Data Processing of RestingState f MRI Part 3








- Slides: 84

Data Processing of Resting-State f. MRI (Part 3) DPARSF Advanced Edition V 2. 2 YAN Chao-Gan 严超赣 Ph. D. ycg. yan@gmail. com Nathan Kline Institute, Child Mind Institute and New York University Child Study Center 1

DPARSF (Yan and Zang, 2010) 2

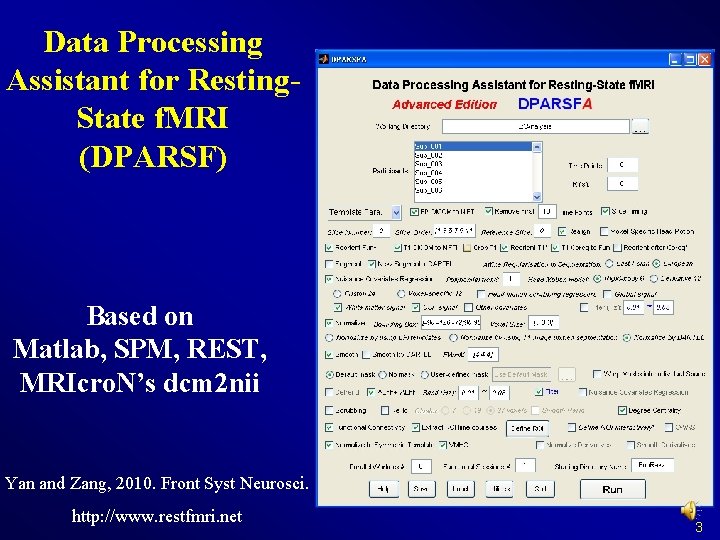
Data Processing Assistant for Resting. State f. MRI (DPARSF) Based on Matlab, SPM, REST, MRIcro. N’s dcm 2 nii Yan and Zang, 2010. Front Syst Neurosci. http: //www. restfmri. net 3

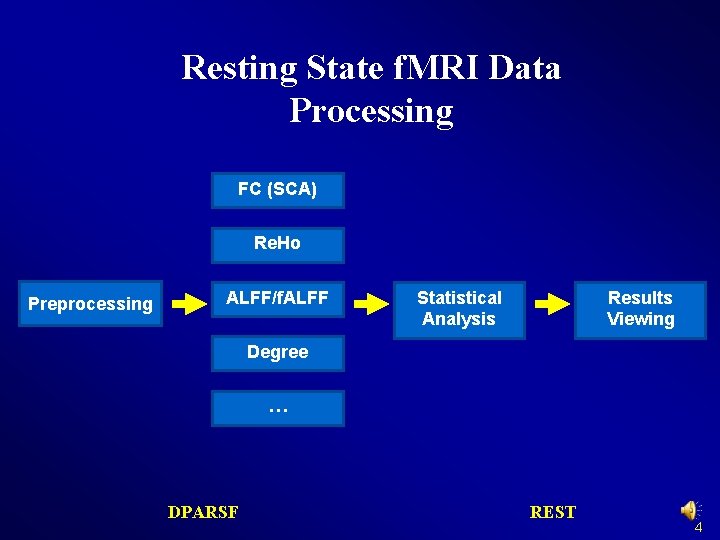
Resting State f. MRI Data Processing FC (SCA) Re. Ho Preprocessing ALFF/f. ALFF Statistical Analysis Results Viewing Degree … DPARSF REST 4

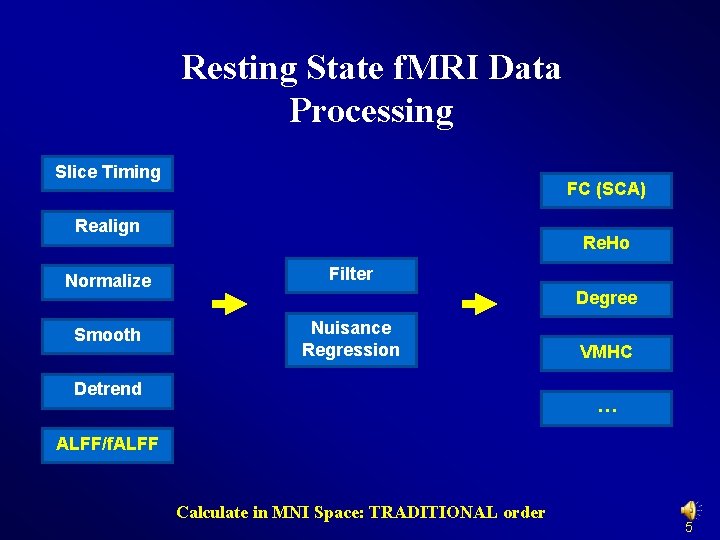
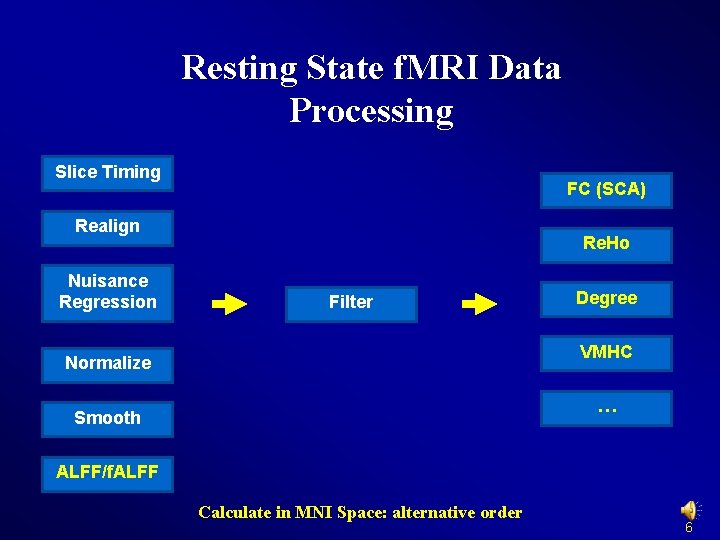
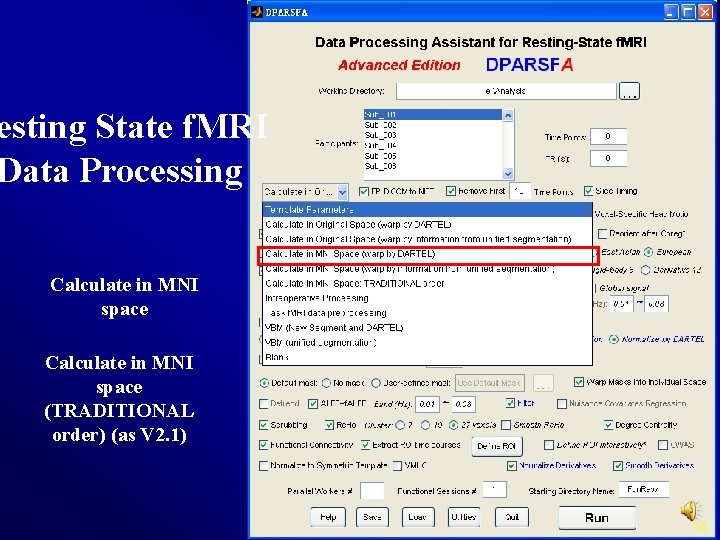
Resting State f. MRI Data Processing Slice Timing FC (SCA) Realign Re. Ho Normalize Filter Smooth Nuisance Regression Degree Detrend VMHC … ALFF/f. ALFF Calculate in MNI Space: TRADITIONAL order 5

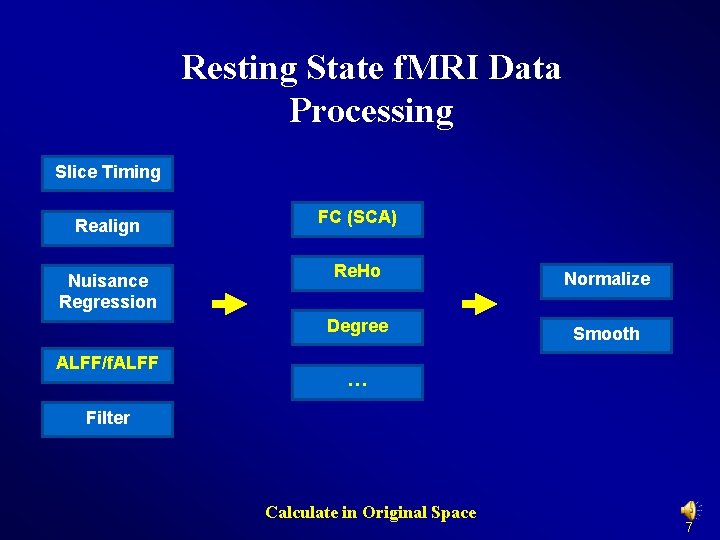
Resting State f. MRI Data Processing Slice Timing FC (SCA) Realign Nuisance Regression Re. Ho Filter Degree VMHC Normalize … Smooth ALFF/f. ALFF Calculate in MNI Space: alternative order 6

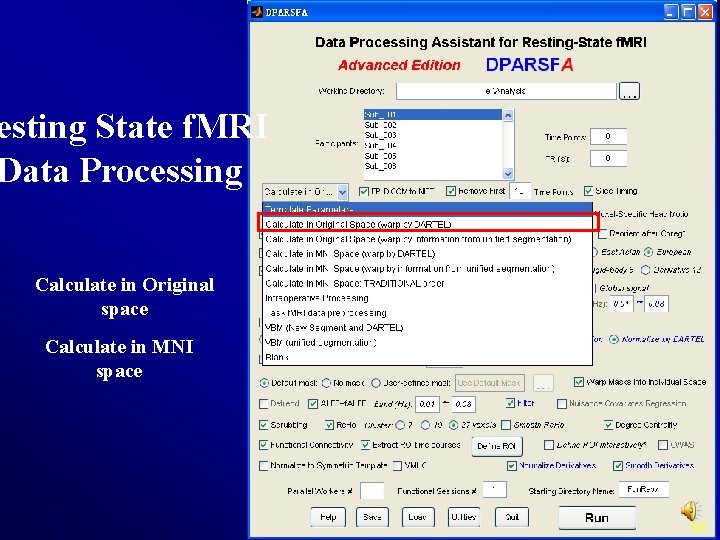
Resting State f. MRI Data Processing Slice Timing Realign Nuisance Regression ALFF/f. ALFF FC (SCA) Re. Ho Normalize Degree Smooth … Filter Calculate in Original Space 7

