Chromosome and genome 11 16 2015 Genome whole



















- Slides: 65

Chromosome and genome 11 -16 -2015

Genome: whole set of DNA to store all genetic information Circular versus linear chromsome


Human Genome • How many chromosomes does each human cell have? – 22 pairs of autosomal chromosome and 1 pair of sex chromosomes • What is the size of the human genome? – The human genome is made up of 3 billion base pairs • How many genes does the Human Genome code for? – Define the meaning of gene first!

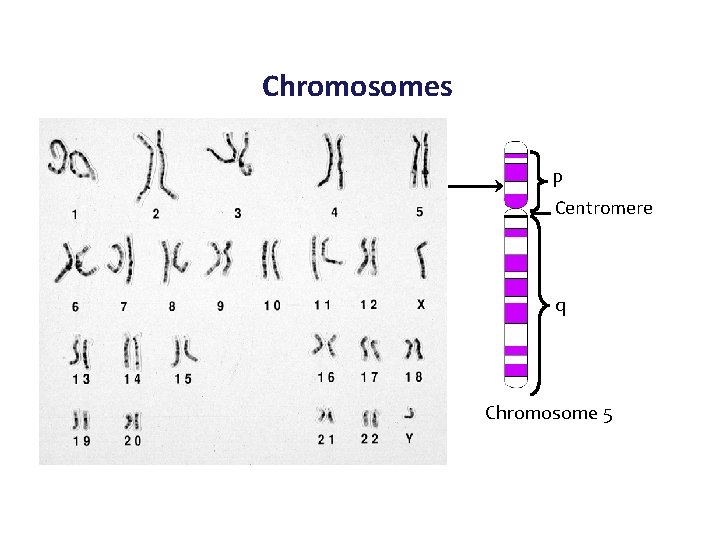
Chromosomes p Centromere q Chromosome 5

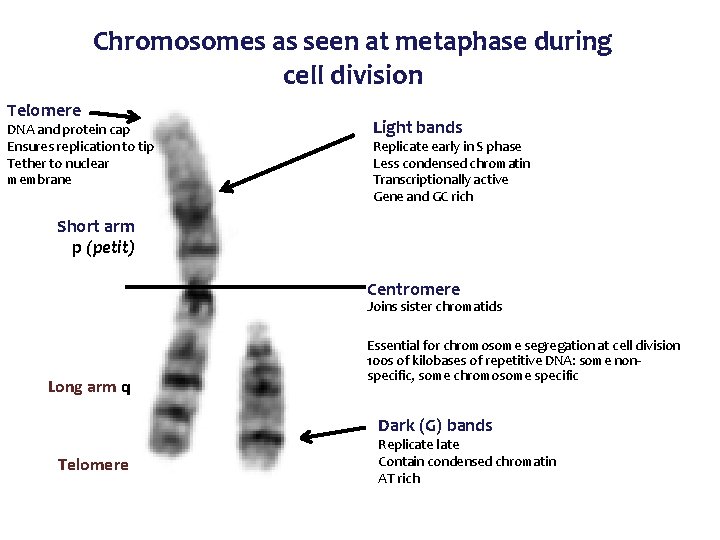
Chromosomes as seen at metaphase during cell division Telomere DNA and protein cap Ensures replication to tip Tether to nuclear membrane Light bands Replicate early in S phase Less condensed chromatin Transcriptionally active Gene and GC rich Short arm p (petit) Centromere Joins sister chromatids Long arm q Essential for chromosome segregation at cell division 100 s of kilobases of repetitive DNA: some nonspecific, some chromosome specific Dark (G) bands Telomere Replicate late Contain condensed chromatin AT rich

