Course Data Processing of RestingState f MRI Part
















































- Slides: 111

Course: Data Processing of Resting-State f. MRI (Part 1) Data Processing Assistant for Resting-State f. MRI: Speed Up Your Data Analysis YAN Chao-Gan 严超赣 Ph. D. ycg. yan@gmail. com State Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University, China 1

Outline • Overview • Data Preparation • Preprocess • Re. Ho, ALFF, f. ALFF Calculation • Functional Connectivity • Utilities 2

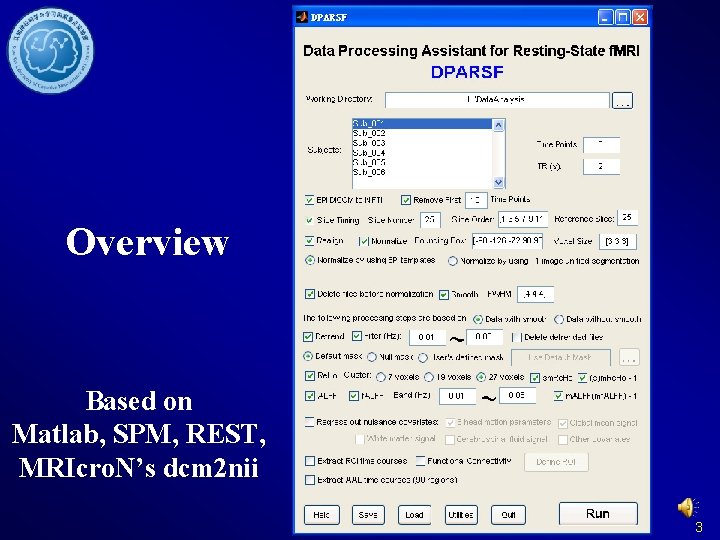
Overview Based on Matlab, SPM, REST, MRIcro. N’s dcm 2 nii 3

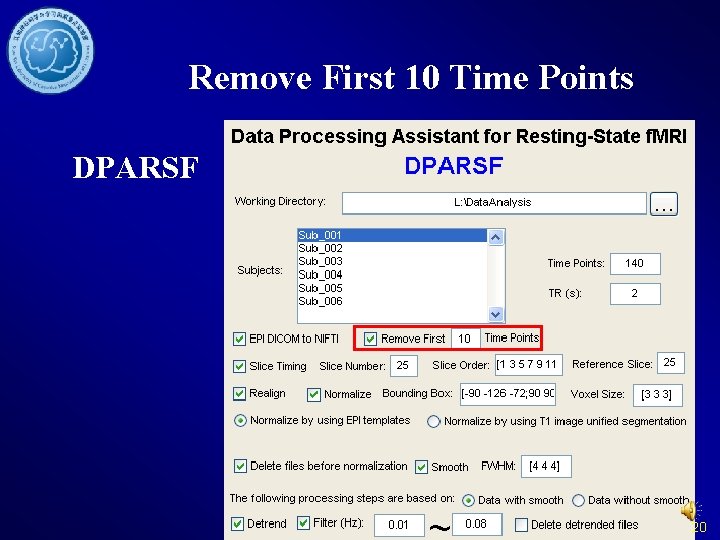
DPARSF's standard procedure l Convert DICOM files to NIFTI images. l Remove First 10 Time Points. l Slice Timing. l Realign. l Normalize. l Smooth (optional). l Detrend. l Filter. l Calculate Re. Ho, ALFF, f. ALFF (optional). l Regress out the Covariables (optional). l Calculate Functional Connectivity (optional). l Extract AAL or ROI time courses for further analysis (optional). 4

Outline • Overview • Data Preparation • Preprocess • Re. Ho, ALFF, f. ALFF Calculation • Functional Connectivity • Utilities 5

Data preparation Arrange the information of the subjects 6

Data preparation Information of subjects 7

Data preparation Arrange the information of the subjects Arrange the MRI data of the subjects Functional MRI data Structural MRI data DTI data 8


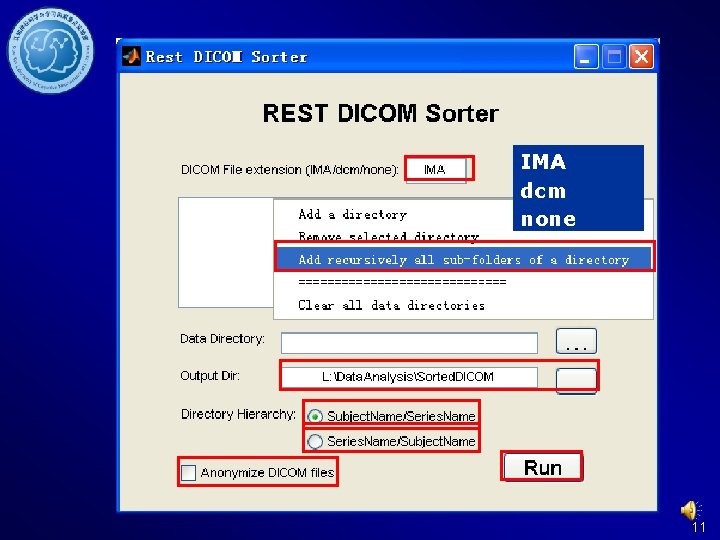
Sort DICOM data 10

IMA dcm none 11

Data preparation Arrange each subject's f. MRI DICOM images in one directory, and then put them in "Fun. Raw" directory under the working directory. Subject 1’s DICOM 1’s Fun. Raw files directory, please name as this Working directory 12

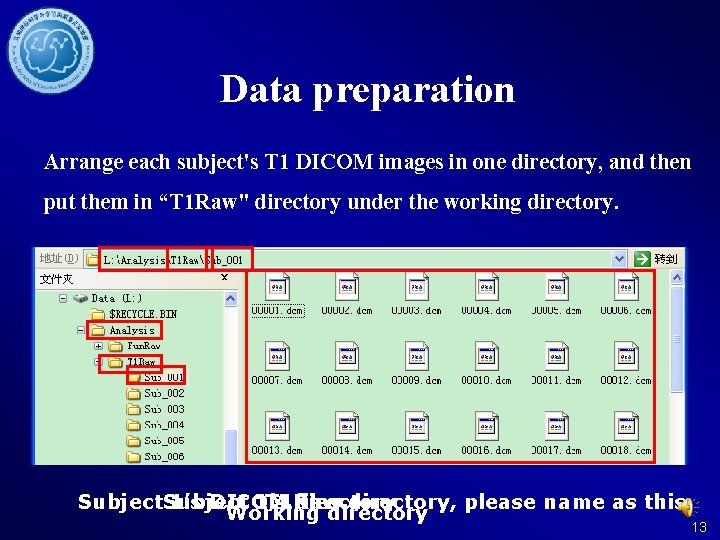
Data preparation Arrange each subject's T 1 DICOM images in one directory, and then put them in “T 1 Raw" directory under the working directory. Subject 1’s DICOM 1’s T 1 Raw files directory, please name as this Working directory 13

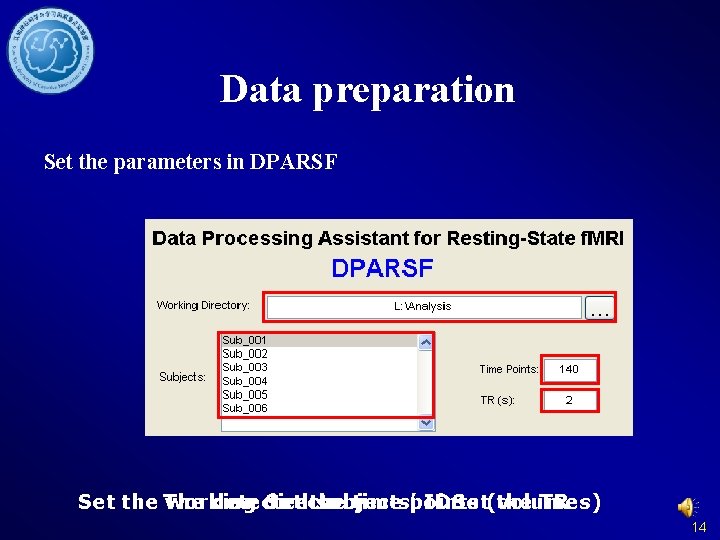
Data preparation Set the parameters in DPARSF Set the The working detected directory Set the subjects’ time points IDSet(volumes) the TR 14

Outline • Overview • Data Preparation • Preprocess • Re. Ho, ALFF, f. ALFF Calculation • Functional Connectivity • Utilities 15

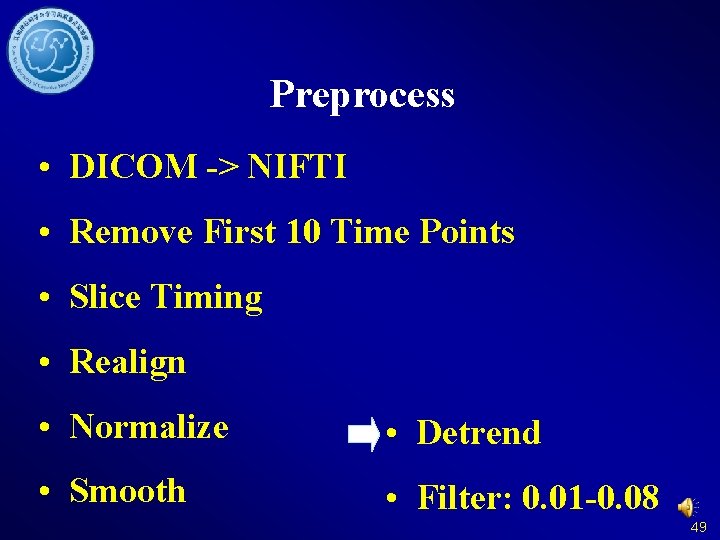
Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 16