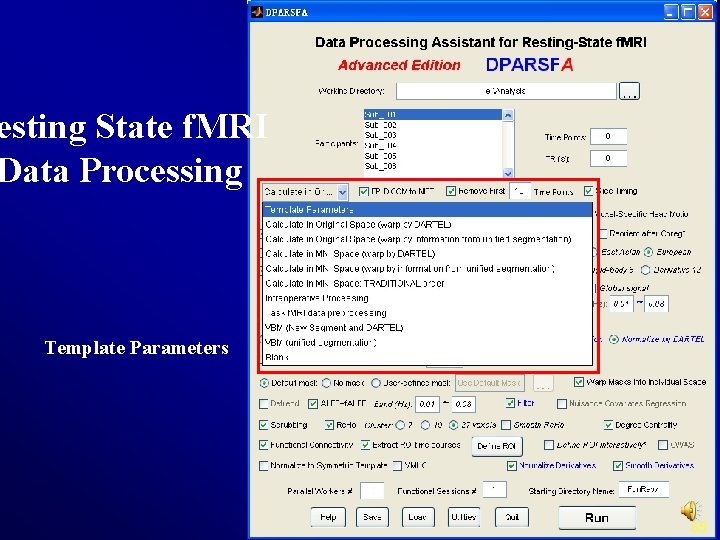
esting State f. MRI Data Processing Template Parameters 8

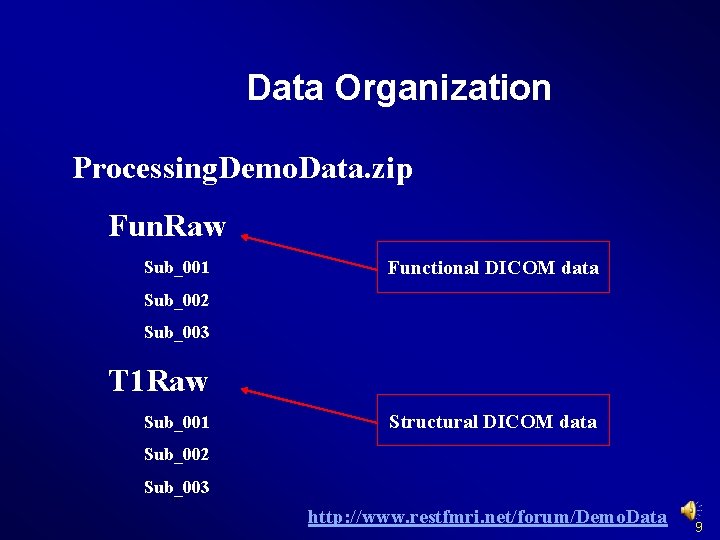
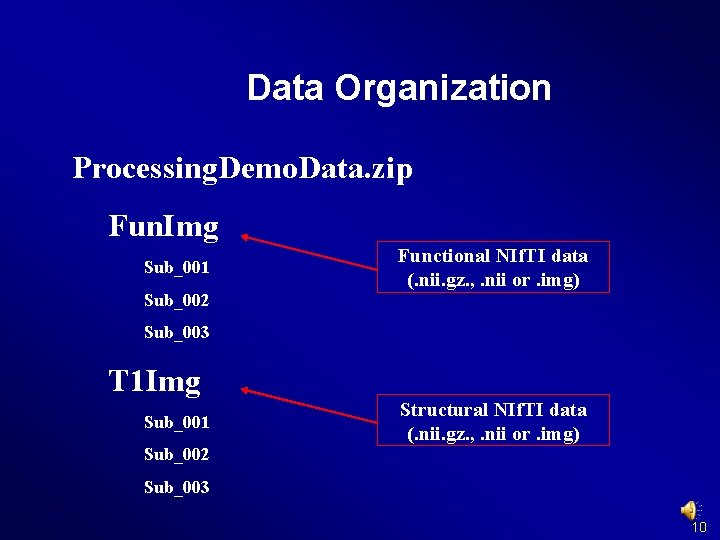
Data Organization Processing. Demo. Data. zip Fun. Raw Sub_001 Functional DICOM data Sub_002 Sub_003 T 1 Raw Sub_001 Structural DICOM data Sub_002 Sub_003 http: //www. restfmri. net/forum/Demo. Data 9

Data Organization Processing. Demo. Data. zip Fun. Img Sub_001 Sub_002 Functional NIf. TI data (. nii. gz. , . nii or. img) Sub_003 T 1 Img Sub_001 Structural NIf. TI data (. nii. gz. , . nii or. img) Sub_002 Sub_003 10

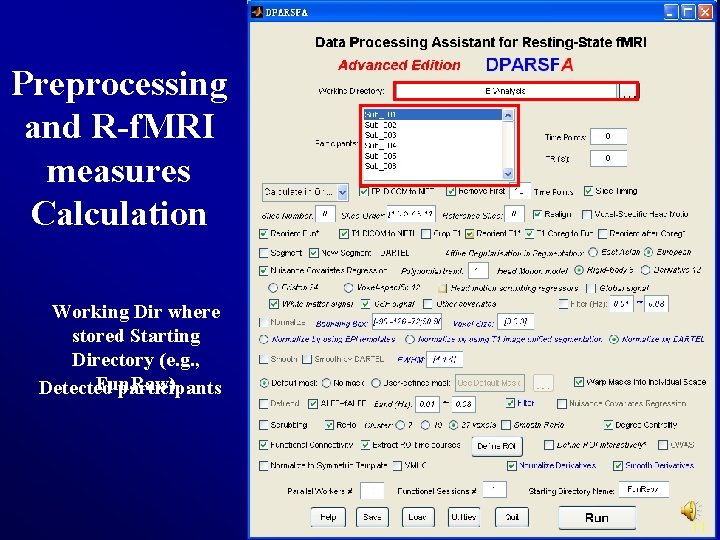
Preprocessing and R-f. MRI measures Calculation Working Dir where stored Starting Directory (e. g. , Fun. Raw) Detected participants 11

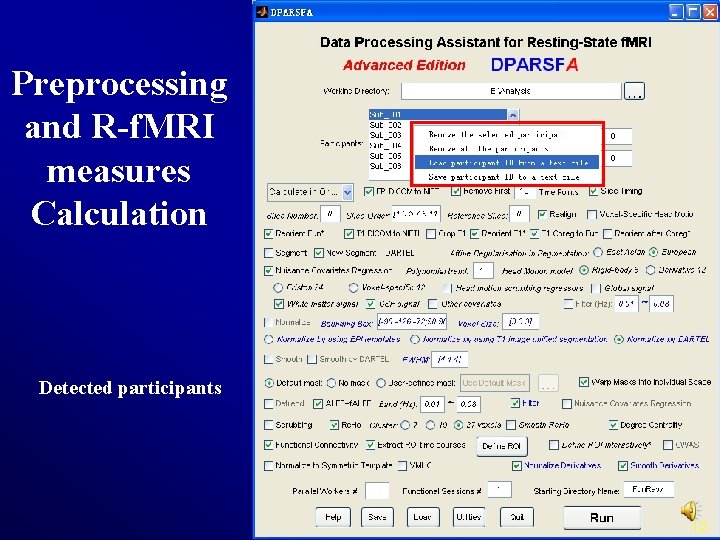
Preprocessing and R-f. MRI measures Calculation Detected participants 12

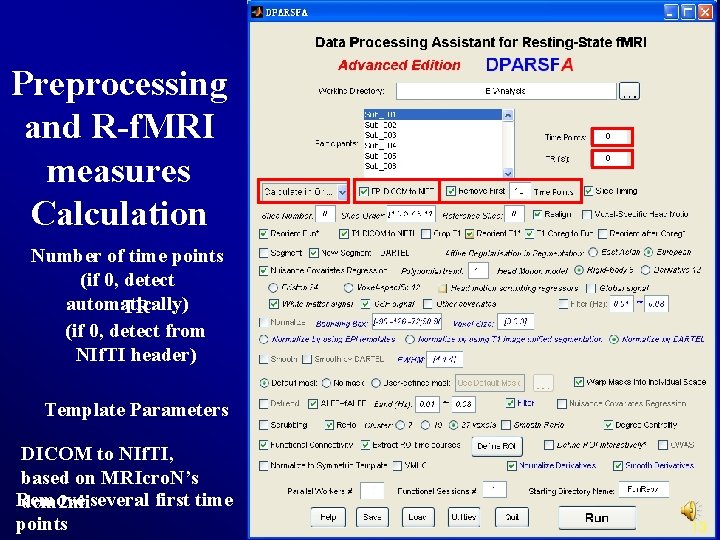
Preprocessing and R-f. MRI measures Calculation Number of time points (if 0, detect automatically) TR (if 0, detect from NIf. TI header) Template Parameters DICOM to NIf. TI, based on MRIcro. N’s Remove dcm 2 niiseveral first time points 13

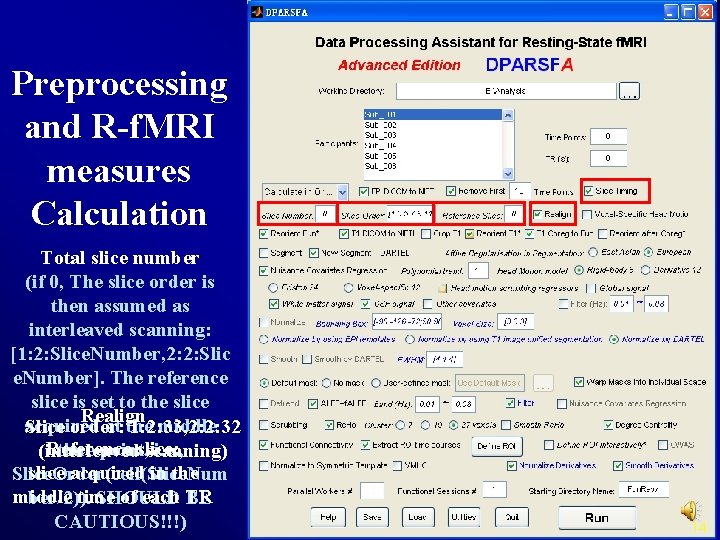
Preprocessing and R-f. MRI measures Calculation Total slice number (if 0, The slice order is then assumed as interleaved scanning: [1: 2: Slice. Number, 2: 2: Slic e. Number]. The reference slice is set to the slice Realign acquired at the middle Slice order: 1: 2: 33, 2: 2: 32 Reference slice: time point, i. e. , (interleaved scanning) slice acquired in the Slice. Order(ceil(Slice. Num middle time of each TR ber/2)). SHOULD BE CAUTIOUS!!!) 14