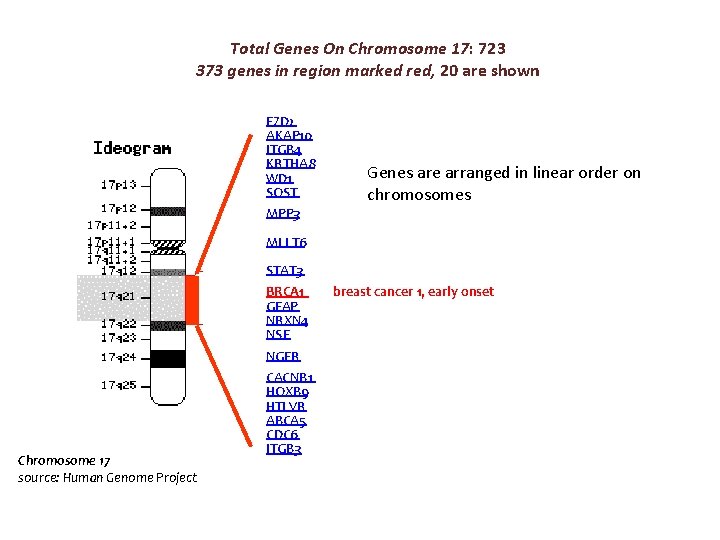
Total Genes On Chromosome 17: 723 373 genes in region marked red, 20 are shown FZD 2 AKAP 10 ITGB 4 KRTHA 8 WD 1 SOST Genes are arranged in linear order on chromosomes MPP 3 MLLT 6 STAT 3 BRCA 1 GFAP NRXN 4 NSF NGFR Chromosome 17 source: Human Genome Project CACNB 1 HOXB 9 HTLVR ABCA 5 CDC 6 ITGB 3 breast cancer 1, early onset

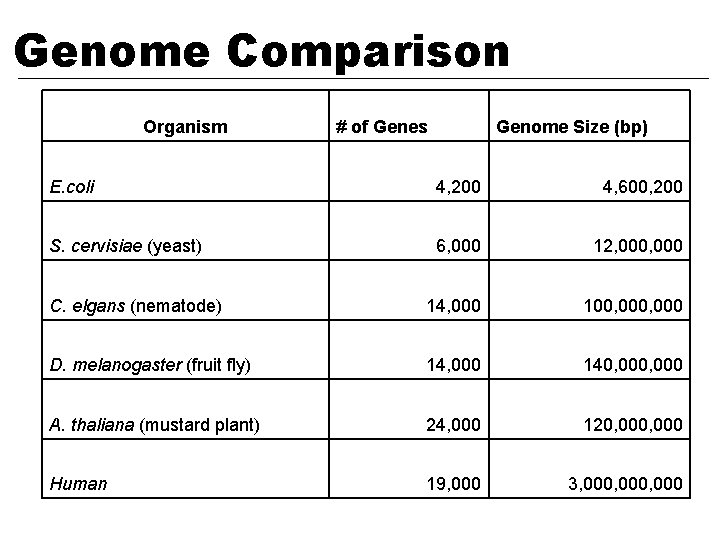
Genome Comparison Organism # of Genes Genome Size (bp) E. coli 4, 200 4, 600, 200 S. cervisiae (yeast) 6, 000 12, 000 C. elgans (nematode) 14, 000 100, 000 D. melanogaster (fruit fly) 14, 000 140, 000 A. thaliana (mustard plant) 24, 000 120, 000 Human 19, 000 3, 000, 000

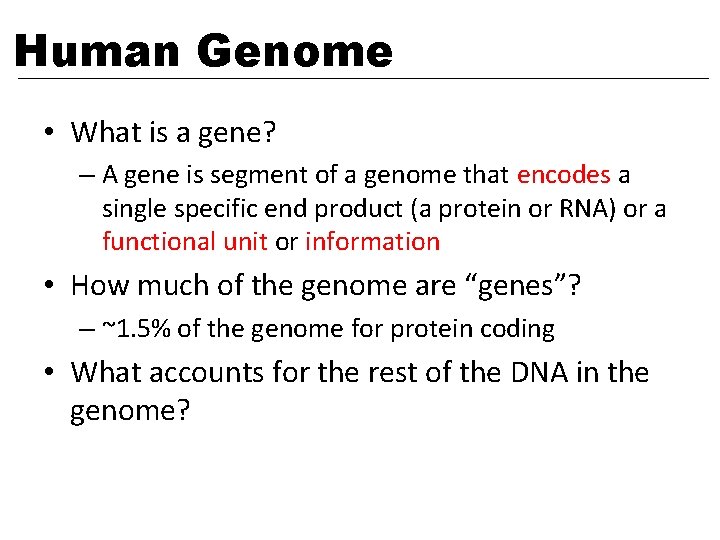
Human Genome • What is a gene? – A gene is segment of a genome that encodes a single specific end product (a protein or RNA) or a functional unit or information • How much of the genome are “genes”? – ~1. 5% of the genome for protein coding • What accounts for the rest of the DNA in the genome?

