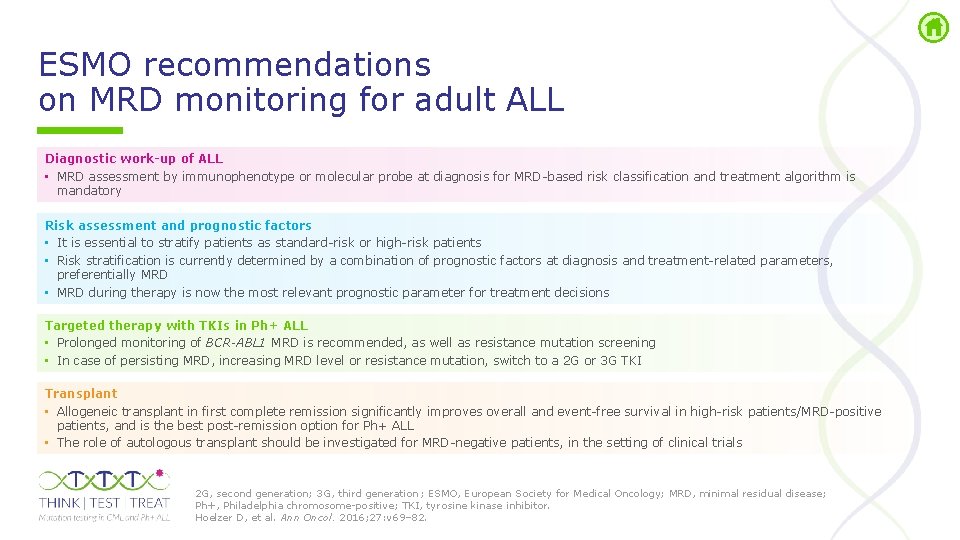
ESMO recommendations on MRD monitoring for adult ALL Diagnostic work-up of ALL • MRD assessment by immunophenotype or molecular probe at diagnosis for MRD based risk classification and treatment algorithm is mandatory Risk assessment and prognostic factors • It is essential to stratify patients as standard risk or high risk patients • Risk stratification is currently determined by a combination of prognostic factors at diagnosis and treatment related parameters, preferentially MRD • MRD during therapy is now the most relevant prognostic parameter for treatment decisions Targeted therapy with TKIs in Ph+ ALL • Prolonged monitoring of BCR-ABL 1 MRD is recommended, as well as resistance mutation screening • In case of persisting MRD, increasing MRD level or resistance mutation, switch to a 2 G or 3 G TKI Transplant • Allogeneic transplant in first complete remission significantly improves overall and event free survival in high risk patients/MRD positive patients, and is the best post remission option for Ph+ ALL • The role of autologous transplant should be investigated for MRD negative patients, in the setting of clinical trials 2 G, second generation; 3 G, third generation ; ESMO, European Society for Medical Oncology; MRD, minimal residual disease; Ph+, Philadelphia chromosome positive; TKI, tyrosine kinase inhibitor. Hoelzer D, et al. Ann Oncol. 2016; 27: v 69– 82.

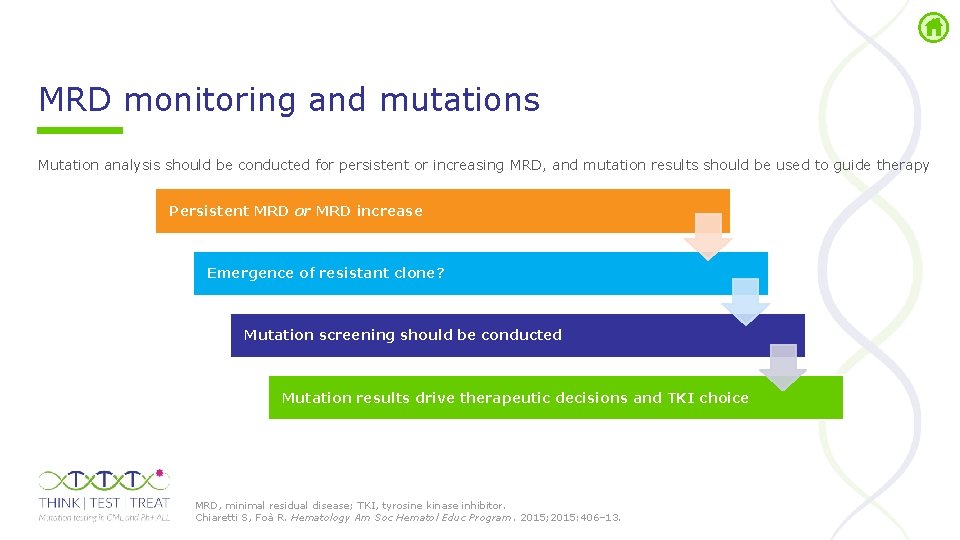
MRD monitoring and mutations Mutation analysis should be conducted for persistent or increasing MRD, and mutation results should be used to guide therapy Persistent MRD or MRD increase Emergence of resistant clone? Mutation screening should be conducted Mutation results drive therapeutic decisions and TKI choice MRD, minimal residual disease; TKI, tyrosine kinase inhibitor. Chiaretti S, Foà R. Hematology Am Soc Hematol Educ Program. 2015; 2015: 406– 13.

BCR-ABL 1 mutation testing in Ph+ ALL

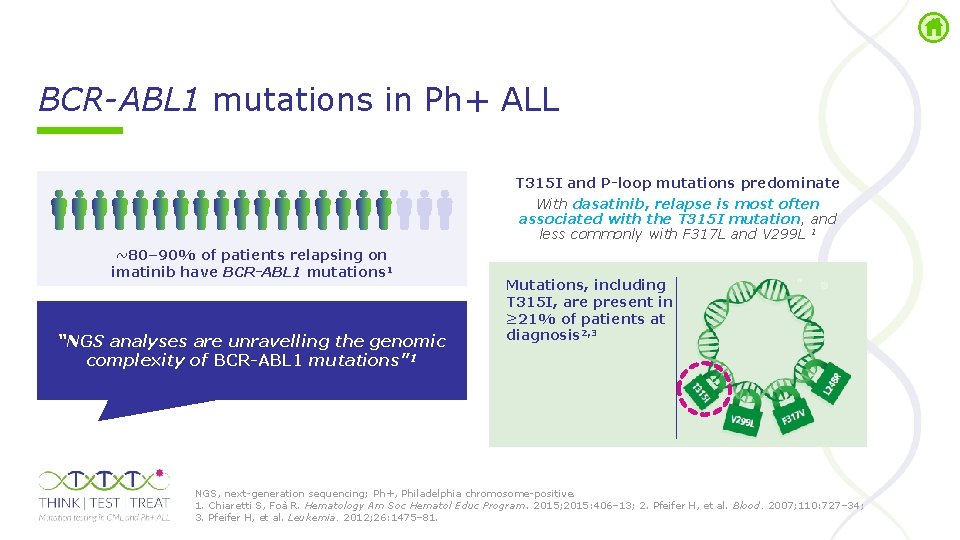
BCR-ABL 1 mutations in Ph+ ALL T 315 I and P-loop mutations predominate With dasatinib, relapse is most often associated with the T 315 I mutation, and less commonly with F 317 L and V 299 L 1 ~80– 90% of patients relapsing on imatinib have BCR-ABL 1 mutations 1 “NGS analyses are unravelling the genomic complexity of BCR-ABL 1 mutations” 1 Mutations, including T 315 I, are present in ≥ 21% of patients at diagnosis 2, 3 NGS, next generation sequencing; Ph+, Philadelphia chromosome positive. 1. Chiaretti S, Foà R. Hematology Am Soc Hematol Educ Program. 2015; 2015: 406– 13; 2. Pfeifer H, et al. Blood. 2007; 110: 727– 34; 3. Pfeifer H, et al. Leukemia. 2012; 26: 1475– 81.

