Use of Toxicological Pathways for Hazard Assessment in












![Validation of Mechanism #1 Calculated HBI ( E# [e. V]) vs. change in RBC Validation of Mechanism #1 Calculated HBI ( E# [e. V]) vs. change in RBC](https://slidetodoc.com/presentation_image/98df2dd4315d47662c9c5ec34cea026e/image-61.jpg)














- Slides: 132

Use of Toxicological Pathways for Hazard Assessment in OECD (Q)SAR Toolbox: LMC, Bourgas University, Bulgaria Chemical Management Center, NITE, Japan Fraunhofer Institute for Toxicology and Experimental Medicine, Germany OECD, Environment Directorate, Paris International QSAR Foundation, USA Mc. Kim Conference September 2008, Duluth, USA

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

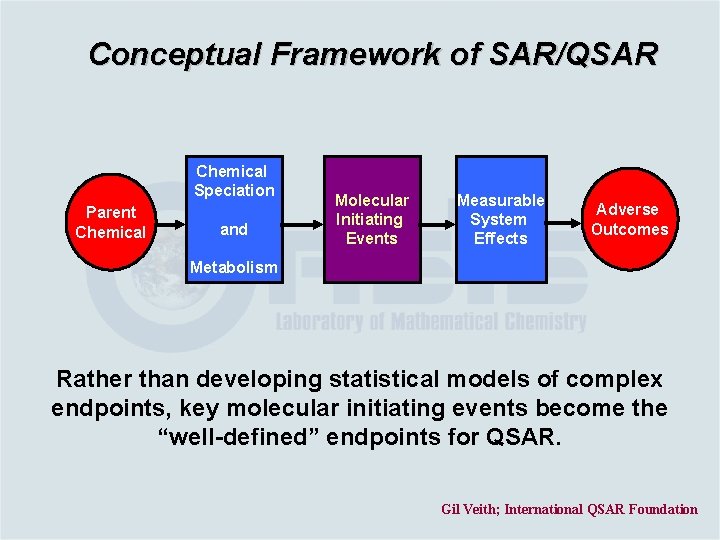
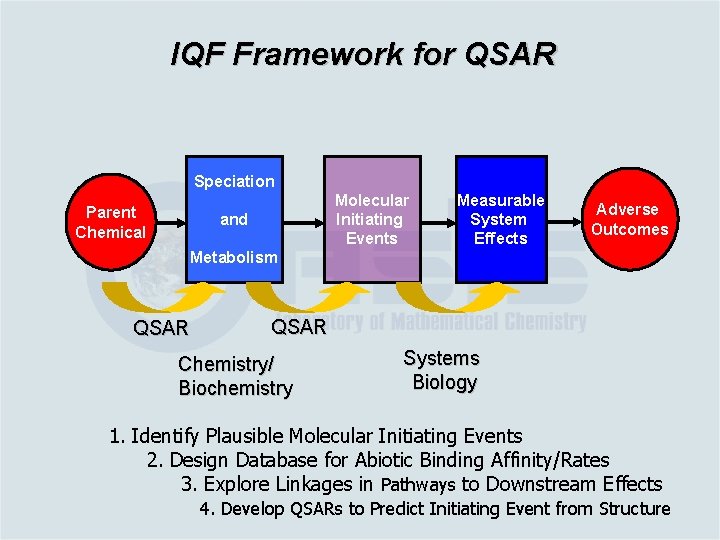
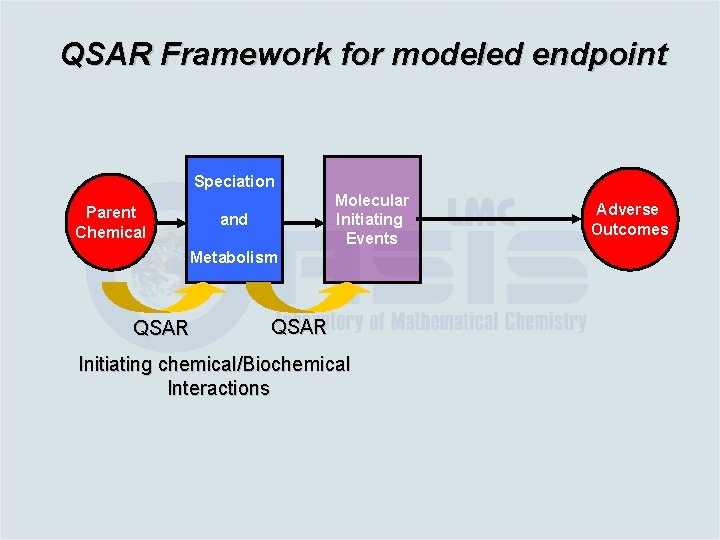
Conceptual Framework of SAR/QSAR Chemical Speciation Parent Chemical and Molecular Initiating Events Measurable System Effects Adverse Outcomes Metabolism Rather than developing statistical models of complex endpoints, key molecular initiating events become the “well-defined” endpoints for QSAR. Gil Veith; International QSAR Foundation

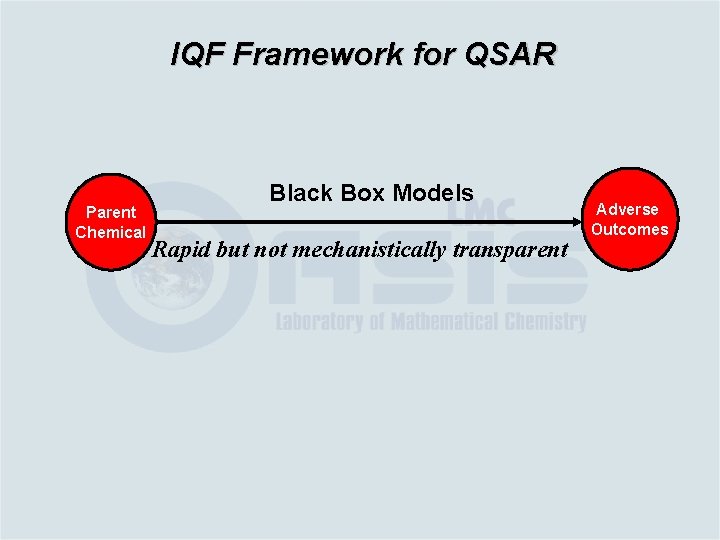
IQF Framework for QSAR Parent Chemical Black Box Models Rapid but not mechanistically transparent Adverse Outcomes

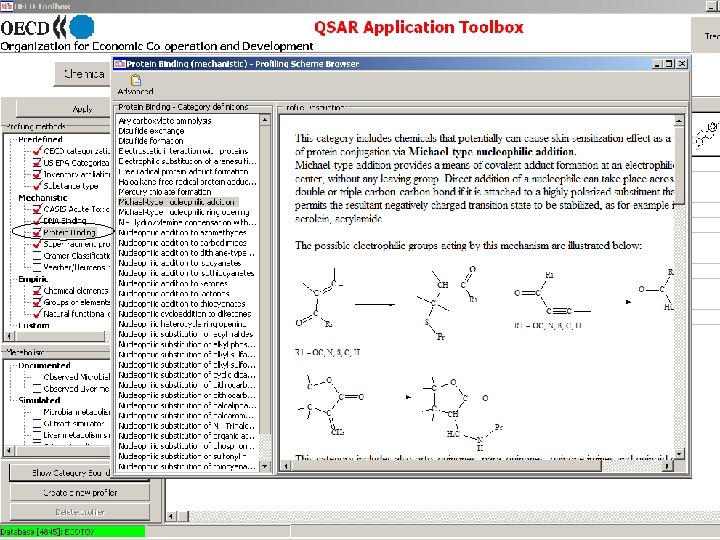
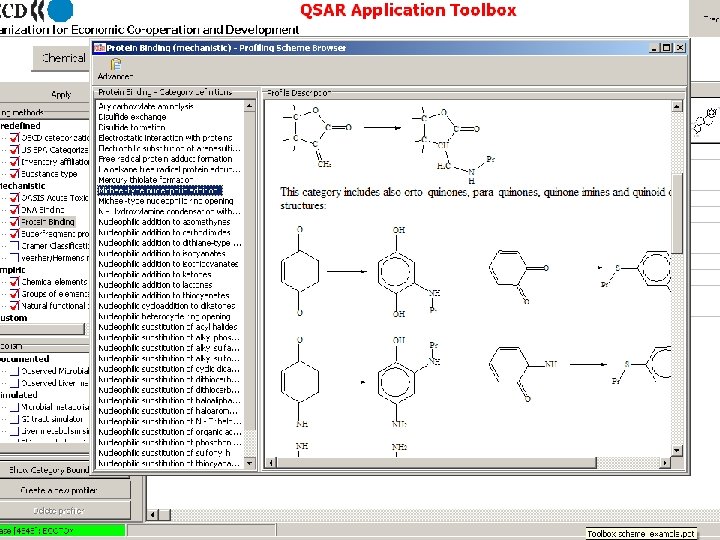
IQF Framework for QSAR Speciation Parent Chemical Molecular Initiating Events and Measurable System Effects Adverse Outcomes Metabolism QSAR Chemistry/ Biochemistry Systems Biology 1. Identify Plausible Molecular Initiating Events 2. Design Database for Abiotic Binding Affinity/Rates 3. Explore Linkages in Pathways to Downstream Effects 4. Develop QSARs to Predict Initiating Event from Structure

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

Categorization and QSAR v The categories concept is part of the historical description of QSARs v QSARs are quantitative models of key mechanistic processes which result in the measured activity

Categorization and QSAR Each QSAR estimate is a result of two predictions: v Qualitative prediction of predominant interaction mechanisms and hazard identification (defined by category) v Quantitative prediction of the intensity (potency) of the specific mechanisms of interaction (predicted by QSAR) Wrong definition for the mechanism of underlying reaction could result in using of a wrong QSAR for the potency estimate

Categorization and QSAR Example v Phenols are polar narcotics, uncouplers or electrophilic chemicals. v QSAR models for predicting acute effects for each mechanism have comparable uncertainty v The potency of the electrophilic mechanism can be orders of magnitude greater than polar narcotics v Wrong categorization of chemicals could cause significant errors in defining the potency

Categorization and QSAR Basic Assumption for Regulatory Acceptance n The logic for selecting a specific model (category) for a specific chemical is the cornerstone of regulatory acceptance OECD QSAR AD-Hoc group meeting, Madrid, April 2007

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

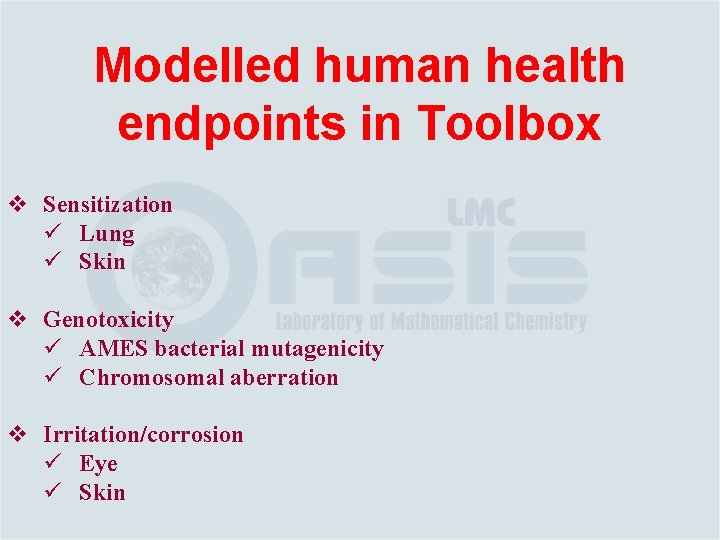
Modelled human health endpoints in Toolbox v Sensitization ü Lung ü Skin v Genotoxicity ü AMES bacterial mutagenicity ü Chromosomal aberration v Irritation/corrosion ü Eye ü Skin

Commonality between the modelled endpoints The effects could be characterized by: v. Single toxicological pathway v. Strong dependency on initiating molecular events (e. g. on molecular structure) v. Small impact of subsequent biological processes (“short” toxicological pathways)

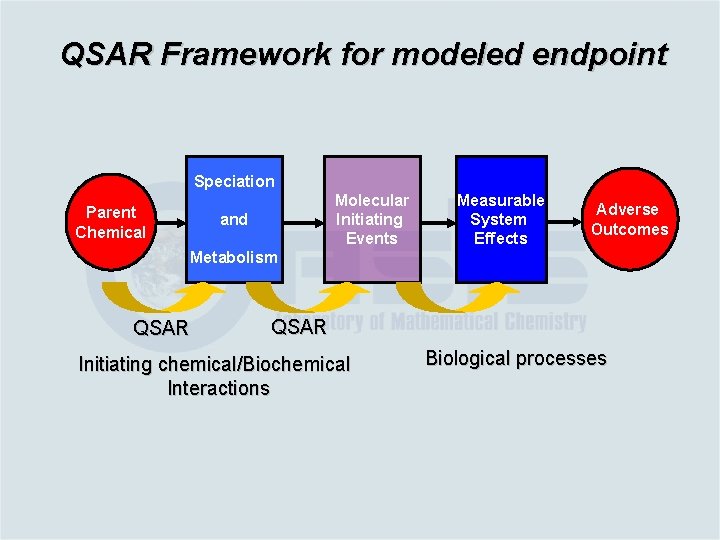
QSAR Framework for modeled endpoint Speciation Parent Chemical Molecular Initiating Events and Measurable System Effects Adverse Outcomes Metabolism QSAR Initiating chemical/Biochemical Interactions Biological processes

QSAR Framework for modeled endpoint Speciation Parent Chemical Molecular Initiating Events and Metabolism QSAR Initiating chemical/Biochemical Interactions Adverse Outcomes

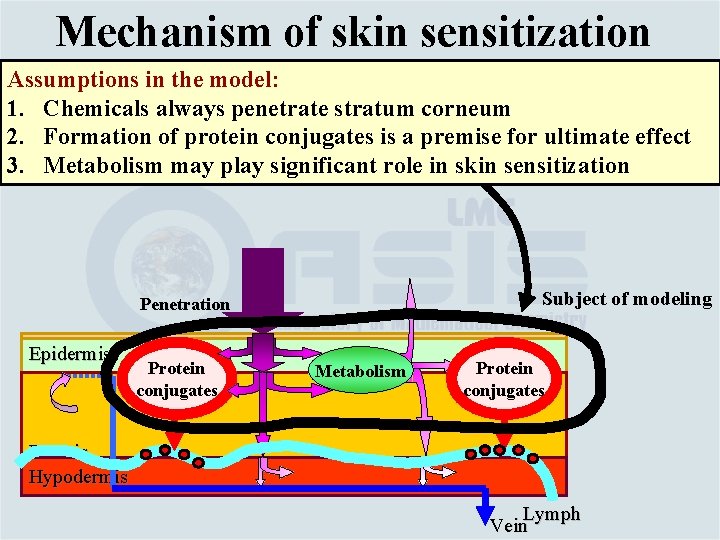
Mechanism of skin sensitization Assumptions in the model: 1. Chemicals always penetrate stratum corneum 2. Formation of protein conjugates is a premise for ultimate effect 3. Metabolism may play significant role in skin sensitization Subject of modeling Penetration Epidermis Protein conjugates Metabolism Protein conjugates Dermis Hypodermis Lymph Vein