Human Genome • Other items in genome – Introns – Repeated sequences (telomeres, centromeres) – Non-coding RNAs • • t. RNA, r. RNA, sn. RNA. mi. RNA Long non-coding RNA Functional or noise? ? – Transposons (SINES and LINES) – Psuedogenes (genes that no longer produce functional products)

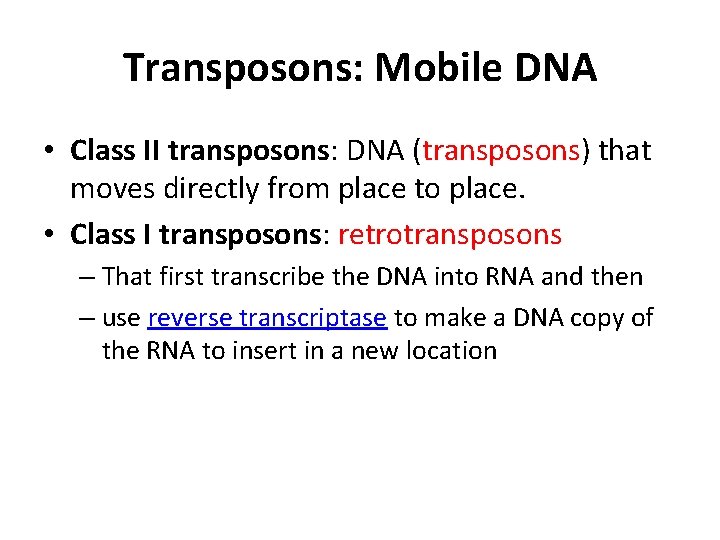
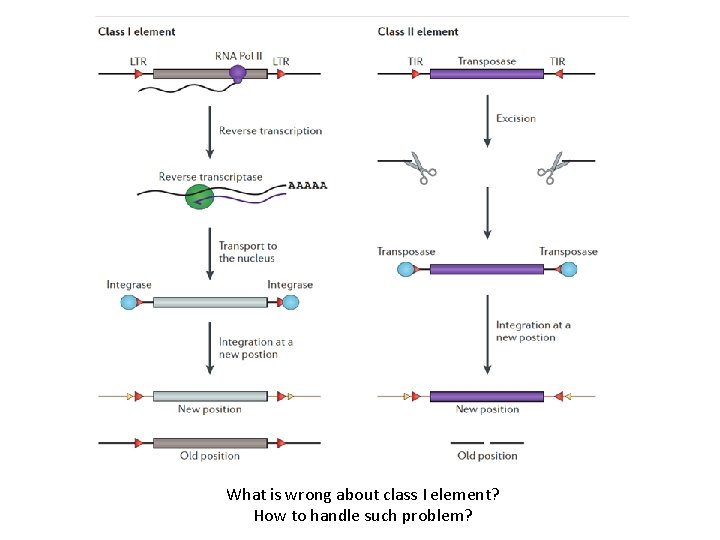
Transposons: Mobile DNA • Class II transposons: DNA (transposons) that moves directly from place to place. • Class I transposons: retrotransposons – That first transcribe the DNA into RNA and then – use reverse transcriptase to make a DNA copy of the RNA to insert in a new location

Discovery of transposons Barbara Mc. Clintock (1902 -1992) Nobel prize in Physiology and Medicine 1983 Mobile genetic elements in maize 1940 -1950

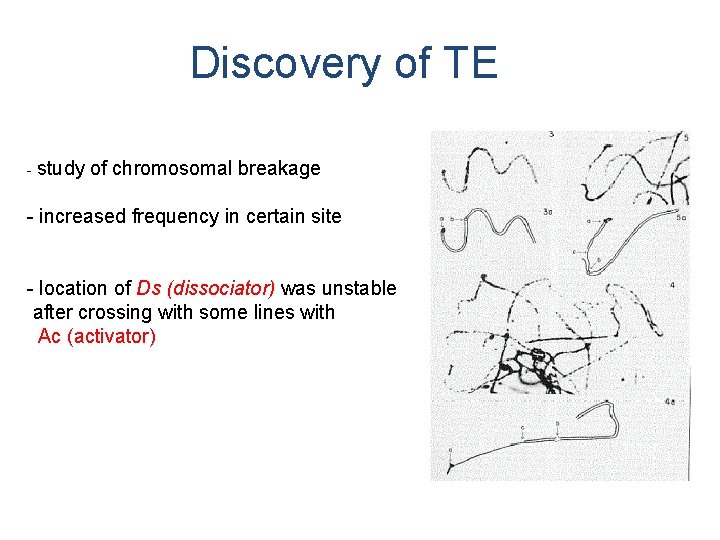
Discovery of TE - study of chromosomal breakage - increased frequency in certain site (= marker „dissociation“ Ds) - location of Ds (dissociator) was unstable after crossing with some lines with Ac (activator) (= line carrying „activator“ Ac)