DICOM->NIFTI MRIcro. N’s dcm 2 niigui SPM 5’s DICOM Import 17

DICOM->NIFTI DPARSF 18

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 19

Remove First 10 Time Points DPARSF 20

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 21

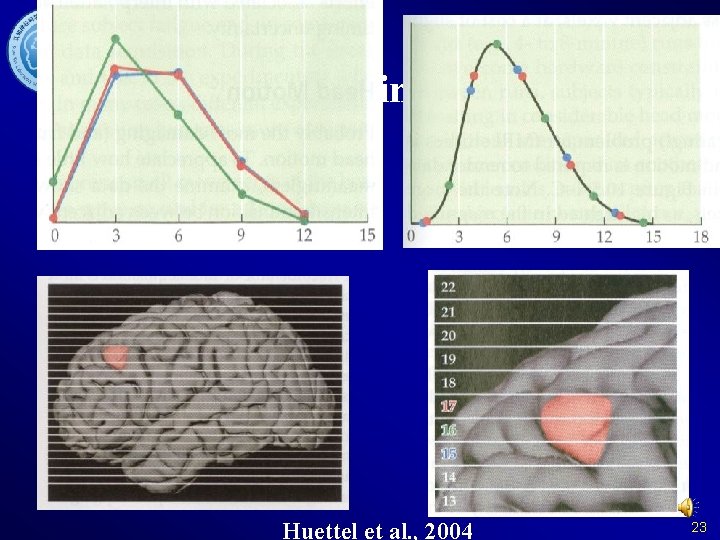
Slice Timing Why? 22

Slice Timing Why? Huettel et al. , 2004 23

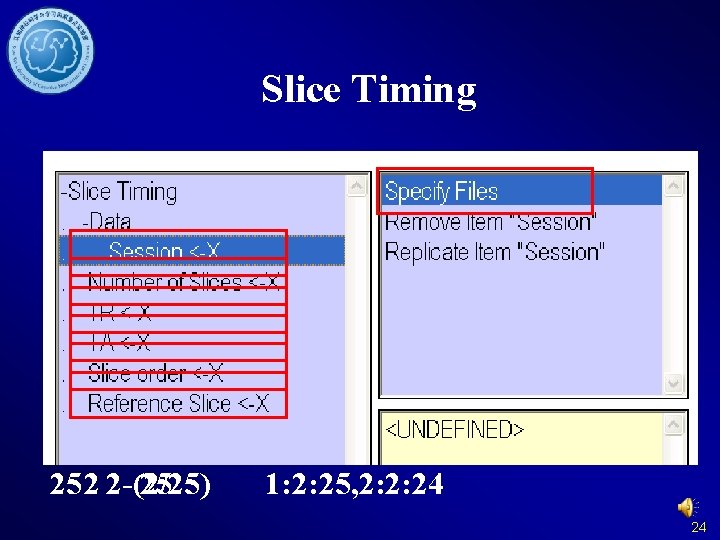
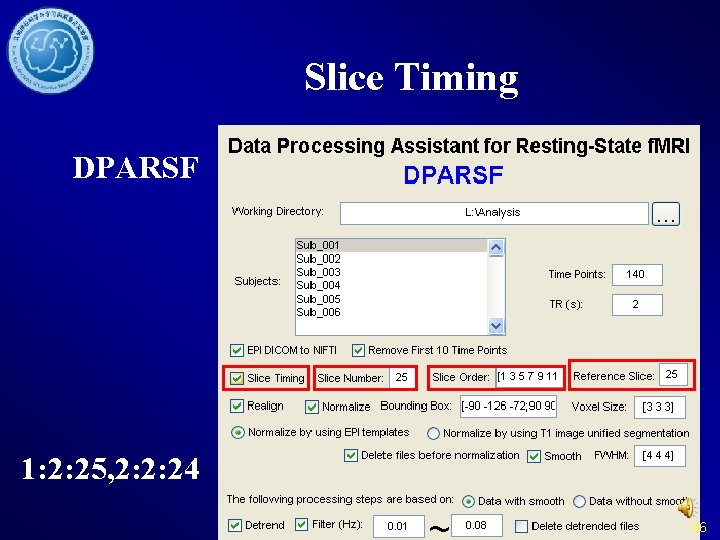
Slice Timing 252 2 -(2/25) 25 1: 2: 25, 2: 2: 24 24

Slice Timing 25

Slice Timing DPARSF 1: 2: 25, 2: 2: 24 26

Slice Timing If you start with NIFTI images (. hdr/. img pairs) before slice timing, you need to arrange each subject's f. MRI NIFTI images in one directory, and then put them in "Fun. Img" directory under the working directory. Fun. Img directory, please name as this 27

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 28

Realign Why? 29

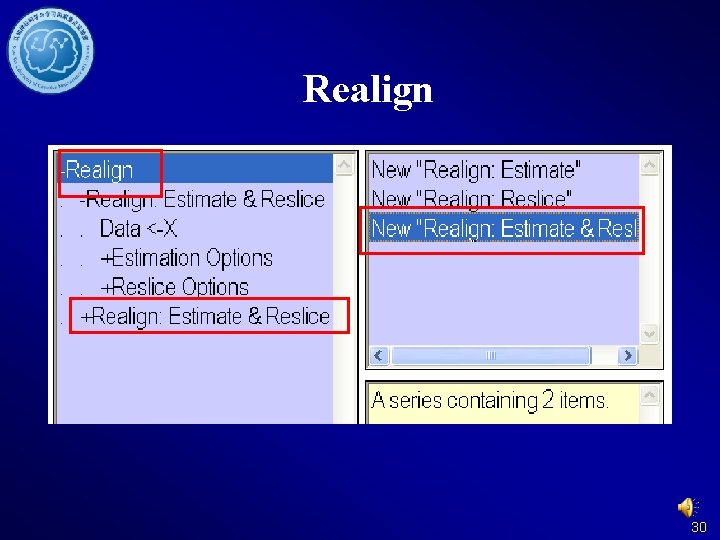
Realign 30

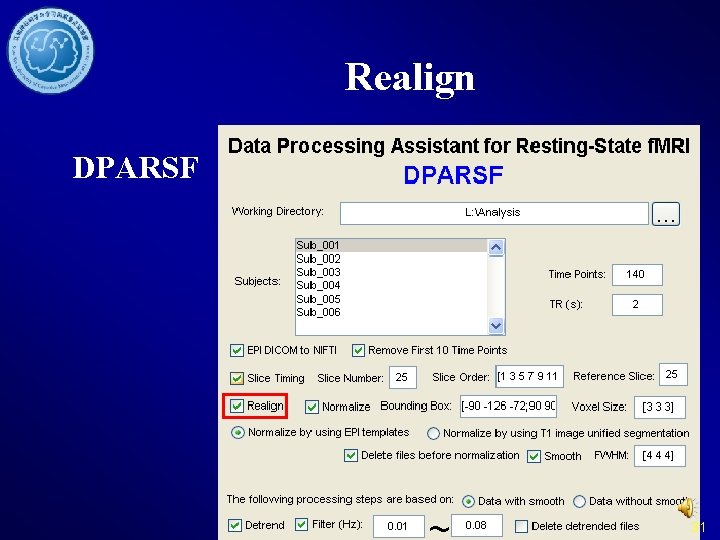
Realign DPARSF 31

Excluding Criteria: 2. 5 mm and 2. 5 degree None Realign Excluding Criteria: 2. 0 mm and 2. 0 degree Check head motion: Sub_013 Excluding Criteria: 1. 5 mm and 1. 5 degree Sub_013 Excluding Criteria: 1. 0 mm and 1. 0 degree Sub_007 Sub_012 Sub_013 Sub_017 Sub_018 32

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 33

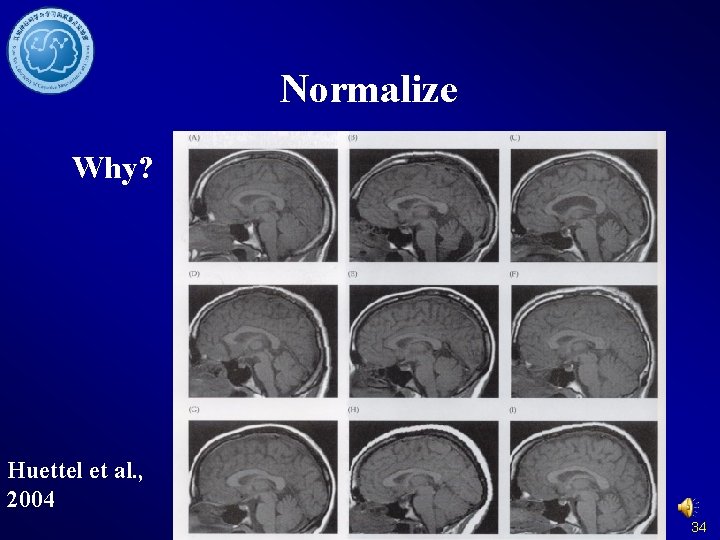
Normalize Why? Huettel et al. , 2004 34

Normalize Methods: I. Normalize by using EPI templates II. Normalize by using T 1 image unified segmentation 35

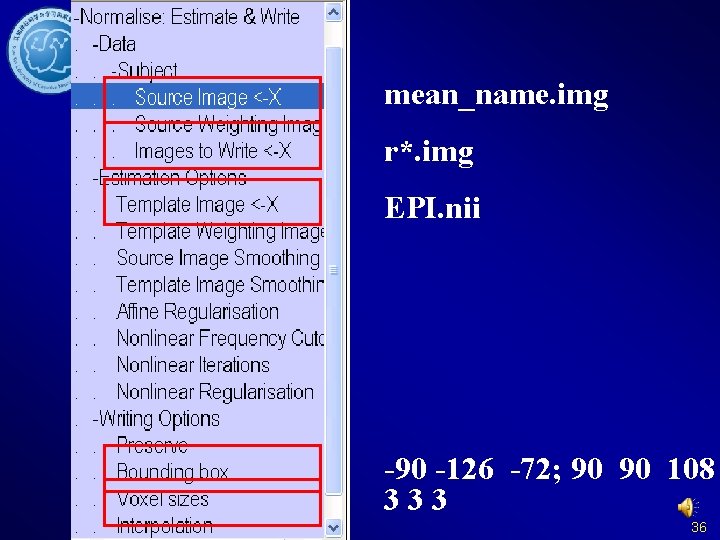
mean_name. img r*. img EPI. nii -90 -126 -72; 90 90 108 333 36

Normalize I 37

Normalize Methods: • Normalize by using EPI templates v Structural image was coregistered to the mean image after the motion • functional Normalize by using T 1 correction image v The transformed structural image was then segmented unified segmentation into gray matter, white matter, cerebrospinal fluid by using a unified segmentation algorithm v Normalize: the motion corrected functional volumes were spatially normalized to the MNI space using the normalization parameters estimated during unified segmentation (*_seg_sn. mat) 38

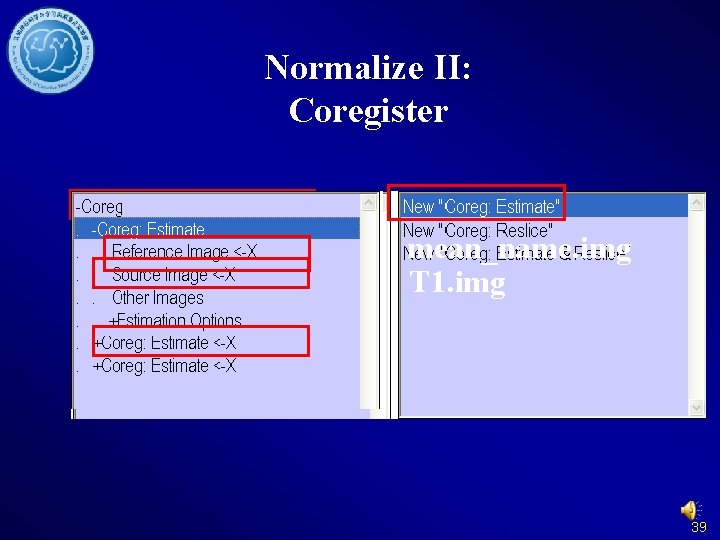
Normalize II: Coregister mean_name. img T 1. img 39