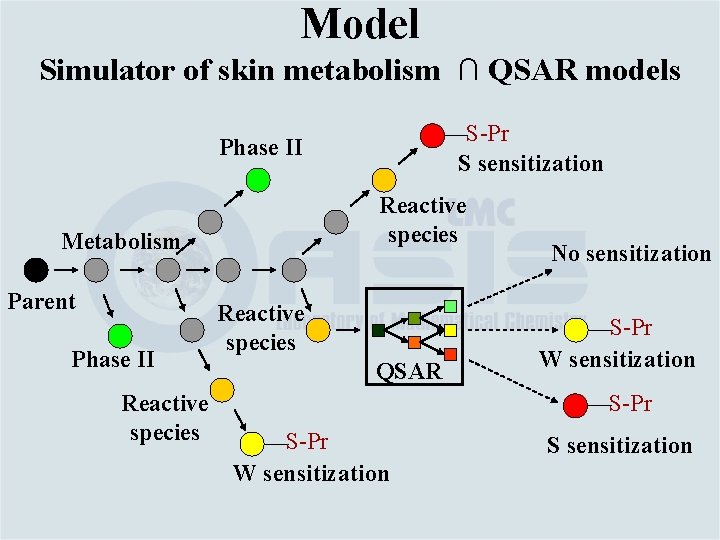
Model Simulator of skin metabolism ∩ QSAR models S-Pr S sensitization Phase II Reactive species Metabolism Parent Phase II Reactive species QSAR No sensitization S-Pr W sensitization S sensitization

Conclusion: The categorization of substances according to chemical mechanisms governing the initiating reaction with protein or DNA is good enough for predicting human health effects resulting from single and “short” toxicological pathways

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

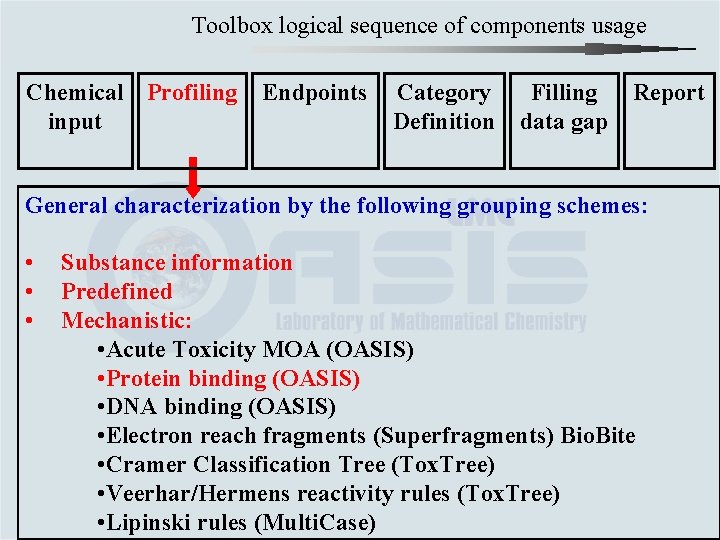
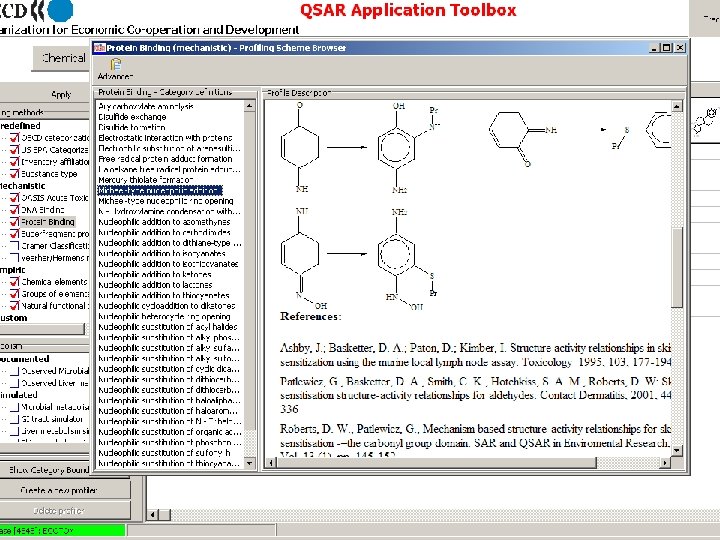
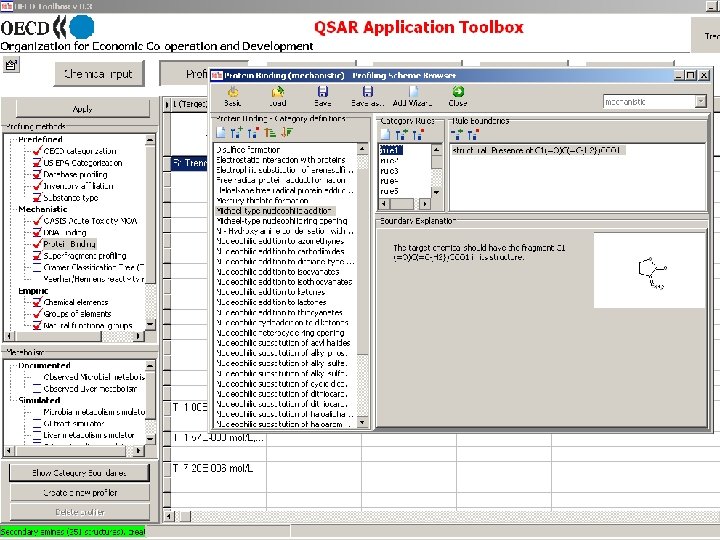
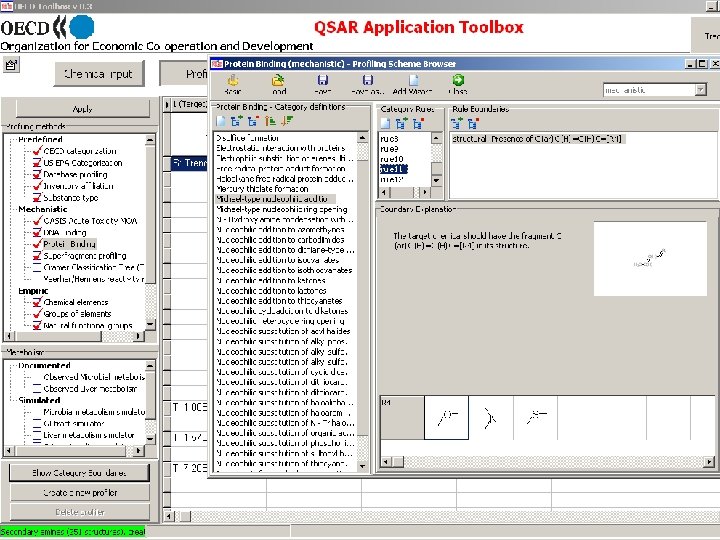
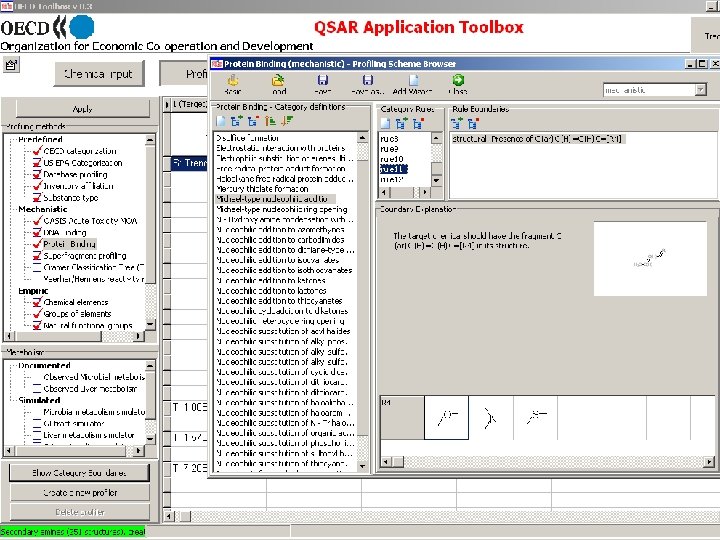
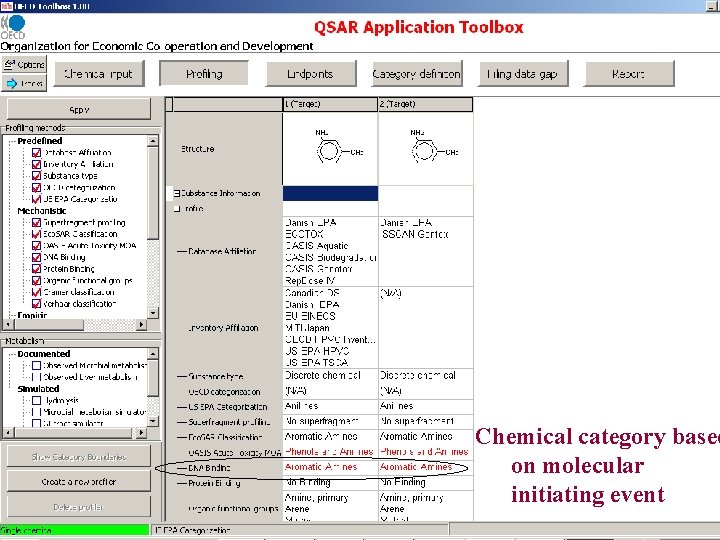
Toolbox logical sequence of components usage Chemical Profiling input Endpoints Category Definition Filling data gap Report General characterization by the following grouping schemes: • • • Substance information Predefined Mechanistic: • Acute Toxicity MOA (OASIS) • Protein binding (OASIS) • DNA binding (OASIS) • Electron reach fragments (Superfragments) Bio. Bite • Cramer Classification Tree (Tox. Tree) • Veerhar/Hermens reactivity rules (Tox. Tree) • Lipinski rules (Multi. Case)








Molecular Initiating Events and Toxicological Pathways General Consideration

Molecular Level Mechanism of chemical interactions

Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 …

Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Distribution in lipid phase Protein binding Arylcarboxylate aminolysis Michael-type addition Schiff base formation … DNA binding Quinones Hydrazines …

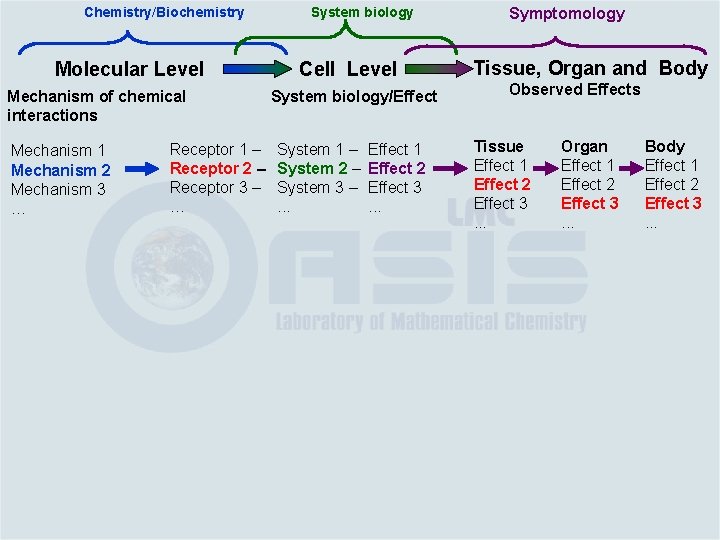
Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … Distribution in lipid phase Protein binding Arylcarboxylate aminolysis Michael-type addition Schiff base formation … DNA binding Quinones Hydrazines …

Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … Initiating event/Receptor • Activation of AP-1、NF-k. B、Ep. RE in hepatocyte →Activation of JNK/AP 1 pathway • Activation of estrogen Signals → Proliferation of bile duct cell and hepatocyte injury • Activation of MAPK Signals Apoptosis • …

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … Initiating event/Receptor • Activation of AP-1、NF-k. B、Ep. RE in hepatocyte →Activation of JNK/AP 1 pathway • Activation of estrogen Signals → Proliferation of bile duct cell and hepatocyte injury • Activation of MAPK Signals Apoptosis • …

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … Cell Level System biology/Effect

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … Cell Level System biology/Effect System 1 – System 2 – System 3 –. . .

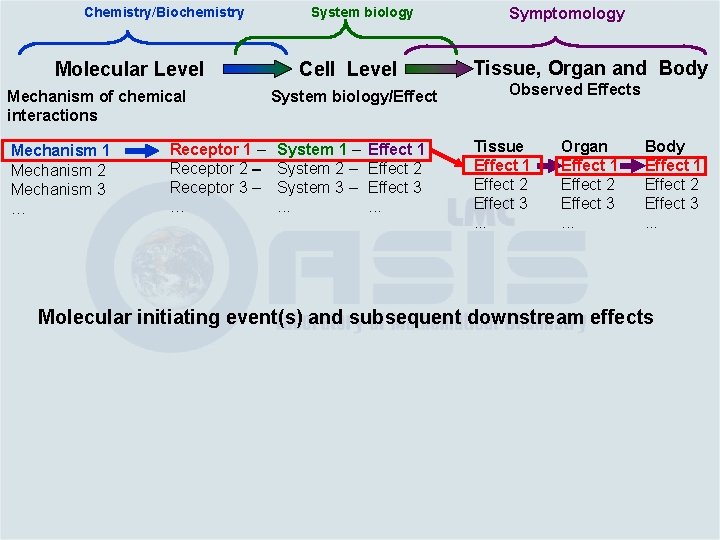
Chemistry/Biochemistry Molecular Level Cell Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … System biology/Effect Receptor 1 – Receptor 2 – Receptor 3 – … System 1 – System 2 – System 3 –. . . System biology Hepatotoxicity mechanism: • Oxidant stress • Mitochondrial damage • Apoptosis • Degradation of membrane phospholipid • Aberration of ion channel • Increase of enzyme activition of drug metabolism • Inflammatory responses • …

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . . Cell Effects • Hepatocyte • Changes in the tubular epithelium • …

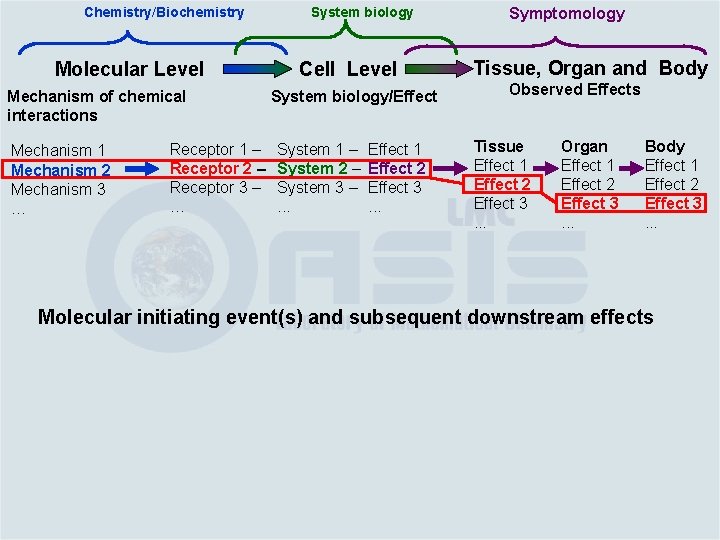
Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … System biology Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . .