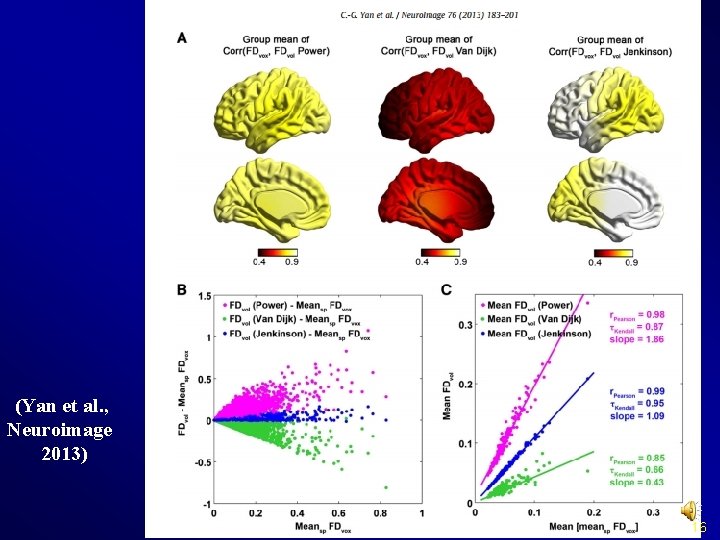
Realign Check head motion: {Working. Dir}Realign. ParameterSub_xxx: rp_*. txt: realign parameters FD_Power_*. txt: Frame-wise Displacement (Power et al. , 2012) FD_Van. Dijk_*. txt: Relative Displacement (Van Dijk et al. , 2012) FD_Jenkinson_*. txt: Relative RMS (Jenkinson et al. , 2002)

Realign (Yan et al. , Neuroimage 2013) 16

Excluding Criteria: 2. 5 mm and 2. 5 degree in max head motion None Realign Excluding Criteria: 2. 0 mm and 2. 0 degree in max head motion Sub_013 Check head motion: {Working. Dir}Realign. Parameter: Excluding Criteria: 1. 5 mm and 1. 5 degree in max head motion Sub_013 Exclude. Subjects. According. To. Max. Head. Motion. txt Excluding Criteria: 1. 0 mm and 1. 0 degree in max head motion Sub_007 Sub_012 Sub_013 Sub_017 Sub_018 17

Realign Check head motion: Head. Motion. csv: head motion characteristics for each subject (e. g. , max or mean motion, mean FD, # or % of FD>0. 2) Threshold: Group mean (mean FD) + 2 * Group SD (mean FD) Yan et al. , in press Neuroimage; Di Martino, in press, Mol Psychiatry 18

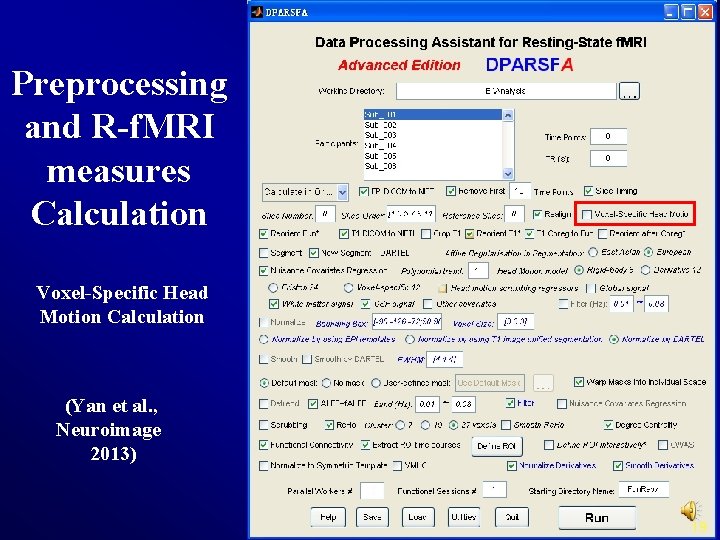
Preprocessing and R-f. MRI measures Calculation Voxel-Specific Head Motion Calculation (Yan et al. , Neuroimage 2013) 19

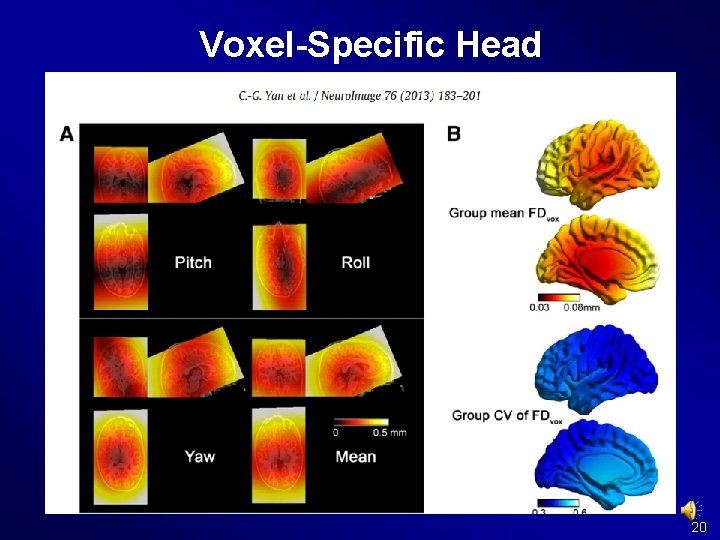
Voxel-Specific Head Motion Calculation {Working. Dir}Voxel. Specific. Head. MotionSub_xxx: HMvox_x_*. nii: voxel specific translation in x axis FDvox_*. nii: Frame-wise Displacement (relative to the previous time point) for each voxel TDvox_*. nii: Total Displacement (relative to the reference time point) for each voxel Mean. FDvox. nii: temporal mean of FDvox for each voxel Mean. TDvox. nii: temporal mean of TDvox for each voxel 20

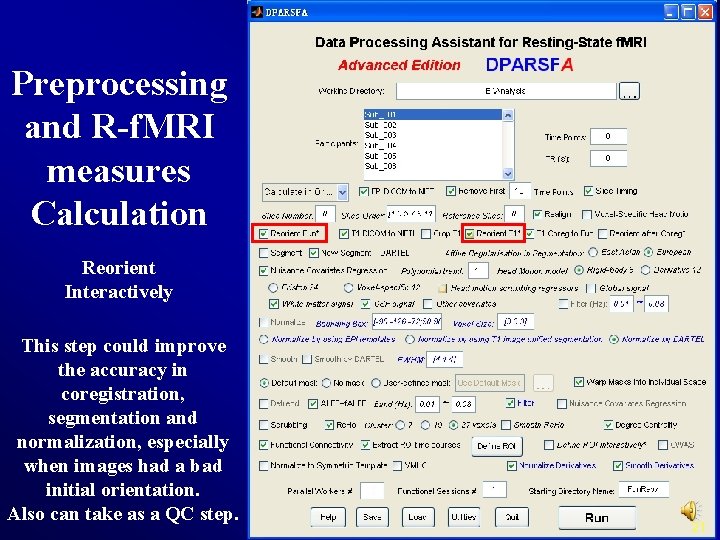
Preprocessing and R-f. MRI measures Calculation Reorient Interactively This step could improve the accuracy in coregistration, segmentation and normalization, especially when images had a bad initial orientation. Also can take as a QC step. 21

22

23

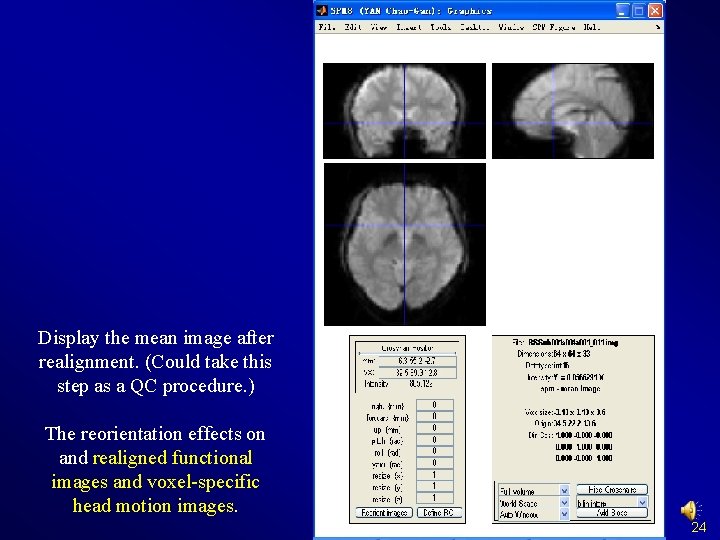
Display the mean image after realignment. (Could take this step as a QC procedure. ) The reorientation effects on and realigned functional images and voxel-specific head motion images. 24

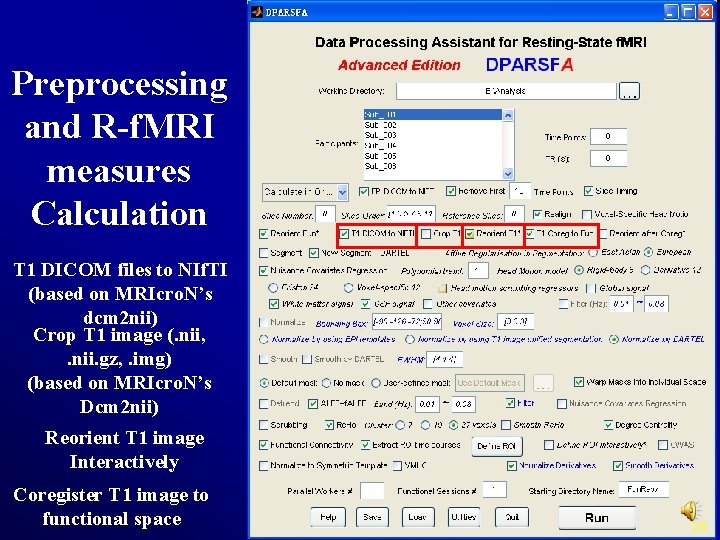
Preprocessing and R-f. MRI measures Calculation T 1 DICOM files to NIf. TI (based on MRIcro. N’s dcm 2 nii) Crop T 1 image (. nii, . nii. gz, . img) (based on MRIcro. N’s Dcm 2 nii) Reorient T 1 image Interactively Coregister T 1 image to functional space 25