Normalize II: T 1_Coregisted. img Light Clean ICBM space template – East Asian brains – European brains 40

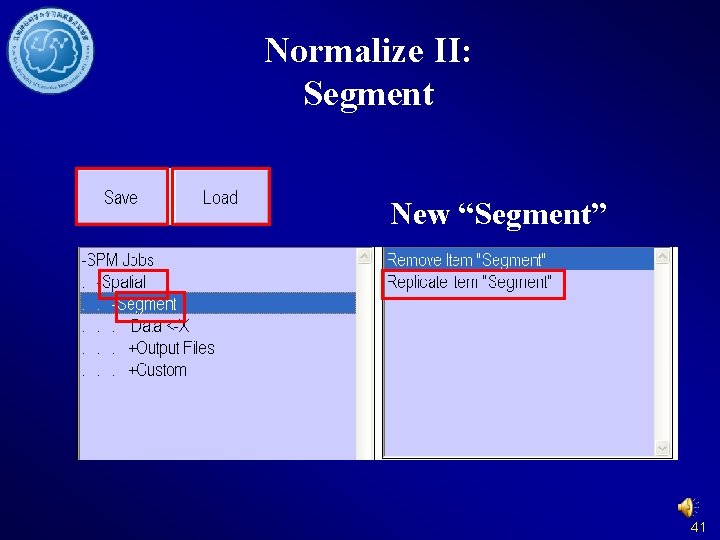
Normalize II: Segment New “Segment” 41

Normalize II: New “Normalize: Write” New “Subject” name_seg_sn. mat r*. img -90 -126 -72; 90 90 108 333 42

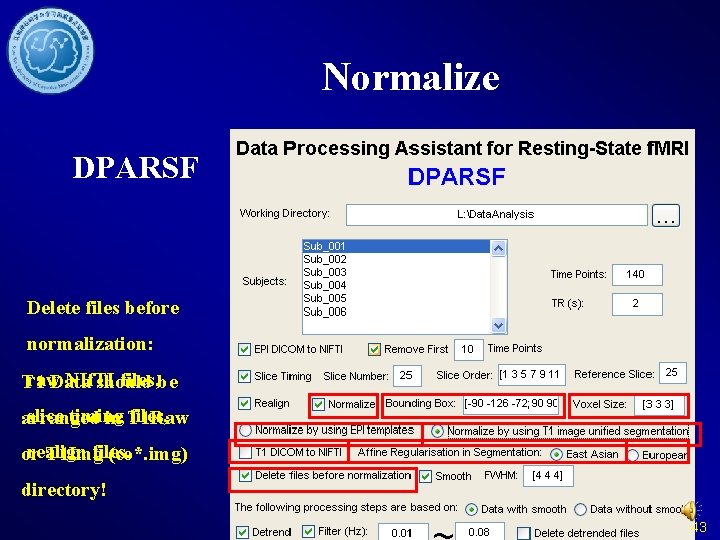
Normalize DPARSF Delete files before normalization: raw NIf. TI files, be T 1 Data should slice timing files, arranged in T 1 Raw realign files. or T 1 Img (co*. img) directory! 43

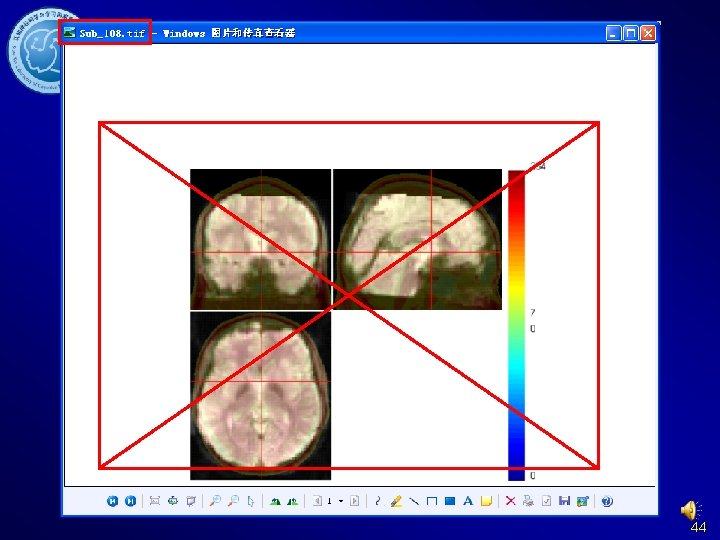
Normalize Check Normalization with DPARSF {WROKDIR}Pictures. For. Chk. Normalization 44

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 45

Smooth Why? • Reduce the effects of the bad normalization • … 46

Smooth w*. img FWHM kernel 47

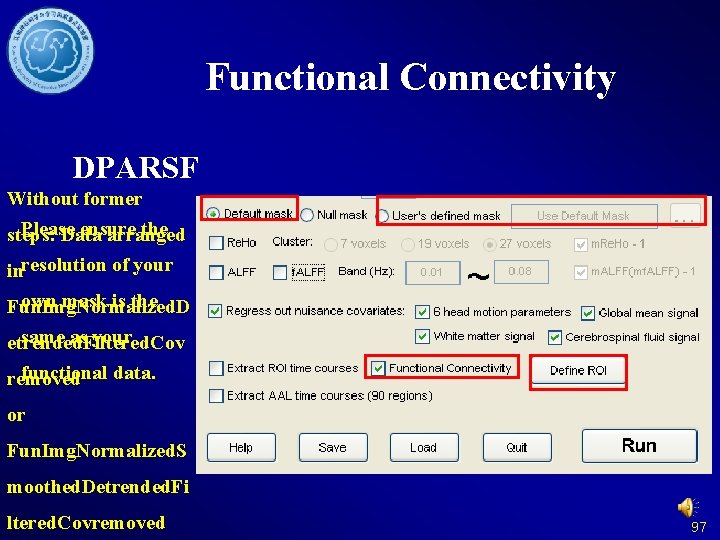
Smooth DPARSF Without former steps: Data arranged in. Re. Ho: Fun. Img. Normalized Data without directory. smoothf. ALFF, Funtional Connectivity: Data with smooth 48

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 49

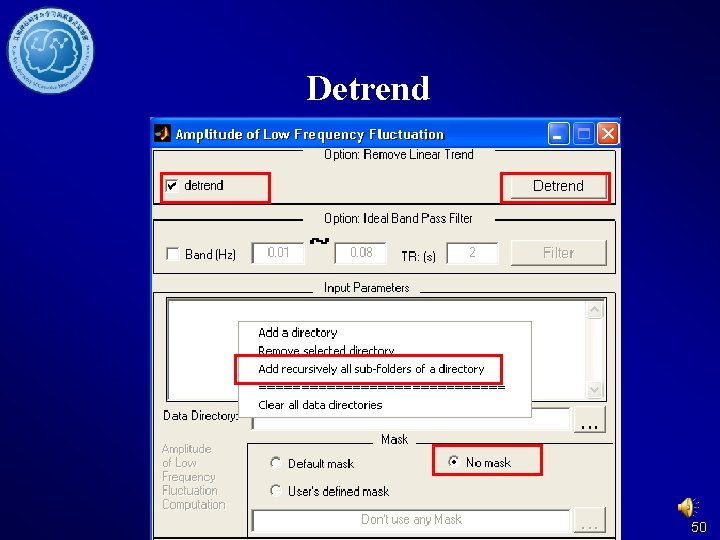
Detrend 50

Preprocess • DICOM -> NIFTI • Remove First 10 Time Points • Slice Timing • Realign • Normalize • Detrend • Smooth • Filter: 0. 01 -0. 08 51

滤波 Why? • Low frequency (0. 01– 0. 08 Hz) fluctuations (LFFs) of the resting-state f. MRI signal were of physiological importance. (Biswal et al. , 2005) • LFFs of resting-state f. MRI signal were suggested to reflect spontaneous neuronal activity (Logothetis et al. , 2001; Lu et al. , 2007). l Biswal B, Yetkin FZ, Haughton VM, Hyde JS (1995) Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34: 537– 541. l Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A (2001) Neurophysiological investigation of the basis of the f. MRI signal. Nature 412: 150– 157. l Lu H, Zuo Y, Gu H, Waltz JA, Zhan W, et al. (2007) Synchronized delta oscillations correlate with the resting-state functional MRI signal. Proc Natl Acad Sci U S A 104: 18265 – 18269. 52

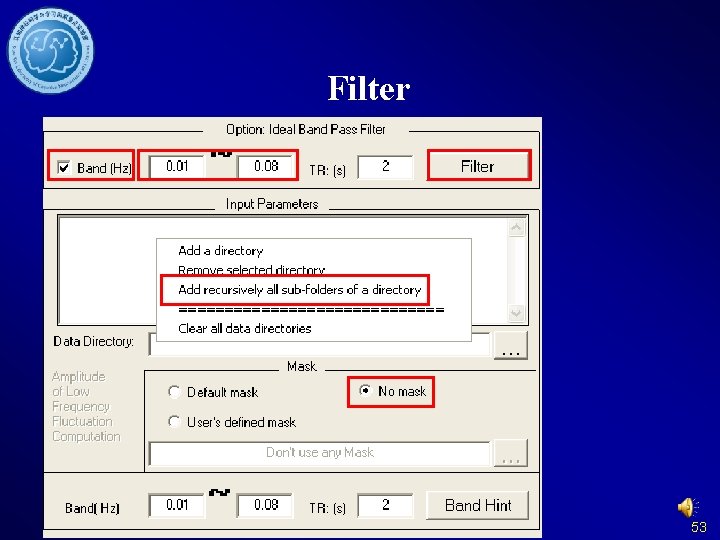
Filter 53

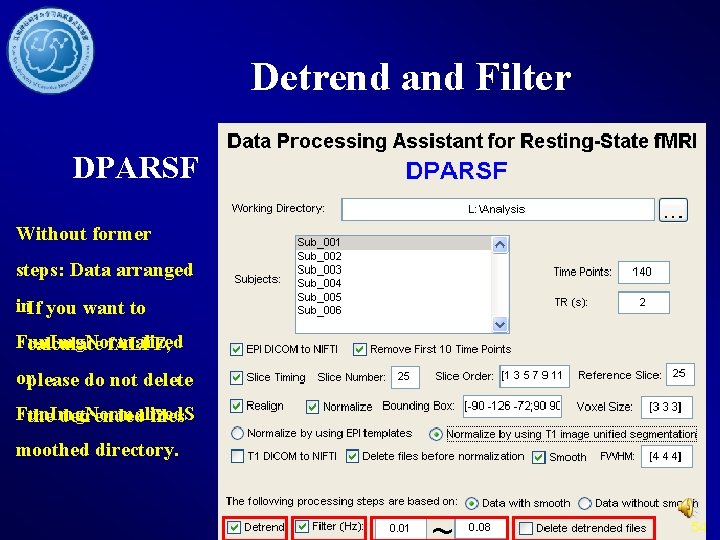
Detrend and Filter DPARSF Without former steps: Data arranged in. If you want to Fun. Img. Normalized calculate f. ALFF, orplease do not delete Fun. Img. Normalized. S the detrended files moothed directory. 54