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … System biology Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . . Symptomology Tissue, Organ and Body Observed Effects

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … System biology Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . . Symptomology Tissue, Organ and Body Observed Effects Tissue Effect 1 Effect 2 Effect 3. . . Organ Effect 1 Effect 2 Effect 3. . . Body Effect 1 Effect 2 Effect 3. . .

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … System biology Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . . Symptomology Tissue, Organ and Body Observed Effects Tissue Effect 1 Effect 2 Effect 3. . . Organ Effect 1 Effect 2 Effect 3. . . Body Effect 1 Effect 2 Effect 3. . . Molecular initiating event(s) and subsequent downstream effects

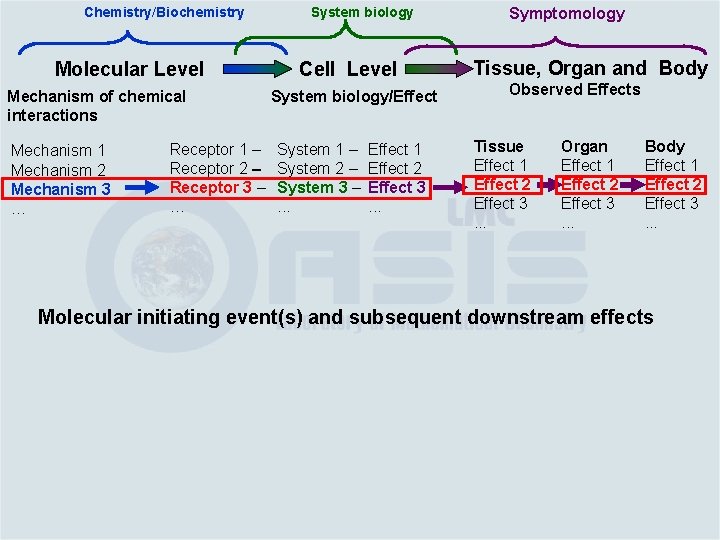
Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … System biology Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . . Symptomology Tissue, Organ and Body Observed Effects Tissue Effect 1 Effect 2 Effect 3. . . Organ Effect 1 Effect 2 Effect 3. . . Body Effect 1 Effect 2 Effect 3. . . Molecular initiating event(s) and subsequent downstream effects

Chemistry/Biochemistry Molecular Level Mechanism of chemical interactions Mechanism 1 Mechanism 2 Mechanism 3 … Receptor 1 – Receptor 2 – Receptor 3 – … System biology Cell Level System biology/Effect System 1 – System 2 – System 3 –. . . Effect 1 Effect 2 Effect 3. . . Symptomology Tissue, Organ and Body Observed Effects Tissue Effect 1 Effect 2 Effect 3. . . Organ Effect 1 Effect 2 Effect 3. . . Body Effect 1 Effect 2 Effect 3. . . Molecular initiating event(s) and subsequent downstream effects

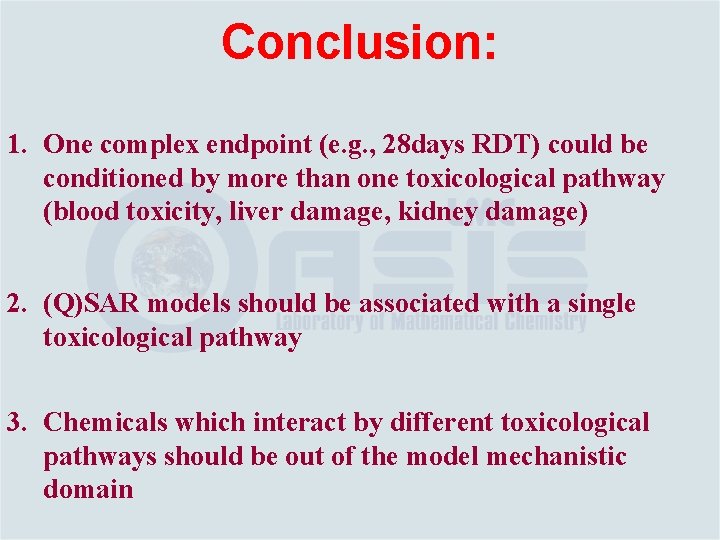
Conclusion: 1. One complex endpoint (e. g. , 28 days RDT) could be conditioned by more than one toxicological pathway (blood toxicity, liver damage, kidney damage) 2. (Q)SAR models should be associated with a single toxicological pathway 3. Chemicals which interact by different toxicological pathways should be out of the model mechanistic domain

Conclusion: 4. The categorization of substances according to chemical mechanisms governing the initiating reactions with protein or DNA is not enough for predicting human health effects resulting from multiple and complex toxicological pathways 5. The link between chemical and toxicological mechanisms and respective categorization schemes needs to be identified

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

Case study: Twenty-eight day repeat dose oral toxicity test of chemicals (28 d RDT) 1. Data produced by: v Safety examination of existing chemicals in NITE- Japan; under Japanese Chemical Substances Control Law; v Fraunhofer Institute for Toxicology and Experimental Medicine, Hanover, Germany 2. Categorization of chemicals for predicting 28 d RDT is based on analysis of data by NITE and LMC

28 -day RDT tests conducted on male rats that tested 14 aromatic amines

Categorization of Anilines 1. Based on their effects on two organs: v Blood v Kidney

Categorization of Anilines 1. Based on their effects on two organs: v Blood v Kidney

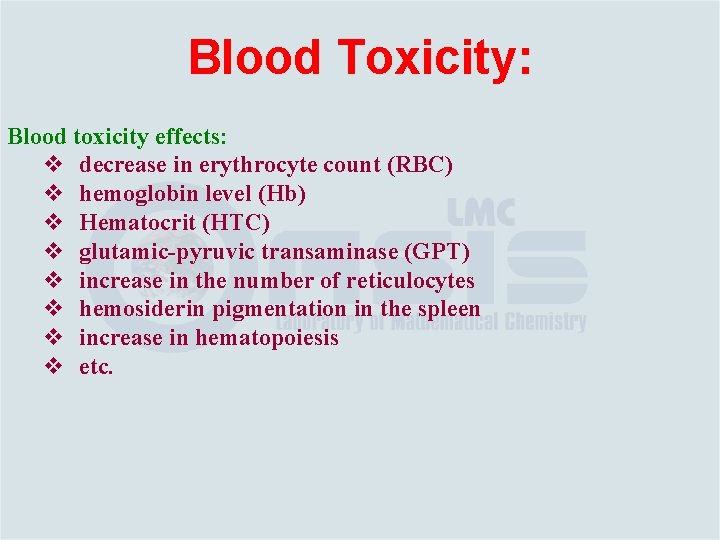
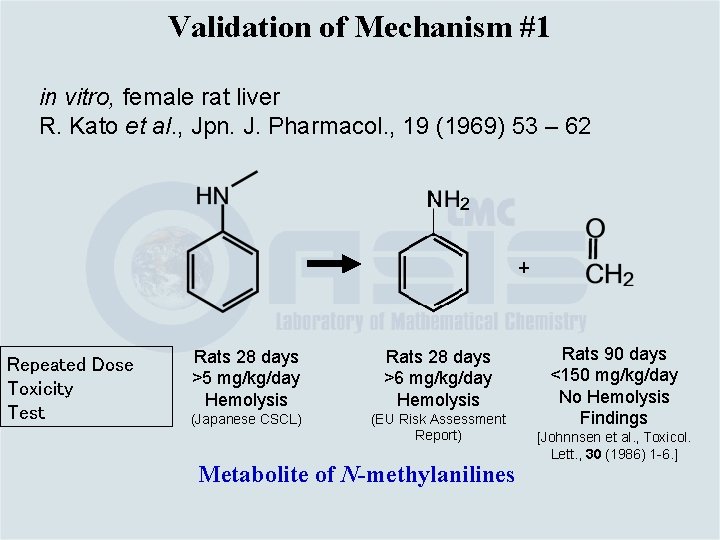
Blood Toxicity: Blood toxicity effects: v decrease in erythrocyte count (RBC) v hemoglobin level (Hb) v Hematocrit (HTC) v glutamic-pyruvic transaminase (GPT) v increase in the number of reticulocytes v hemosiderin pigmentation in the spleen v increase in hematopoiesis v etc.

Toxicity scale of Intensity scale: basis of the number of effects indicative of toxicity v strong v medium v weak v non

Toxicity scale of Intensity Example: determining LOEL of N-ethylaniline: v Test doses - 0, 5, 25, 125 mg/kg/day v Decrease in RBC only has been observed at 5 mg/kg v Decrease in RBC, Hb and HTC–at 25 and 125 mg/kg v Hence, LOEL for hemolysis is 5 mg/kg/day

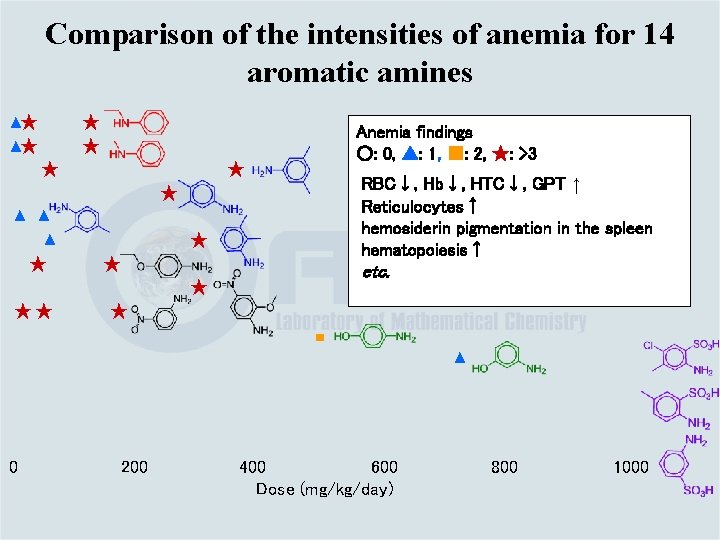
Comparison of the intensities of anemia for 14 aromatic amines ▲★ ▲★ ★ Anemia findings ○: 0, ▲: 1, ■: 2, ★: >3 ★ RBC↓, Hb↓, HTC↓, GPT ↑ Reticulocytes↑ hemosiderin pigmentation in the spleen hematopoiesis↑ ★ ▲ ▲ ▲ ★ ★ ★ etc. ★ ★★ ★ ■ ▲ 0 200 400 600 Dose (mg/kg/day) 800 1000

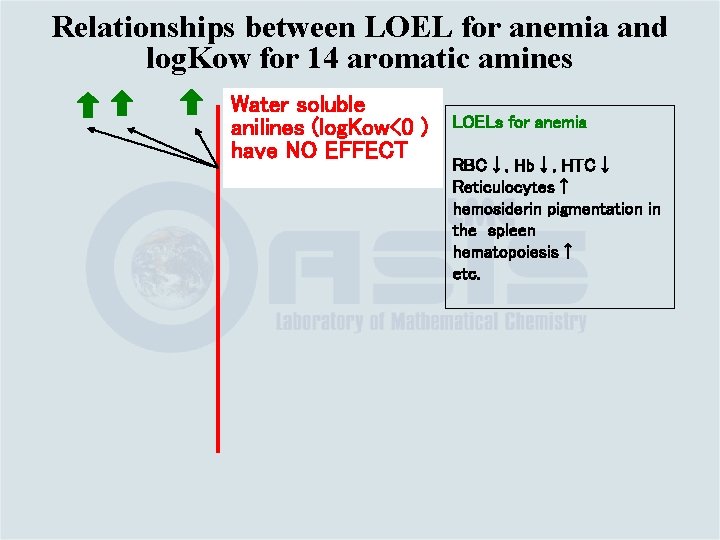
Relationships between LOEL for anemia and log. Kow for 14 aromatic amines Water soluble anilines (log. Kow<0 ) have NO EFFECT LOELs for anemia RBC↓, Hb↓, HTC↓ Reticulocytes↑ hemosiderin pigmentation in the spleen hematopoiesis↑ etc.