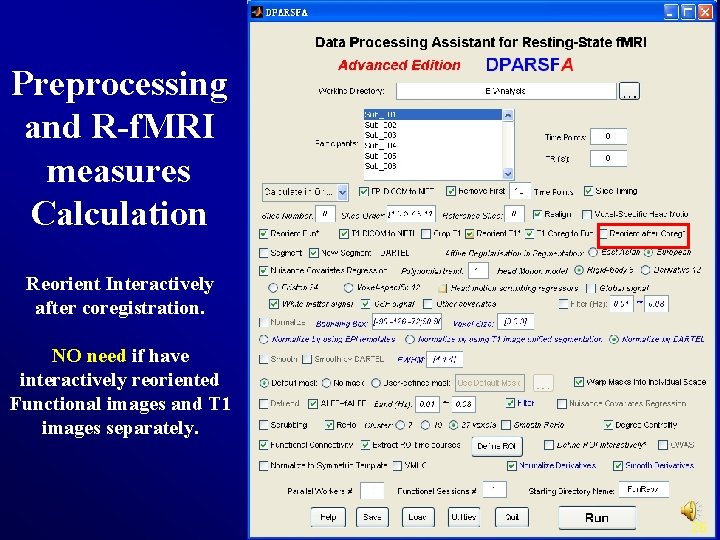
Preprocessing and R-f. MRI measures Calculation Reorient Interactively after coregistration. NO need if have interactively reoriented Functional images and T 1 images separately. 26

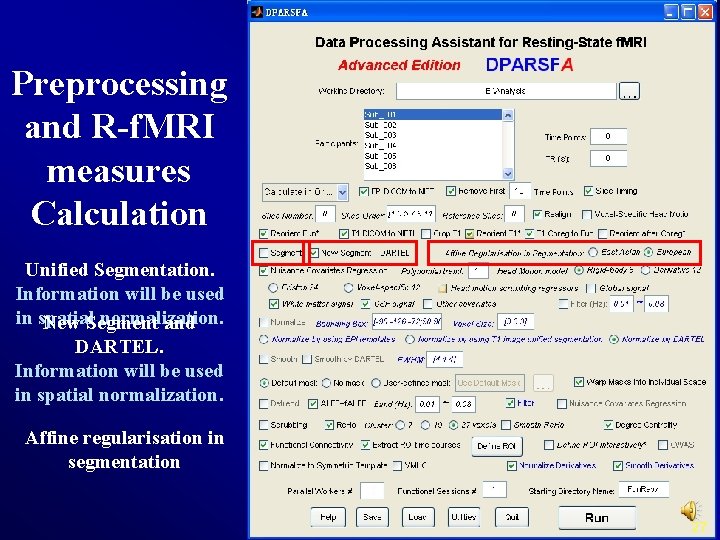
Preprocessing and R-f. MRI measures Calculation Unified Segmentation. Information will be used in spatial normalization. New Segment and DARTEL. Information will be used in spatial normalization. Affine regularisation in segmentation 27

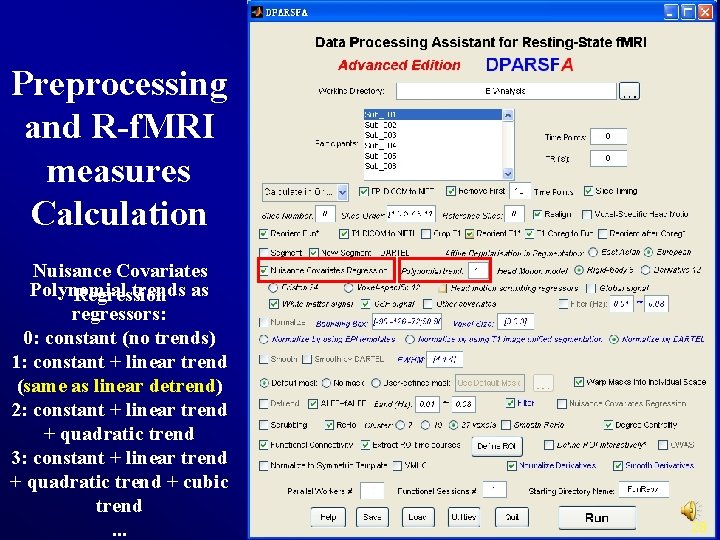
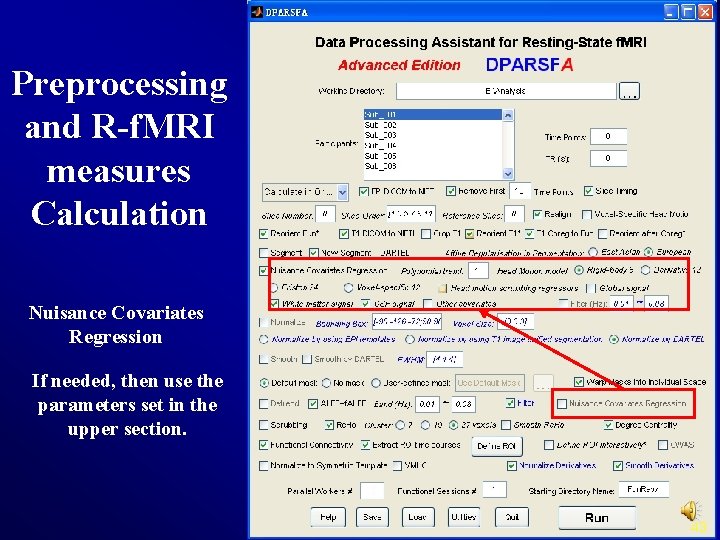
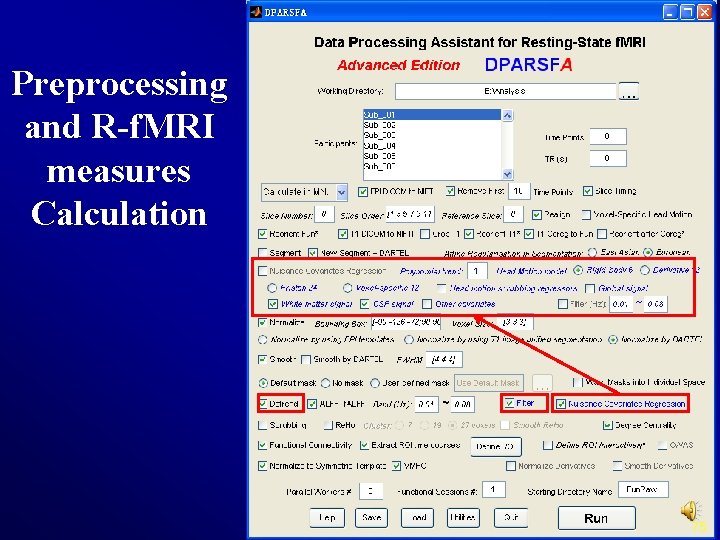
Preprocessing and R-f. MRI measures Calculation Nuisance Covariates Polynomial trends as Regression regressors: 0: constant (no trends) 1: constant + linear trend (same as linear detrend) 2: constant + linear trend + quadratic trend 3: constant + linear trend + quadratic trend + cubic trend. . . 28

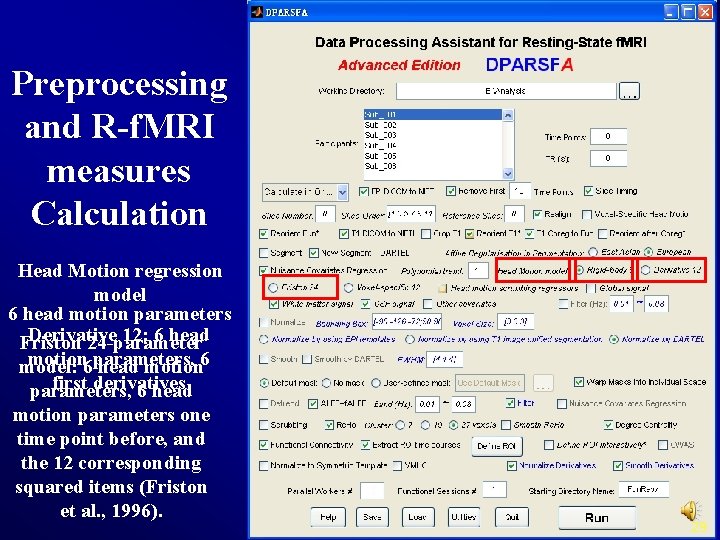
Preprocessing and R-f. MRI measures Calculation Head Motion regression model 6 head motion parameters Derivative 12: 6 head Friston 24 -parameter motion 6 parameters, model: head motion 6 first derivatives parameters, 6 head motion parameters one time point before, and the 12 corresponding squared items (Friston et al. , 1996). 29

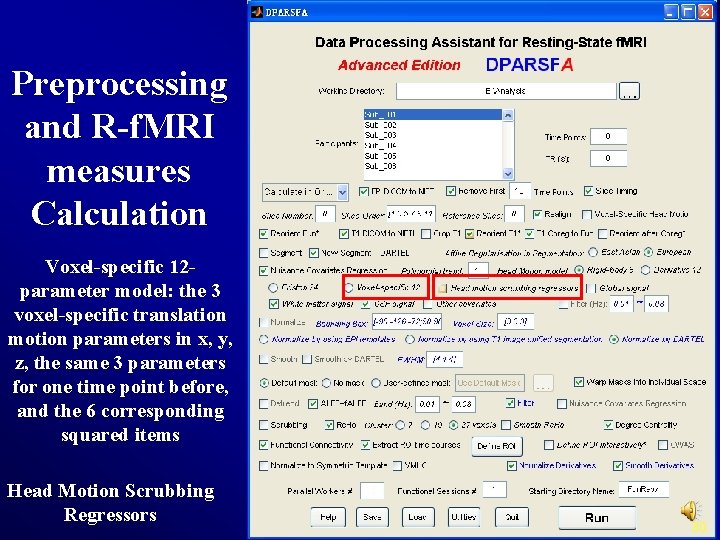
Preprocessing and R-f. MRI measures Calculation Voxel-specific 12 parameter model: the 3 voxel-specific translation motion parameters in x, y, z, the same 3 parameters for one time point before, and the 6 corresponding squared items Head Motion Scrubbing Regressors 30

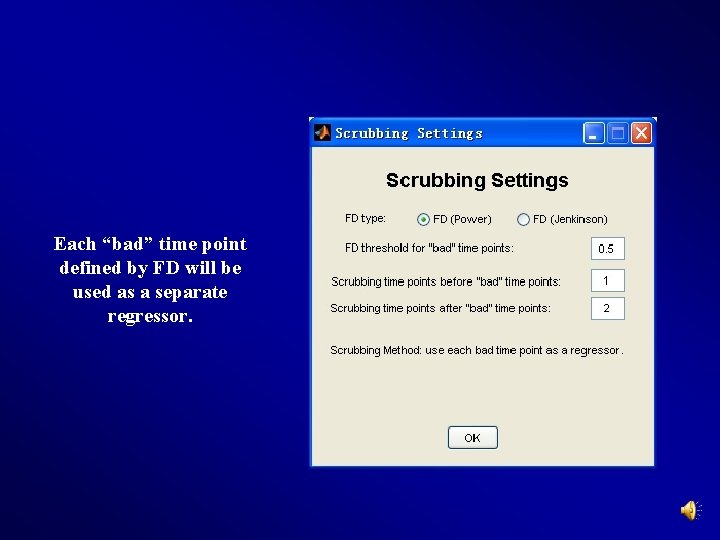
Each “bad” time point defined by FD will be used as a separate regressor.