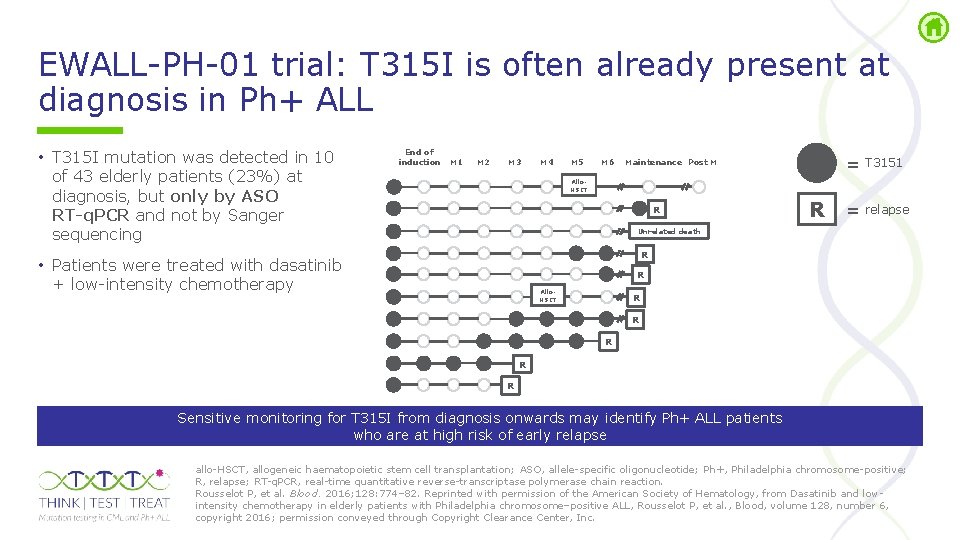
EWALL PH 01 trial: T 315 I is often already present at diagnosis in Ph+ ALL • T 315 I mutation was detected in 10 of 43 elderly patients (23%) at diagnosis, but only by ASO RT-q. PCR and not by Sanger sequencing End of induction M 1 M 2 M 3 M 4 M 5 M 6 = T 3151 Maintenance Post M Allo. HSCT R R = relapse Unrelated death R • Patients were treated with dasatinib + low intensity chemotherapy R Allo. HSCT R R R Sensitive monitoring for T 315 I from diagnosis onwards may identify Ph+ ALL patients who are at high risk of early relapse allo HSCT, allogeneic haematopoietic stem cell transplantation; ASO, allele specific oligonucleotide; Ph+, Philadelphia chromosome positive; R, relapse; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. Rousselot P, et al. Blood. 2016; 128: 774– 82. Reprinted with permission of the American Society of Hematology, from Dasatinib and low intensity chemotherapy in elderly patients with Philadelphia chromosome–positive ALL, Rousselot P, et al. , Blood, volume 128, number 6, copyright 2016; permission conveyed through Copyright Clearance Center, Inc.

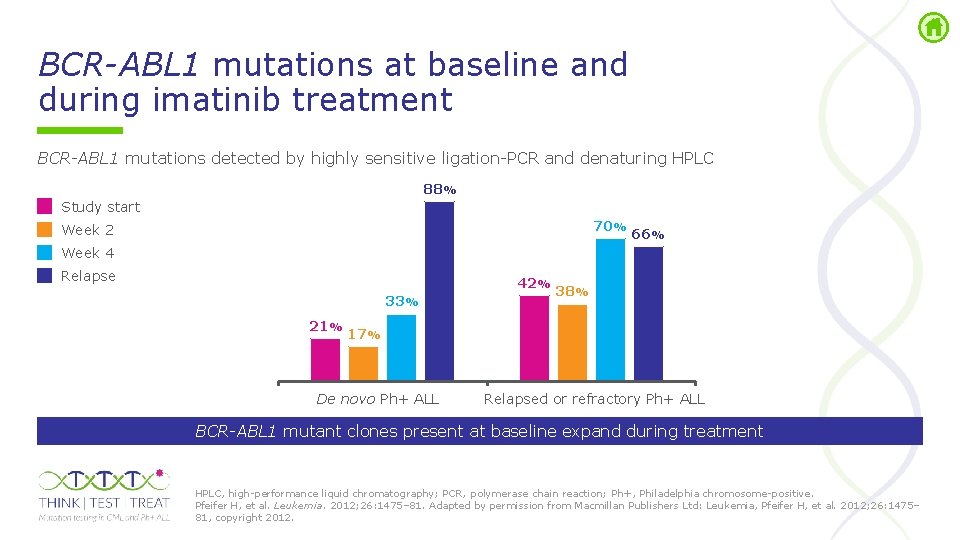
BCR-ABL 1 mutations at baseline and during imatinib treatment BCR-ABL 1 mutations detected by highly sensitive ligation PCR and denaturing HPLC 88% Study start 70% Week 2 66% Week 4 Relapse 42% 33% 21% 38% 17% De novo Ph+ ALL Relapsed or refractory Ph+ ALL BCR-ABL 1 mutant clones present at baseline expand during treatment HPLC, high performance liquid chromatography; PCR, polymerase chain reaction; Ph+, Philadelphia chromosome positive. Pfeifer H, et al. Leukemia. 2012; 26: 1475– 81. Adapted by permission from Macmillan Publishers Ltd: Leukemia, Pfeifer H, et al. 2012; 26: 1475– 81, copyright 2012.

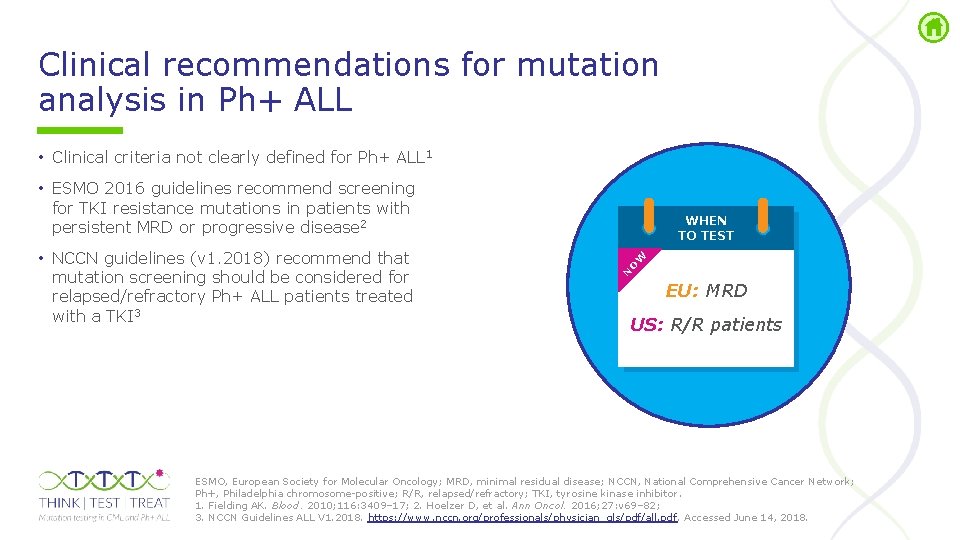
Clinical recommendations for mutation analysis in Ph+ ALL • Clinical criteria not clearly defined for Ph+ ALL 1 • ESMO 2016 guidelines recommend screening for TKI resistance mutations in patients with persistent MRD or progressive disease 2 W O N • NCCN guidelines (v 1. 2018) recommend that mutation screening should be considered for relapsed/refractory Ph+ ALL patients treated with a TKI 3 WHEN TO TEST EU: MRD US: R/R patients ESMO, European Society for Molecular Oncology; MRD, minimal residual disease; NCCN, National Comprehensive Cancer Network; Ph+, Philadelphia chromosome positive; R/R, relapsed/refractory; TKI, tyrosine kinase inhibitor. 1. Fielding AK. Blood. 2010; 116: 3409– 17; 2. Hoelzer D, et al. Ann Oncol. 2016; 27: v 69– 82; 3. NCCN Guidelines ALL V 1. 2018. https: //www. nccn. org/professionals/physician_gls/pdf/all. pdf. Accessed June 14, 2018.