Outline • Overview • Data Preparation • Preprocess • Re. Ho, ALFF, f. ALFF Calculation • Functional Connectivity • Utilities 55

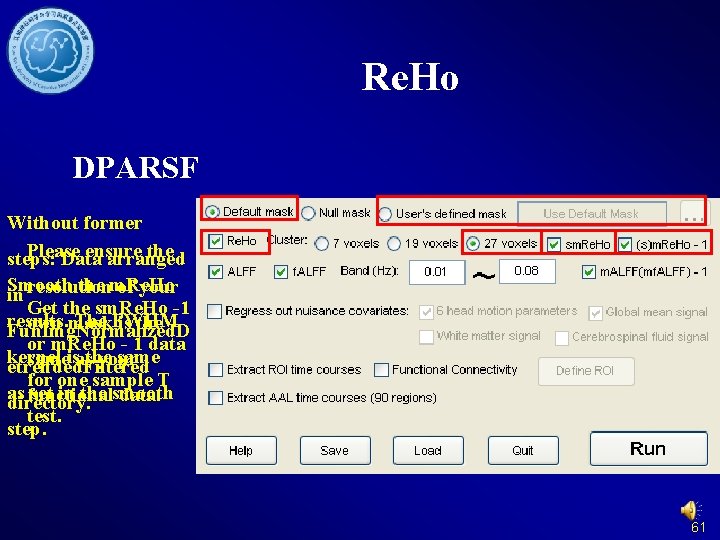
Re. Ho (Regional Homogeneity) Note: Please do not smooth your data in preprocessing, just smooth your data after Re. Ho calculation. Zang et al. , 2004 Zang YF, Jiang TZ, Lu YL, He Y, Tian LX (2004) Regional homogeneity approach to f. MRI data analysis. Neuroimage 22: 394– 400. 56

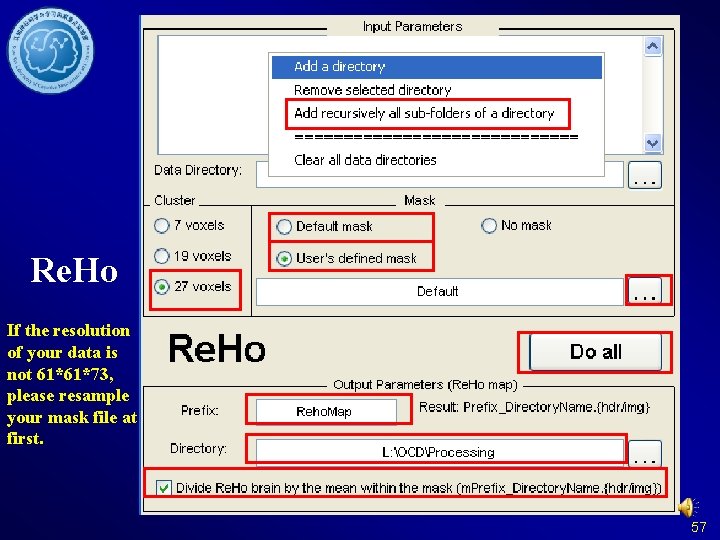
Re. Ho If the resolution of your data is not 61*61*73, please resample your mask file at first. 57

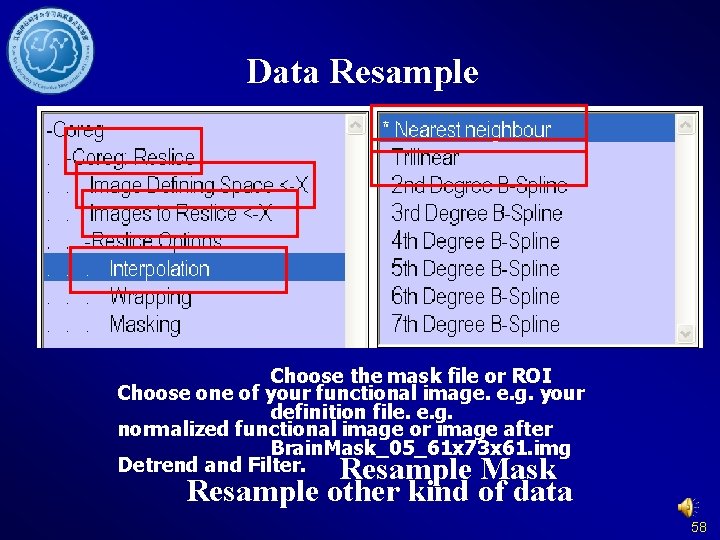
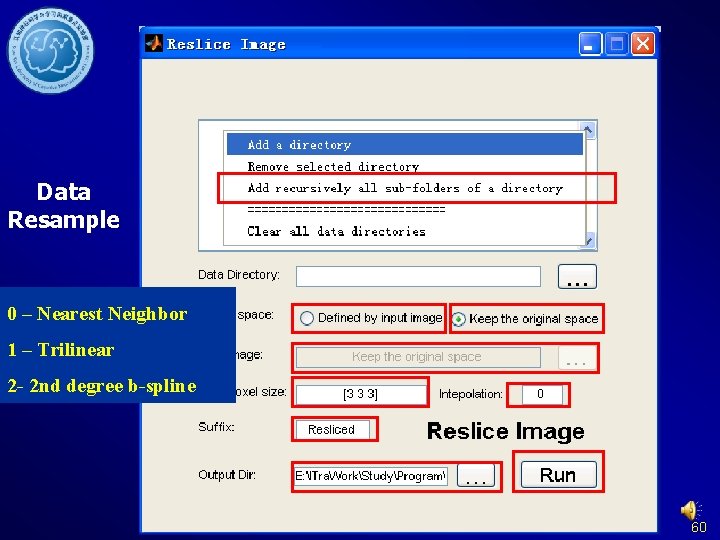
Data Resample Choose the mask file or ROI Choose one of your functional image. e. g. your definition file. e. g. normalized functional image or image after Brain. Mask_05_61 x 73 x 61. img Detrend and Filter. Resample Mask Resample other kind of data 58

Data Resample 59

Data Resample 0 – Nearest Neighbor 1 – Trilinear 2 - 2 nd degree b-spline 60

Re. Ho DPARSF Without former Please ensure the steps: Data arranged Smooth the m. Re. Ho of your in resolution Get the sm. Re. Ho -1 results. The FWHM own mask is the Fun. Img. Normalized. D or m. Re. Ho - 1 data kernel sameisasthe your etrended. Filtered for one sample T as set in the smooth functional data. directory. test. step. 61

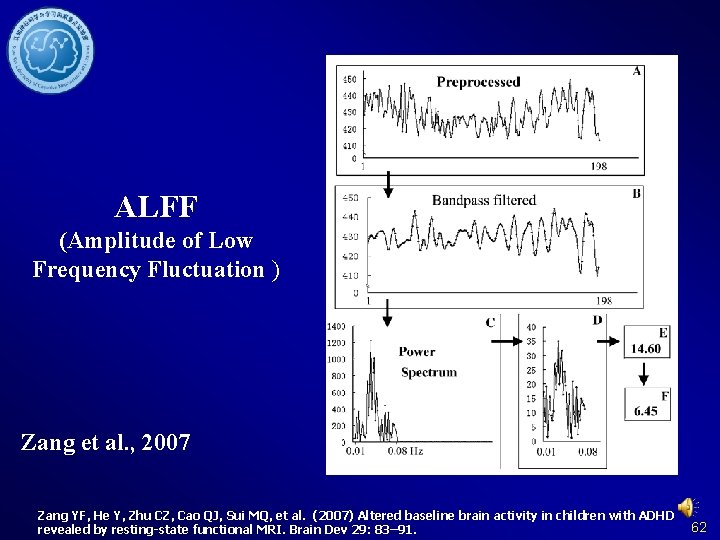
ALFF (Amplitude of Low Frequency Fluctuation ) Zang et al. , 2007 Zang YF, He Y, Zhu CZ, Cao QJ, Sui MQ, et al. (2007) Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev 29: 83– 91. 62

f. ALFF (fractional ALFF ) PCC: posterior cingulate cortex SC: suprasellar cistern Zou et al. , 2008 Zou QH, Zhu CZ, Yang Y, Zuo XN, Long XY, et al. (2008) An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state f. MRI: fractional ALFF. J Neurosci Methods 172: 137 -141. 63

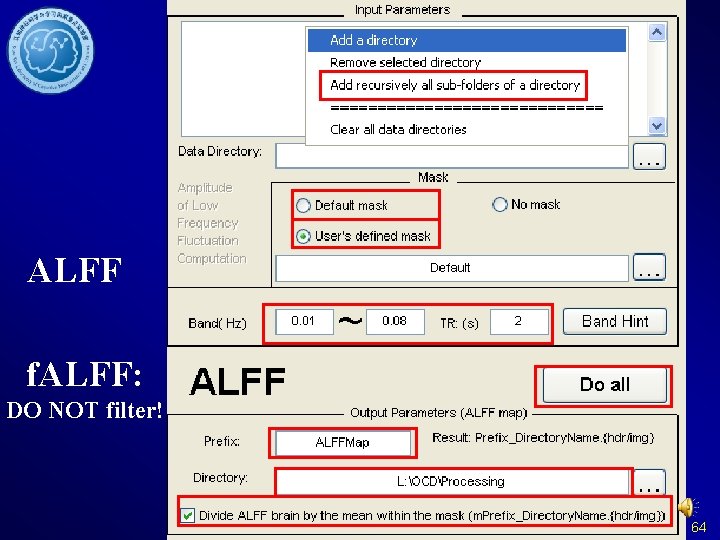
ALFF f. ALFF: DO NOT filter! 64

ALFF and f. ALFF DPARSF Without formerthe Please ensure steps: Data arranged resolution your Please DO of NOT inown mask is the delete the detrended Fun. Img. Normalized. S same as your files before filter. moothed. Detrended. Fi functional data. DPARSF will Get the m. ALFF - 1 ltered calculated the oror (mf. ALFF - 1) f. ALFF based on data for one sample Fun. Img. Normalized. S data before filter. T test. moothed. Detrended 65

Outline • Overview • Data Preparation • Preprocess • Re. Ho, ALFF, f. ALFF Calculation • Functional Connectivity • Utilities 66