Yan et al. , 2013, Neuroimage

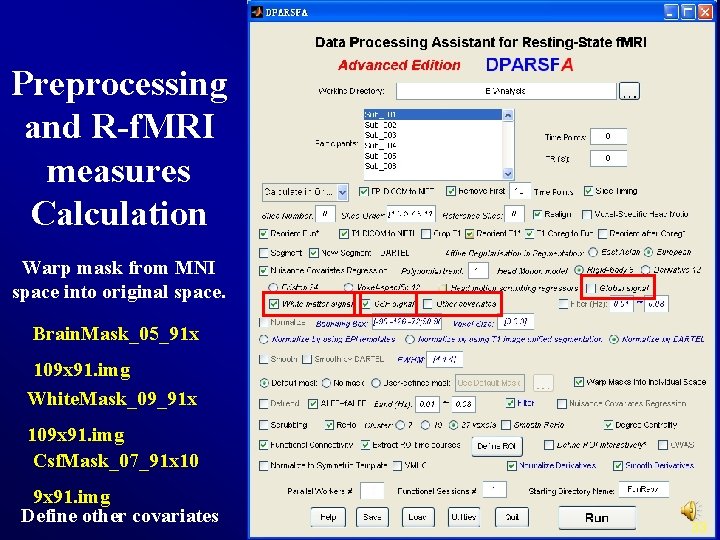
Preprocessing and R-f. MRI measures Calculation Warp mask from MNI space into original space. Brain. Mask_05_91 x 109 x 91. img White. Mask_09_91 x 109 x 91. img Csf. Mask_07_91 x 10 9 x 91. img Define other covariates 33

Preprocessing and R-f. MRI measures Calculation Plan to implement Comp. Cor (Behzadi et al. , 2007), ANATICOR (Jo et al. , 2010) in the next version. Also use the eroded mask from segmentation as well. 34

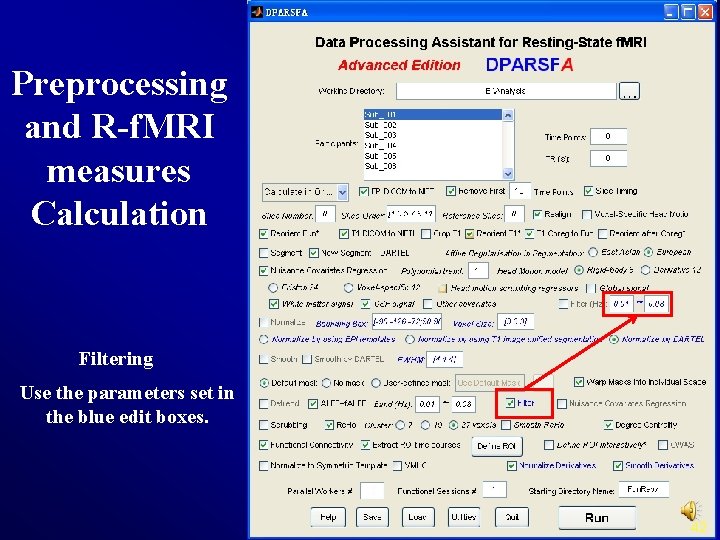
Preprocessing and R-f. MRI measures Calculation Filtering The filtering parameters will be used later (Blue checkbox). 35

Preprocessing and R-f. MRI measures Calculation Spatial Normalize Calculate in original space, the normalization parameters will be used in normalizing the R-f. MRI metrics calculated in original space (derivatives) (Blue checkbox). 36

Preprocessing and R-f. MRI measures Calculation Spatial Smooth Calculate in original space, the smooth parameters will be used in smoothing the R-f. MRI metrics calculated in original space (derivatives) (Blue checkbox). 37

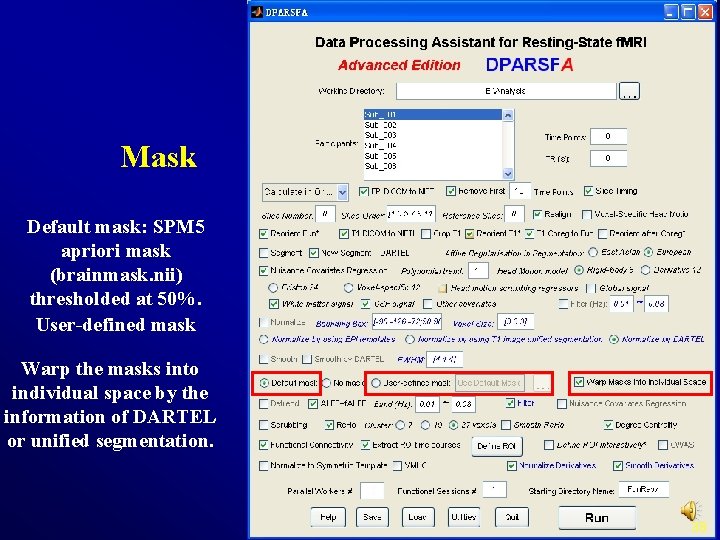
Mask Default mask: SPM 5 apriori mask (brainmask. nii) thresholded at 50%. User-defined mask Warp the masks into individual space by the information of DARTEL or unified segmentation. 38

Preprocessing and R-f. MRI measures Calculation Linear detrend (NO need since included in nuisance covariate regression) 39

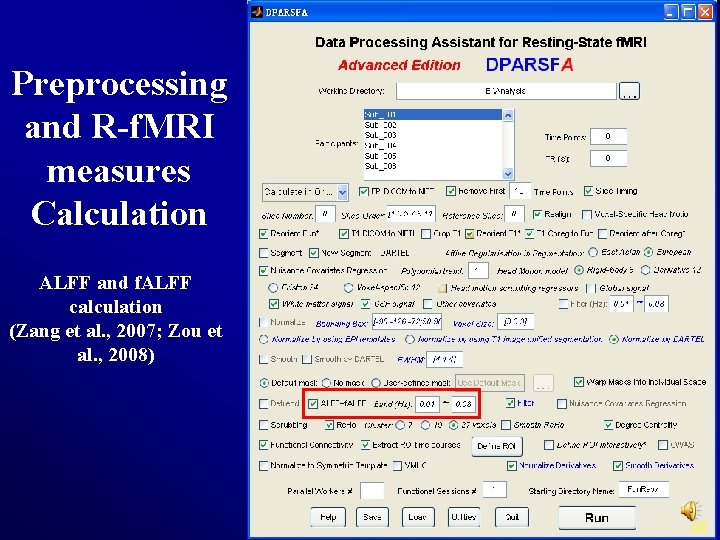
Preprocessing and R-f. MRI measures Calculation ALFF and f. ALFF calculation (Zang et al. , 2007; Zou et al. , 2008) 40

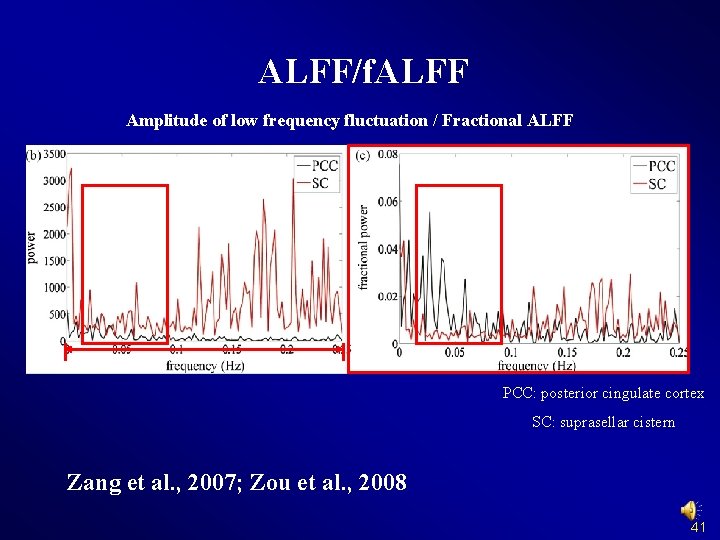
ALFF/f. ALFF Amplitude of low frequency fluctuation / Fractional ALFF PCC: posterior cingulate cortex SC: suprasellar cistern Zang et al. , 2007; Zou et al. , 2008 41

Preprocessing and R-f. MRI measures Calculation Filtering Use the parameters set in the blue edit boxes. 42

Preprocessing and R-f. MRI measures Calculation Nuisance Covariates Regression If needed, then use the parameters set in the upper section. 43

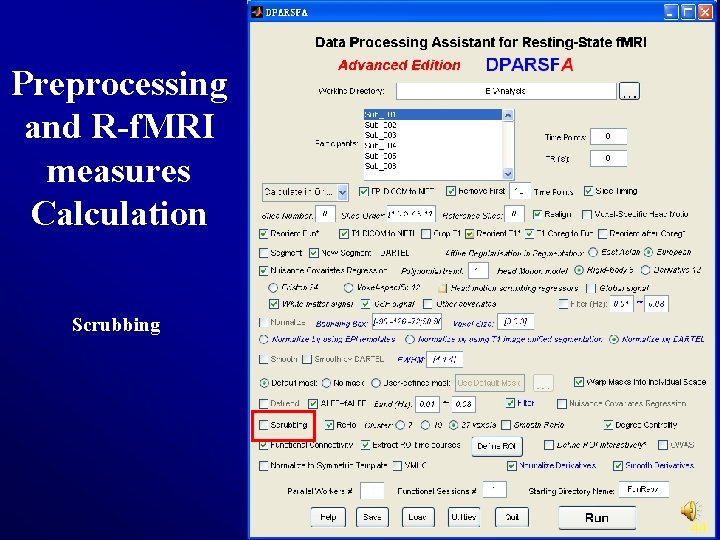
Preprocessing and R-f. MRI measures Calculation Scrubbing 44

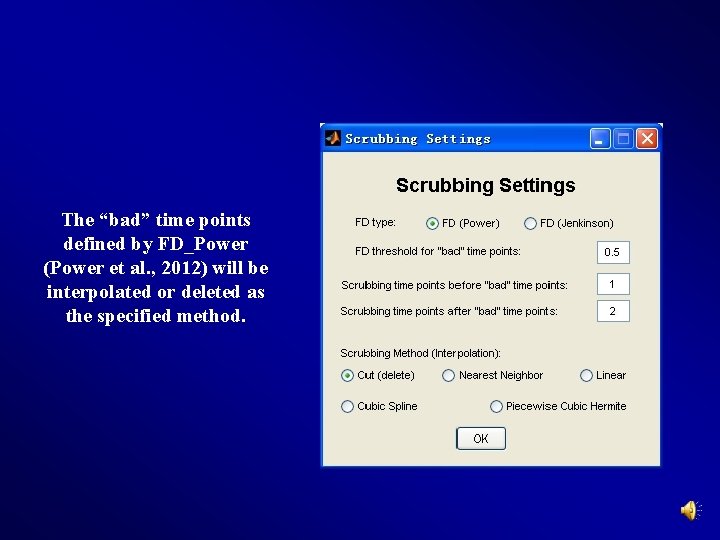
The “bad” time points defined by FD_Power (Power et al. , 2012) will be interpolated or deleted as the specified method.