Challenges in MRD monitoring and mutational analysis in Ph+ ALL • Multiple and regular MRD analyses are required in a typical treatment schedule for patients with Ph+ ALL 1, 2 • In Ph+ ALL, BCR-ABL 1 RT q. PCR of BM is the best approach for MRD monitoring, 3, 4 and this material can be used for mutation analysis if indicated by MRD results, but: - Adequate yield of cells for analysis may be a problem at the time of haematopoietic stem cell transplantation 4 - BM sampling is an invasive technique that is painful for most patients 5 BM, bone marrow; MRD, minimal residual disease; Ph+, Philadelphia chromosome positive; RT q. PCR, real time quantitative reverse transcriptase polymerase chain reaction. 1. Cazzaniga G, et al. Haematologica. 2018; 103: 107– 15; 2. Hoelzer D, et al. Ann Oncol. 2016; 27: v 69– 82; 3. Bruggemann M, et al. Leukemia. 2010; 24: 521– 35; 4. Van Dongen JJM, et al. Blood. 2015; 125: 3996– 4009; 5. Hjortholm N, et al. Ann Hematol. 2013; 92: 145– 9.

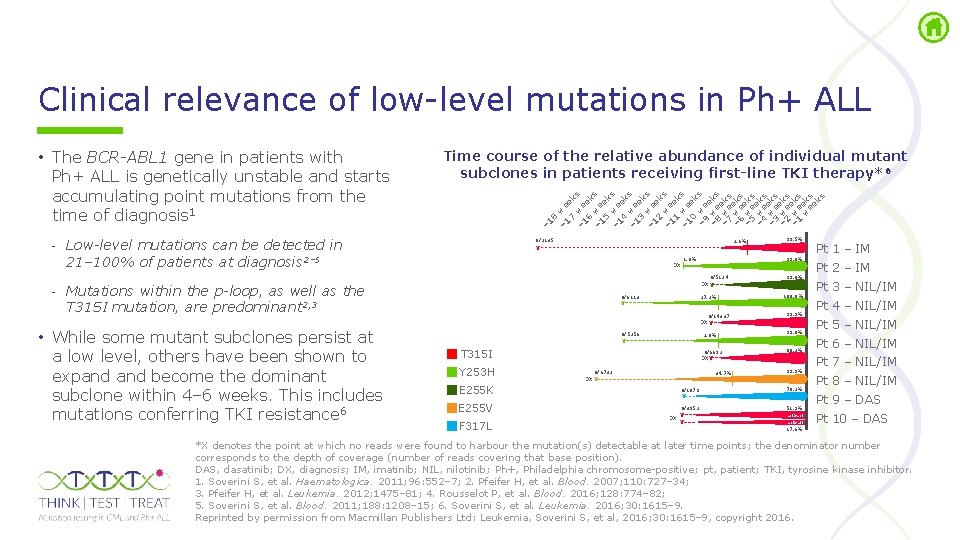
Clinical relevance of low level mutations in Ph+ ALL 8 – 1 we 7 ek – 1 we s 6 ek – 1 we s 5 ek – 1 we s 4 ek – 1 we s 3 ek – 1 we s 2 ek – 1 we s 1 ek – 1 we s 0 ek – 9 we s e – 8 we ks e w – 7 e ks – 6 weeks – 5 weeks – 4 weeks – 3 weeks – 2 weeks – 1 weeks w ek ee s ks Time course of the relative abundance of individual mutant subclones in patients receiving first-line TKI therapy*6 – 1 • The BCR-ABL 1 gene in patients with Ph+ ALL is genetically unstable and starts accumulating point mutations from the time of diagnosis 1 - Low-level mutations can be detected in 21– 100% of patients at diagnosis 2– 5 0/2135 2. 6% 99. 8% 1. 8% DX 0/5124 DX X - Mutations within the p-loop, as well as the T 315 I mutation, are predominant 2, 3 • While some mutant subclones persist at a low level, others have been shown to expand become the dominant subclone within 4– 6 weeks. This includes mutations conferring TKI resistance 6 99. 5% X 0/6113 X 27. 2% 0/14337 DX X 0/5256 X 2. 0% T 315 I Y 253 H E 255 K E 255 V F 317 L 0/6692 DX X 0/6732 DX X 34. 7% 99. 0% 100. 0% 99. 9% 99. 8% 88. 2% 99. 9% 0/1079 X 70. 2% 0/3952 X DX X 51. 9% act>act act>att Pt 1 – IM Pt 2 – IM Pt 3 – NIL/IM Pt 4 – NIL/IM Pt 5 – NIL/IM Pt 6 – NIL/IM Pt 7 – NIL/IM Pt 8 – NIL/IM Pt 9 – DAS Pt 10 – DAS 17. 6% *X denotes the point at which no reads were found to harbour the mutation(s) detectable at later time points; the denominator number corresponds to the depth of coverage (number of reads covering that base position). DAS, dasatinib; DX, diagnosis; IM, imatinib; NIL, nilotinib; Ph+, Philadelphia chromosome positive; pt, patient; TKI, tyrosine kinase inhibitor. 1. Soverini S, et al. Haematologica. 2011; 96: 552– 7; 2. Pfeifer H, et al. Blood. 2007; 110: 727– 34; 3. Pfeifer H, et al. Leukemia. 2012; 1475– 81; 4. Rousselot P, et al. Blood. 2016; 128: 774– 82; 5. Soverini S, et al. Blood. 2011; 188: 1208– 15; 6. Soverini S, et al. Leukemia. 2016; 30: 1615– 9. Reprinted by permission from Macmillan Publishers Ltd: Leukemia, Soverini S, et al, 2016; 30: 1615– 9, copyright 2016.