Regress out nuisance covariates • Head motion parameters: rp_name. txt • Global mean signal • White matter signal • Cerebrospinal fluid signal 67

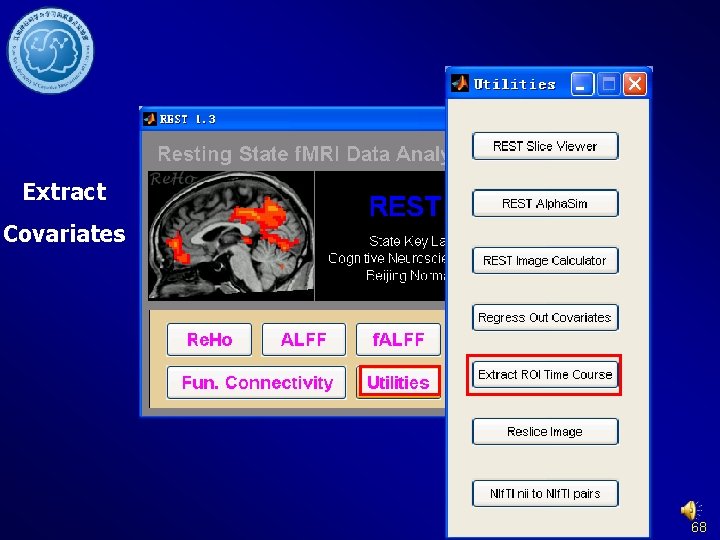
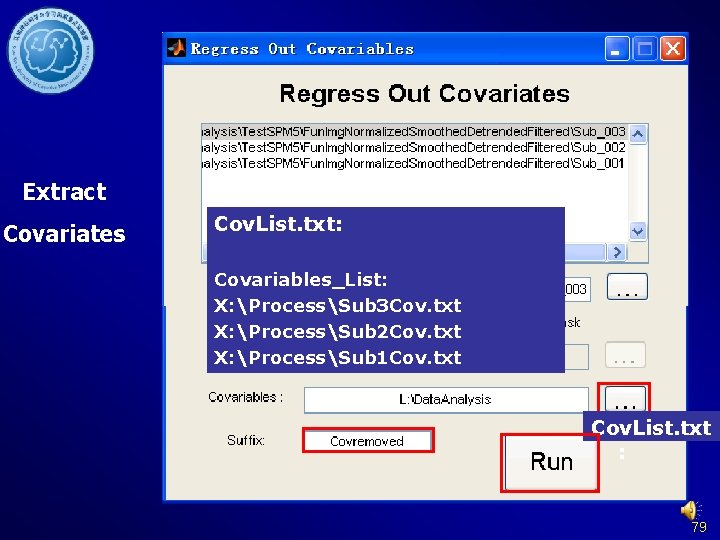
Extract Covariates 68

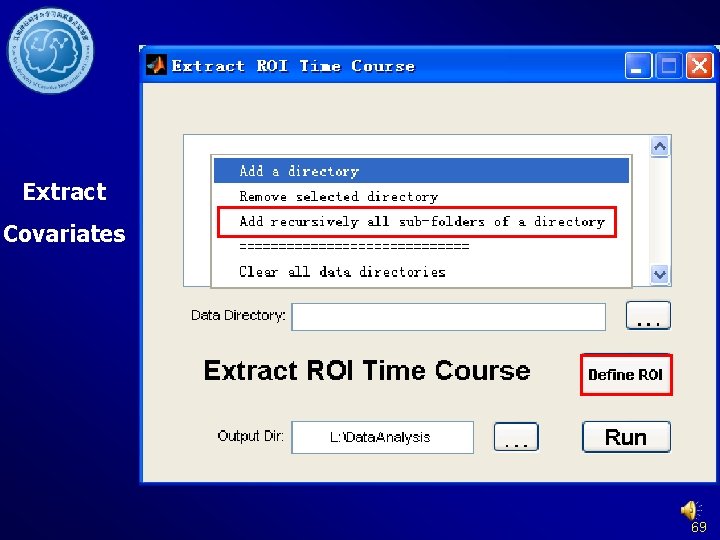
Extract Covariates 69

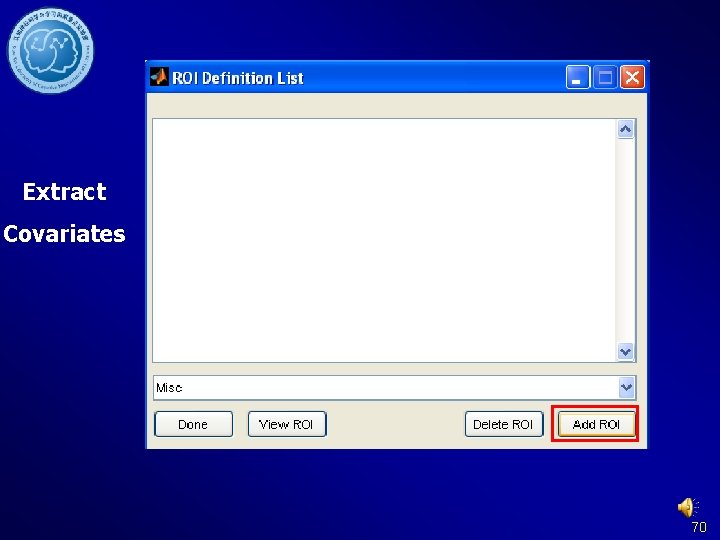
Extract Covariates 70

Extract Covariates 71

Extract Covariates 72

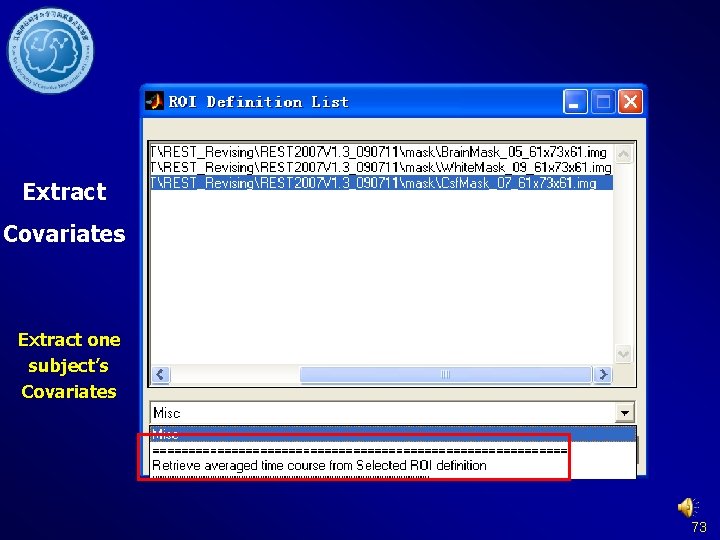
Extract Covariates Extract one subject’s Covariates 73

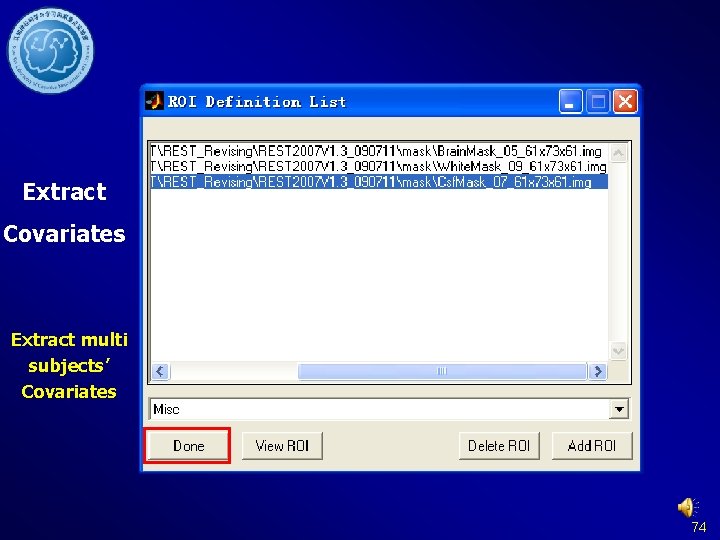
Extract Covariates Extract multi subjects’ Covariates 74

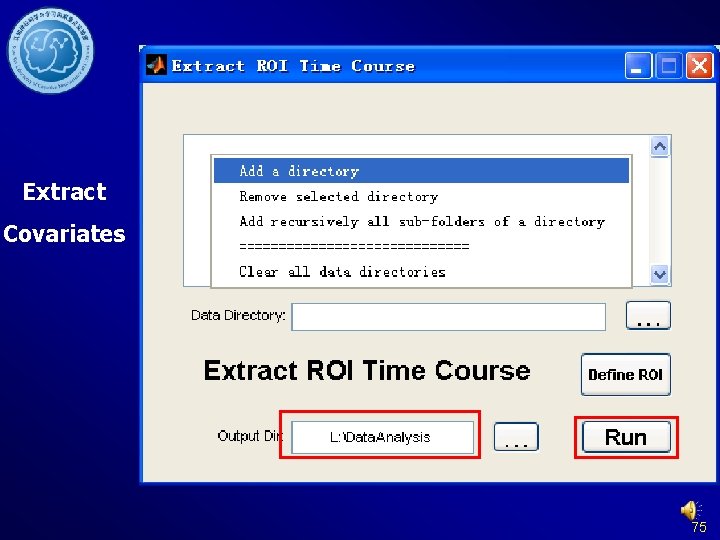
Extract Covariates 75

Extract Covariates 76

Regress out nuisance Covariates Extract Covariates • • Head motion parameters: rp_name. txt Global mean signal White matter signal Cerebrospinal fluid signal • Combine the covariates for future using in REST v v RPCov=load('rp_name. txt'); BCWCov=load('ROI_FCMap_name. txt'); Cov=[RPCov, BCWCov]; save('Cov. txt', 'Cov', '-ASCII', '-DOUBLE', 'TABS'); 77

Regress out Covariates 78

Extract Covariates Cov. List. txt: Covariables_List: X: ProcessSub 3 Cov. txt X: ProcessSub 2 Cov. txt X: ProcessSub 1 Cov. txt Cov. List. txt : 79