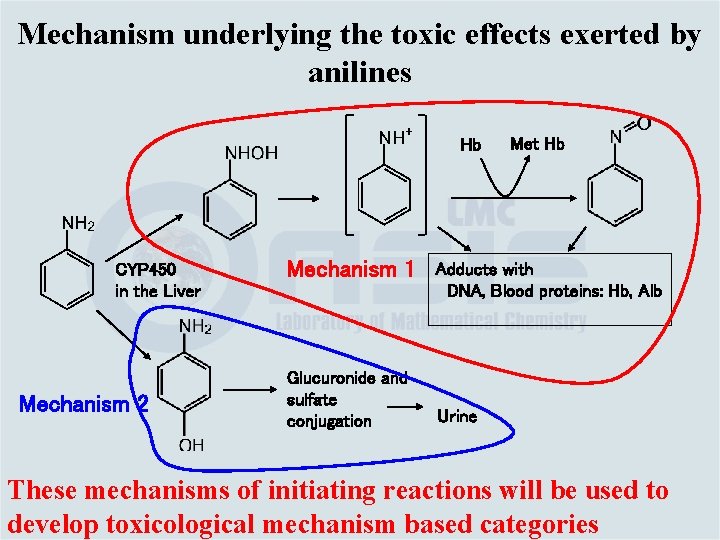
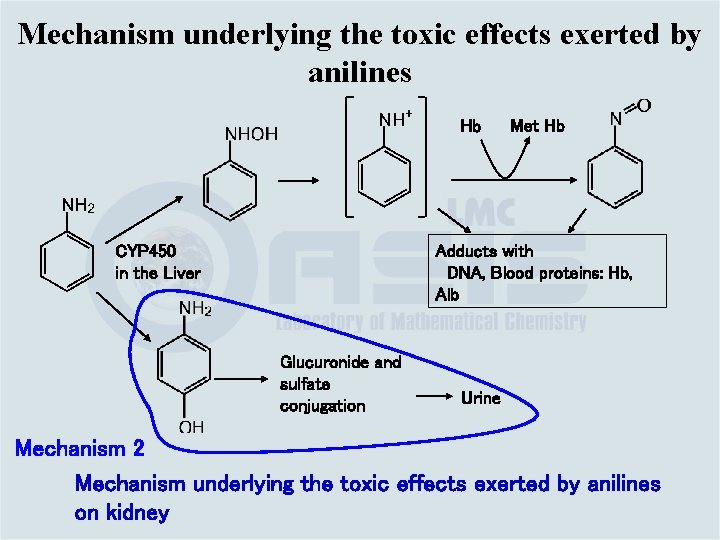
Mechanism underlying the toxic effects exerted by anilines Hb CYP 450 in the Liver Mechanism 2 Mechanism 1 Glucuronide and sulfate conjugation Met Hb Adducts with DNA, Blood proteins: Hb, Alb Urine These mechanisms of initiating reactions will be used to develop toxicological mechanism based categories

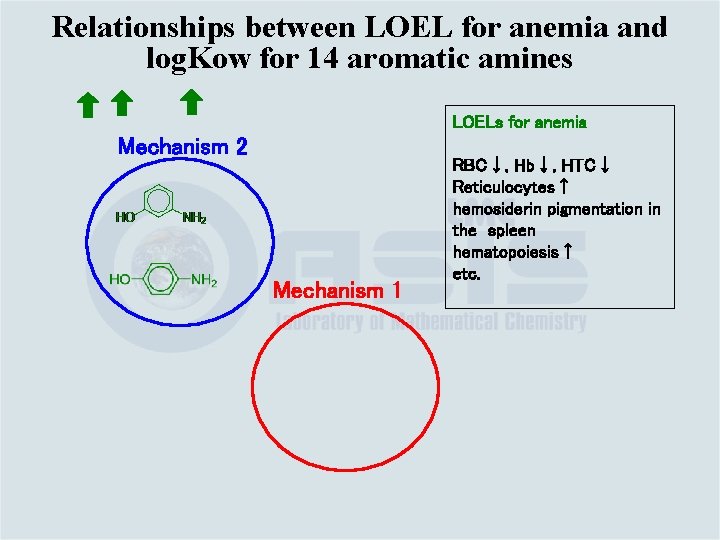
Relationships between LOEL for anemia and log. Kow for 14 aromatic amines LOELs for anemia Mechanism 2 Mechanism 1 RBC↓, Hb↓, HTC↓ Reticulocytes↑ hemosiderin pigmentation in the spleen hematopoiesis↑ etc.

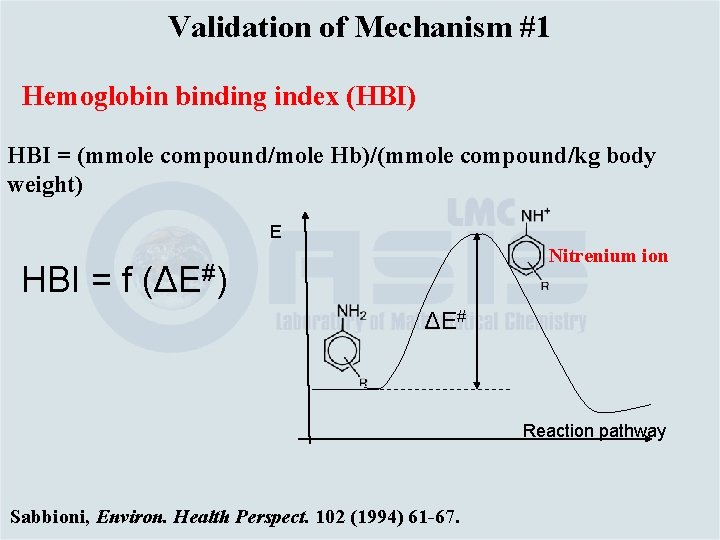
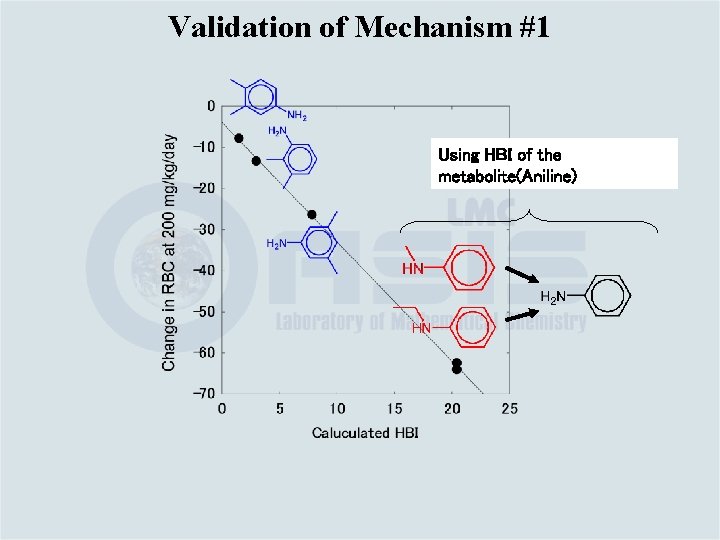
Validation of Mechanism #1 Hemoglobin binding index (HBI) HBI = (mmole compound/mole Hb)/(mmole compound/kg body weight) E Nitrenium ion HBI = f (ΔE#) ΔE# Reaction pathway Sabbioni, Environ. Health Perspect. 102 (1994) 61 -67.
![Validation of Mechanism 1 Calculated HBI E e V vs change in RBC Validation of Mechanism #1 Calculated HBI ( E# [e. V]) vs. change in RBC](https://slidetodoc.com/presentation_image/98df2dd4315d47662c9c5ec34cea026e/image-61.jpg)
Validation of Mechanism #1 Calculated HBI ( E# [e. V]) vs. change in RBC in RDT test

Validation of Mechanism #1 in vitro, female rat liver R. Kato et al. , Jpn. J. Pharmacol. , 19 (1969) 53 – 62 + Repeated Dose Toxicity Test Rats 28 days >5 mg/kg/day Hemolysis Rats 28 days >6 mg/kg/day Hemolysis (Japanese CSCL) (EU Risk Assessment Report) Metabolite of N-methylanilines Rats 90 days <150 mg/kg/day No Hemolysis Findings [Johnnsen et al. , Toxicol. Lett. , 30 (1986) 1 -6. ]

Validation of Mechanism #1 Using HBI of the metabolite(Aniline)

Categorization of Anilines 1. Based on their effects on two organs: v Blood v Kidney

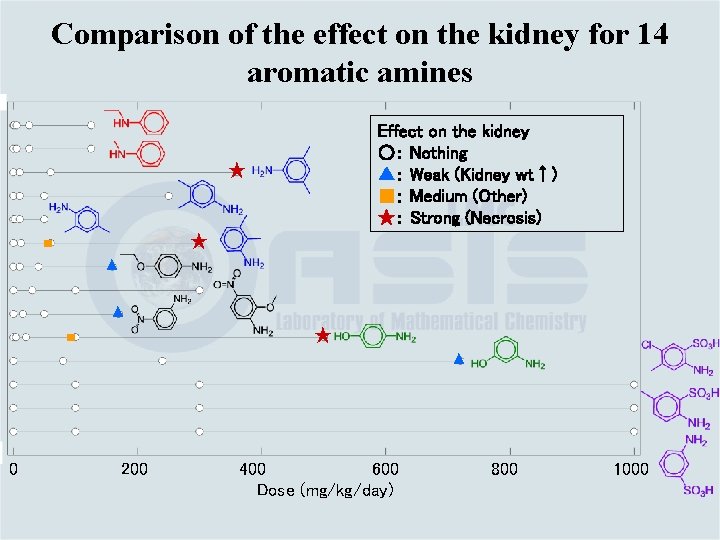
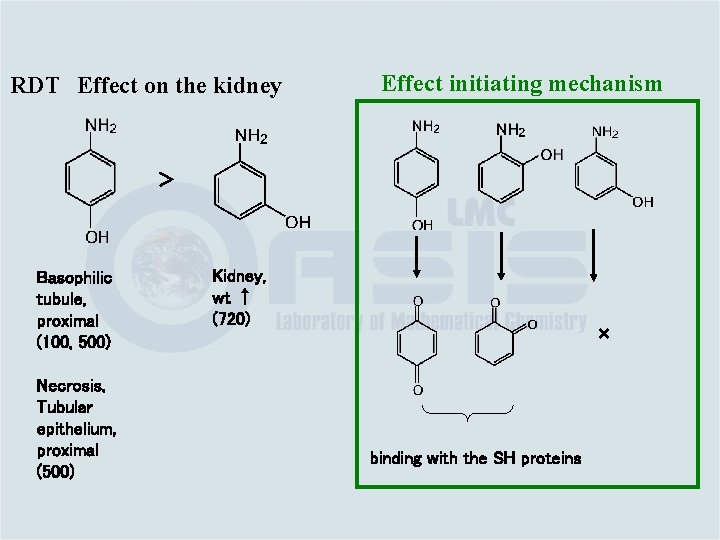
Comparison of the effect on the kidney for 14 aromatic amines Effect on the kidney ○: Nothing ▲: Weak (Kidney wt↑) ■: Medium (Other) ★: Strong (Necrosis) ★ ★ ■ ▲ ▲ ★ ■ ▲ 0 200 400 600 Dose (mg/kg/day) 800 1000

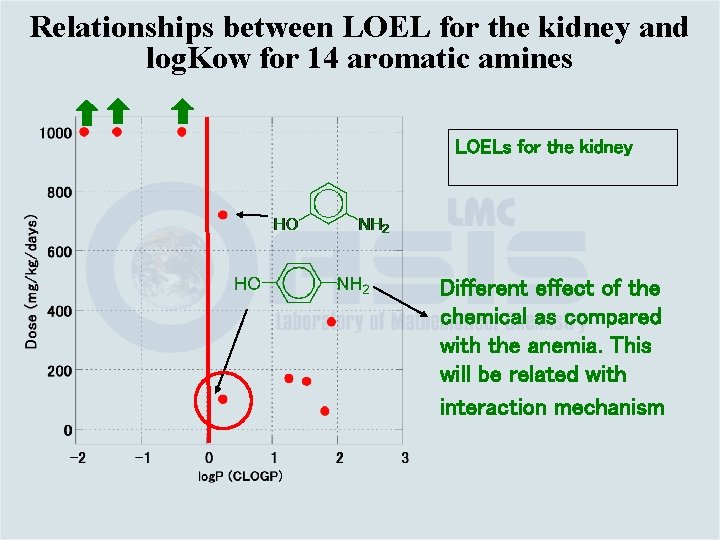
Relationships between LOEL for the kidney and log. Kow for 14 aromatic amines LOELs for the kidney Different effect of the chemical as compared with the anemia. This will be related with interaction mechanism

Mechanism underlying the toxic effects exerted by anilines Hb Met Hb Adducts with DNA, Blood proteins: Hb, Alb CYP 450 in the Liver Glucuronide and sulfate conjugation Urine Mechanism 2 Mechanism underlying the toxic effects exerted by anilines on kidney

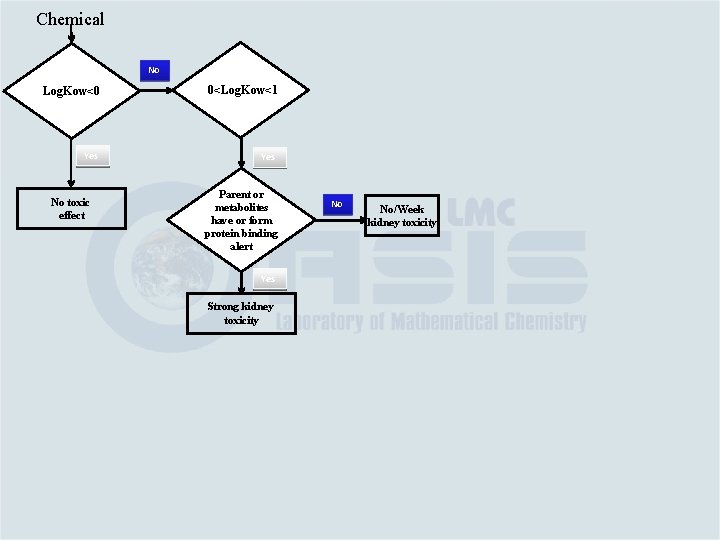
RDT Effect on the kidney Effect initiating mechanism > Basophilic tubule, proximal (100, 500) Necrosis, Tubular epithelium, proximal (500) Kidney, wt ↑ (720) × binding with the SH proteins

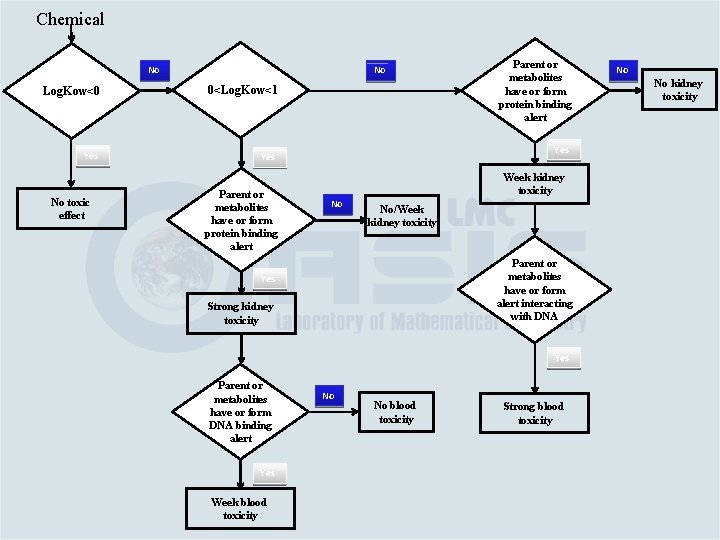
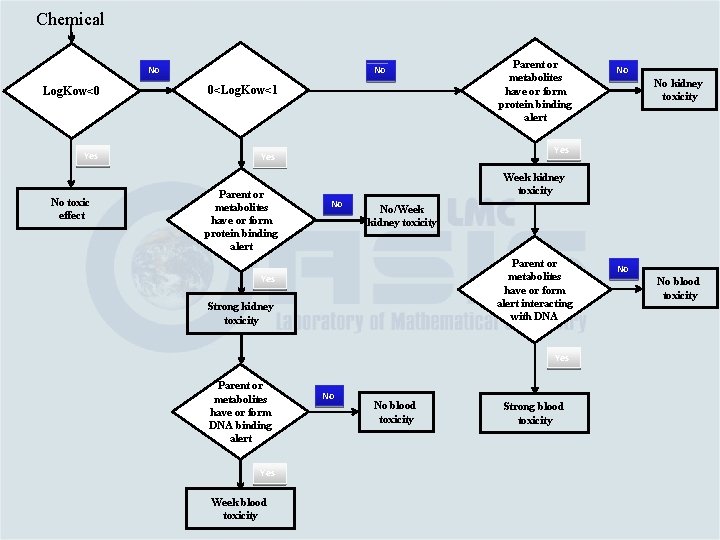
Category building based on the link between chemical and toxicological mechanisms