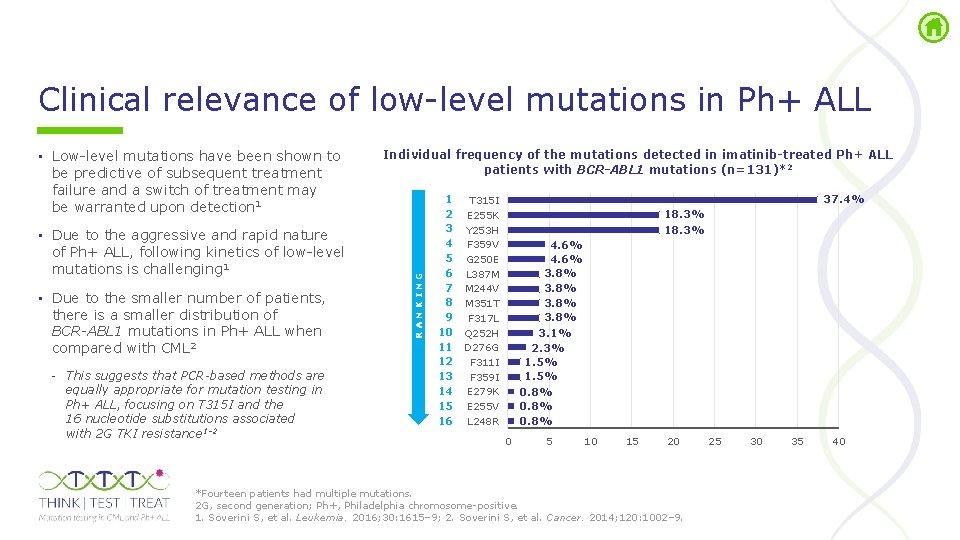
Clinical relevance of low level mutations in Ph+ ALL • Due to the aggressive and rapid nature of Ph+ ALL, following kinetics of low level mutations is challenging 1 • Due to the smaller number of patients, there is a smaller distribution of BCR-ABL 1 mutations in Ph+ ALL when compared with CML 2 - This suggests that PCR-based methods are equally appropriate for mutation testing in Ph+ ALL, focusing on T 315 I and the 16 nucleotide substitutions associated with 2 G TKI resistance 1– 2 Individual frequency of the mutations detected in imatinib-treated Ph+ ALL patients with BCR-ABL 1 mutations (n=131)*2 RANKING • Low level mutations have been shown to be predictive of subsequent treatment failure and a switch of treatment may be warranted upon detection 1 1 T 315 I 2 E 255 K 3 Y 253 H 4 F 359 V 5 G 250 E 6 L 387 M 7 M 244 V 8 M 351 T 9 F 317 L 10 Q 252 H 11 D 276 G 12 F 311 I 13 F 359 I 14 E 279 K 15 E 255 V 16 L 248 R 37. 4% 18. 3% 4. 6% 3. 8% 3. 1% 2. 3% 1. 5% 0. 8% 0 5 10 15 20 *Fourteen patients had multiple mutations. 2 G, second generation; Ph+, Philadelphia chromosome positive. 1. Soverini S, et al. Leukemia. 2016; 30: 1615– 9; 2. Soverini S, et al. Cancer. 2014; 120: 1002– 9. 25 30 35 40

Methods of BCR-ABL 1 mutation testing

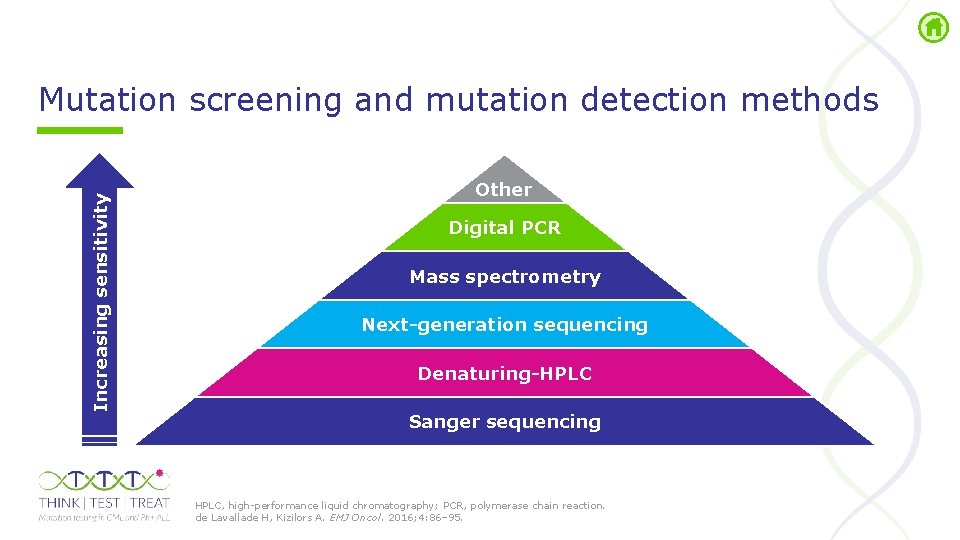
Increasing sensitivity Mutation screening and mutation detection methods Other Digital PCR Mass spectrometry Next-generation sequencing Denaturing-HPLC Sanger sequencing HPLC, high performance liquid chromatography; PCR, polymerase chain reaction. de Lavallade H, Kizilors A. EMJ Oncol. 2016; 4: 86– 95.

Sanger (direct) sequencing Currently the most used mutational analysis method a. DNA polymerase binds to a single stranded DNA template (blue) and synthesises a complementary strand of DNA (red) b. The reaction contains a mixture of normal d. NTPs and fluorescently labelled dd. NTPs c. When DNA polymerase randomly incorporates a dd. NTP base, synthesis terminates - The dd. NTP molecules lack a 3' hydroxyl group, which is required to bind to the next nucleotide in the chain. - This step produces a mixture of newly synthesised DNA strands that differ in length by a single nucleotide. Each strand is labelled at the 3′ end with a fluorescently labelled dd. NTP base d. The DNA mixture is separated by electrophoresis e. The electropherogram results show peaks representing the colour and signal intensity of each DNA band From these data, the sequence of the newly synthesised DNA strand is determined, as shown above the peaks d. NTP, deoxynucleotide triphosphate; dd. NTP, dideoxynucleotide base. Chial, H. Nature Education. 2008; 1: 219. Chial H, Nature Education, published 2008, © 2003 Macmillan Publishers, reproduced with permission of SNCSC.

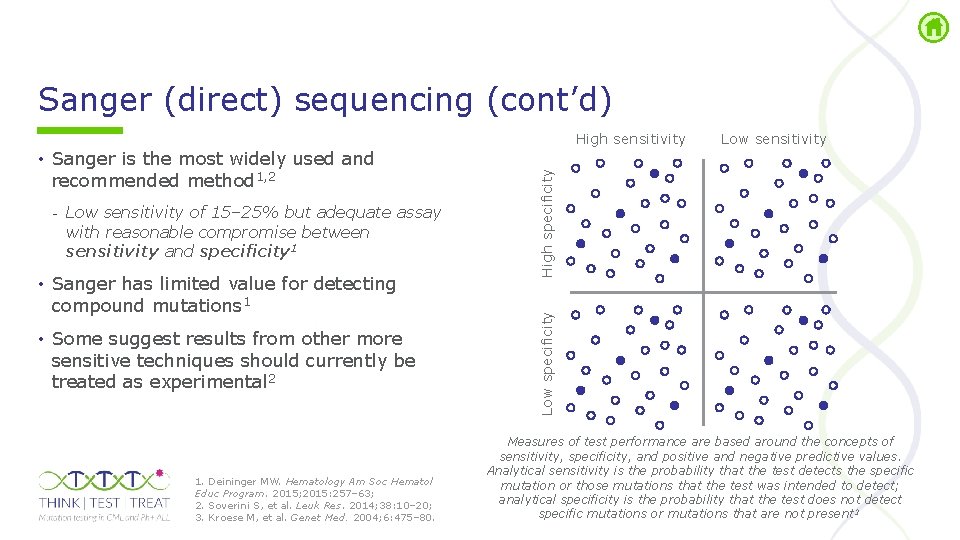
Sanger (direct) sequencing (cont’d) - Low sensitivity of 15– 25% but adequate assay with reasonable compromise between sensitivity and specificity 1 • Sanger has limited value for detecting compound mutations 1 • Some suggest results from other more sensitive techniques should currently be treated as experimental 2 1. Deininger MW. Hematology Am Soc Hematol Educ Program. 2015; 2015: 257– 63; 2. Soverini S, et al. Leuk Res. 2014; 38: 10– 20; 3. Kroese M, et al. Genet Med. 2004; 6: 475– 80. Low specificity • Sanger is the most widely used and recommended method 1, 2 Low sensitivity High specificity High sensitivity Measures of test performance are based around the concepts of sensitivity, specificity, and positive and negative predictive values. Analytical sensitivity is the probability that the test detects the specific mutation or those mutations that the test was intended to detect; analytical specificity is the probability that the test does not detect specific mutations or mutations that are not present 3

