Loglinear Models HRP 261 030304 LogLinear Models for












- Slides: 47

Log-linear Models HRP 261 03/03/04

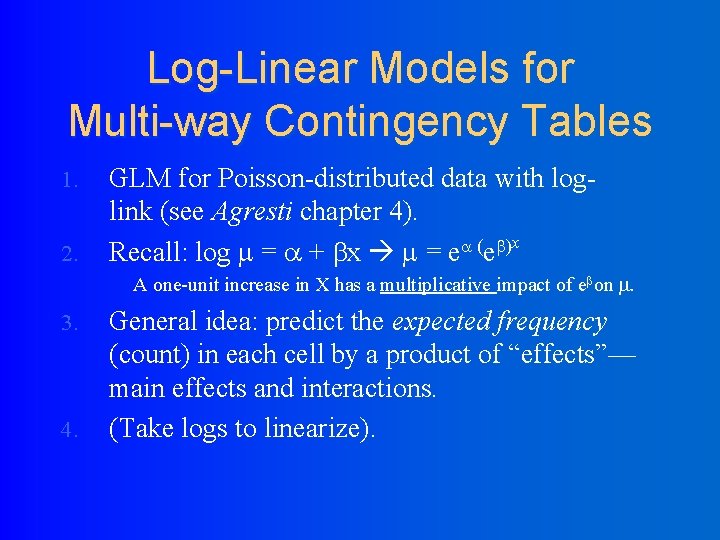
Log-Linear Models for Multi-way Contingency Tables 1. 2. GLM for Poisson-distributed data with loglink (see Agresti chapter 4). x ( ) Recall: log = + x = e e A one-unit increase in X has a multiplicative impact of e on . 3. 4. General idea: predict the expected frequency (count) in each cell by a product of “effects”— main effects and interactions. (Take logs to linearize).

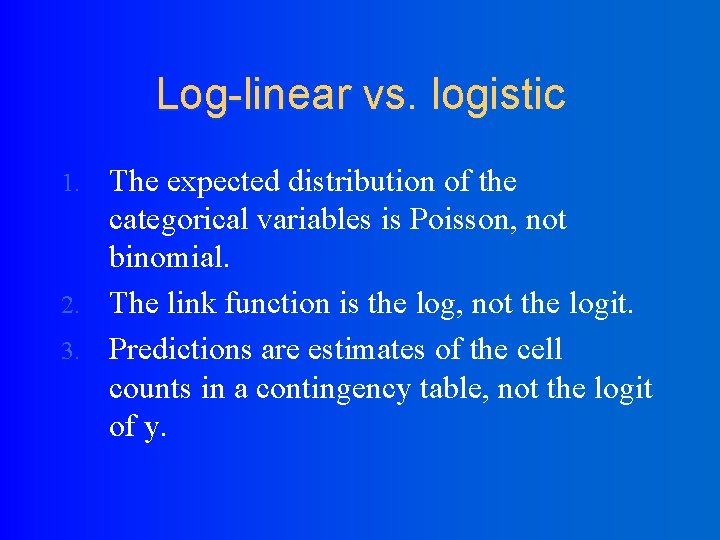
Log-linear vs. logistic The expected distribution of the categorical variables is Poisson, not binomial. 2. The link function is the log, not the logit. 3. Predictions are estimates of the cell counts in a contingency table, not the logit of y. 1.

Log-linear vs. logistic The variables investigated by log linear models are all treated as “response variables. ” l Therefore, loglinear models only demonstrate association between variables (like chi-square or correlation coefficient). l If clear explanatory and response variables exist, then logistic regression should be used instead. l Also, if the variables are continuous and cannot be broken down into discrete categories, logistic regression is preferable. l

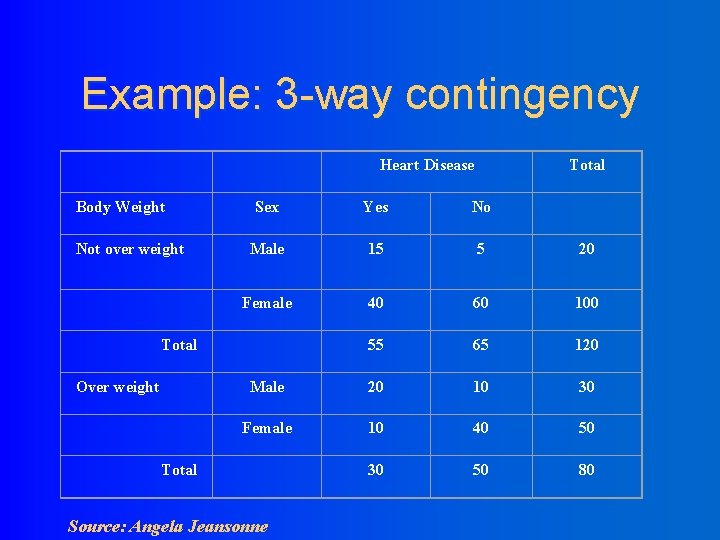
Example: 3 -way contingency Heart Disease Body Weight Not over weight Sex Yes No Male 15 5 20 Female 40 60 100 55 65 120 Male 20 10 30 Female 10 40 50 30 50 80 Total Over weight Total Source: Angela Jeansonne

l In class exercise: Analyze these data using methods we have already learned. l Is gender related to heart disease and is this effect modified or confounded by weight? l What’s the relationship between overweight and gender (controlled for chd) and overweight and heart disease (controlled for gender)? l

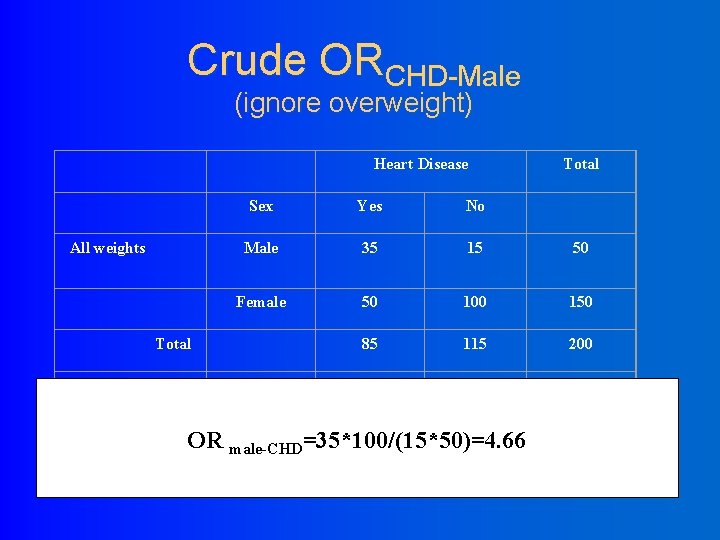
Crude ORCHD-Male (ignore overweight) Heart Disease All weights Sex Yes No Male 35 15 50 Female 50 100 150 85 115 200 Male 20 10 30 Female 10 40 50 30 50 80 Total Over weight OR male-CHD=35*100/(15*50)=4. 66 Total