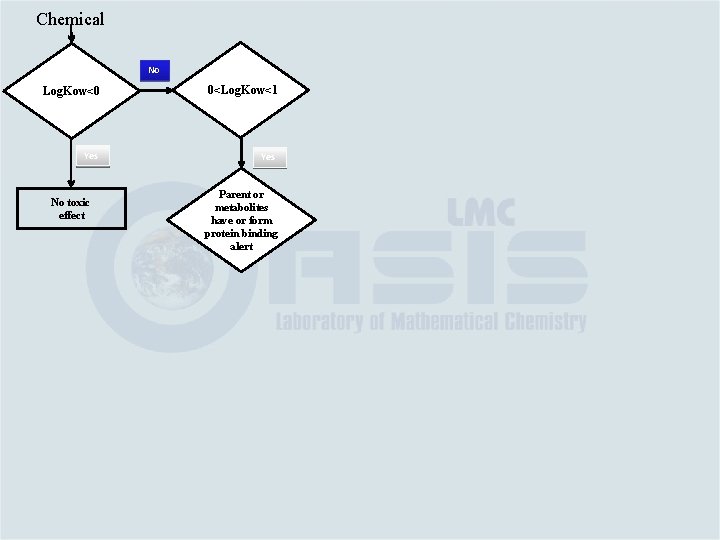
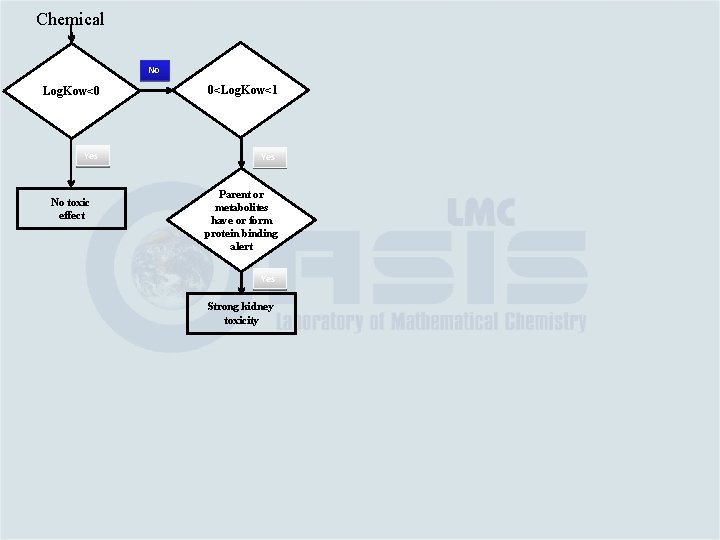
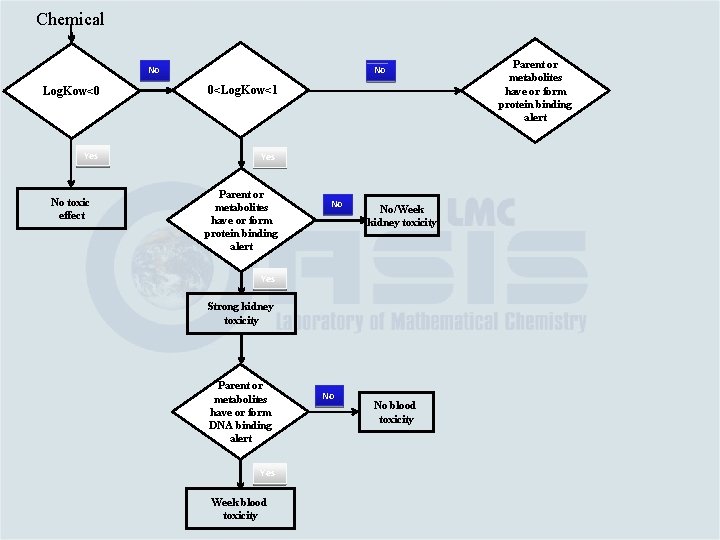
Category Building Based on the link between chemical and toxicological mechanisms Category #1. Water soluble aromatic amines – log. Kow ≤ 0 – no effect Category #2. If 0<log. Kow ≤ 1; it is eliminated by mechanism #2 and as parent or metabolite has alerting groups interacting with: v. Proteins (such as quinone imines; Mechanisms #2) – strong kidney toxicity and weak blood toxicity (Category #2 a) v. DNA or blood proteins (Mechanisms #1) – week blood toxicity (Category #2 b)

Category Building Based on the link between chemical and toxicological mechanisms Category #3. If log. Kow > 1 and as parent or metabolite has alerting groups interacting with: v. Proteins (Mechanisms #2) – week kidney toxicity (Category #3 a) v. DNA (Mechanisms #1) – strong blood toxicity (Category #3 b)

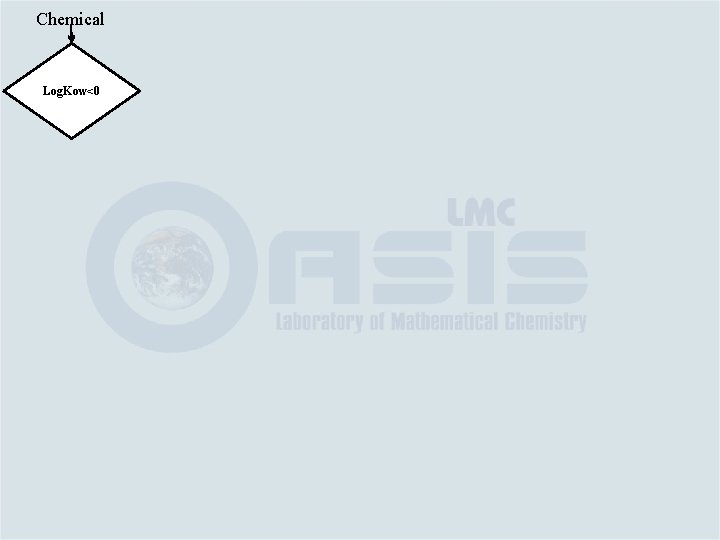
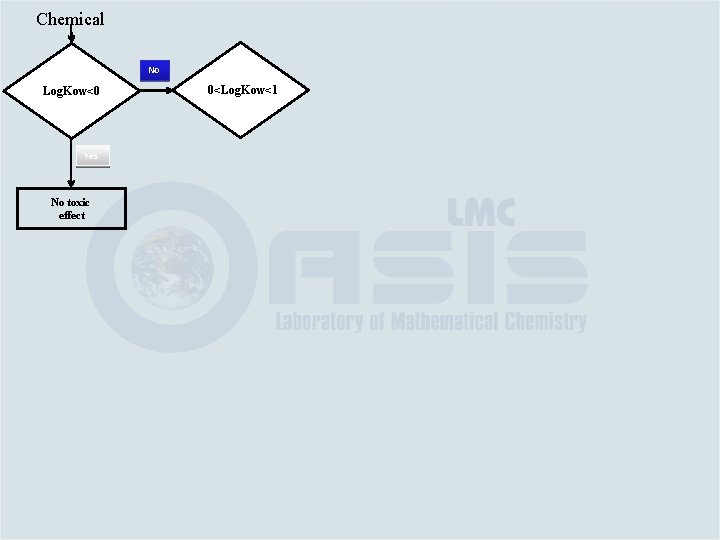
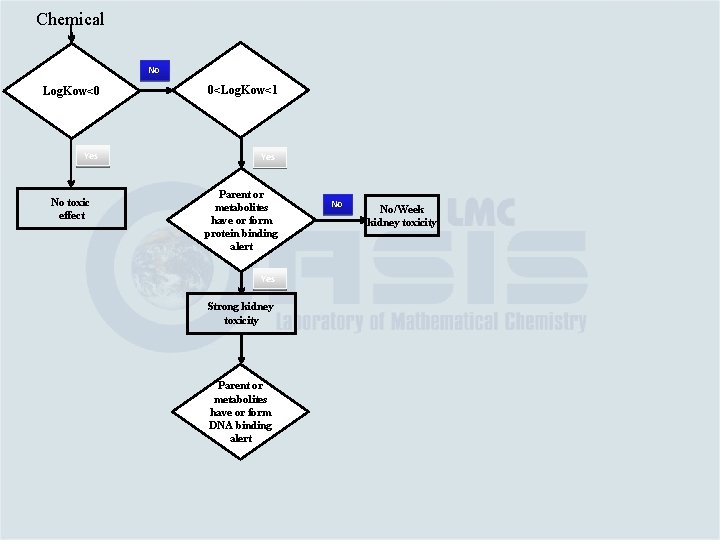
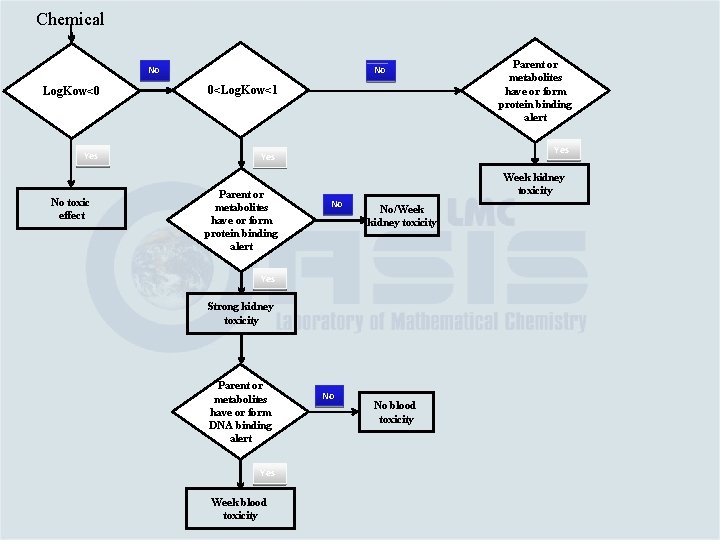
Chemical Log. Kow<0

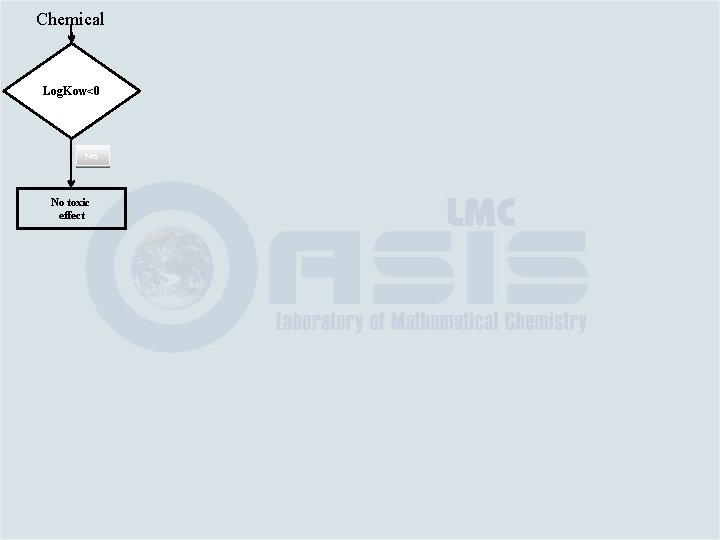
Chemical Log. Kow<0 Yes No toxic effect

Chemical No Log. Kow<0 Yes No toxic effect 0<Log. Kow<1

Chemical No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert

Chemical No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Strong kidney toxicity

Chemical No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Strong kidney toxicity No No/Week kidney toxicity

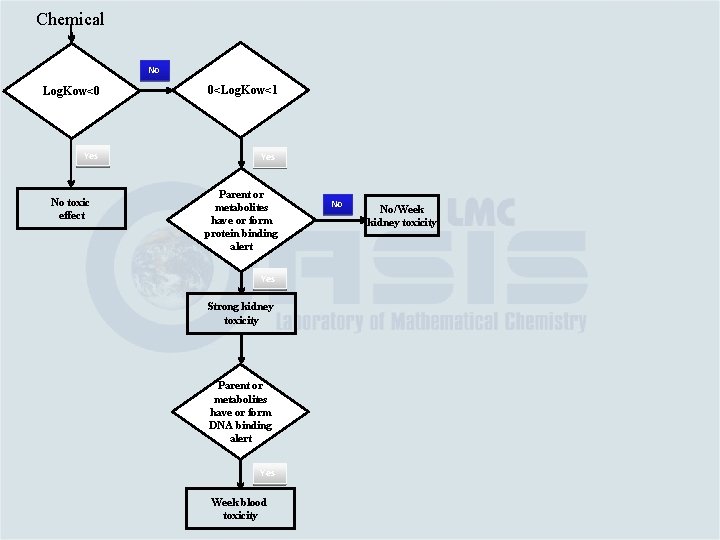
Chemical No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert No No/Week kidney toxicity

Chemical No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert Yes Week blood toxicity No No/Week kidney toxicity

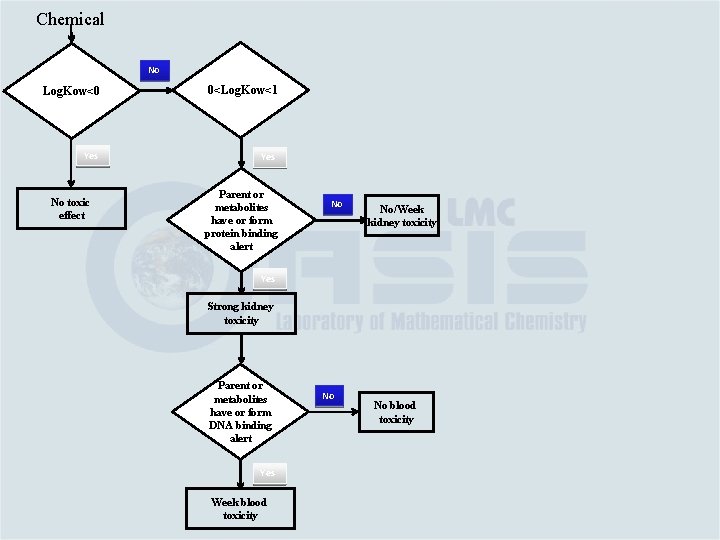
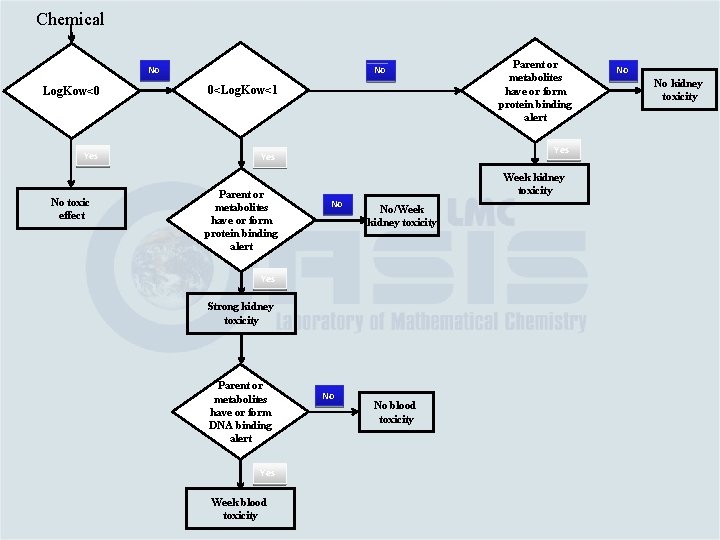
Chemical No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert No No/Week kidney toxicity Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert Yes Week blood toxicity No No blood toxicity