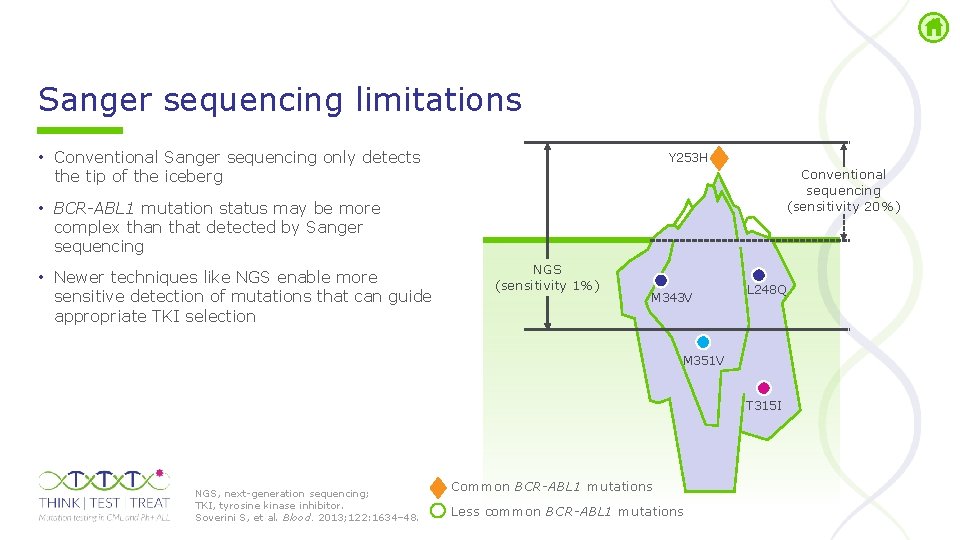
Sanger sequencing limitations • Conventional Sanger sequencing only detects the tip of the iceberg Y 253 H Conventional sequencing (sensitivity 20%) • BCR-ABL 1 mutation status may be more complex than that detected by Sanger sequencing • Newer techniques like NGS enable more sensitive detection of mutations that can guide appropriate TKI selection NGS (sensitivity 1%) M 343 V L 248 Q M 351 V T 315 I NGS, next generation sequencing; TKI, tyrosine kinase inhibitor. Soverini S, et al. Blood. 2013; 122: 1634– 48. Common BCR-ABL 1 mutations Less common BCR-ABL 1 mutations

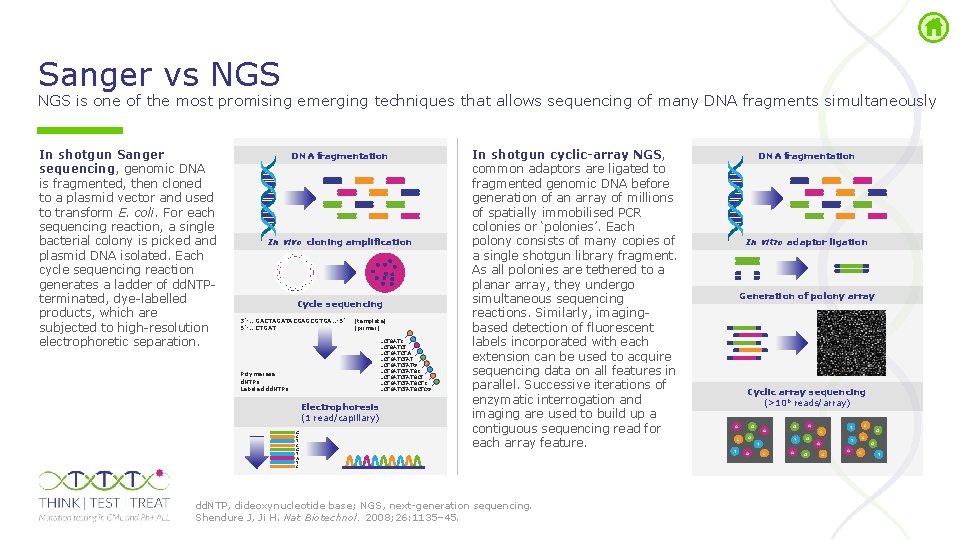
Sanger vs NGS is one of the most promising emerging techniques that allows sequencing of many DNA fragments simultaneously In shotgun Sanger sequencing, genomic DNA is fragmented, then cloned to a plasmid vector and used to transform E. coli. For each sequencing reaction, a single bacterial colony is picked and plasmid DNA isolated. Each cycle sequencing reaction generates a ladder of dd. NTP terminated, dye labelled products, which are subjected to high resolution electrophoretic separation. DNA fragmentation In vivo cloning amplification Cycle sequencing 3’ … GACTAGATACGAGCGTGA… 5’ 5’ … CTGAT (template) (primer) …CTGATCTAT …CTGATCTATGCTC …CTGATCTATGCTCG Polymerase d. NTPs Labeled dd. NTPs Electrophoresis (1 read/capillary) G C T C G T A T C In shotgun cyclic-array NGS, common adaptors are ligated to fragmented genomic DNA before generation of an array of millions of spatially immobilised PCR colonies or ‘polonies’. Each polony consists of many copies of a single shotgun library fragment. As all polonies are tethered to a planar array, they undergo simultaneous sequencing reactions. Similarly, imaging based detection of fluorescent labels incorporated with each extension can be used to acquire sequencing data on all features in parallel. Successive iterations of enzymatic interrogation and imaging are used to build up a contiguous sequencing read for each array feature. dd. NTP, dideoxynucleotide base; NGS, next generation sequencing. Shendure J, Ji H. Nat Biotechnol. 2008; 26: 1135– 45. DNA fragmentation In vitro adaptor ligation Generation of polony array Cyclic array sequencing (>106 reads/array) A C T G A T T A C A G G C T A C C T A G C T

NGS can be used to detect low level mutations in Ph+ ALL and compound mutations Compound mutations are an emergent clinical problem 1 • Among 68 patients with Ph+ ALL relapsing on second line dasatinib, 30 (44%) had multiple mutations, including 23 (34%) with a complex clonal architecture comprising both compound and polyclonal mutations 2 Analysis of relapsed patients who have no evidence of mutations by conventional Sanger sequencing shows that clinically relevant low-level mutations may be present in almost half of these patients 3 • Detection of low-level mutations by deep sequencing can predict treatment failure in Ph+ ALL 3 Compound mutations can be detected by NGS if these mutations are on the same read 1. Chiaretti S, Foà R. Hematology Am Soc Hematol Educ Program. 2015; 2015: 406– 13; 2. Soverini S, et al. Cancer. 2014; 120: 1002– 9; 3. Soverini S, et al. Leukemia. 2016; 30: 1615– 9.