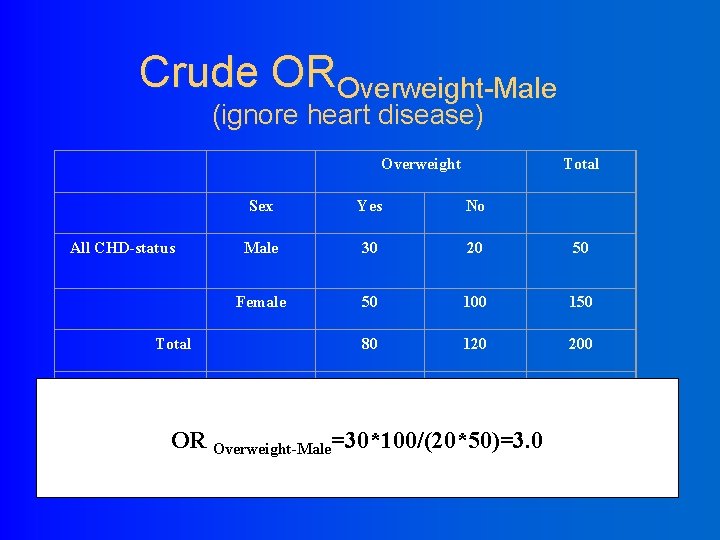
Crude OROverweight-Male (ignore heart disease) Overweight All CHD-status Sex Yes No Male 30 20 50 Female 50 100 150 80 120 200 Male 20 10 30 Female 10 40 50 30 50 80 Total Over weight Total OR Overweight-Male=30*100/(20*50)=3. 0 Total

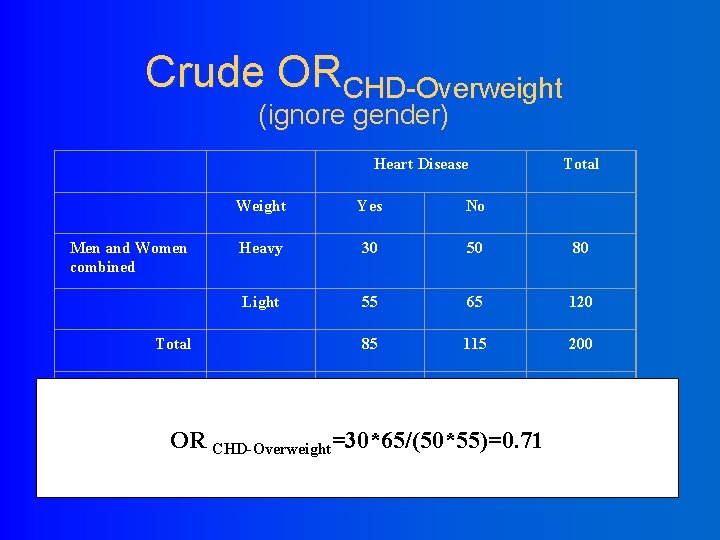
Crude ORCHD-Overweight (ignore gender) Heart Disease Men and Women combined Weight Yes No Heavy 30 50 80 Light 55 65 120 85 115 200 Male 20 10 30 Female 10 40 50 30 50 80 Total Over weight OR CHD-Overweight=30*65/(50*55)=0. 71 Total

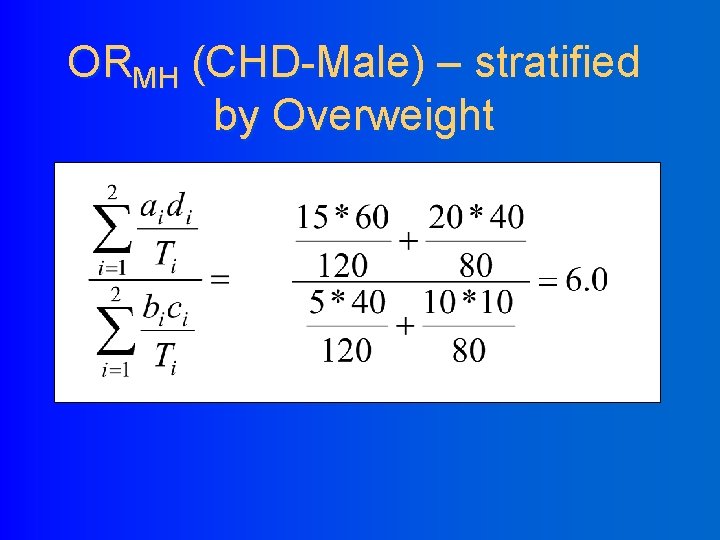
ORMH (CHD-Male) – stratified by Overweight

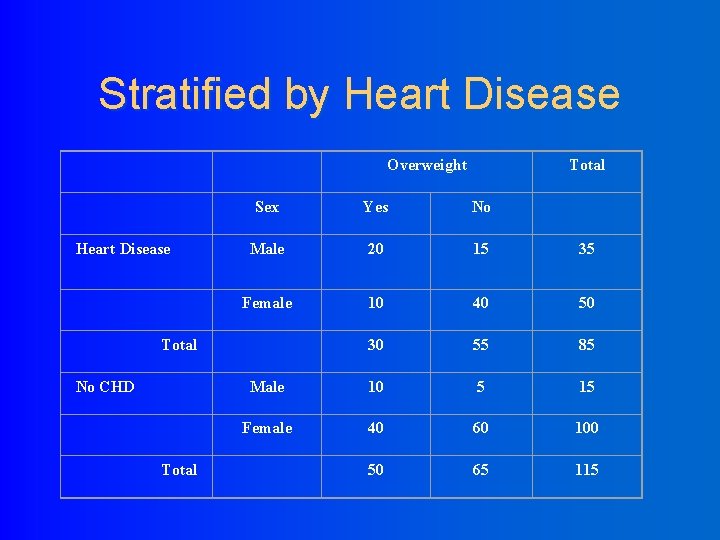
Stratified by Heart Disease Overweight Heart Disease Sex Yes No Male 20 15 35 Female 10 40 50 30 55 85 Male 10 5 15 Female 40 60 100 50 65 115 Total No CHD Total

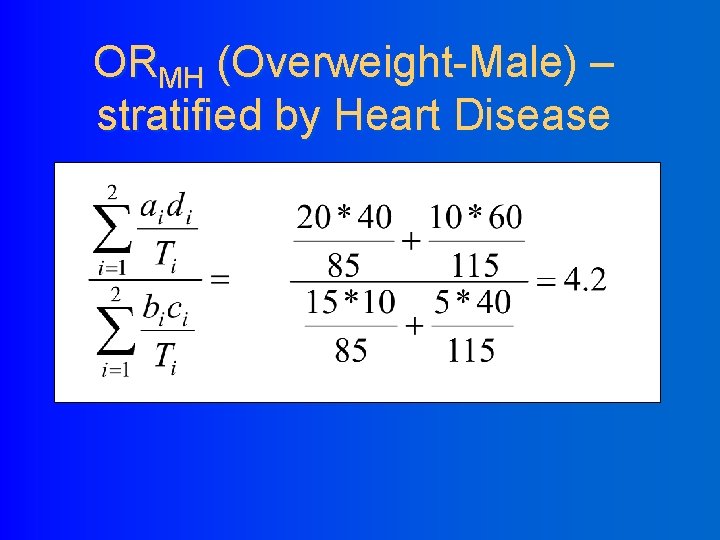
ORMH (Overweight-Male) – stratified by Heart Disease