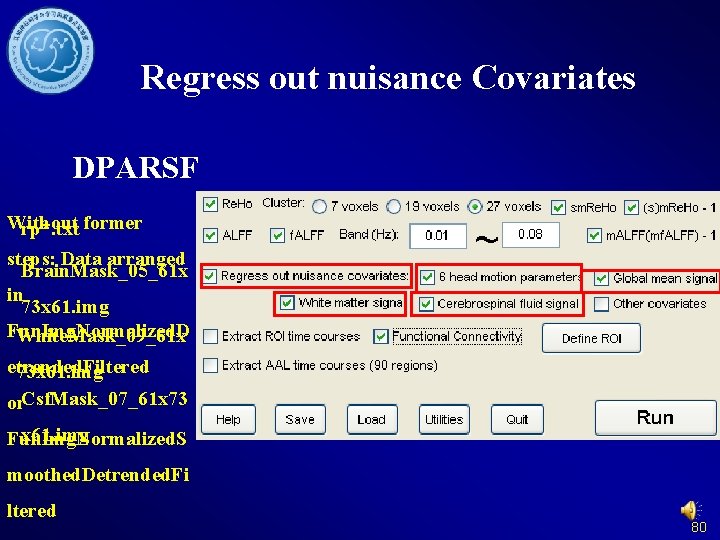
Regress out nuisance Covariates DPARSF Without rp*. txt former steps: Data arranged Brain. Mask_05_61 x in 73 x 61. img Fun. Img. Normalized. D White. Mask_09_61 x etrended. Filtered 73 x 61. img or. Csf. Mask_07_61 x 73 x 61. img Fun. Img. Normalized. S moothed. Detrended. Fi ltered 80

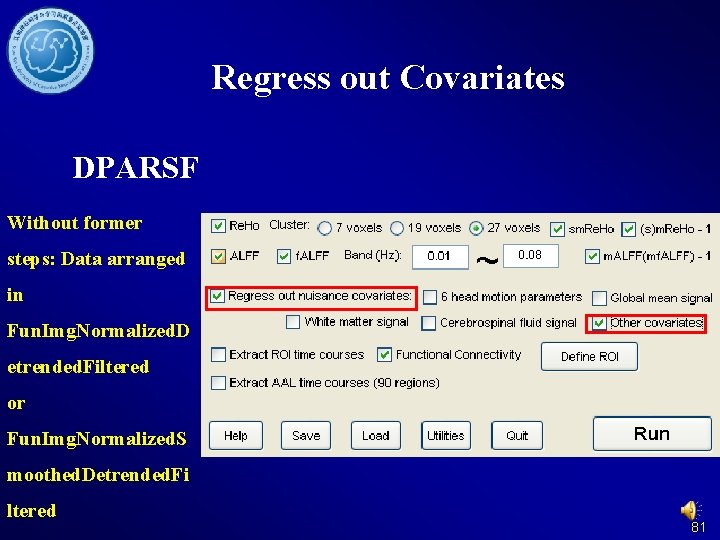
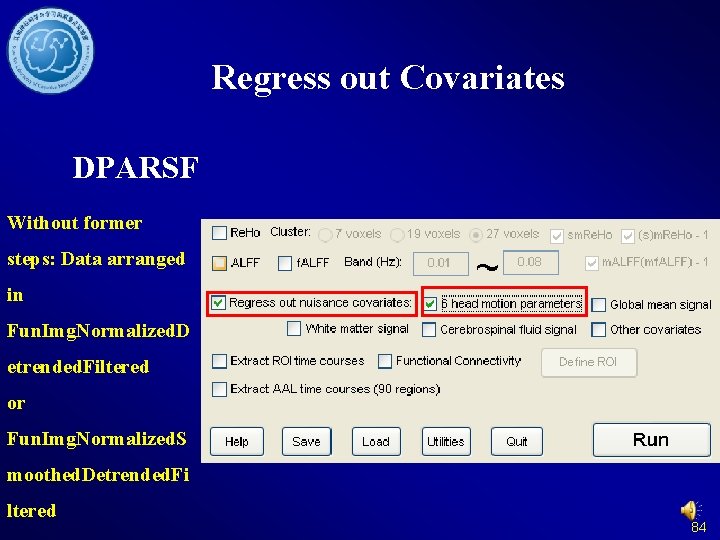
Regress out Covariates DPARSF Without former steps: Data arranged in Fun. Img. Normalized. D etrended. Filtered or Fun. Img. Normalized. S moothed. Detrended. Fi ltered 81

Regress out Covariates 82

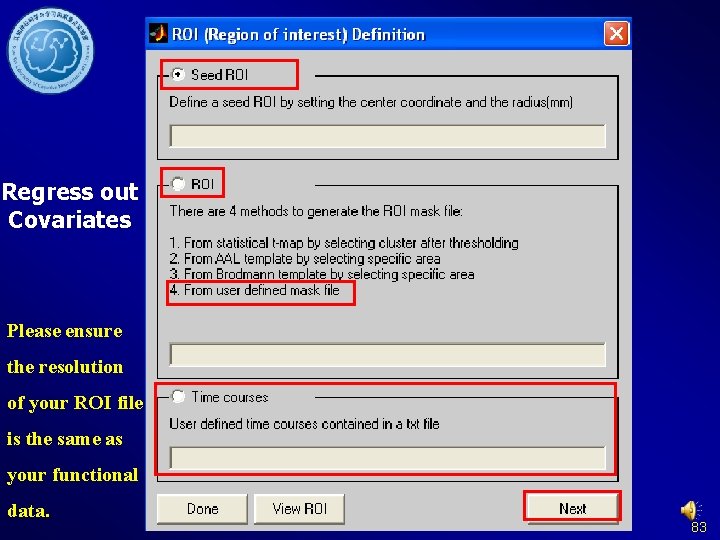
Regress out Covariates Please ensure the resolution of your ROI file is the same as your functional data. 83

Regress out Covariates DPARSF Without former steps: Data arranged in Fun. Img. Normalized. D etrended. Filtered or Fun. Img. Normalized. S moothed. Detrended. Fi ltered 84

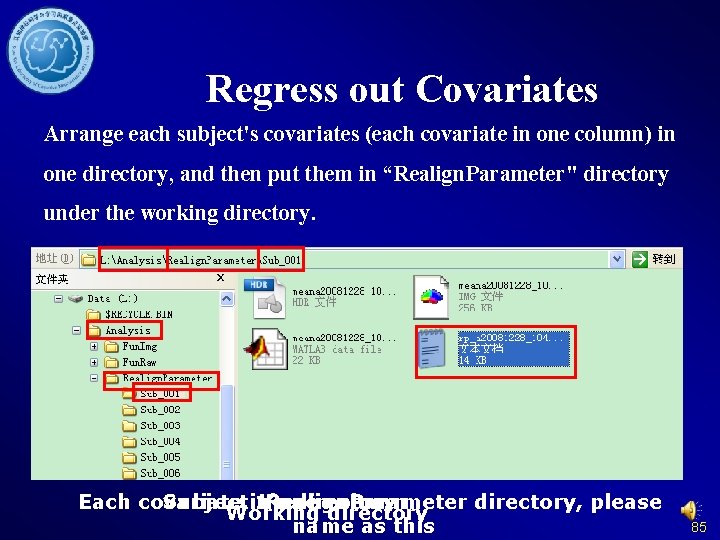
Regress out Covariates Arrange each subject's covariates (each covariate in one column) in one directory, and then put them in “Realign. Parameter" directory under the working directory. Each covariate Subject in 1’s Realign. Parameter one directory column directory, please Working directory name as this 85

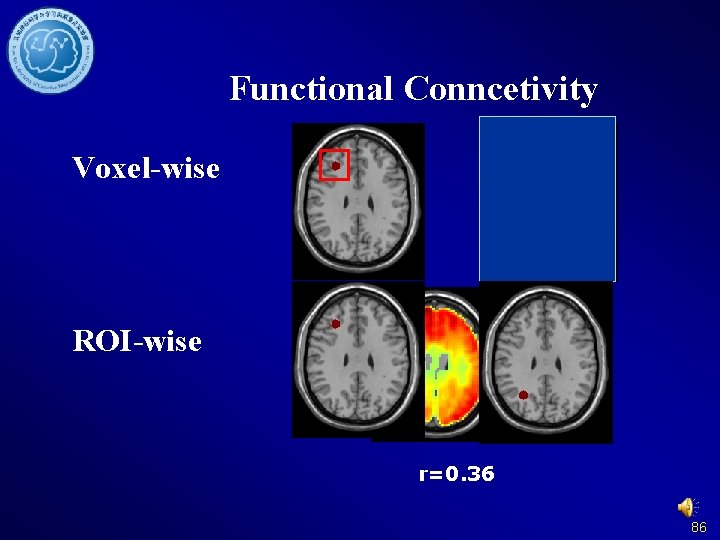
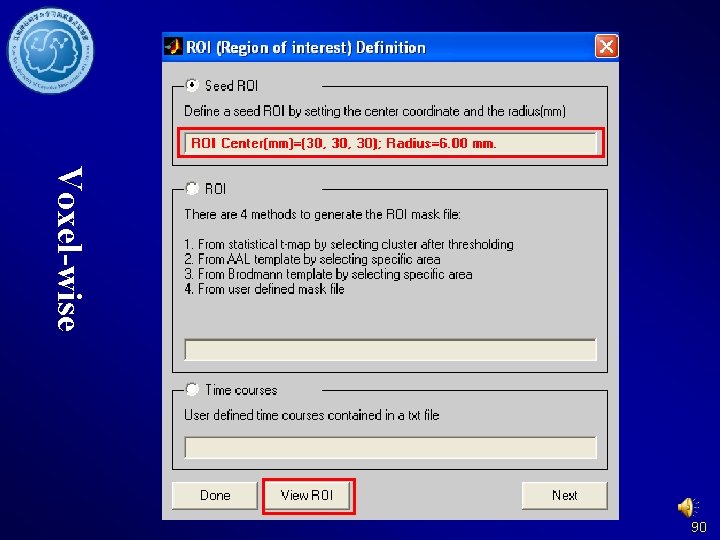
Functional Conncetivity Voxel-wise ROI-wise r=0. 36 86