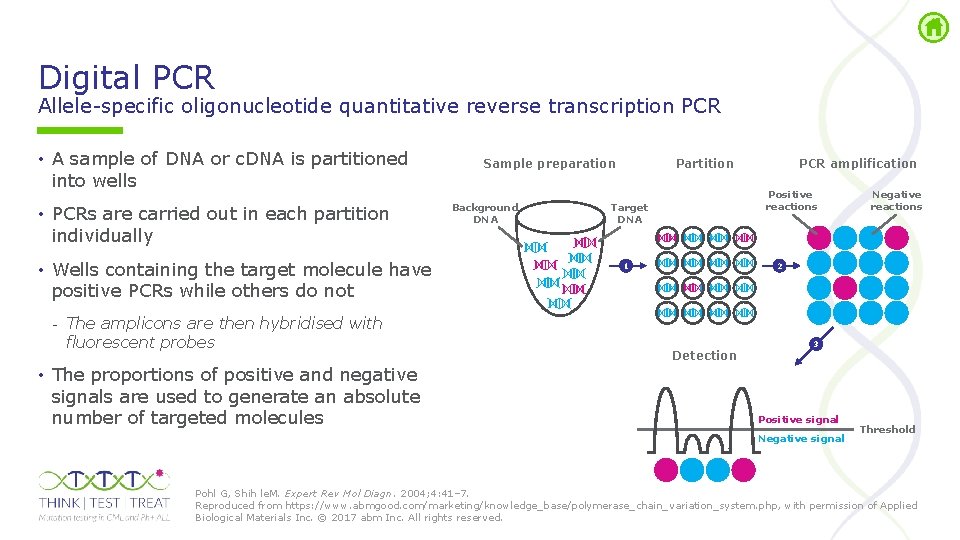
Digital PCR Allele specific oligonucleotide quantitative reverse transcription PCR • A sample of DNA or c. DNA is partitioned into wells • PCRs are carried out in each partition individually • Wells containing the target molecule have positive PCRs while others do not - The amplicons are then hybridised with fluorescent probes • The proportions of positive and negative signals are used to generate an absolute number of targeted molecules Sample preparation Background DNA Partition PCR amplification Positive reactions Target DNA 1 Negative reactions 2 Detection 3 Positive signal Negative signal Threshold Pohl G, Shih le. M. Expert Rev Mol Diagn. 2004; 4: 41– 7. Reproduced from https: //www. abmgood. com/marketing/knowledge_base/polymerase_chain_variation_system. php, with permission of Applied Biological Materials Inc. © 2017 abm Inc. All rights reserved.

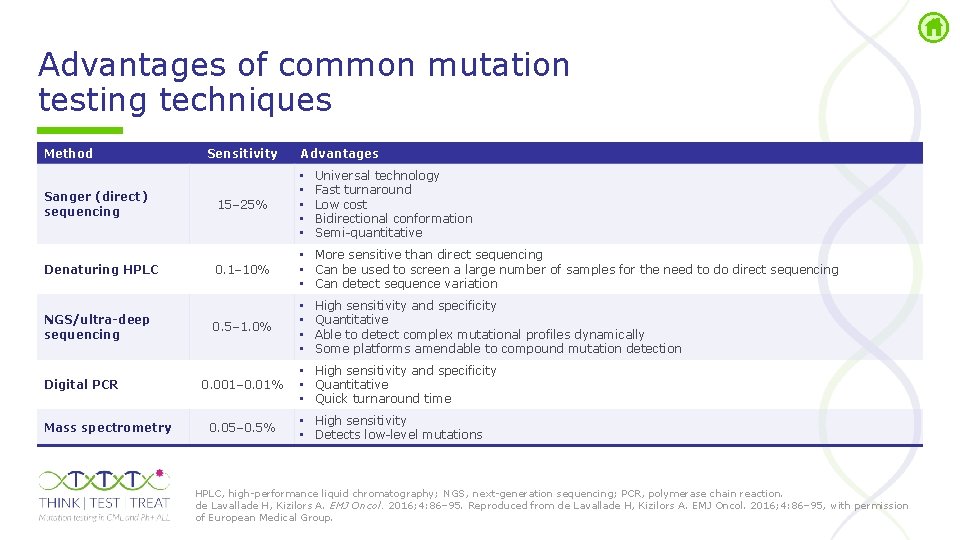
Advantages of common mutation testing techniques Method Sanger (direct) sequencing Denaturing HPLC NGS/ultra-deep sequencing Digital PCR Mass spectrometry Sensitivity Advantages Universal technology Fast turnaround Low cost Bidirectional conformation Semi quantitative 15– 25% • • • 0. 1– 10% • More sensitive than direct sequencing • Can be used to screen a large number of samples for the need to do direct sequencing • Can detect sequence variation 0. 5– 1. 0% • • 0. 001– 0. 01% 0. 05– 0. 5% High sensitivity and specificity Quantitative Able to detect complex mutational profiles dynamically Some platforms amendable to compound mutation detection • High sensitivity and specificity • Quantitative • Quick turnaround time • High sensitivity • Detects low level mutations HPLC, high performance liquid chromatography; NGS, next generation sequencing; PCR, polymerase chain reaction. de Lavallade H, Kizilors A. EMJ Oncol. 2016; 4: 86– 95. Reproduced from de Lavallade H, Kizilors A. EMJ Oncol. 2016; 4: 86– 95, with permission of European Medical Group.

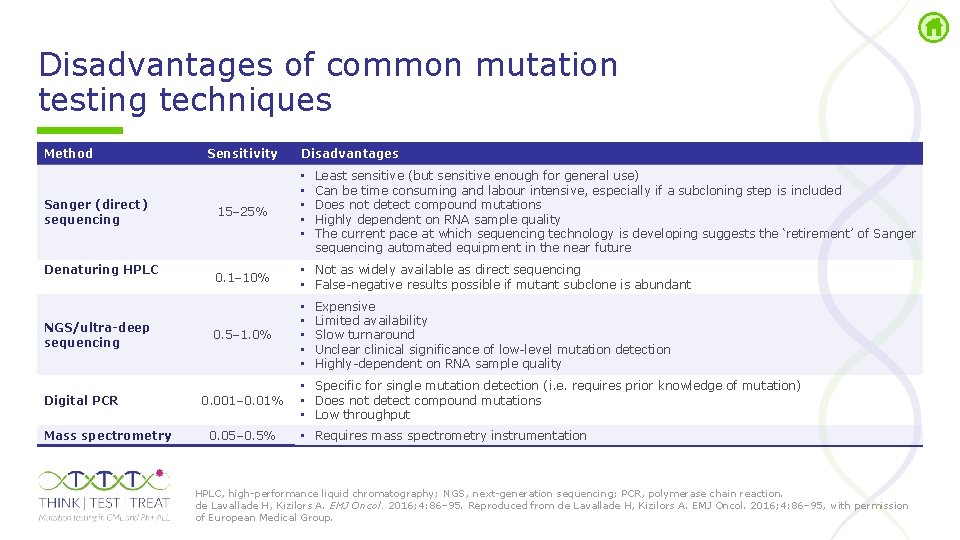
Disadvantages of common mutation testing techniques Method Sanger (direct) sequencing Denaturing HPLC NGS/ultra-deep sequencing Digital PCR Mass spectrometry Sensitivity 15– 25% Disadvantages • • • Least sensitive (but sensitive enough for general use) Can be time consuming and labour intensive, especially if a subcloning step is included Does not detect compound mutations Highly dependent on RNA sample quality The current pace at which sequencing technology is developing suggests the ‘retirement’ of Sanger sequencing automated equipment in the near future 0. 1– 10% • Not as widely available as direct sequencing • False negative results possible if mutant subclone is abundant 0. 5– 1. 0% • • • 0. 001– 0. 01% 0. 05– 0. 5% Expensive Limited availability Slow turnaround Unclear clinical significance of low level mutation detection Highly dependent on RNA sample quality • Specific for single mutation detection (i. e. requires prior knowledge of mutation) • Does not detect compound mutations • Low throughput • Requires mass spectrometry instrumentation HPLC, high performance liquid chromatography; NGS, next generation sequencing; PCR, polymerase chain reaction. de Lavallade H, Kizilors A. EMJ Oncol. 2016; 4: 86– 95. Reproduced from de Lavallade H, Kizilors A. EMJ Oncol. 2016; 4: 86– 95, with permission of European Medical Group.