Chemical No No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert No No/Week kidney toxicity Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert Yes Week blood toxicity No No blood toxicity Parent or metabolites have or form protein binding alert

Chemical No No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Week kidney toxicity No No/Week kidney toxicity Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert Yes Week blood toxicity Parent or metabolites have or form protein binding alert No No blood toxicity

Chemical No No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Week kidney toxicity No No/Week kidney toxicity Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert Yes Week blood toxicity Parent or metabolites have or form protein binding alert No No blood toxicity No No kidney toxicity

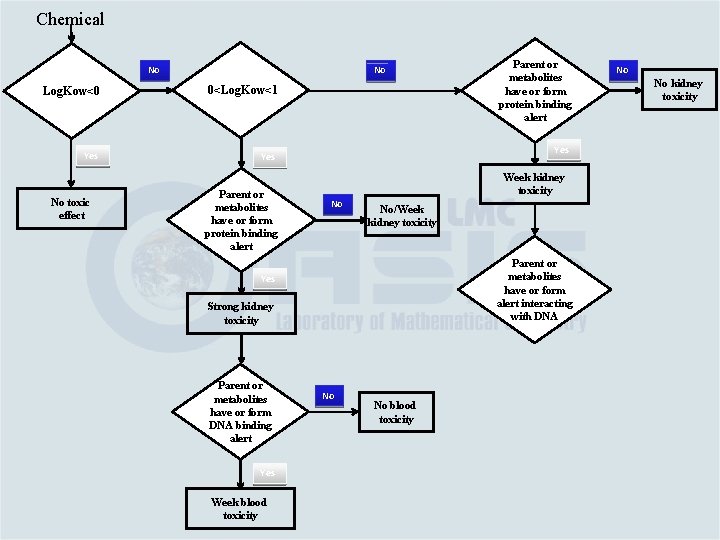
Chemical No No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Week kidney toxicity No No/Week kidney toxicity Parent or metabolites have or form alert interacting with DNA Yes Strong kidney toxicity Parent or metabolites have or form DNA binding alert Yes Week blood toxicity Parent or metabolites have or form protein binding alert No No blood toxicity No No kidney toxicity

Chemical No No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Yes Week kidney toxicity No No/Week kidney toxicity Parent or metabolites have or form alert interacting with DNA Yes Strong kidney toxicity Yes Parent or metabolites have or form DNA binding alert Yes Week blood toxicity No No blood toxicity Strong blood toxicity No No kidney toxicity

Chemical No No Log. Kow<0 0<Log. Kow<1 Yes No toxic effect Parent or metabolites have or form protein binding alert Week kidney toxicity No No/Week kidney toxicity Parent or metabolites have or form alert interacting with DNA Strong kidney toxicity Yes Week blood toxicity No kidney toxicity Yes Parent or metabolites have or form DNA binding alert No No No blood toxicity Strong blood toxicity No No blood toxicity

New Pilot Functionalities in Toolbox Demonstrated by using RDT database, Fraunhover ITEM, Hanover, Germany

Input chemical

Chemical category based on molecular initiating event

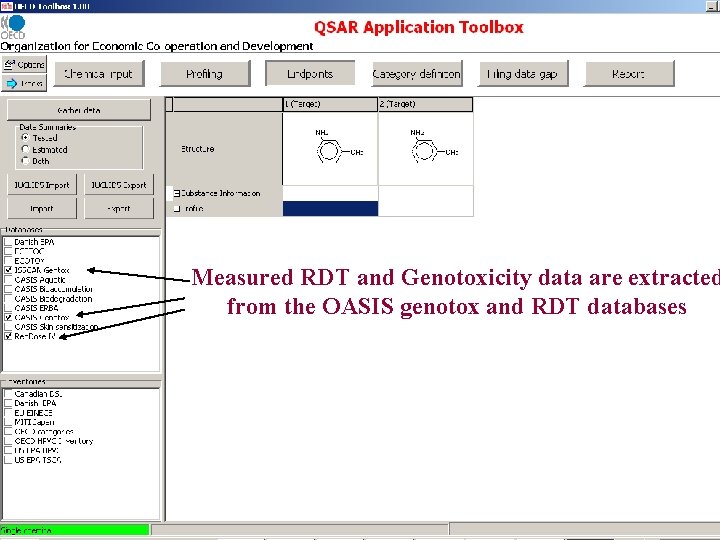
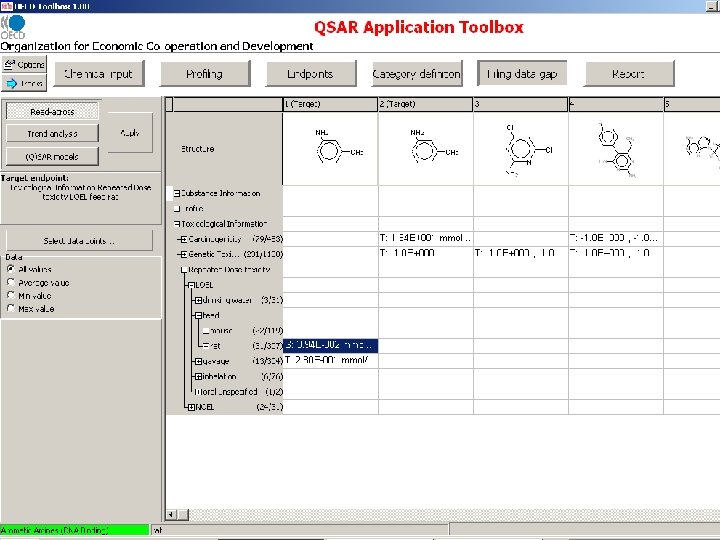
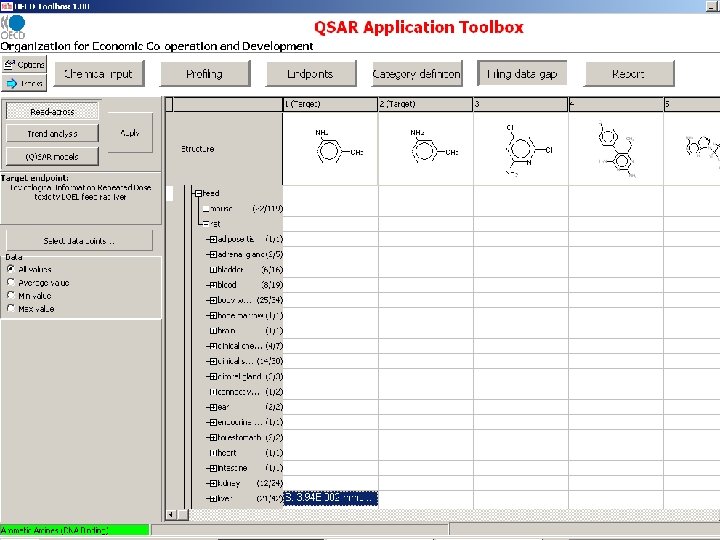
Measured RDT and Genotoxicity data are extracted from the OASIS genotox and RDT databases

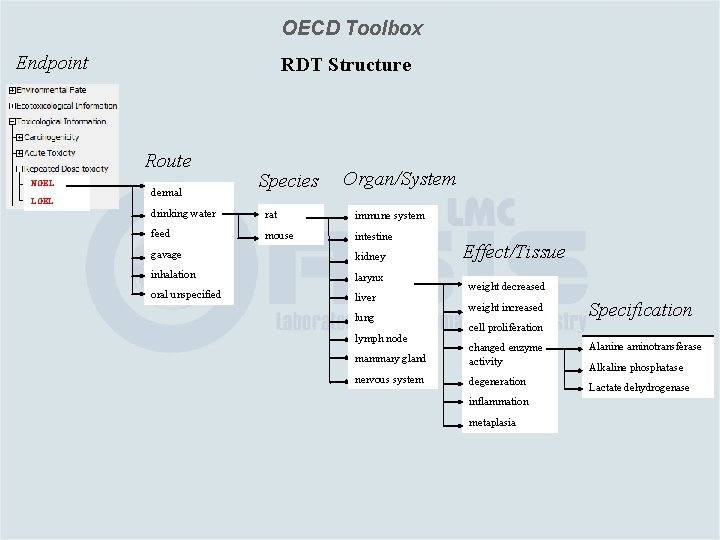
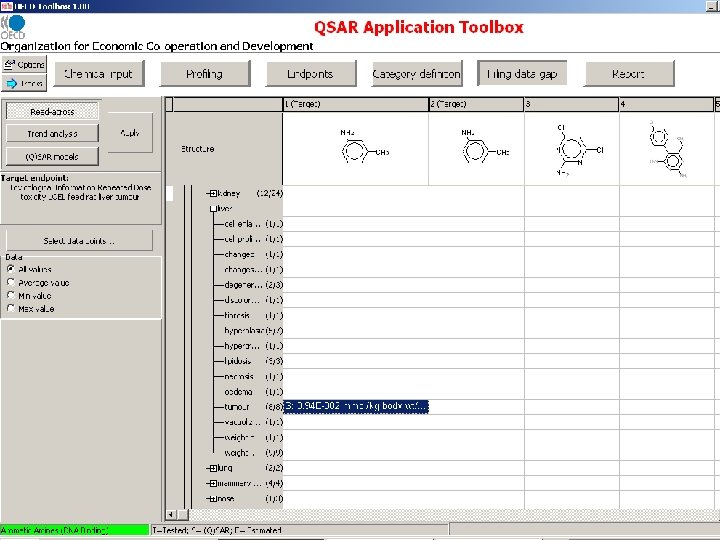
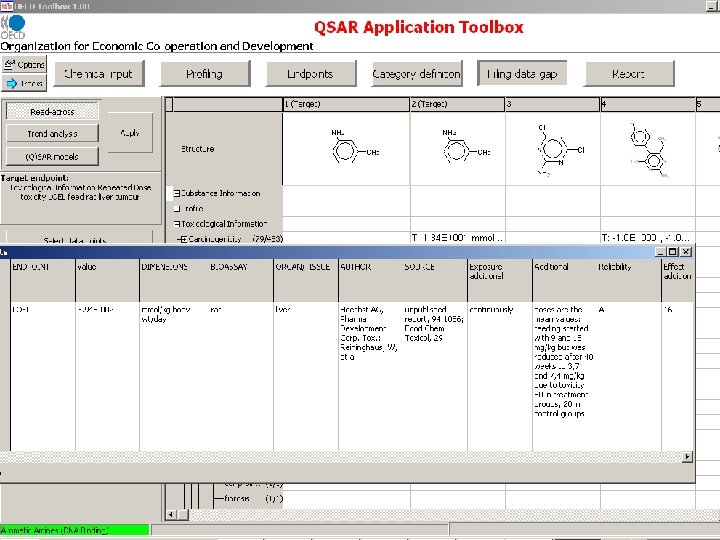
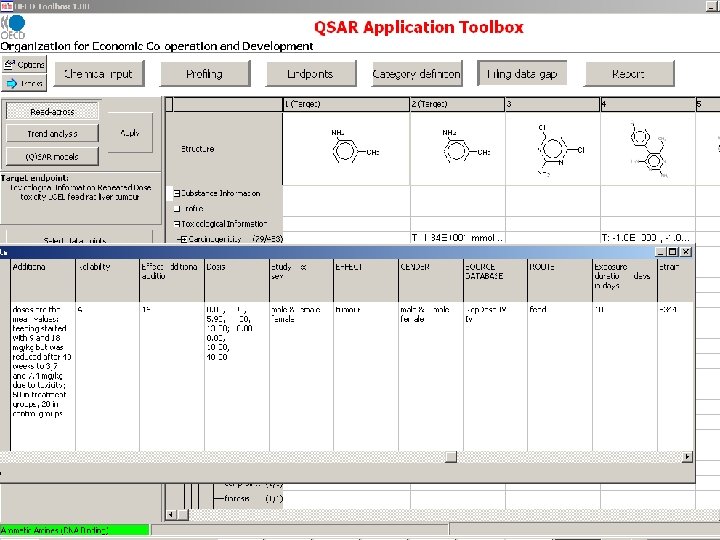
OECD Toolbox Endpoint RDT Structure Route NOEL LOEL dermal Species Organ/System drinking water rat immune system feed mouse intestine gavage kidney inhalation larynx oral unspecified liver lung lymph node Effect/Tissue weight decreased weight increased Specification cell proliferation mammary gland changed enzyme activity nervous system degeneration inflammation metaplasia Alanine aminotransferase Alkaline phosphatase Lactate dehydrogenase