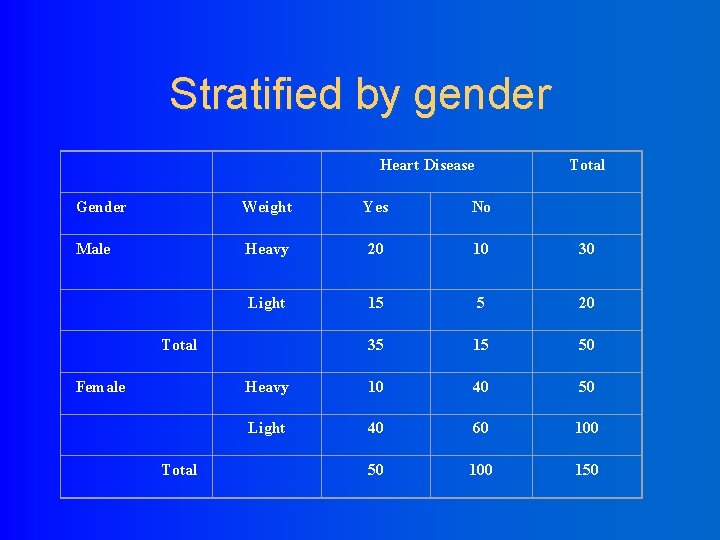
Stratified by gender Heart Disease Total Gender Weight Yes No Male Heavy 20 10 30 Light 15 5 20 35 15 50 Heavy 10 40 50 Light 40 60 100 50 100 150 Total Female Total

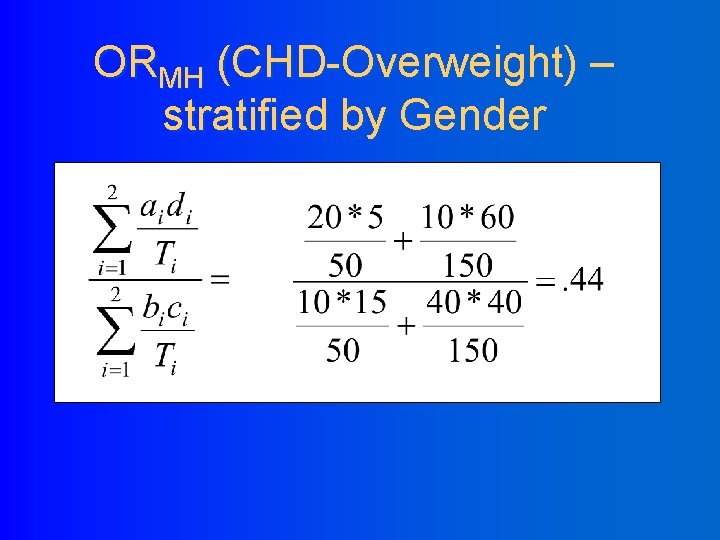
ORMH (CHD-Overweight) – stratified by Gender

Model with log-linear models

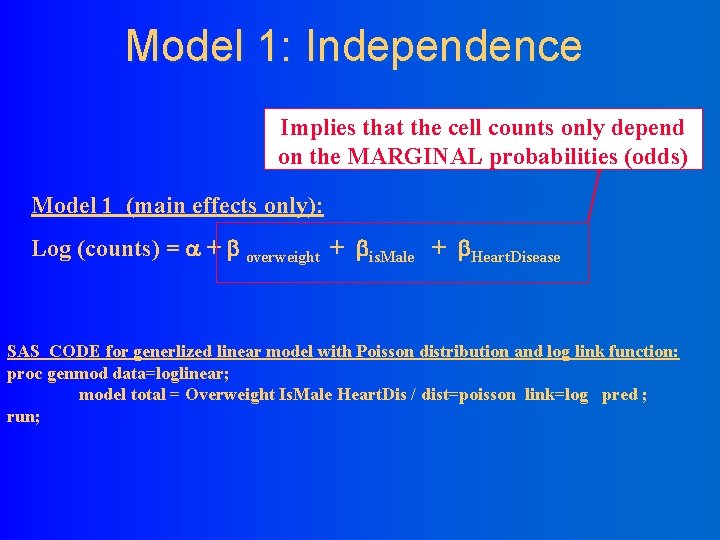
Model 1: Independence Implies that the cell counts only depend on the MARGINAL probabilities (odds) Model 1 (main effects only): Log (counts) = + overweight + is. Male + Heart. Disease SAS CODE for generlized linear model with Poisson distribution and log link function: proc genmod data=loglinear; model total = Overweight Is. Male Heart. Dis / dist=poisson link=log pred ; run;

Independence model: parameters Parameter Intercept Overweight Is. Male Heart. Dis DF Estimate Standard Error 1 1 3. 9464 -0. 4055 -1. 0986 -0. 3023 0. 1170 0. 1443 0. 1633 0. 1430 Parameter Intercept Overweight Is. Male Heart. Dis Wald 95% Confidence Limits 3. 7171 -0. 6884 -1. 4187 -0. 5826 4. 1758 -0. 1226 -0. 7786 -0. 0219 Chi. Square 1137. 17 7. 89 45. 26 4. 47 Pr > Chi. Sq <. 0001 0. 0050 <. 0001 0. 0346 Model 1: Log (counts) = 3. 95 -. 41 (weight) – 1. 1 (male) -. 30 (heart disease)

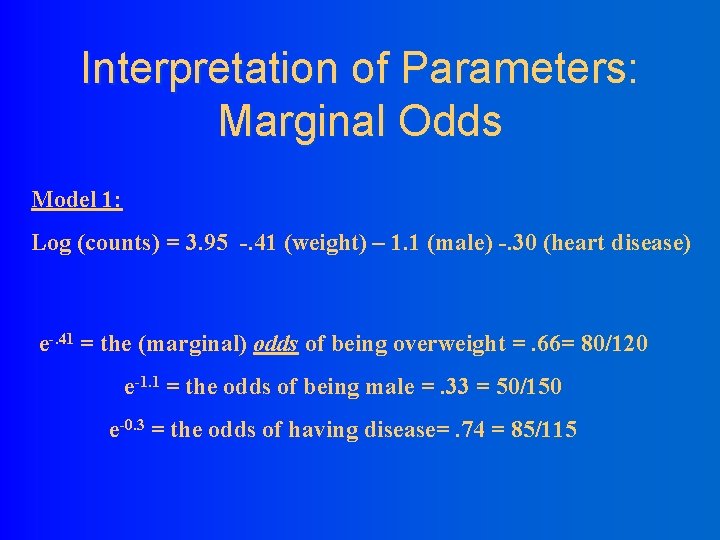
Interpretation of Parameters: Marginal Odds Model 1: Log (counts) = 3. 95 -. 41 (weight) – 1. 1 (male) -. 30 (heart disease) e-. 41 = the (marginal) odds of being overweight =. 66= 80/120 e-1. 1 = the odds of being male =. 33 = 50/150 e-0. 3 = the odds of having disease=. 74 = 85/115