Interpretation of mutation testing results

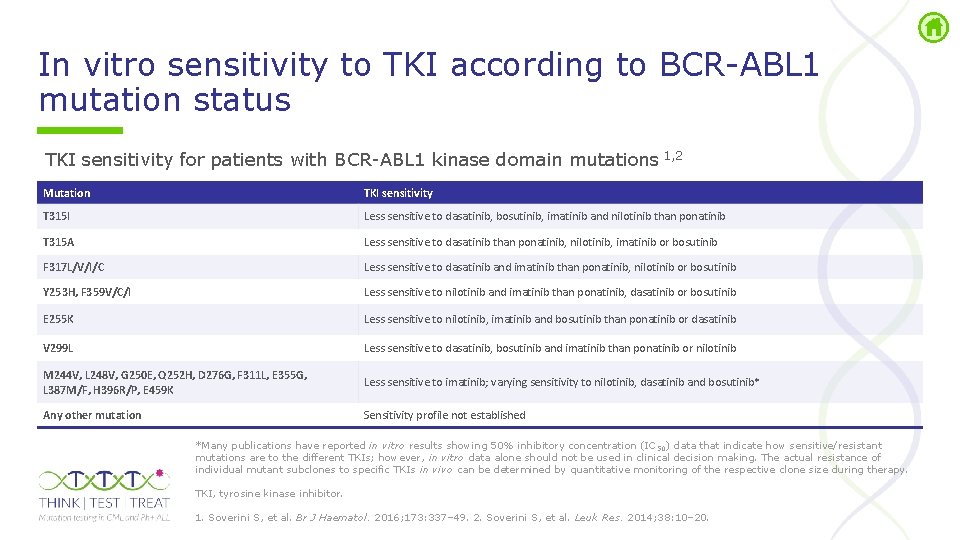
In vitro sensitivity to TKI according to BCR ABL 1 mutation status TKI sensitivity for patients with BCR ABL 1 kinase domain mutations 1, 2 Mutation TKI sensitivity T 315 I Less sensitive to dasatinib, bosutinib, imatinib and nilotinib than ponatinib T 315 A Less sensitive to dasatinib than ponatinib, nilotinib, imatinib or bosutinib F 317 L/V/I/C Less sensitive to dasatinib and imatinib than ponatinib, nilotinib or bosutinib Y 253 H, F 359 V/C/I Less sensitive to nilotinib and imatinib than ponatinib, dasatinib or bosutinib E 255 K Less sensitive to nilotinib, imatinib and bosutinib than ponatinib or dasatinib V 299 L Less sensitive to dasatinib, bosutinib and imatinib than ponatinib or nilotinib M 244 V, L 248 V, G 250 E, Q 252 H, D 276 G, F 311 L, E 355 G, L 387 M/F, H 396 R/P, E 459 K Less sensitive to imatinib; varying sensitivity to nilotinib, dasatinib and bosutinib* Any other mutation Sensitivity profile not established *Many publications have reported in vitro results showing 50% inhibitory concentration (IC 50) data that indicate how sensitive/resistant mutations are to the different TKIs; however, in vitro data alone should not be used in clinical decision making. The actual resistance of individual mutant subclones to specific TKIs in vivo can be determined by quantitative monitoring of the respective clone size during therapy. TKI, tyrosine kinase inhibitor. 1. Soverini S, et al. Br J Haematol. 2016; 173: 337– 49. 2. Soverini S, et al. Leuk Res. 2014; 38: 10– 20.

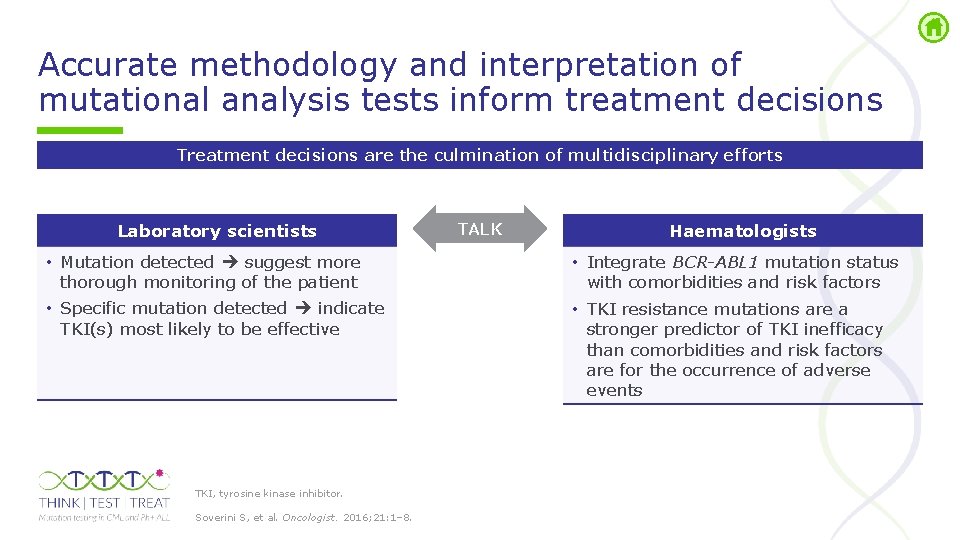
Accurate methodology and interpretation of mutational analysis tests inform treatment decisions Treatment decisions are the culmination of multidisciplinary efforts Laboratory scientists • Mutation detected suggest more thorough monitoring of the patient • Specific mutation detected indicate TKI(s) most likely to be effective TKI, tyrosine kinase inhibitor. Soverini S, et al. Oncologist. 2016; 21: 1– 8. TALK Haematologists • Integrate BCR-ABL 1 mutation status with comorbidities and risk factors • TKI resistance mutations are a stronger predictor of TKI inefficacy than comorbidities and risk factors are for the occurrence of adverse events

Further information

Steering Committee of European experts Simona Soverini University of Bologna, Italy Hugues de Lavallade King’s College Hospital, London, UK Lucia Cavelier Uppsala University, Sweden Torsten Haferlach Munich Haematology Medical Practice & Munich Leukaemia Laboratory, Germany Joaquin Martinez Complutense University de Madrid; Hospital 12 de Octubre & Spanish Center of Oncology, Spain Jean-Michel Cayuela Saint-Louis Hospital Paris, France Katerina Machova Charles University, Prague, Czech Republic Nick Cross University of Southampton & Wessex Regional Genetics Laboratory, Salisbury, UK Pascal Vannuffel Institute of Pathology and Genetics, Gosselies, Belgium Thomas Lion Lab. Dia Labordiagnostik & Children’s Cancer Research Institute, Austria Thomas Ernst University Hospital Jena, Germany