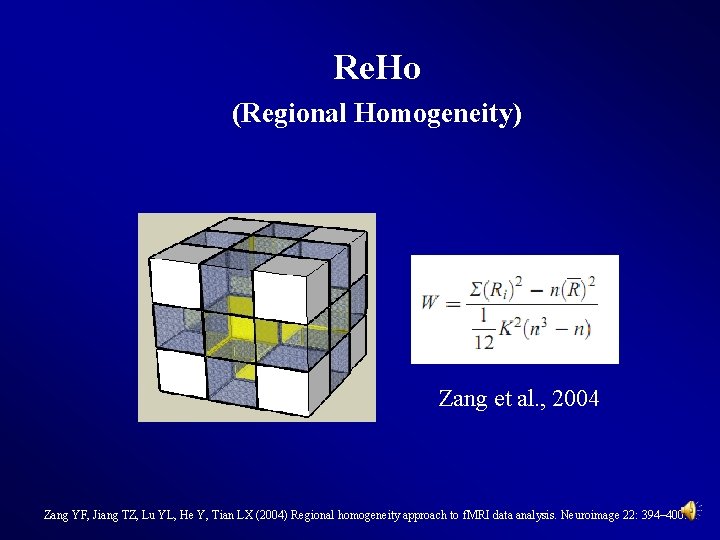
Preprocessing and R-f. MRI measures Calculation Regional Homogeneity (Re. Ho) Calculation (Zang et al. , 2004) 46

Re. Ho (Regional Homogeneity) Zang et al. , 2004 Zang YF, Jiang TZ, Lu YL, He Y, Tian LX (2004) Regional homogeneity approach to f. MRI data analysis. Neuroimage 22: 394– 400.

Preprocessing and R-f. MRI measures Calculation Regional Homogeneity (Re. Ho) Calculation (Zang et al. , 2004) 48

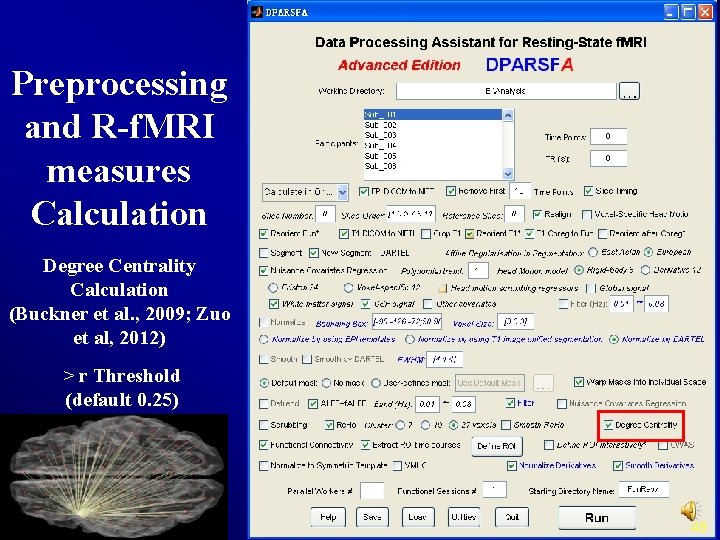
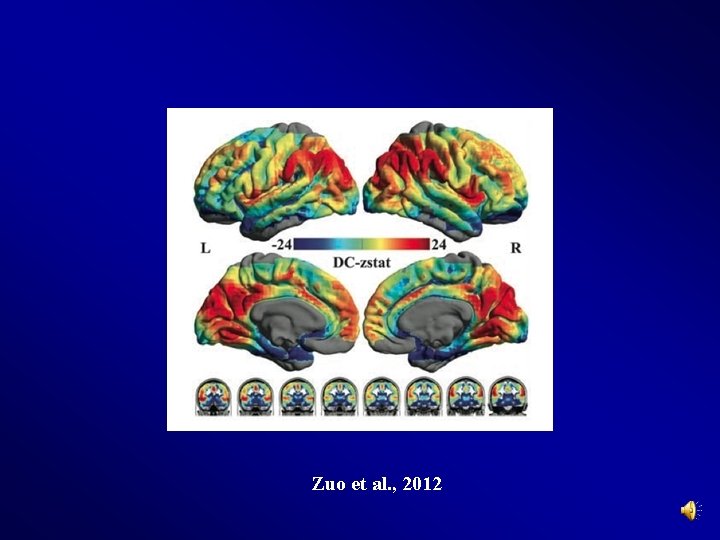
Preprocessing and R-f. MRI measures Calculation Degree Centrality Calculation (Buckner et al. , 2009; Zuo et al, 2012) > r Threshold (default 0. 25) 49

Zuo et al. , 2012

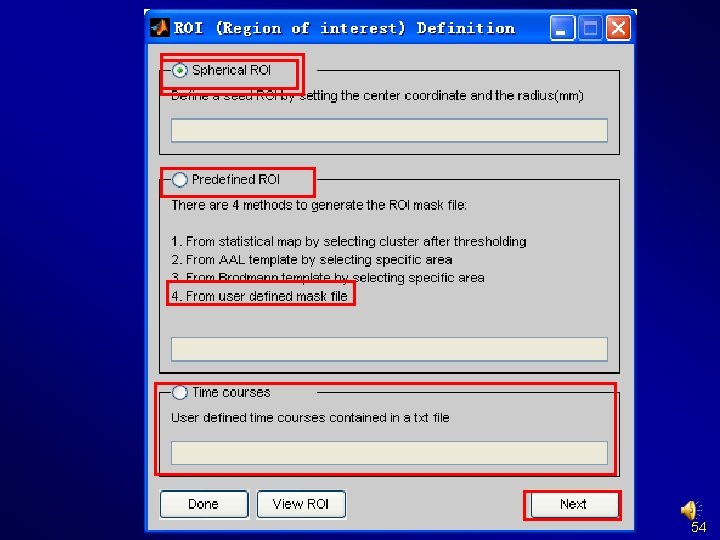
Preprocessing and R-f. MRI measures Calculation Functional Connectivity (voxel-wise seed based correlation analysis) Extract ROI time courses (also for ROI-wise Functional Connectivity) Define ROI 51

Define ROI Multiple labels in mask file: each label is considered as one ROI Dosenbach et al. , 2010 Andrews-Hanna et al. , 2010 Craddock et al. , 2011 Define other ROIs 52

Define ROI 53

54

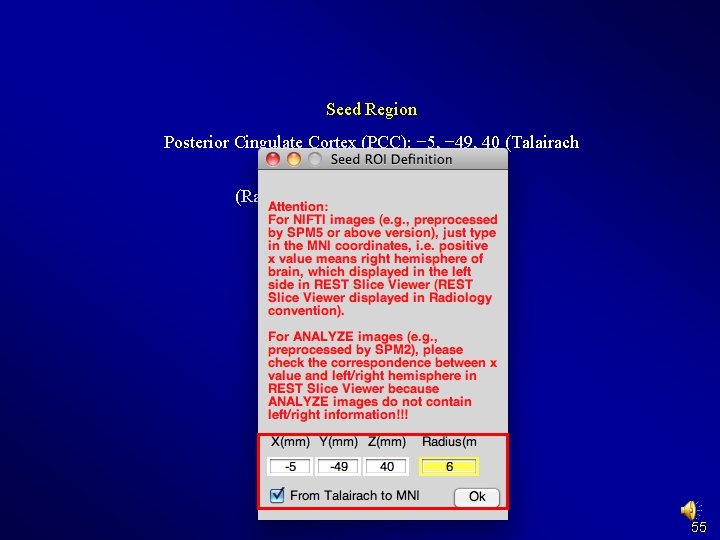
Seed Region Posterior Cingulate Cortex (PCC): − 5, − 49, 40 (Talairach coordinates) (Raichle et al. , 2001; Fox et al. , 2005) 55

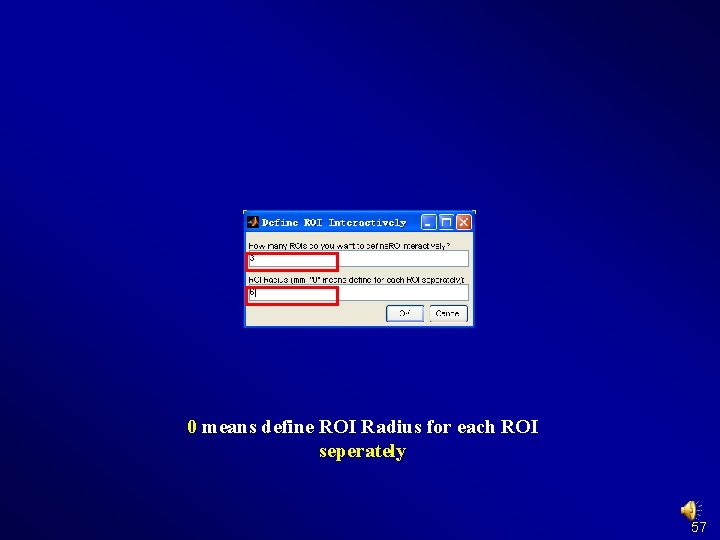
Preprocessing and R-f. MRI measures Calculation Define ROI Interactively 56