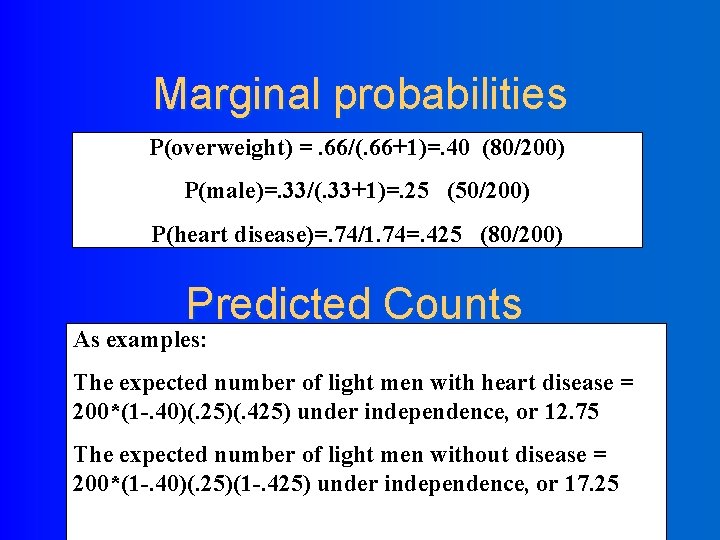
Marginal probabilities P(overweight) =. 66/(. 66+1)=. 40 (80/200) P(male)=. 33/(. 33+1)=. 25 (50/200) P(heart disease)=. 74/1. 74=. 425 (80/200) Predicted Counts As examples: The expected number of light men with heart disease = 200*(1 -. 40)(. 25)(. 425) under independence, or 12. 75 The expected number of light men without disease = 200*(1 -. 40)(. 25)(1 -. 425) under independence, or 17. 25

Independence model: goodness-of-fit Cells light/male/disease Observed 15 Pred 12. 75 light/male/no disease 5 17. 25 light/female/disease 40 38. 25 light/female/no disease 60 51. 75 heavy/male/disease 20 8. 5 heavy/male/no disease 10 11. 5 heavy/female/disease 10 25. 5 heavy/female/no disease 40 34. 5 df = cells – parameters in model=8 -4 Suggests independen ce model is a poor fit!!

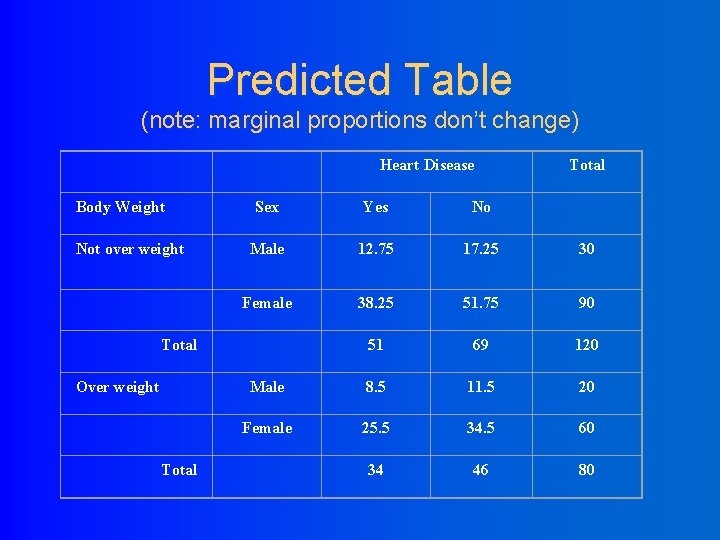
Predicted Table (note: marginal proportions don’t change) Heart Disease Body Weight Not over weight Sex Yes No Male 12. 75 17. 25 30 Female 38. 25 51. 75 90 51 69 120 Male 8. 5 11. 5 20 Female 25. 5 34. 5 60 34 46 80 Total Over weight Total

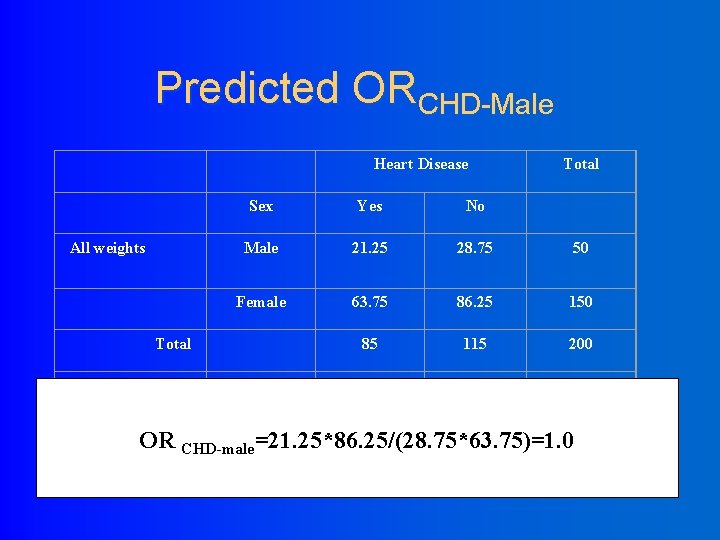
Predicted ORCHD-Male Heart Disease All weights Sex Yes No Male 21. 25 28. 75 50 Female 63. 75 86. 25 150 85 115 200 Male 20 10 30 Female 10 40 50 30 50 80 Total Over weight Total OR CHD-male=21. 25*86. 25/(28. 75*63. 75)=1. 0 Total

The model coefficients have an odds ratio interpretation…

Coefficients represent predicted counts in each cell Coefficients have a direct odds ratio interpretation Calculate OR CHD-Male in each Weight stratum This interpretation becomes more interesting/useful when interaction terms occur!

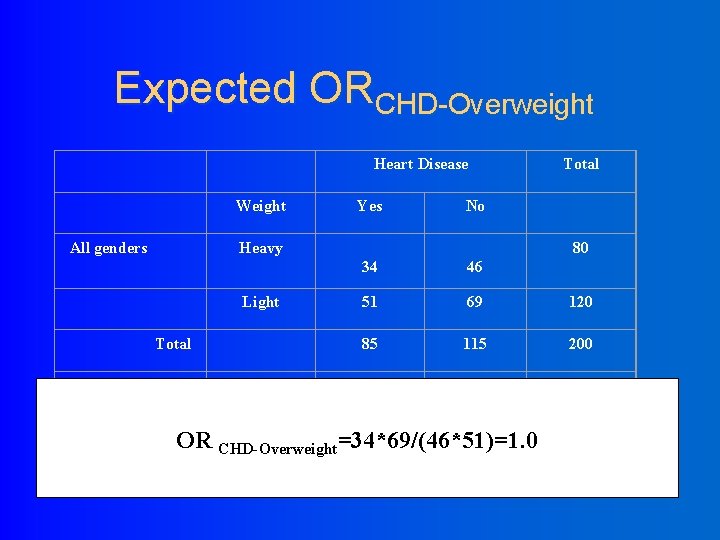
Expected ORCHD-Overweight Heart Disease Weight All genders Yes No Heavy 80 34 46 51 69 120 85 115 200 Male 20 10 30 Female 10 40 50 30 50 80 Light Total Over weight OR CHD-Overweight=34*69/(46*51)=1. 0 Total