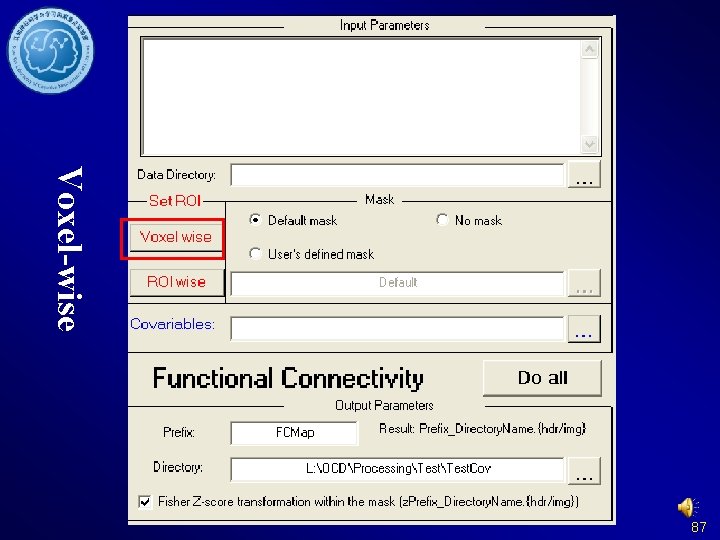
Voxel-wise 87

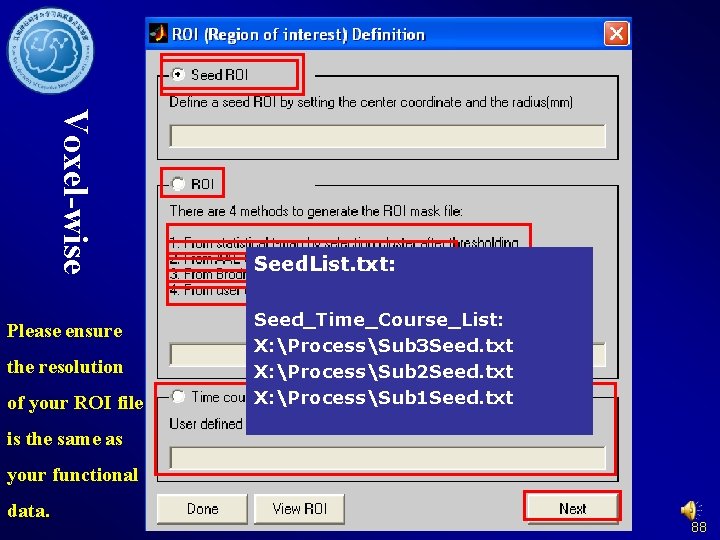
Voxel-wise Please ensure the resolution of your ROI file Seed. List. txt: Seed_Time_Course_List: X: ProcessSub 3 Seed. txt X: ProcessSub 2 Seed. txt X: ProcessSub 1 Seed. txt is the same as your functional data. 88

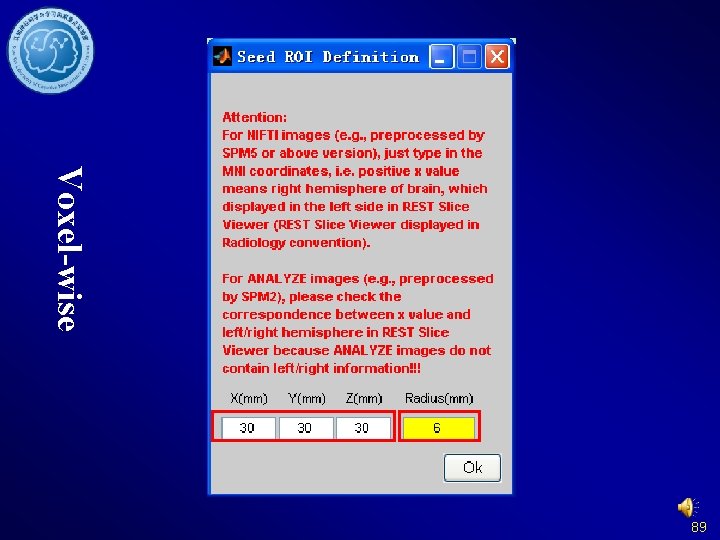
Voxel-wise 89

Voxel-wise 90

Voxel-wise 91

Voxel-wise 92

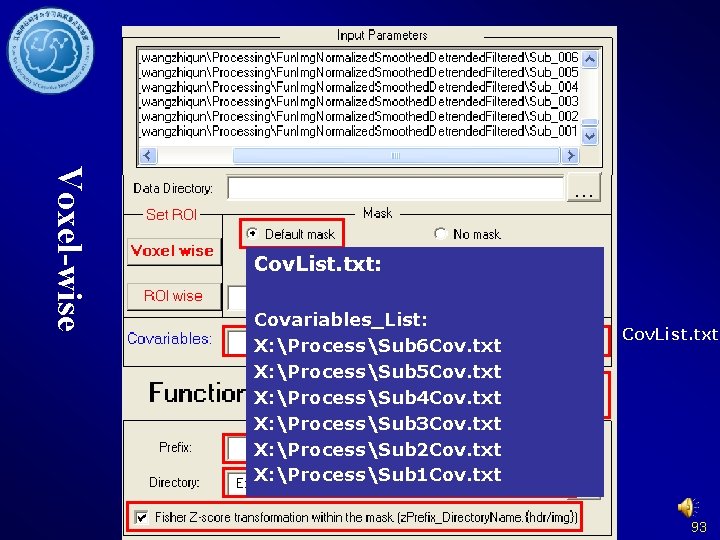
Voxel-wise Cov. List. txt: Covariables_List: X: ProcessSub 6 Cov. txt X: ProcessSub 5 Cov. txt X: ProcessSub 4 Cov. txt X: ProcessSub 3 Cov. txt X: ProcessSub 2 Cov. txt X: ProcessSub 1 Cov. txt Cov. List. txt 93

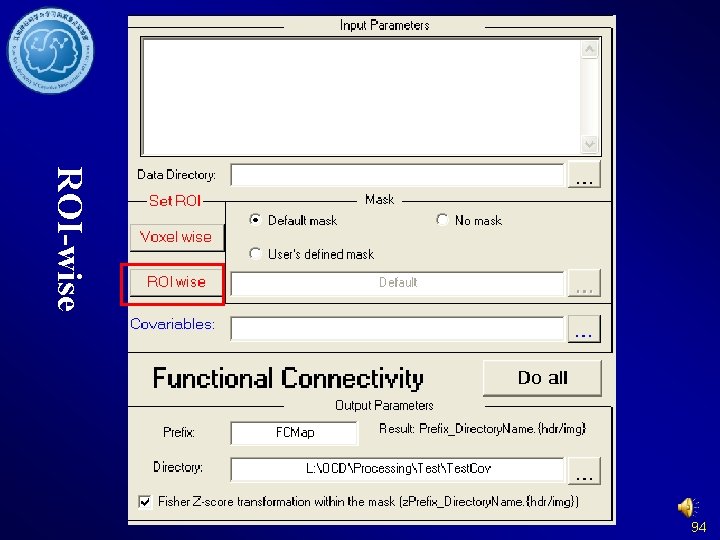
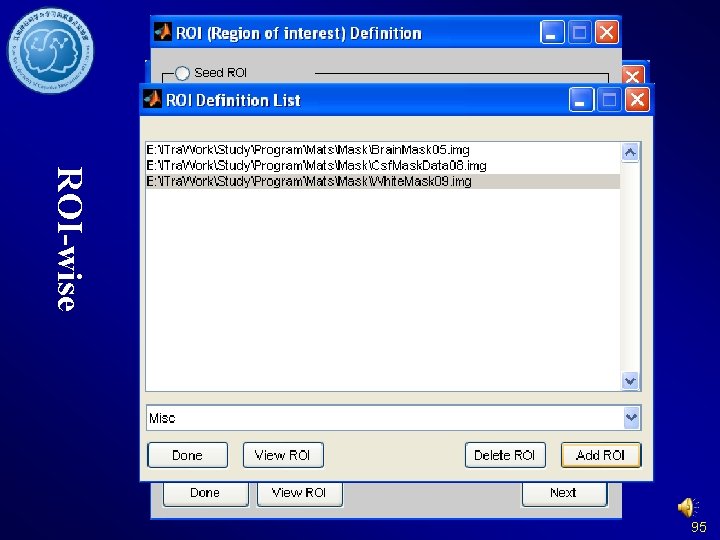
ROI-wise 94

ROI-wise 95

ROI-wise Cov. List. txt: Covariables_List: X: ProcessSub 6 Cov. txt X: ProcessSub 5 Cov. txt X: ProcessSub 4 Cov. txt X: ProcessSub 3 Cov. txt X: ProcessSub 2 Cov. txt X: ProcessSub 1 Cov. txt Cov. List. txt 96

Functional Connectivity DPARSF Without former Please ensure the steps: Data arranged inresolution of your own mask is the Fun. Img. Normalized. D same as your etrended. Filtered. Cov functional data. removed or Fun. Img. Normalized. S moothed. Detrended. Fi ltered. Covremoved 97

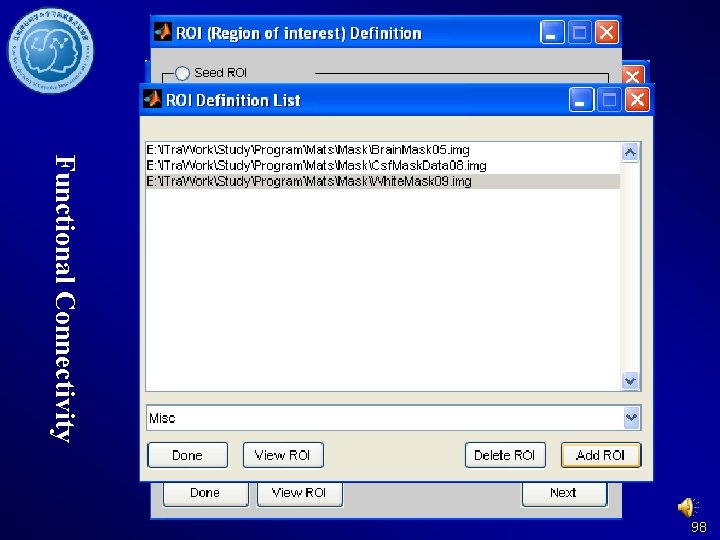
Functional Connectivity 98

Functional Connectivity DPARSF You will get the Voxel-wise functional connectivity results of each ROI in {working directory}ResultsFC: z. ROI 1 FCMap_Sub_001. img z. ROI 2 FCMap_Sub_001. img For ROI-wise results, please see Part Utilities: Extract ROI time courses. 99

Outline • Overview • Data Preparation • Preprocess • Re. Ho, ALFF, f. ALFF Calculation • Functional Connectivity • Utilities 100

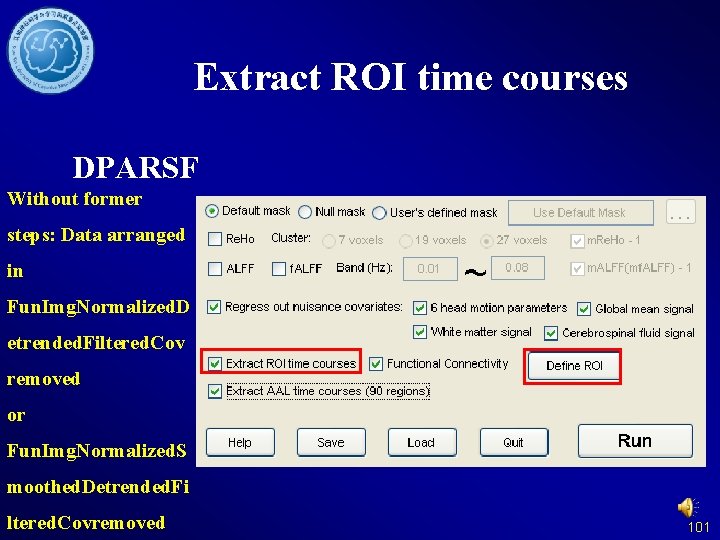
Extract ROI time courses DPARSF Without former steps: Data arranged in Fun. Img. Normalized. D etrended. Filtered. Cov removed or Fun. Img. Normalized. S moothed. Detrended. Fi ltered. Covremoved 101