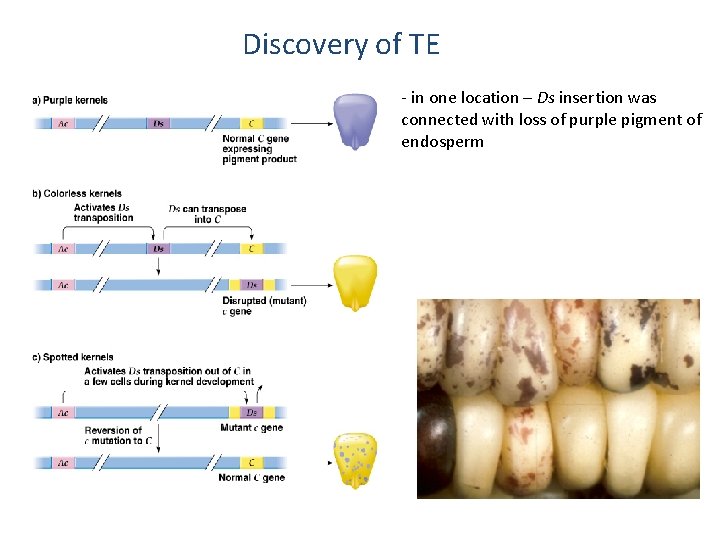
Discovery of TE - in one location – Ds insertion was connected with loss of purple pigment of endosperm - after crossing with activator line pigment synthesis was recovered in some cells

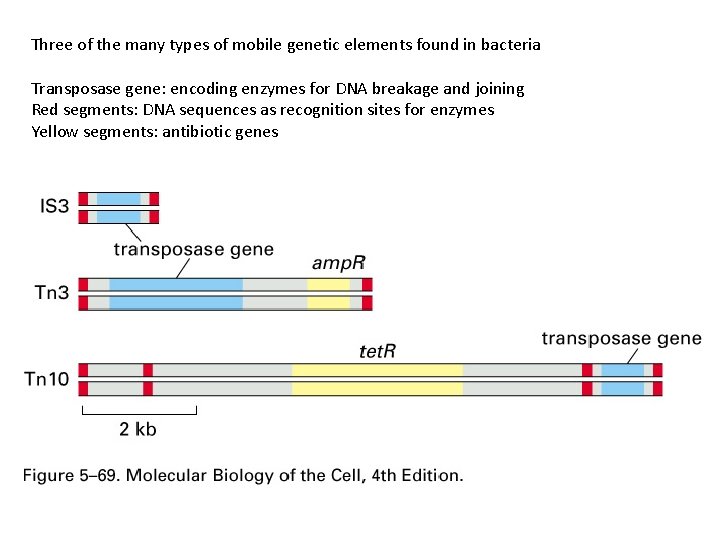
Three of the many types of mobile genetic elements found in bacteria Transposase gene: encoding enzymes for DNA breakage and joining Red segments: DNA sequences as recognition sites for enzymes Yellow segments: antibiotic genes

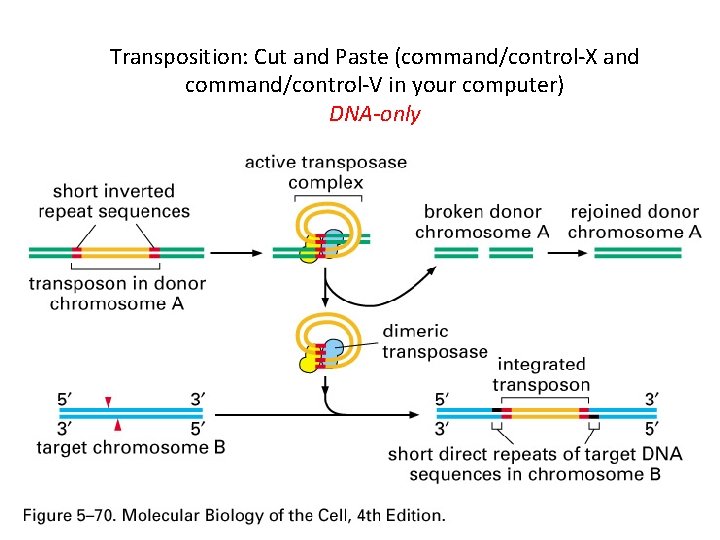
Transposition: Cut and Paste (command/control-X and command/control-V in your computer) DNA-only

What is wrong about class I element? How to handle such problem?