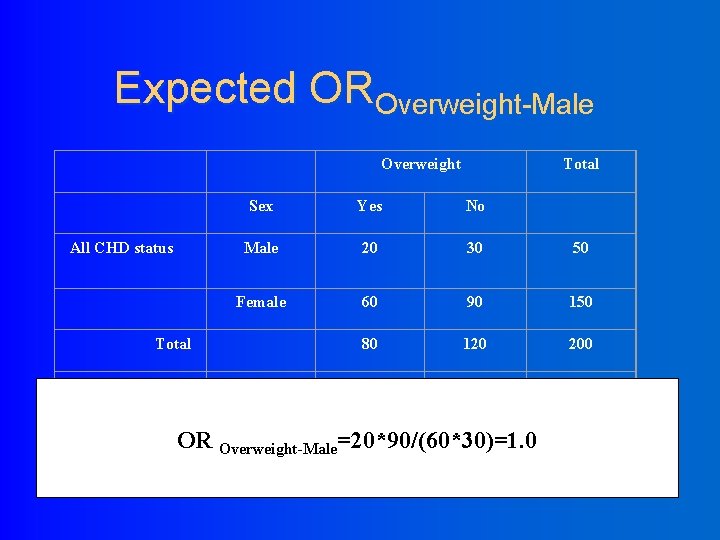
Expected OROverweight-Male Overweight All CHD status Sex Yes No Male 20 30 50 Female 60 90 150 80 120 200 Male 20 10 30 Female 10 40 50 30 50 80 Total Over weight Total OR Overweight-Male=20*90/(60*30)=1. 0 Total

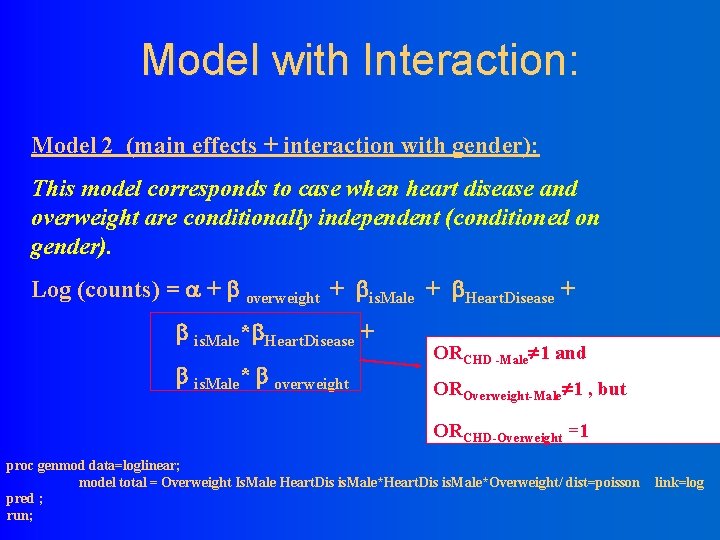
Model with Interaction: Model 2 (main effects + interaction with gender): This model corresponds to case when heart disease and overweight are conditionally independent (conditioned on gender). Log (counts) = + overweight + is. Male + Heart. Disease + is. Male* overweight Implies that 1 gender OR andis associated with CHD -Male heart disease and with overweight but and 1 heart disease are ORoverweight , but Overweight-Male independent. ORCHD-Overweight =1 proc genmod data=loglinear; model total = Overweight Is. Male Heart. Dis is. Male*Overweight/ dist=poisson pred ; run; link=log

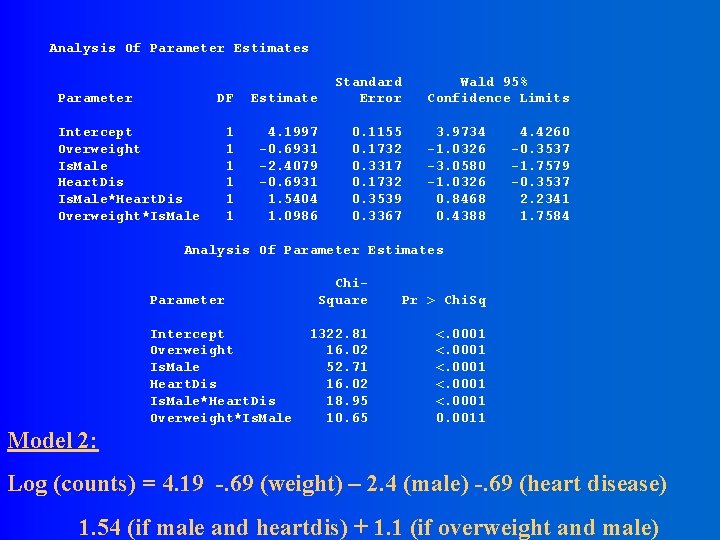
Analysis Of Parameter Estimates Parameter DF Estimate Standard Error 1 1 1 4. 1997 -0. 6931 -2. 4079 -0. 6931 1. 5404 1. 0986 0. 1155 0. 1732 0. 3317 0. 1732 0. 3539 0. 3367 Intercept Overweight Is. Male Heart. Dis Is. Male*Heart. Dis Overweight*Is. Male Wald 95% Confidence Limits 3. 9734 -1. 0326 -3. 0580 -1. 0326 0. 8468 0. 4388 4. 4260 -0. 3537 -1. 7579 -0. 3537 2. 2341 1. 7584 Analysis Of Parameter Estimates Parameter Intercept Overweight Is. Male Heart. Dis Is. Male*Heart. Dis Overweight*Is. Male Chi. Square Pr > Chi. Sq 1322. 81 16. 02 52. 71 16. 02 18. 95 10. 65 <. 0001 0. 0011 Model 2: Log (counts) = 4. 19 -. 69 (weight) – 2. 4 (male) -. 69 (heart disease) 1. 54 (if male and heartdis) + 1. 1 (if overweight and male)

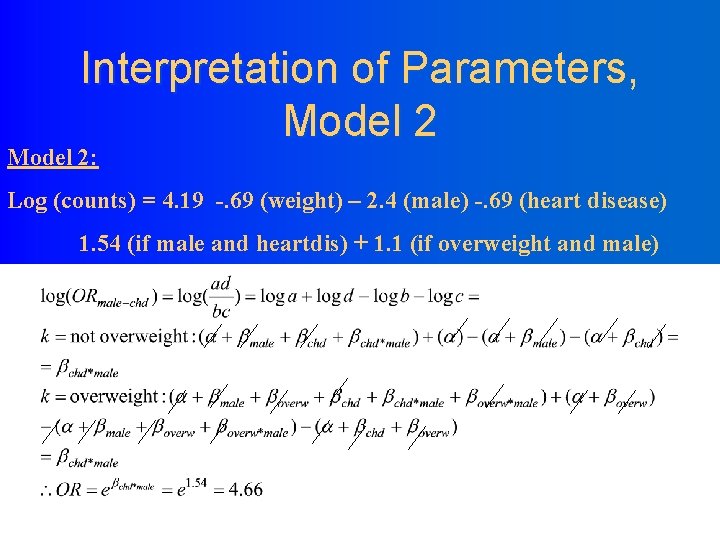
Interpretation of Parameters, Model 2: Log (counts) = 4. 19 -. 69 (weight) – 2. 4 (male) -. 69 (heart disease) 1. 54 (if male and heartdis) + 1. 1 (if overweight and male)