Where to find more information Source Link Description Educational brochure for HCPs • www. thinktesttreat. com A brochure summarising practical aspects of BCR-ABL 1 mutation testing, clinical guidelines on when to perform testing and TKI resistance profiles European Leukemia. Net Website • https: //www. leukemia-net. org Official Website of the European Leukemia. Net research network with information for physicians and patients ESMO guidelines for CML • Hochhaus A, et al. Ann Oncol. 2017; 28(4): iv 41– Guidelines for diagnosis, treatment and follow-up of CML in adult patients 51 ELN recommendations for CML Pocket guide ELN recommendations • Baccarani M, et al. Blood. 2013; 122: 872– 84 • Pocket guide ELN recommendations European Leukemia. Net recommendations for the management of chronic myeloid leukaemia ESMO recommendations for Ph+ ALL • Hoelzer D, et al. Ann Oncol. 2016; 27: v 69– 82. Acute lymphoblastic leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up NCCN Clinical Practice Guidelines – CML V 4. 2018 • https: //www. nccn. org/professionals/physician National Comprehensive Cancer Network clinical practice guidelines for the _gls/pdf/cml. pdf management of chronic myeloid leukaemia NCCN Clinical Practice Guidelines – ALL V 1. 2018 • https: //www. nccn. org/professionals/physician National Comprehensive Cancer Network clinical practice guidelines for the _gls/pdf/all. pdf management of Philadelphia-positive acute lymphoblastic leukaemia ELN, European Leukemia. Net; ESMO, European Society for Molecular Oncology; HCP, healthcare professional; Ph+, Philadelphia chromosome positive; TKI, tyrosine kinase inhibitor. Soverini S, et al. Oncologist. 2016; 21: 1– 8.
 Molecular response
Molecular response Cml
Cml Cml 100
Cml 100 Chronic myeloid leukemia treatment
Chronic myeloid leukemia treatment Cml equation
Cml equation Diagnosi cml
Diagnosi cml Procesadores cml microcircuits
Procesadores cml microcircuits Cml vs sml
Cml vs sml Números naturales
Números naturales Obm cml
Obm cml Cml awareness day
Cml awareness day Rumus cml
Rumus cml Cml
Cml Cml
Cml 20115n-cml
20115n-cml Slope of cml
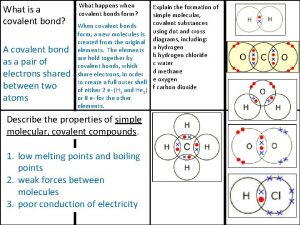
Slope of cml Covalent bond boiling point
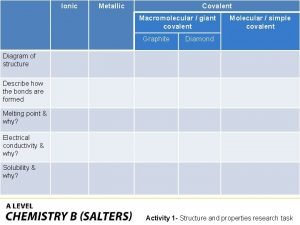
Covalent bond boiling point Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Giant molecular structure vs simple molecular structure
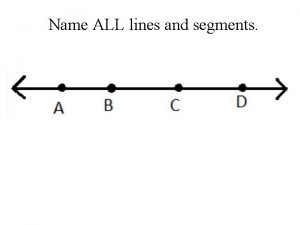
Giant molecular structure vs simple molecular structure Name all the rays
Name all the rays She's all states and all princes i
She's all states and all princes i What is educational psychology discuss its nature and scope
What is educational psychology discuss its nature and scope Love all serve all
Love all serve all Interventi sociali rivolti alla persona
Interventi sociali rivolti alla persona Above all powers
Above all powers I work all night i work all day to pay the bills
I work all night i work all day to pay the bills All-to-all personalized communication
All-to-all personalized communication Sistem all in all out
Sistem all in all out Carpi volare
Carpi volare Silent night holy night all is calm
Silent night holy night all is calm 馮定華神父2020
馮定華神父2020 All of you is more than enough for all of me
All of you is more than enough for all of me What height of love what depth of peace
What height of love what depth of peace Above all powers
Above all powers Pregnancy and infant cohort monitoring and evaluation
Pregnancy and infant cohort monitoring and evaluation Covalent molecular and covalent network
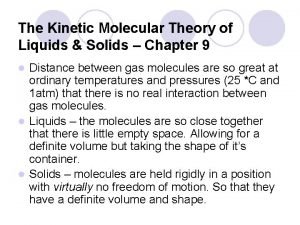
Covalent molecular and covalent network Kinetic molecular theory of solids
Kinetic molecular theory of solids Empirical formula to percent composition
Empirical formula to percent composition Naming and writing formulas for molecular compounds
Naming and writing formulas for molecular compounds Relationship between bond dipoles and molecular dipoles
Relationship between bond dipoles and molecular dipoles Nh3 shape and polarity
Nh3 shape and polarity Axe notation
Axe notation Lewis structure of pf3
Lewis structure of pf3 Molecular geometry and bonding theories
Molecular geometry and bonding theories Bent lewis dot structure
Bent lewis dot structure Molecular theory of gases and liquids
Molecular theory of gases and liquids Covalent molecular and covalent network
Covalent molecular and covalent network Covalent molecular and covalent network
Covalent molecular and covalent network Silicon carbide covalent bond
Silicon carbide covalent bond Empirical and molecular formula worksheet
Empirical and molecular formula worksheet Molecular formula from empirical formula
Molecular formula from empirical formula How to determine empirical formula
How to determine empirical formula Emprical formula
Emprical formula Dipole moment polarity
Dipole moment polarity Chemical formula covalent compounds
Chemical formula covalent compounds What electronegativity difference is polar
What electronegativity difference is polar Ibuprofen percent composition
Ibuprofen percent composition How are atomic and molecular orbitals related
How are atomic and molecular orbitals related Cellular and molecular immunology
Cellular and molecular immunology Kinetic molecular theory of liquids and solids
Kinetic molecular theory of liquids and solids Difference between atomic and molecular spectroscopy
Difference between atomic and molecular spectroscopy Pcr lab setup
Pcr lab setup Difference between atomic and molecular spectroscopy
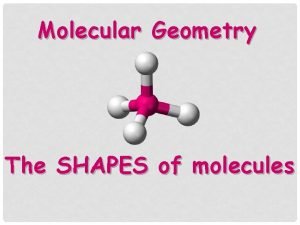
Difference between atomic and molecular spectroscopy Molecular shapes
Molecular shapes Ch6n lewis structure
Ch6n lewis structure Chapter 7 chemical formulas and chemical compounds test
Chapter 7 chemical formulas and chemical compounds test Vbt vs mot
Vbt vs mot Basic principle of valence bond theory
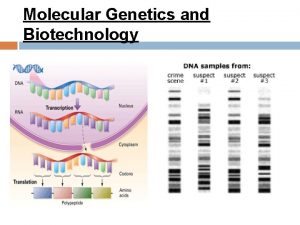
Basic principle of valence bond theory Molecular genetics and biotechnology
Molecular genetics and biotechnology Valence bond theory and molecular orbital theory
Valence bond theory and molecular orbital theory Bond order examples
Bond order examples Valence bond theory and molecular orbital theory
Valence bond theory and molecular orbital theory Valence bond theory example problems
Valence bond theory example problems