0 means define ROI Radius for each ROI seperately 57

58

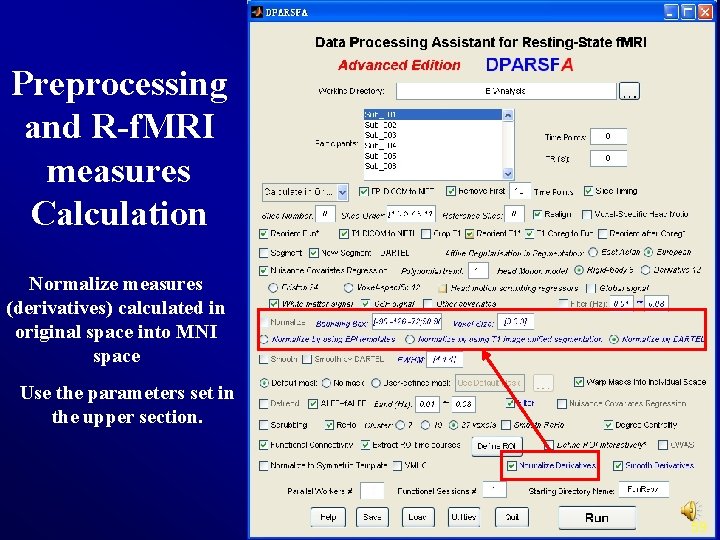
Preprocessing and R-f. MRI measures Calculation Normalize measures (derivatives) calculated in original space into MNI space Use the parameters set in the upper section. 59

Normalize Check Normalization with DPARSF {WORKINGDIR}Pictures. For. Chk. Normalization 60

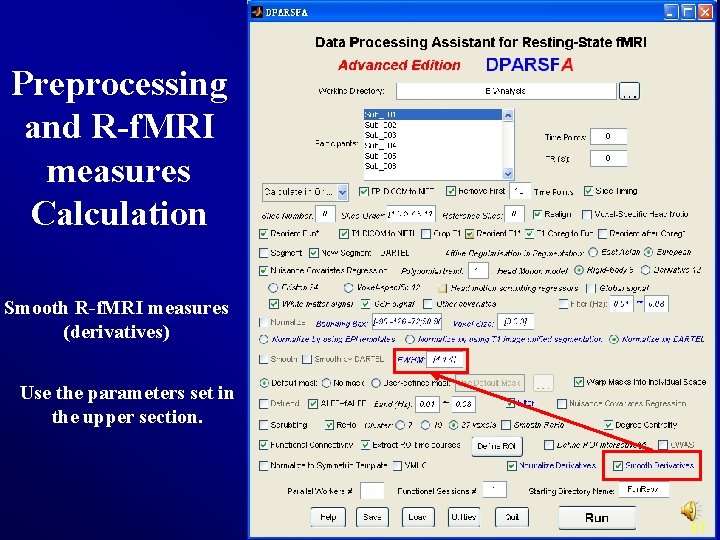
Preprocessing and R-f. MRI measures Calculation Smooth R-f. MRI measures (derivatives) Use the parameters set in the upper section. 61

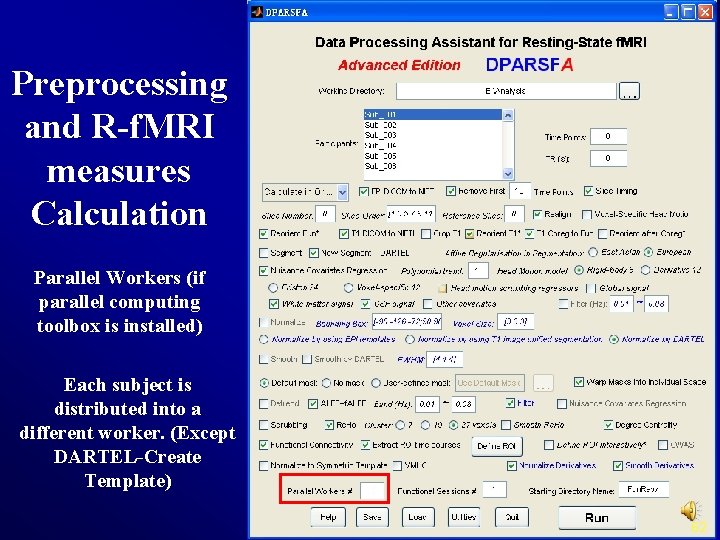
Preprocessing and R-f. MRI measures Calculation Parallel Workers (if parallel computing toolbox is installed) Each subject is distributed into a different worker. (Except DARTEL-Create Template) 62

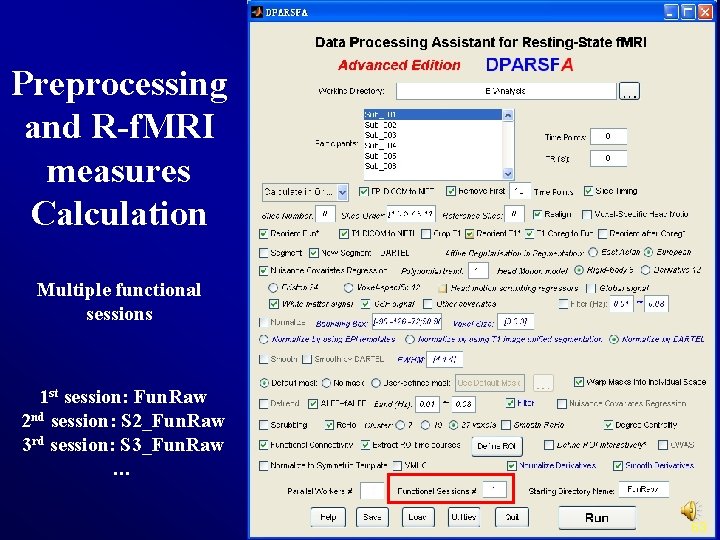
Preprocessing and R-f. MRI measures Calculation Multiple functional sessions 1 st session: Fun. Raw 2 nd session: S 2_Fun. Raw 3 rd session: S 3_Fun. Raw … 63

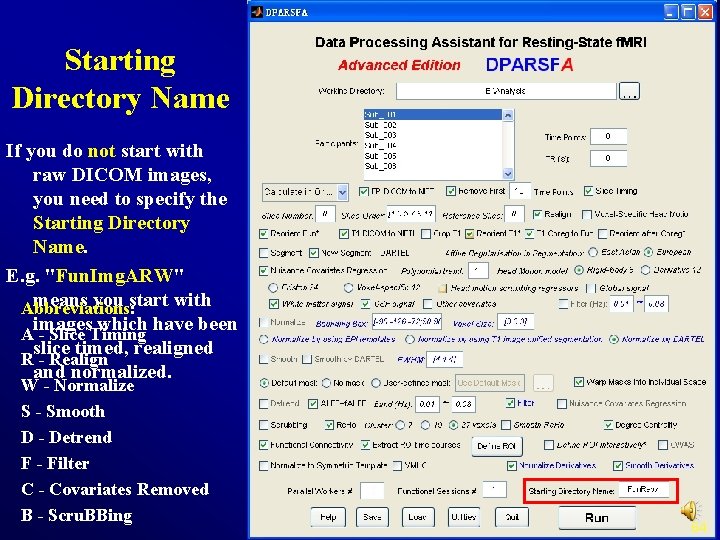
Starting Directory Name If you do not start with raw DICOM images, you need to specify the Starting Directory Name. E. g. "Fun. Img. ARW" means you start with Abbreviations: images which have been A - Slice Timing slice timed, realigned R - Realign and normalized. W - Normalize S - Smooth D - Detrend F - Filter C - Covariates Removed B - Scru. BBing 64

esting State f. MRI Data Processing Template Parameters 65

esting State f. MRI Data Processing Calculate in Original space Calculate in MNI space 66

Preprocessing and R-f. MRI measures Calculation 67

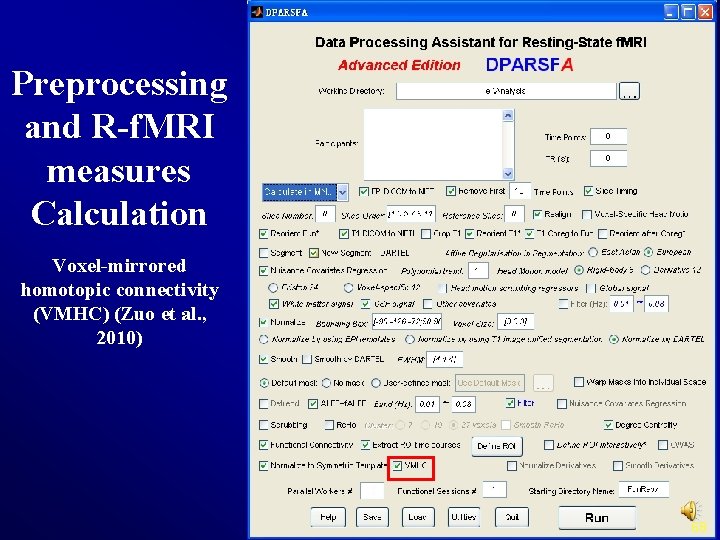
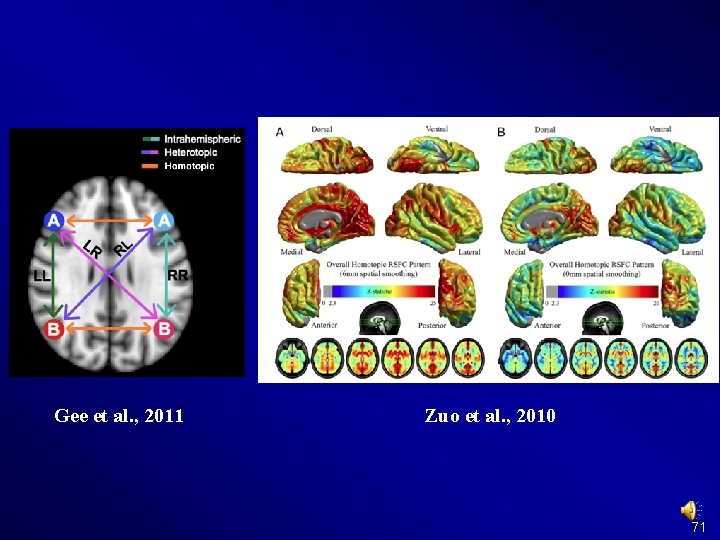
Preprocessing and R-f. MRI measures Calculation Voxel-mirrored homotopic connectivity (VMHC) (Zuo et al. , 2010) 68

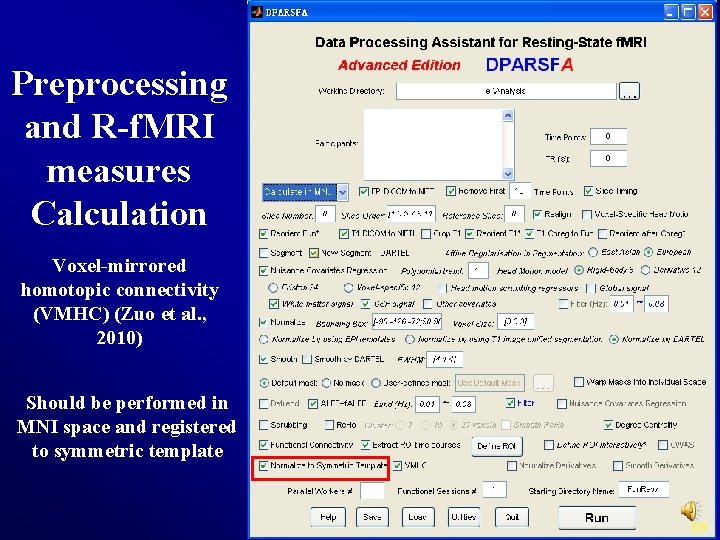
Preprocessing and R-f. MRI measures Calculation Voxel-mirrored homotopic connectivity (VMHC) (Zuo et al. , 2010) Should be performed in MNI space and registered to symmetric template 69