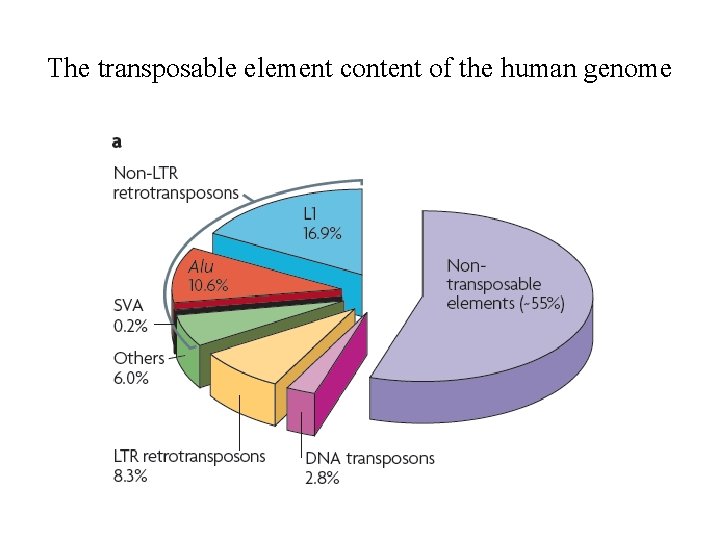
The transposable element content of the human genome

What we should know about Alu element! • All Alus are approximately 300 bp in length. • A single recognition site for the restriction enzyme Alu. I located near the middle of the Alu element. • Alu elements are found only in primates. • Human chromosomes contain about 1, 000 Alu copies (10% of the total genome). • Alu is a "jumping gene" – a transposable DNA sequence that "reproduces" by copying itself and inserting into new chromosome locations.

What we should know about Alu element! • No evidence that it is ever excised or lost from a chromosome locus. Each Alu insertion is the "fossil" of a unique transposition event that occurred only once in primate evolution. • Each Alu element has an internal promoter for RNA polymerase III. • However, Alu lacks the enzyme functions to produce a DNA copy of itself and to integrate into a new chromosome position

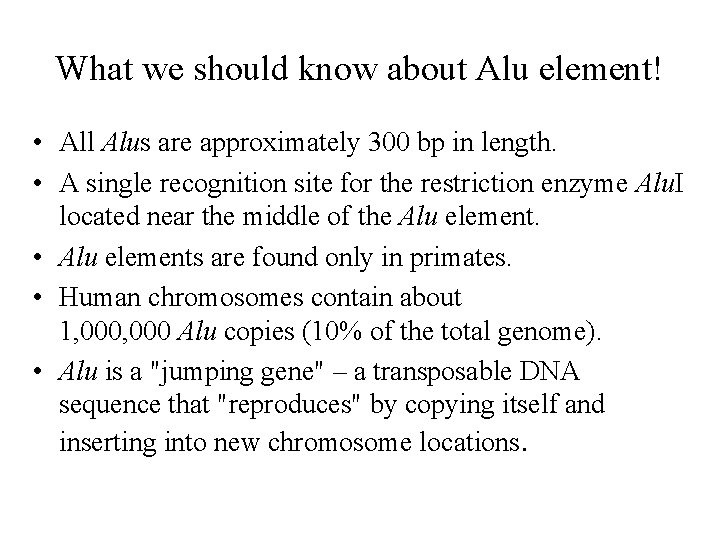
Alu transposition via an RNA intermediate: L 1 provides RT for reverse transcription and a nick on the host chromosome! Alu is a parasite of L 1, which, in turn, is a relic of a retrovirus ancestor.

What we should know about Alu element! • The rate of Alu transposition is about one in every 200 newborns today. • The vast majority of Alu insertions occur in noncoding regions and are thought to be neutral. • An Alu insertion in the NF-1 gene is responsible for neurofibromatosis I. • Alu insertions in introns of genes for tissue plasminogen activator (TPA) and angiotensin converter enzyme (ACE) are associated with heart disease.