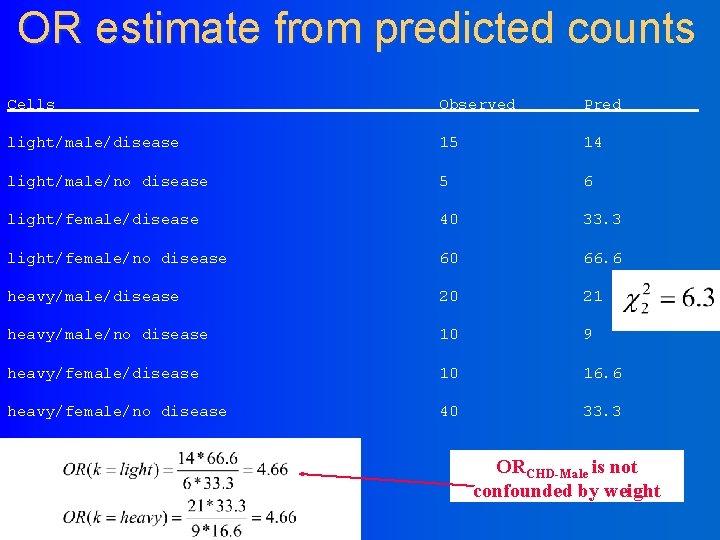
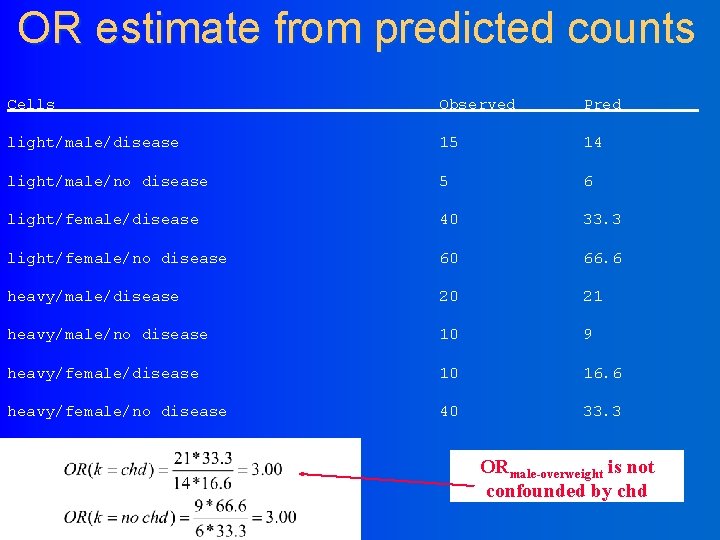
OR estimate from predicted counts Cells Observed Pred light/male/disease 15 14 light/male/no disease 5 6 light/female/disease 40 33. 3 light/female/no disease 60 66. 6 heavy/male/disease 20 21 heavy/male/no disease 10 9 heavy/female/disease 10 16. 6 heavy/female/no disease 40 33. 3 ORCHD-Male is not confounded by weight

OROverweight-Male Model 2: Log (counts) = 4. 19 -. 69 (weight) – 2. 4 (male) -. 69 (heart disease) 1. 54 (if male and heartdis) + 1. 1 (if overweight and male)

OR estimate from predicted counts Cells Observed Pred light/male/disease 15 14 light/male/no disease 5 6 light/female/disease 40 33. 3 light/female/no disease 60 66. 6 heavy/male/disease 20 21 heavy/male/no disease 10 9 heavy/female/disease 10 16. 6 heavy/female/no disease 40 33. 3 ORmale-overweight is not confounded by chd

ORCHD-OVerweight Model 2: Log (counts) = 4. 19 -. 69 (weight) – 2. 4 (male) -. 69 (heart disease) 1. 54 (if male and heartdis) + 1. 1 (if overweight and male)

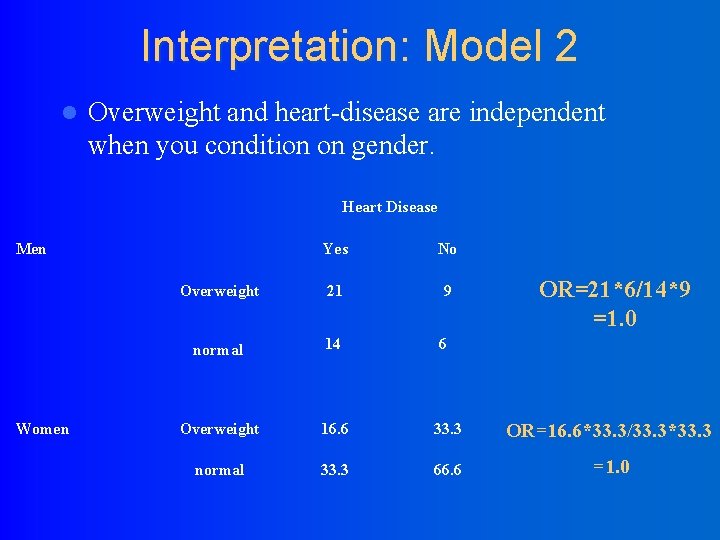
Interpretation: Model 2 l Overweight and heart-disease are independent when you condition on gender. Heart Disease Men Women Yes No Overweight 21 9 normal 14 6 Overweight 16. 6 33. 3 OR=16. 6*33. 3/33. 3*33. 3 normal 33. 3 66. 6 =1. 0 OR=21*6/14*9 =1. 0

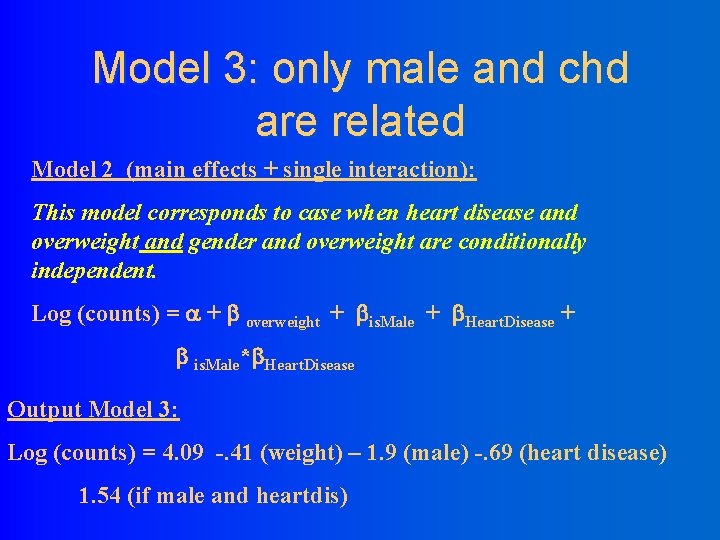
Model 3: only male and chd are related Model 2 (main effects + single interaction): This model corresponds to case when heart disease and overweight and gender and overweight are conditionally independent. Log (counts) = + overweight + is. Male + Heart. Disease + is. Male* Heart. Disease Output Model 3: Log (counts) = 4. 09 -. 41 (weight) – 1. 9 (male) -. 69 (heart disease) 1. 54 (if male and heartdis)