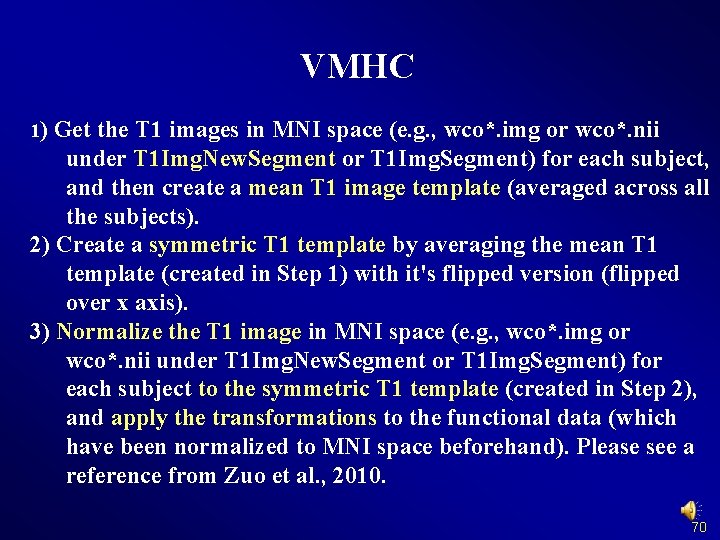
VMHC 1) Get the T 1 images in MNI space (e. g. , wco*. img or wco*. nii under T 1 Img. New. Segment or T 1 Img. Segment) for each subject, and then create a mean T 1 image template (averaged across all the subjects). 2) Create a symmetric T 1 template by averaging the mean T 1 template (created in Step 1) with it's flipped version (flipped over x axis). 3) Normalize the T 1 image in MNI space (e. g. , wco*. img or wco*. nii under T 1 Img. New. Segment or T 1 Img. Segment) for each subject to the symmetric T 1 template (created in Step 2), and apply the transformations to the functional data (which have been normalized to MNI space beforehand). Please see a reference from Zuo et al. , 2010. 70

Gee et al. , 2011 Zuo et al. , 2010 71

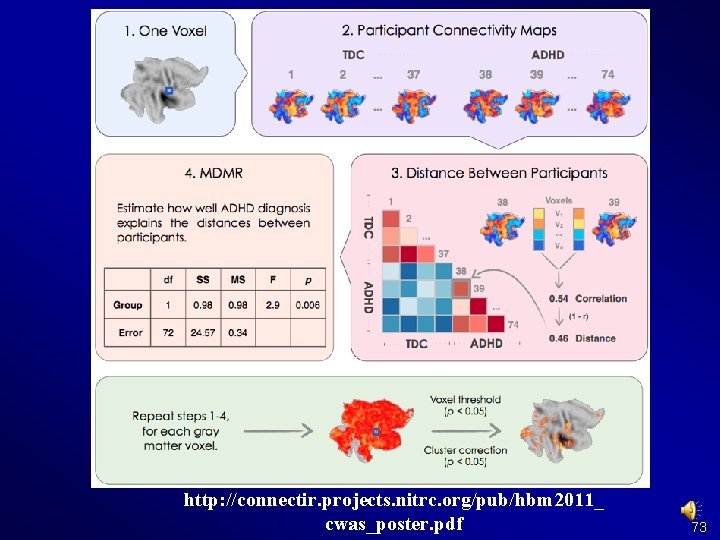
Preprocessing and R-f. MRI measures Calculation Connectome-wide association studies based on multivariate distance matrix regression (Shehzad et al. , 2011) Resource consuming as compared to other measures 72

http: //connectir. projects. nitrc. org/pub/hbm 2011_ cwas_poster. pdf 73

esting State f. MRI Data Processing Calculate in MNI space (TRADITIONAL order) (as V 2. 1) 74

Preprocessing and R-f. MRI measures Calculation 75

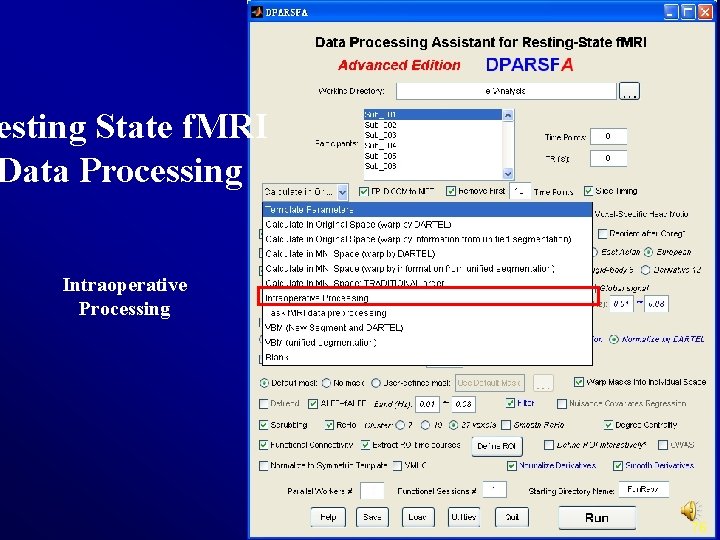
esting State f. MRI Data Processing Intraoperative Processing 76

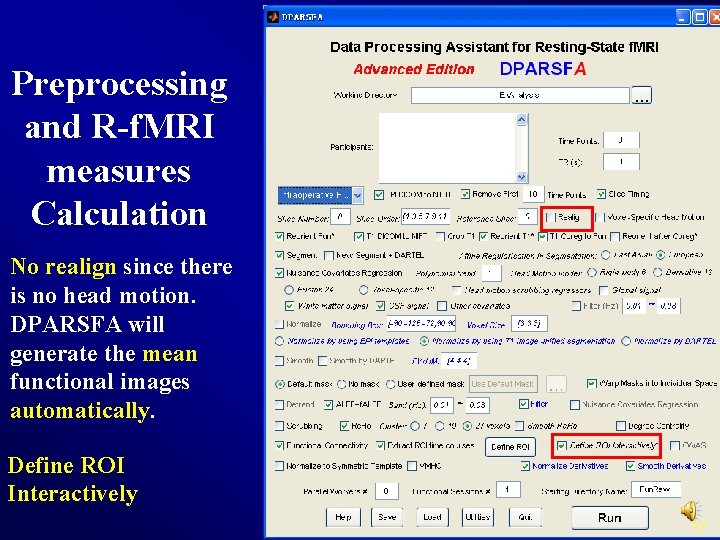
Preprocessing and R-f. MRI measures Calculation No realign since there is no head motion. DPARSFA will generate the mean functional images automatically. Define ROI Interactively 77

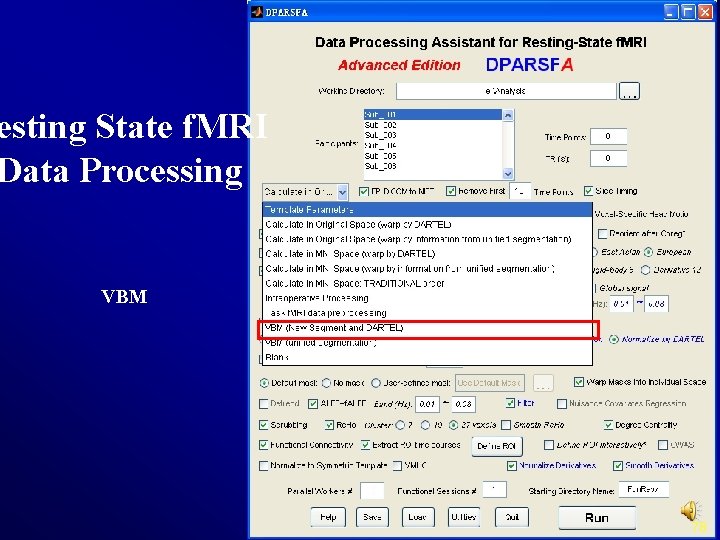
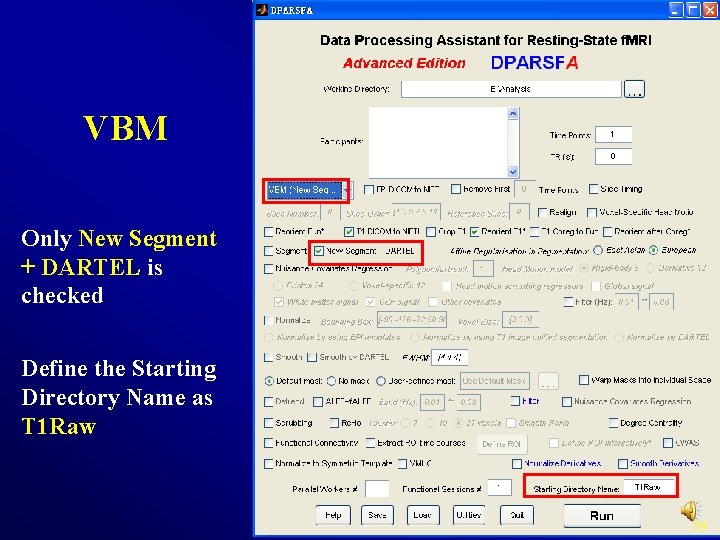
esting State f. MRI Data Processing VBM 78

VBM Only New Segment + DARTEL is checked Define the Starting Directory Name as T 1 Raw 79

esting State f. MRI Data Processing Blank 80

Blank 81

Further Help Further questions: www. restfmri. net 82

Thanks to DONG Zhang-Ye GUO Xiao-Juan HE Yong LONG Xiang-Yu SONG Xiao-Wei WANG Xin-Di YAO Li ZANG Yu-Feng ZANG Zhen-Xiang ZHANG Han ZHU Chao-Zhe ZOU Qi-Hong ZUO Xi-Nian …… Brian CHEUNG Qingyang LI Michael MILHAM …… SPM Team: Wellcome Department of Imaging Neuroscience, UCL MRIcro. N Team: Chris RORDEN Xjview Team: CUI Xu …… 83

Thanks for your attention! 84