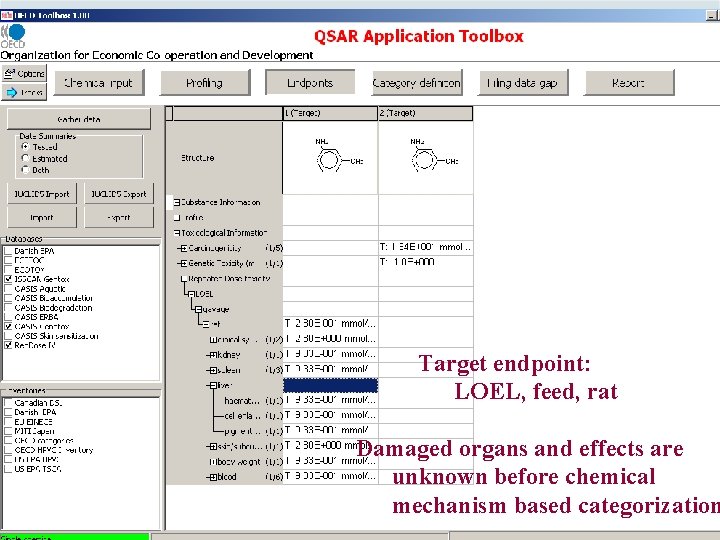
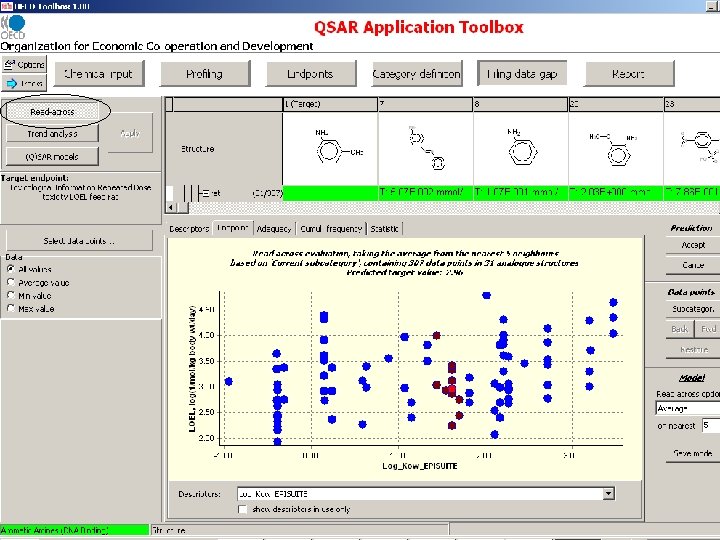
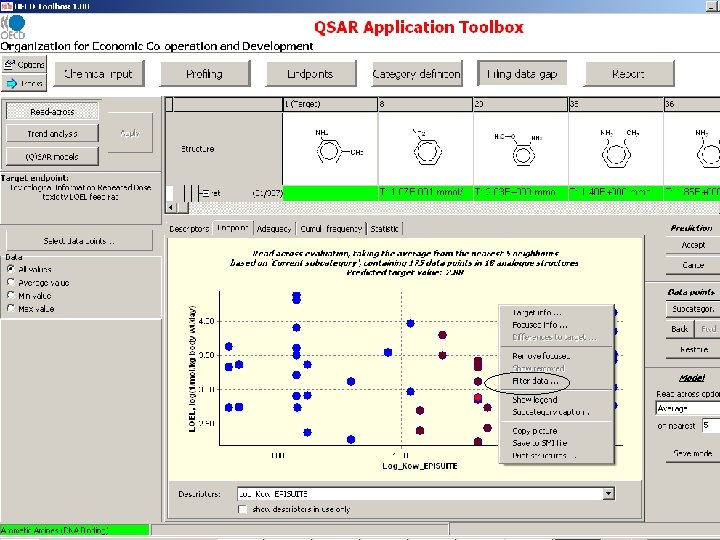
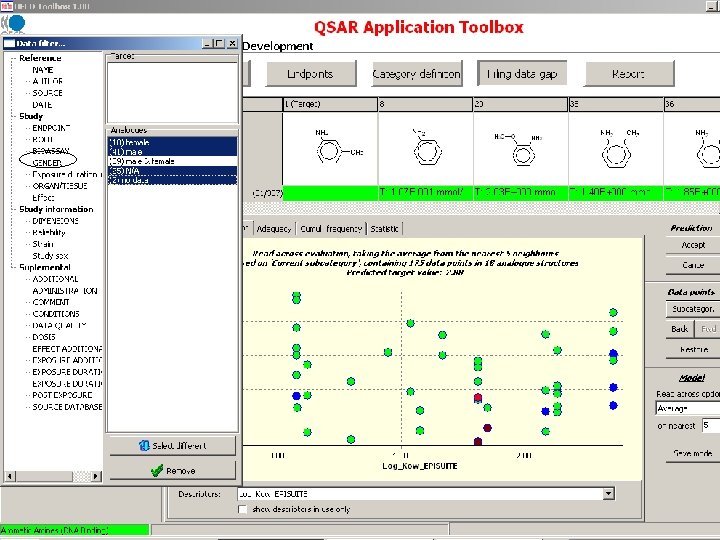
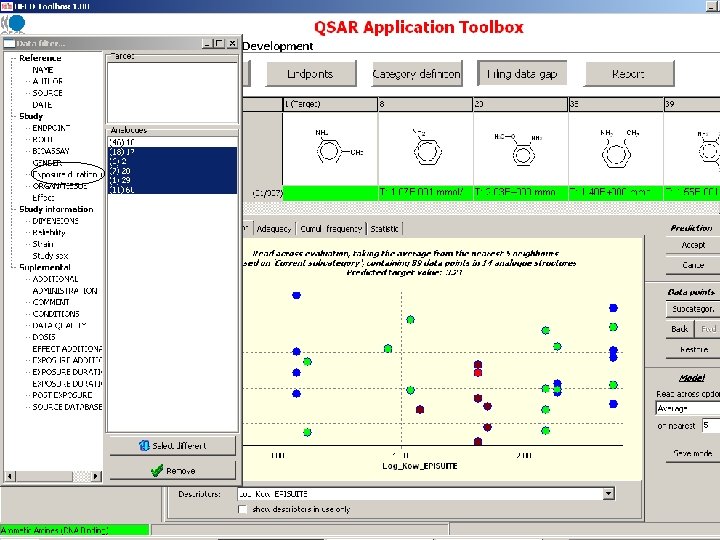
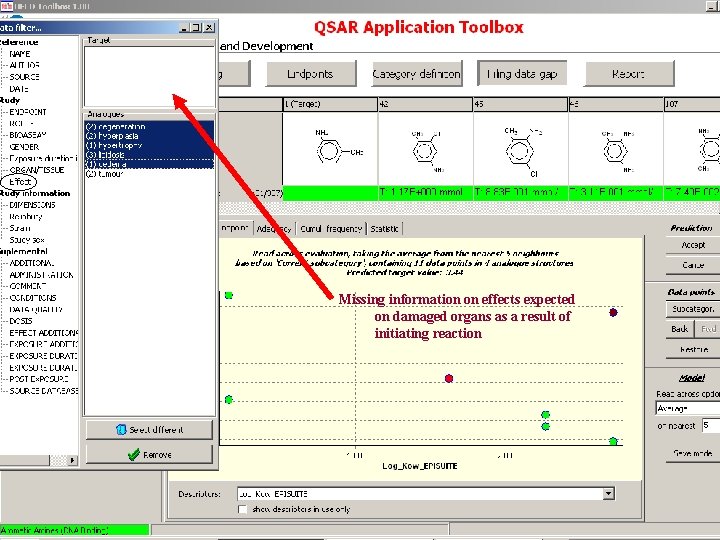
Target endpoint: LOEL, feed, rat Damaged organs and effects are unknown before chemical mechanism based categorization

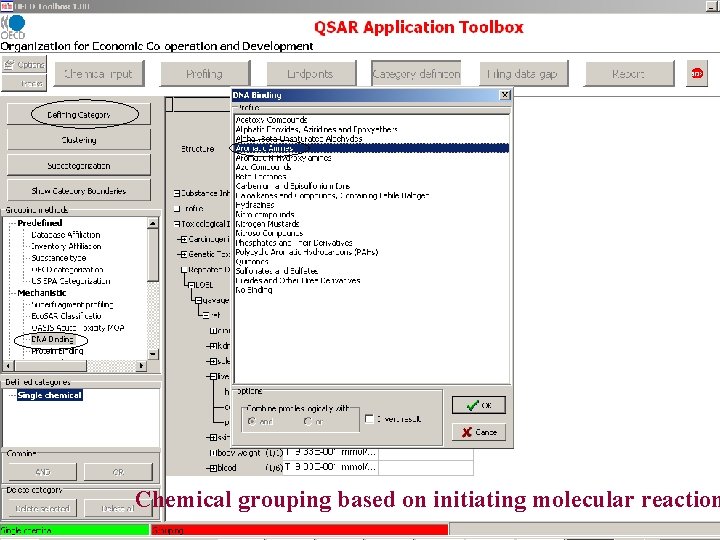
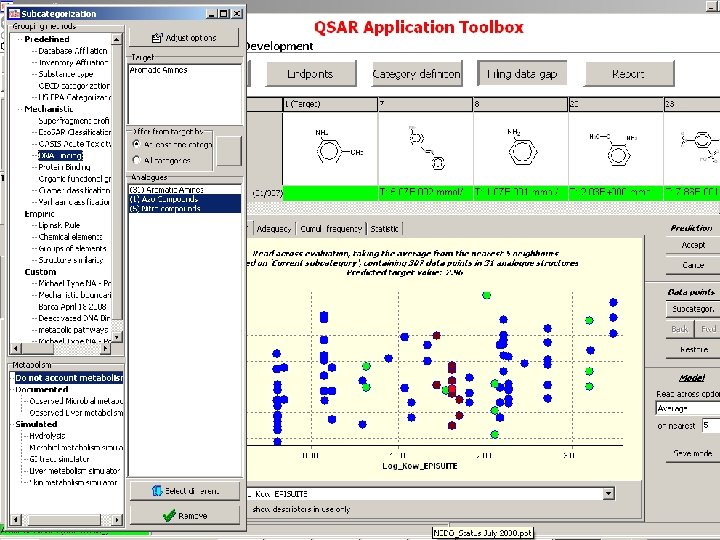
Chemical grouping based on initiating molecular reaction

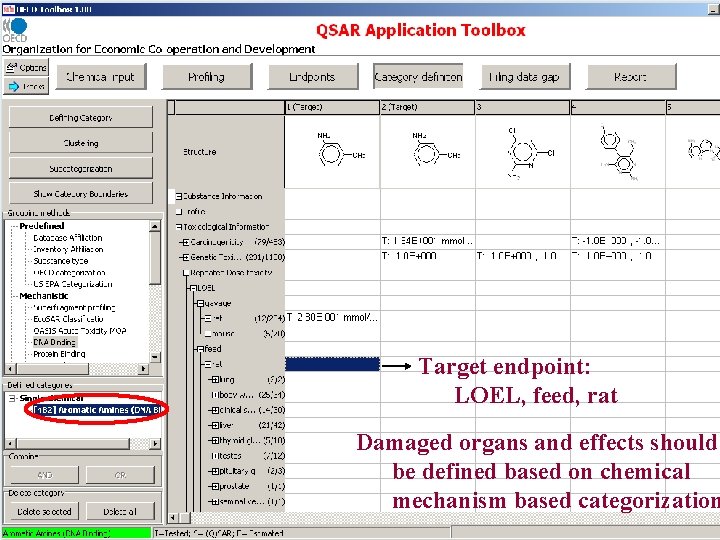
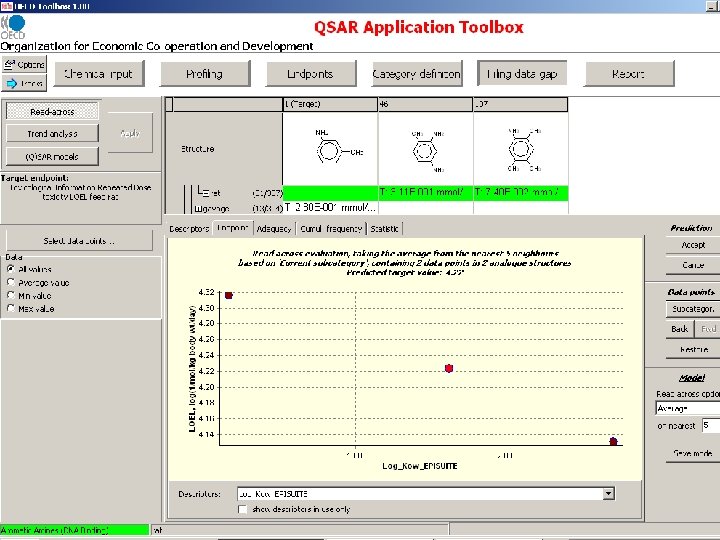
Target endpoint: LOEL, feed, rat Damaged organs and effects should be defined based on chemical mechanism based categorization


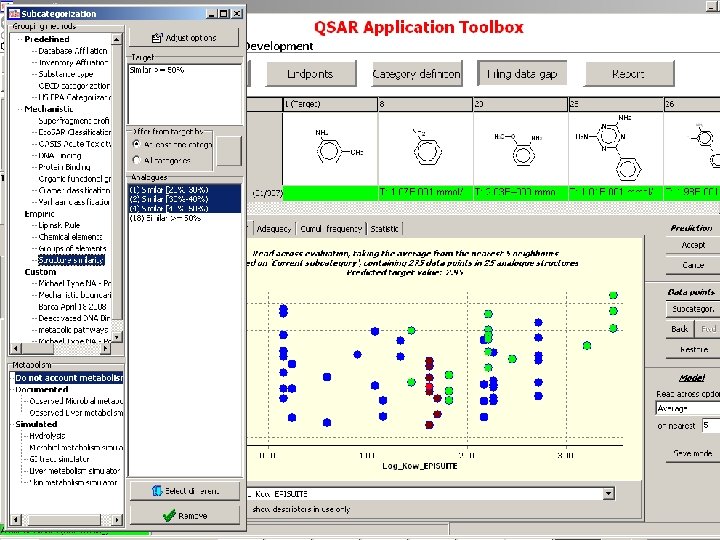
Subcategorization based on chemical interaction mechanisms and/or structure based similarity



Subcategorization based on toxicological mechanisms resulting from underlying chemical interaction mechanisms




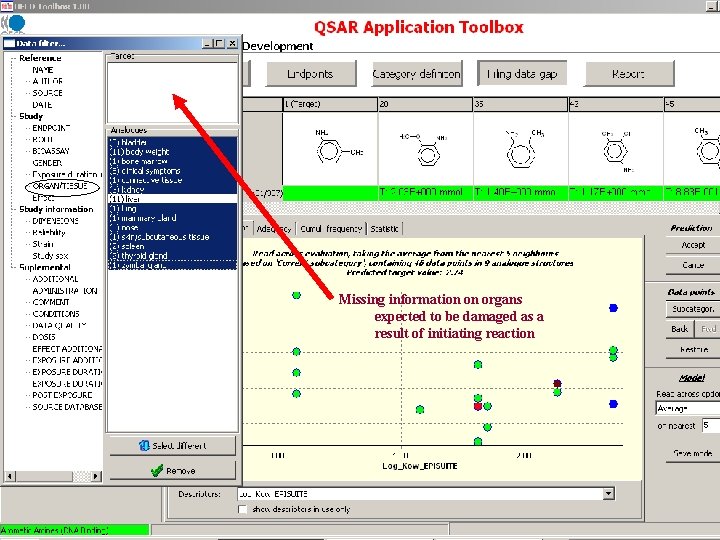
Missing information on organs expected to be damaged as a result of initiating reaction

Missing information on effects expected on damaged organs as a result of initiating reaction