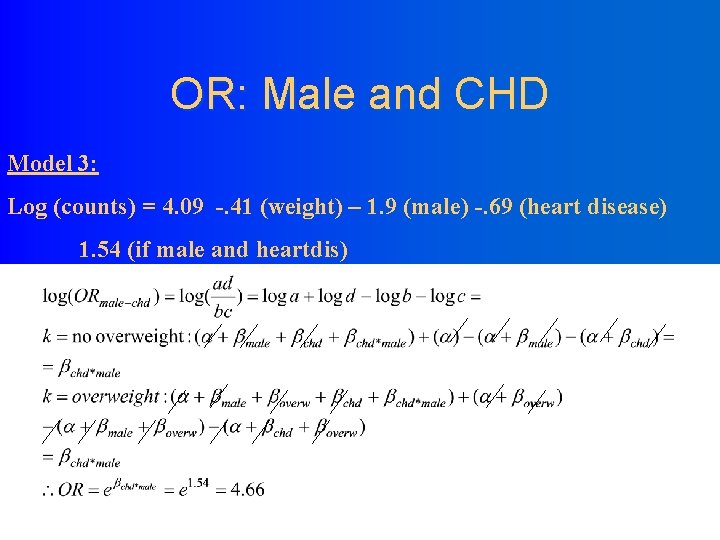
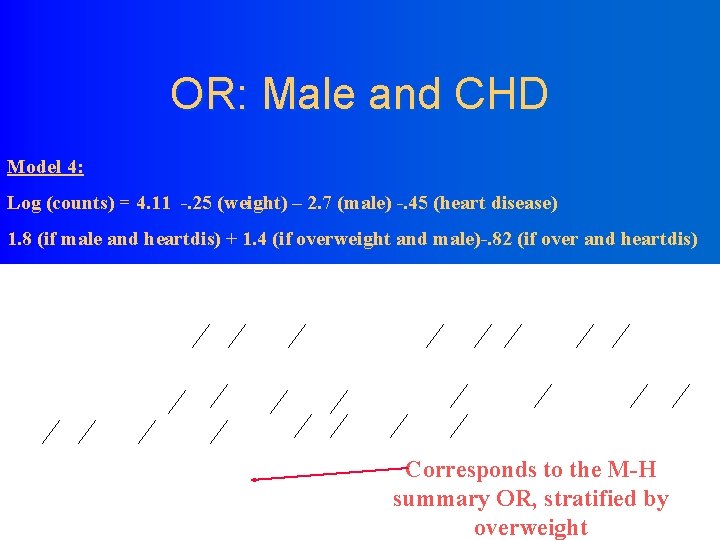
OR: Male and CHD Model 3: Log (counts) = 4. 09 -. 41 (weight) – 1. 9 (male) -. 69 (heart disease) 1. 54 (if male and heartdis)

Model 3: only male and chd are related Cells Observed Pred light/male/disease 15 21 light/male/no disease 5 9 light/female/disease 40 30 light/female/no disease 60 60 heavy/male/disease 20 14 heavy/male/no disease 10 6 heavy/female/disease 10 20 heavy/female/no disease 40 40

Collapses to… Male Female CHD 35 50 No CHD 15 100

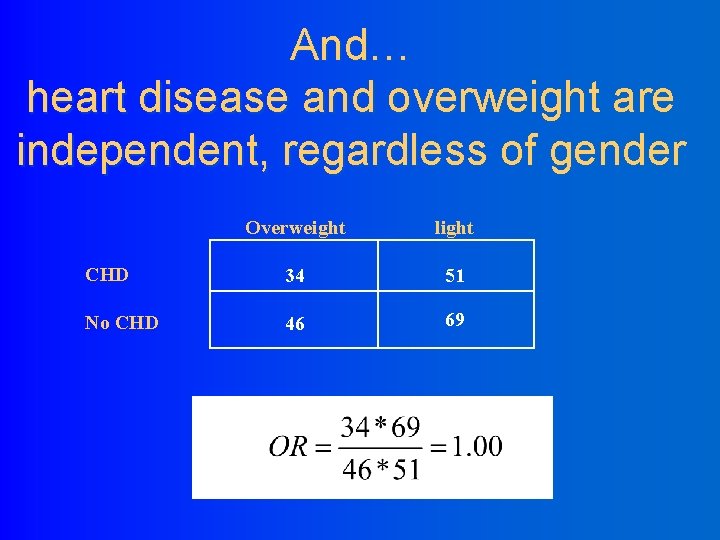
And… heart disease and overweight are independent, regardless of gender Overweight light CHD 34 51 No CHD 46 69

And… overweight and gender are independent, regardless of disease Overweight light Male 20 30 Female 60 90

M 4: All pair-wise interactions Model 4 (main effects +all pairwise interactions): No pair of variables is conditionally independent. Log (counts) = + overweight + is. Male + Heart. Disease is. Male* Heart. Disease + is. Male* overweight + Heart. Dis* overweight proc genmod data=loglinear; model total = Overweight Is. Male Heart. Dis is. Male*Overweight*Heart. Dis / dist=poisson link=log pred ; run;

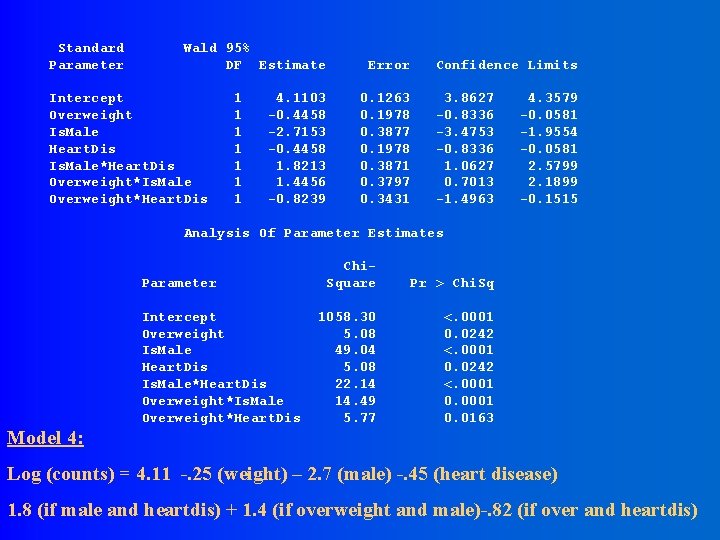
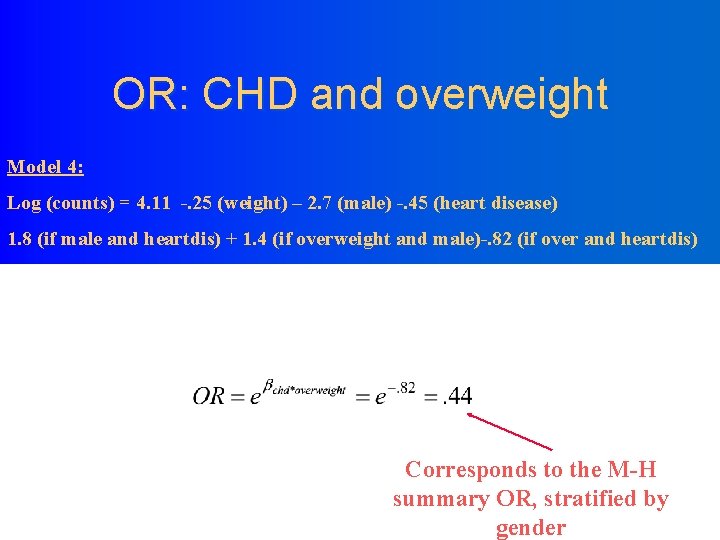
Standard Parameter Wald 95% DF Estimate Intercept Overweight Is. Male Heart. Dis Is. Male*Heart. Dis Overweight*Is. Male Overweight*Heart. Dis 1 1 1 1 Error 4. 1103 -0. 4458 -2. 7153 -0. 4458 1. 8213 1. 4456 -0. 8239 Confidence Limits 0. 1263 0. 1978 0. 3877 0. 1978 0. 3871 0. 3797 0. 3431 3. 8627 -0. 8336 -3. 4753 -0. 8336 1. 0627 0. 7013 -1. 4963 4. 3579 -0. 0581 -1. 9554 -0. 0581 2. 5799 2. 1899 -0. 1515 Analysis Of Parameter Estimates Parameter Intercept Overweight Is. Male Heart. Dis Is. Male*Heart. Dis Overweight*Is. Male Overweight*Heart. Dis Chi. Square Pr > Chi. Sq 1058. 30 5. 08 49. 04 5. 08 22. 14 14. 49 5. 77 <. 0001 0. 0242 <. 0001 0. 0163 Model 4: Log (counts) = 4. 11 -. 25 (weight) – 2. 7 (male) -. 45 (heart disease) 1. 8 (if male and heartdis) + 1. 4 (if overweight and male)-. 82 (if over and heartdis)