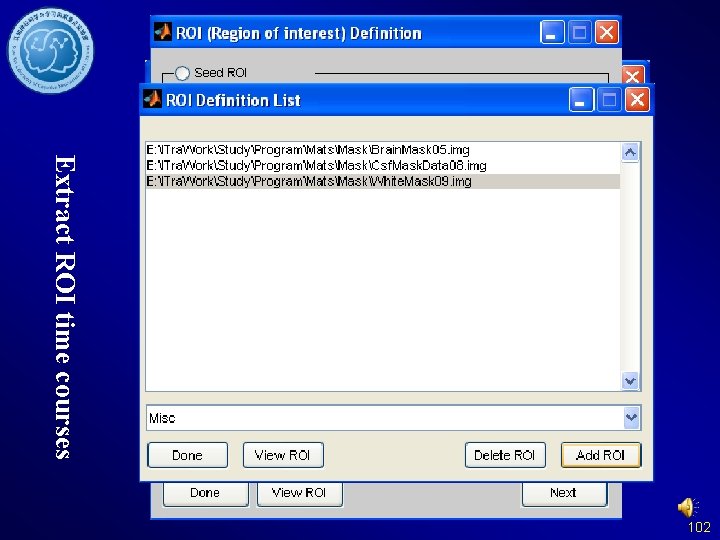
Extract ROI time courses 102

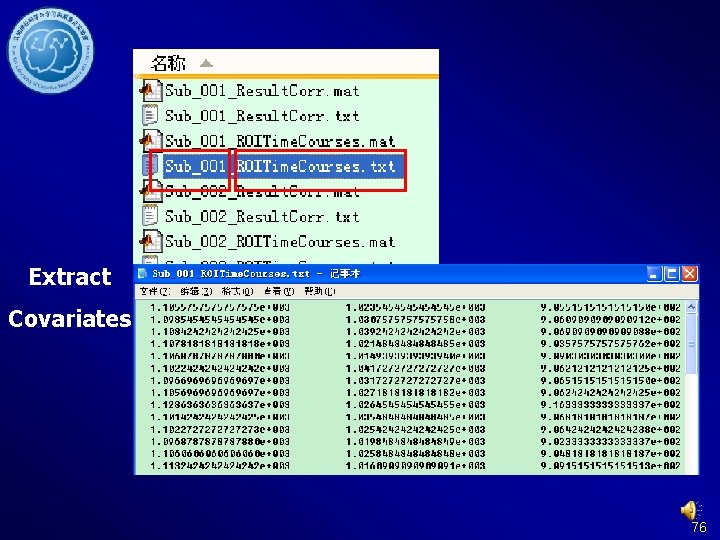
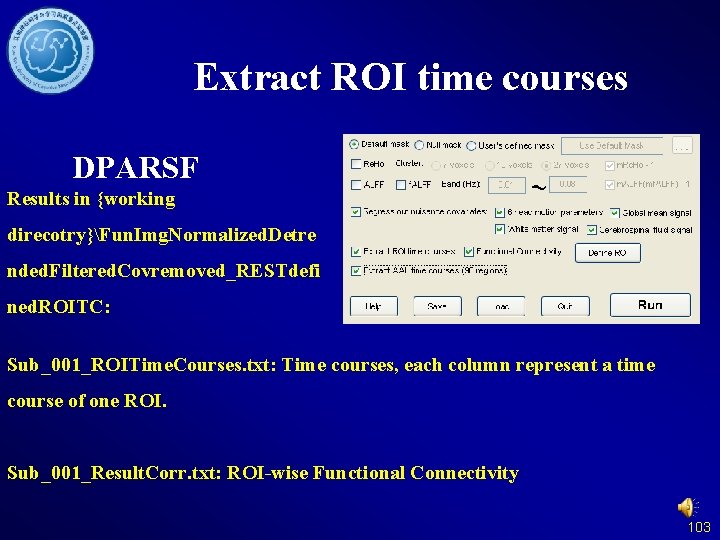
Extract ROI time courses DPARSF Results in {working direcotry}Fun. Img. Normalized. Detre nded. Filtered. Covremoved_RESTdefi ned. ROITC: Sub_001_ROITime. Courses. txt: Time courses, each column represent a time course of one ROI. Sub_001_Result. Corr. txt: ROI-wise Functional Connectivity 103

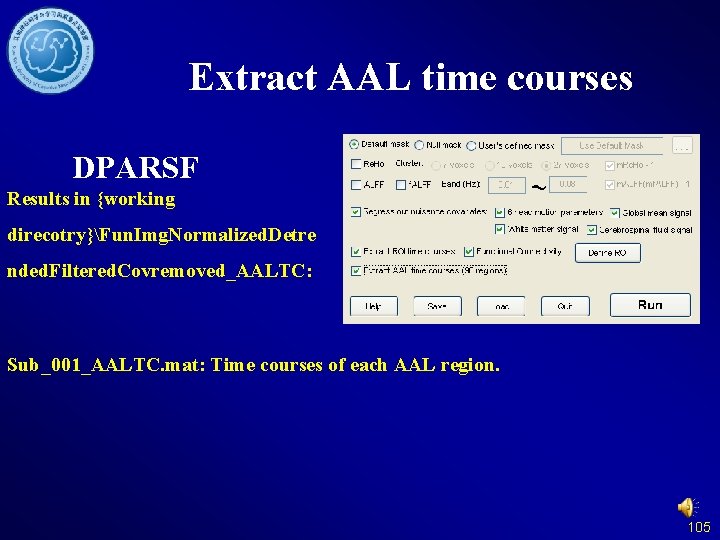
Extract AAL time courses DPARSF Without former steps: Data arranged in Fun. Img. Normalized. D etrended. Filtered. Cov removed or Fun. Img. Normalized. S moothed. Detrended. Fi ltered. Covremoved 104

Extract AAL time courses DPARSF Results in {working direcotry}Fun. Img. Normalized. Detre nded. Filtered. Covremoved_AALTC: Sub_001_AALTC. mat: Time courses of each AAL region. 105

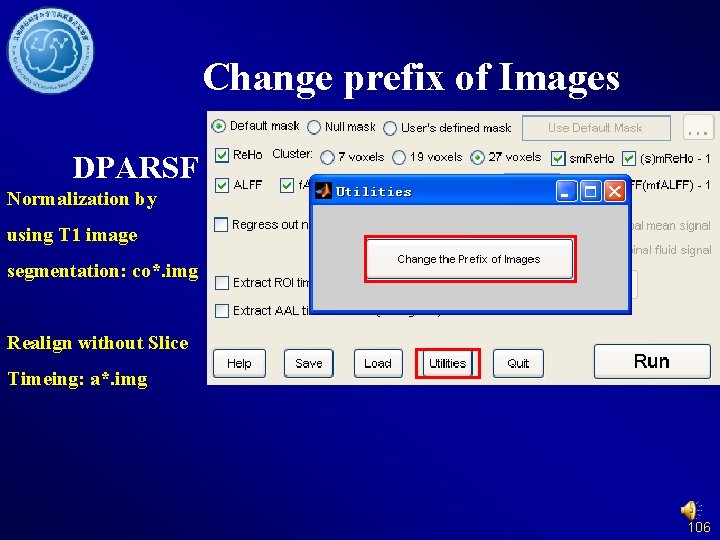
Change prefix of Images DPARSF Normalization by using T 1 image segmentation: co*. img Realign without Slice Timeing: a*. img 106

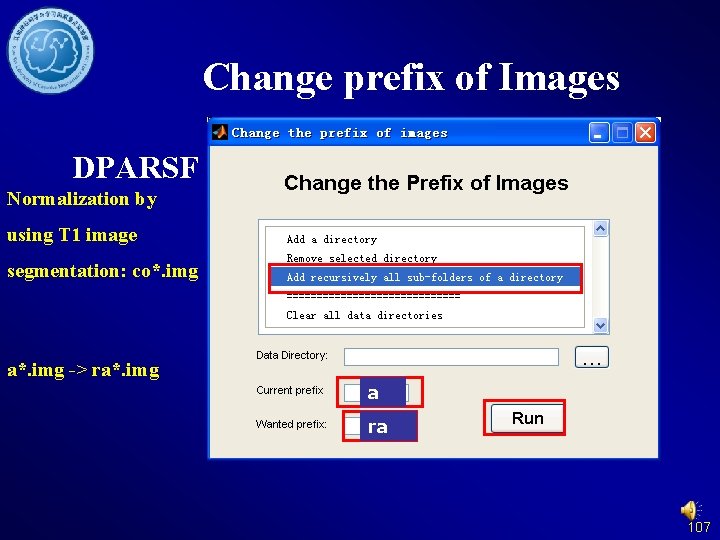
Change prefix of Images DPARSF Normalization by using T 1 image segmentation: co*. img a*. img -> ra*. img a ra 107

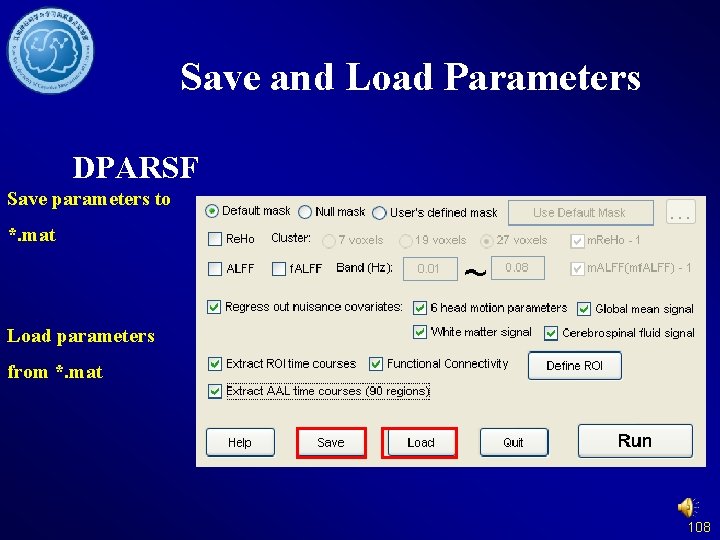
Save and Load Parameters DPARSF Save parameters to *. mat Load parameters from *. mat 108

Further Help Further questions: www. restfmri. net Further professional data analysis service: Brain Imaging Data Analysis and Consultation Section (BIDACS) bidacs@gmail. com 109

Thanks to DONG Zhang-Ye GUO Xiao-Juan HE Yong LONG Xiang-Yu SONG Xiao-Wei YAO Li ZANG Yu-Feng ZHANG Han ZHU Chao-Zhe ZOU Qi-Hong ZUO Xi-Nian …… SPM Team: Wellcome Department of Imaging Neuroscience, UCL MRIcro. N Team: Chris Rorden …… All the group members! 110

Thanks for your attention! 111