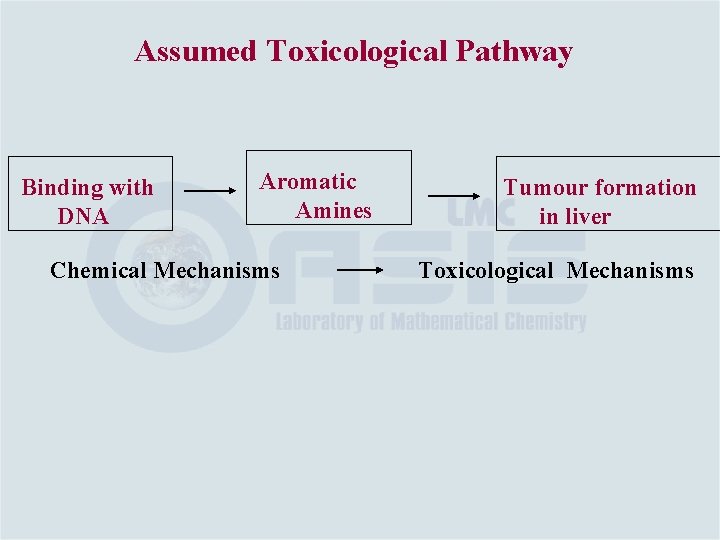
Assumed Toxicological Pathway Binding with DNA Aromatic Amines Chemical Mechanisms Tumour formation in liver Toxicological Mechanisms

Defining toxicological pathway and building the mechanism data base is critical for the Toolbox project Chemical Mechanisms Toxicological Mechanisms

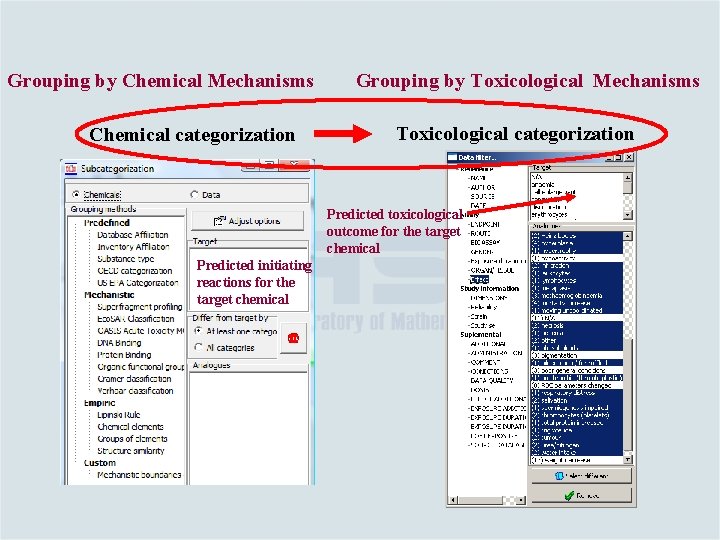
Grouping by Chemical Mechanisms Chemical categorization Grouping by Toxicological Mechanisms Toxicological categorization Predicted toxicological outcome for the target chemical Predicted initiating reactions for the target chemical

Outline v Conceptual framework of QSAR v Categorization and QSAR v Predicting human health endpoints in Toolbox v Molecular initiating events and toxicological pathways v Case study with 28 d RDT v Mechanism database in Toolbox

The Mechanism Database in the Toolbox Project

Chemistry/Biochemistry System biology Symptomology

Chemistry/Biochemistry Molecular Level System biology Symptomology Cell Level Tissue, Organ and Body

Chemistry/Biochemistry Molecular Level System biology Symptomology Cell Level Tissue, Organ and Body Search / Collection of in vitro Test Information

Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level Search / Collection of in vitro Test Information • • • Functions Pathways Processes

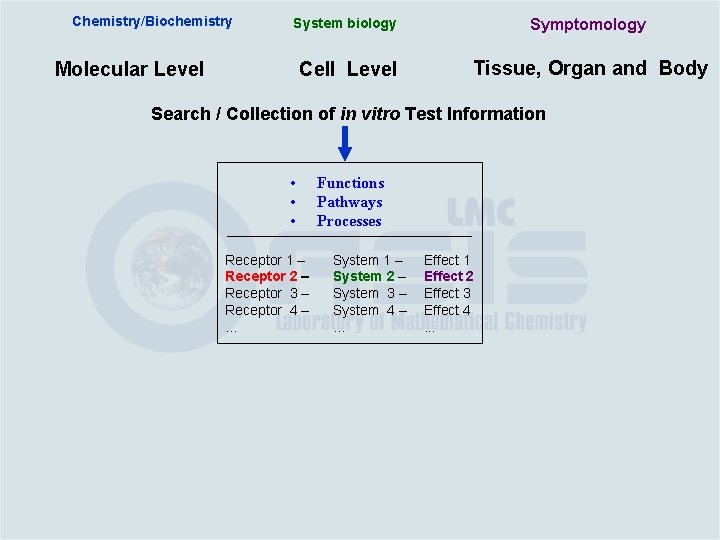
Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level Search / Collection of in vitro Test Information • • • Receptor 1 – Receptor 2 – Receptor 3 – Receptor 4 –. . . Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . .

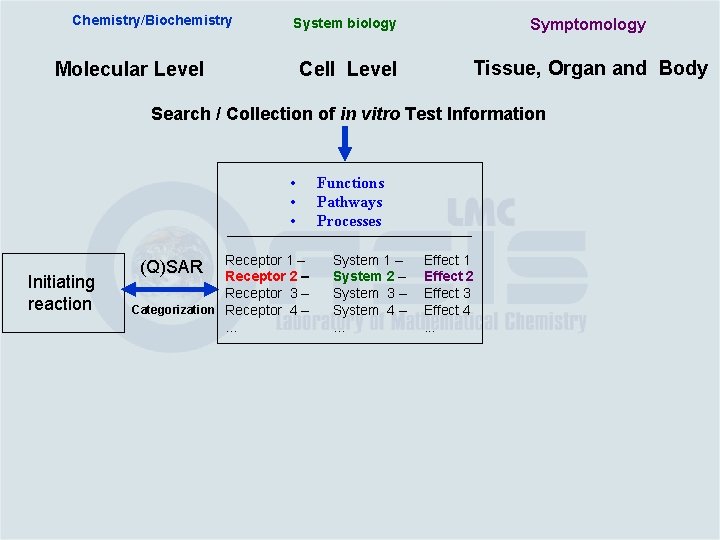
Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level Search / Collection of in vitro Test Information • • • Initiating reaction Receptor 1 – Receptor 2 – Receptor 3 – Categorization Receptor 4 –. . . (Q)SAR Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . .

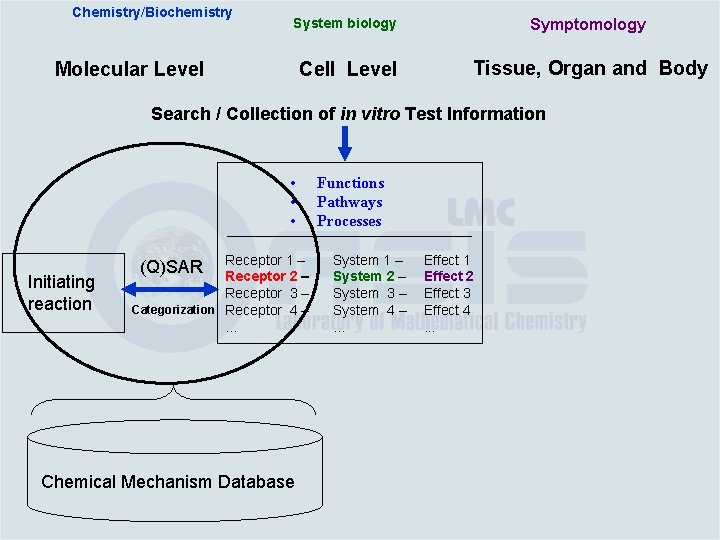
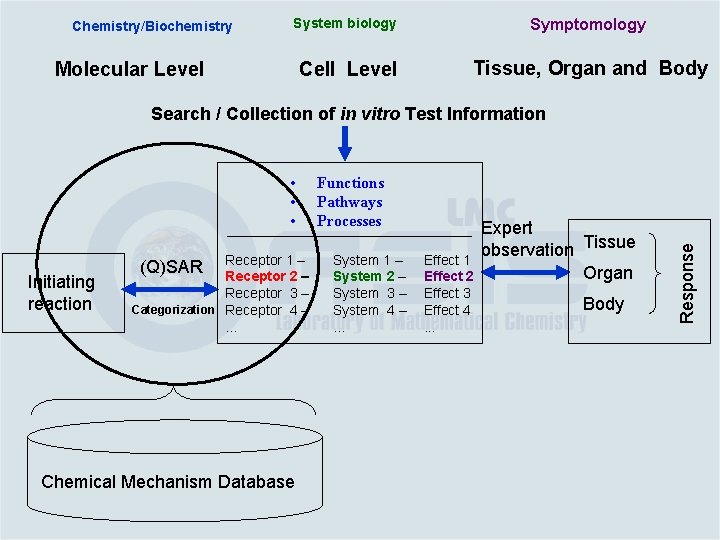
Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level Search / Collection of in vitro Test Information • • • Initiating reaction Receptor 1 – Receptor 2 – Receptor 3 – Categorization Receptor 4 –. . . (Q)SAR Chemical Mechanism Database Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . .

Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level • • • Initiating reaction Receptor 1 – Receptor 2 – Receptor 3 – Categorization Receptor 4 –. . . (Q)SAR Chemical Mechanism Database Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . . Expert observation Tissue Organ Body Response Search / Collection of in vitro Test Information

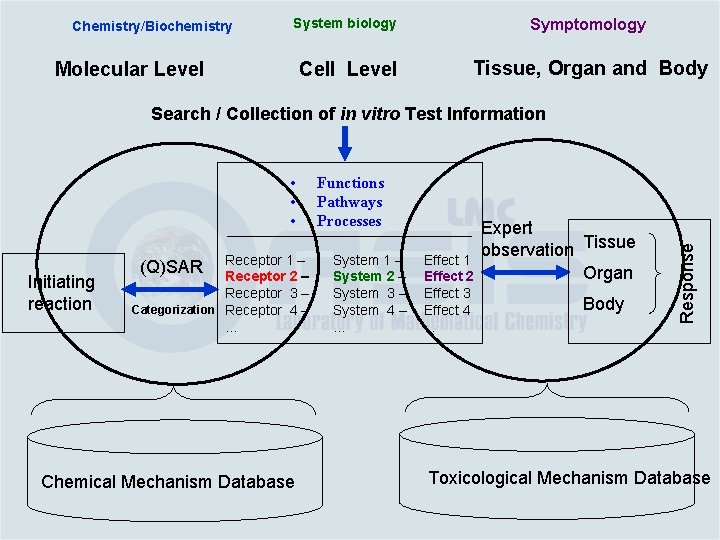
Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level • • • Initiating reaction Receptor 1 – Receptor 2 – Receptor 3 – Categorization Receptor 4 –. . . (Q)SAR Chemical Mechanism Database Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . . Expert observation Tissue Organ Body Response Search / Collection of in vitro Test Information Toxicological Mechanism Database

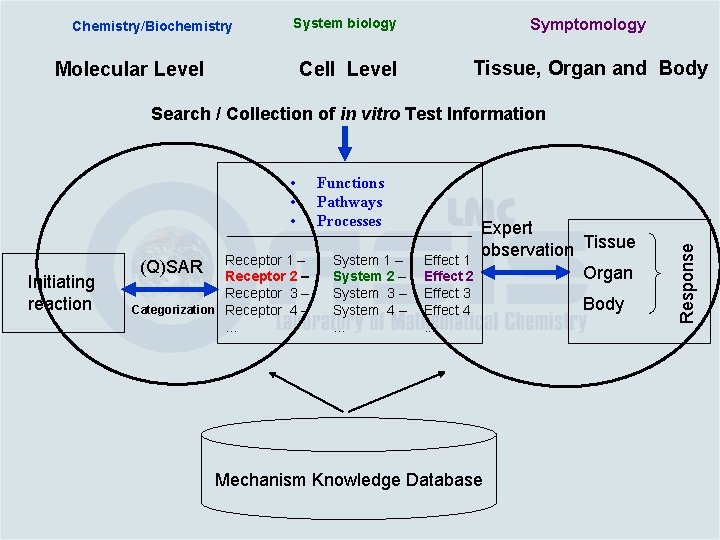
Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level • • • Initiating reaction Receptor 1 – Receptor 2 – Receptor 3 – Categorization Receptor 4 –. . . (Q)SAR Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . . Expert observation Tissue Organ Mechanism Knowledge Database Body Response Search / Collection of in vitro Test Information

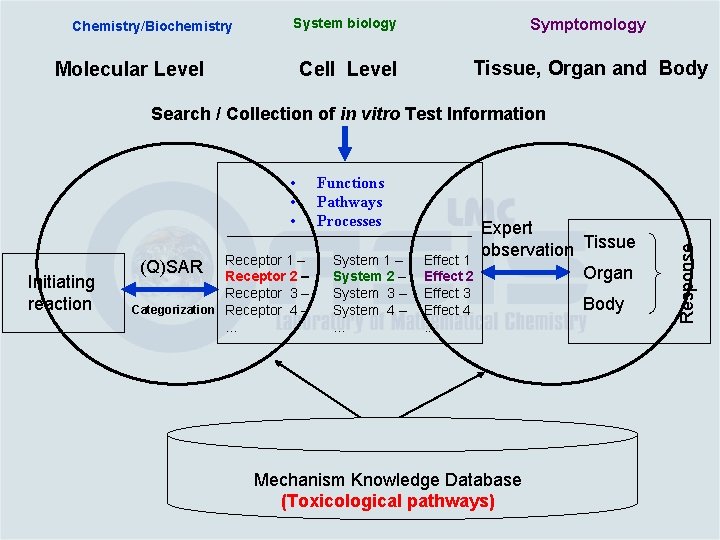
Chemistry/Biochemistry System biology Symptomology Cell Level Tissue, Organ and Body Molecular Level • • • Initiating reaction Receptor 1 – Receptor 2 – Receptor 3 – Categorization Receptor 4 –. . . (Q)SAR Functions Pathways Processes System 1 – System 2 – System 3 – System 4 –. . . Effect 1 Effect 2 Effect 3 Effect 4. . . Expert observation Tissue Organ Mechanism Knowledge Database (Toxicological pathways) Body Response Search / Collection of in vitro Test Information

Contributors:

Laboratory of mathematical Chemistry, Bourgas, Bulgaria O. Mekenyan S. Dimitrov T. Pavlov G. Chankov A. Chapkanov

Chemical Management Center, NITE, Japan Case study on RDT of aromatic amines Chemical vs. toxicological mechanisms (Project of NEDO Japan) Jun Yamada Yuki Sakuratani

Fraunhofer Institute for Toxicology and Experimental Medicine, Department Chemical Risk Assessment, Hanover, Germany Data from REPDOSE Database (Project of CEFIC LRI) Inge Mangelsdorf Sylvia Escher Annette Bitsch

Environment Directorate, OECD, Paris Bob Diderich Terry Schultz

International QSAR Foundation, USA Gilman Veith