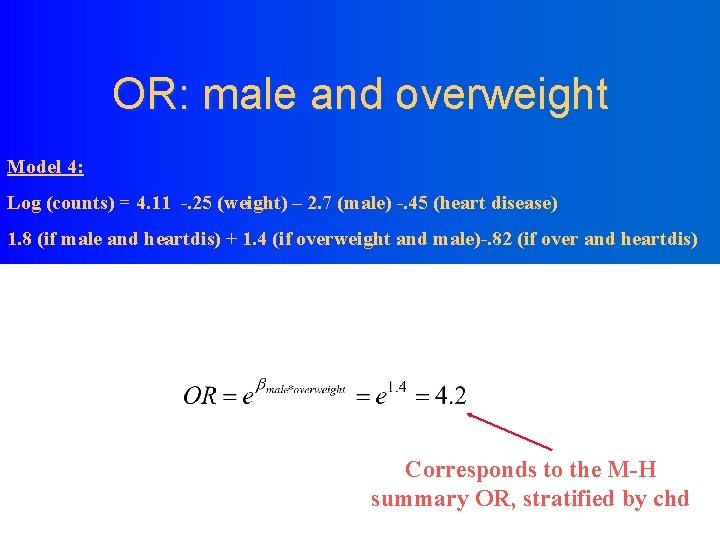
OR: Male and CHD Model 4: Log (counts) = 4. 11 -. 25 (weight) – 2. 7 (male) -. 45 (heart disease) 1. 8 (if male and heartdis) + 1. 4 (if overweight and male)-. 82 (if over and heartdis) Corresponds to the M-H summary OR, stratified by overweight

OR: CHD and overweight Model 4: Log (counts) = 4. 11 -. 25 (weight) – 2. 7 (male) -. 45 (heart disease) 1. 8 (if male and heartdis) + 1. 4 (if overweight and male)-. 82 (if over and heartdis) Corresponds to the M-H summary OR, stratified by gender

OR: male and overweight Model 4: Log (counts) = 4. 11 -. 25 (weight) – 2. 7 (male) -. 45 (heart disease) 1. 8 (if male and heartdis) + 1. 4 (if overweight and male)-. 82 (if over and heartdis) Corresponds to the M-H summary OR, stratified by chd

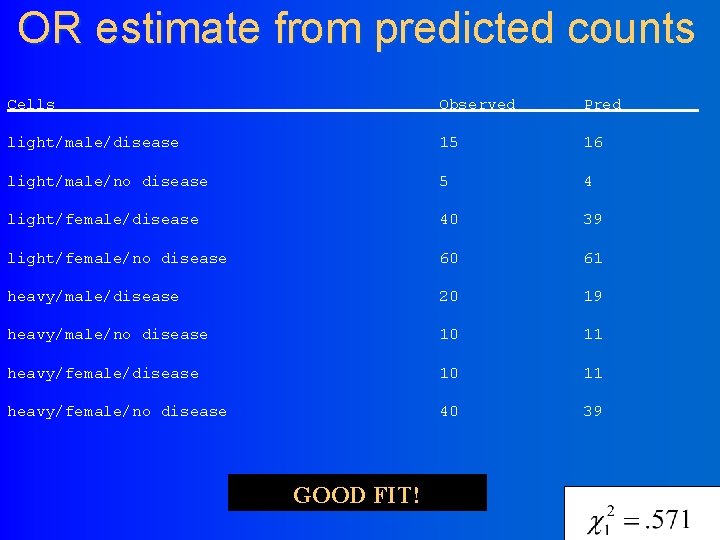
OR estimate from predicted counts Cells Observed Pred light/male/disease 15 16 light/male/no disease 5 4 light/female/disease 40 39 light/female/no disease 60 61 heavy/male/disease 20 19 heavy/male/no disease 10 11 heavy/female/no disease 40 39 GOOD FIT!

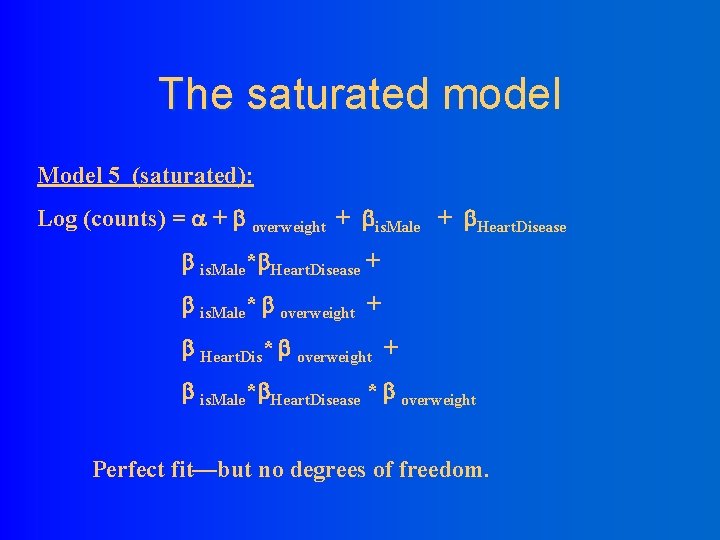
The saturated model Model 5 (saturated): Log (counts) = + overweight + is. Male + Heart. Disease is. Male* Heart. Disease + is. Male* overweight + Heart. Dis* overweight + is. Male* Heart. Disease * overweight Perfect fit—but no degrees of freedom.