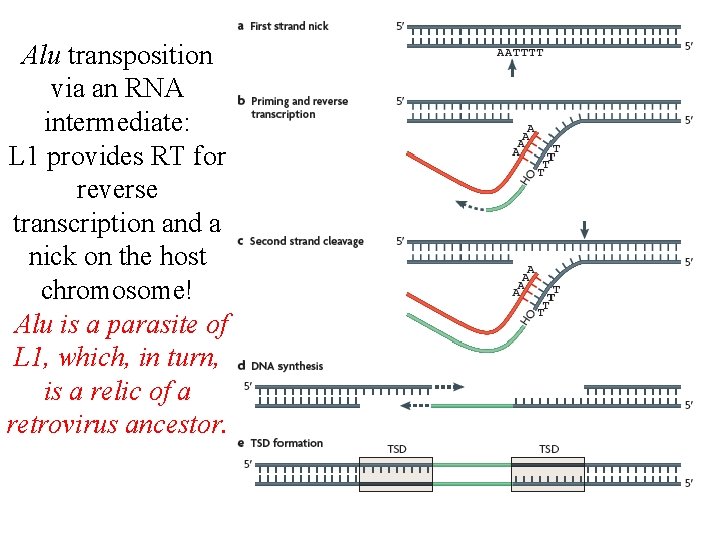
PV 92, a human-specific Alu insertion on chromosome 16

How Alu jump! http: //labcenter. dnalc. org/labs/dnafinge rprintalu/dnafingerprintalu_d. html

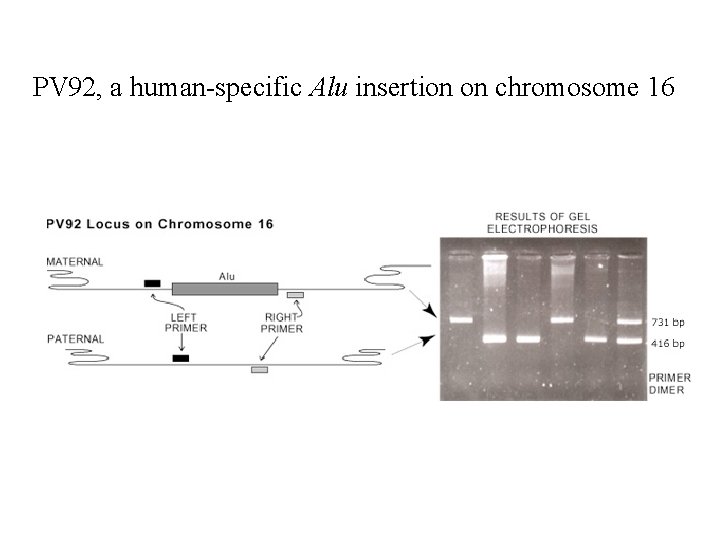
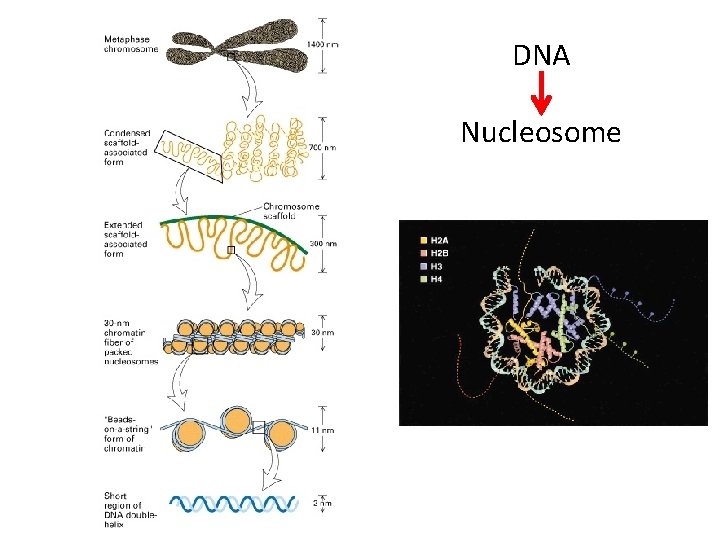
DNA Nucleosome


細胞所有的遺傳資訊都完全登 錄在DNA上的ATGC序列嗎? Epigenetics: the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence (Riggs et al. 1996)

Epigenetic chromatin regulation A. Modification at the DNA level 1. cytosine methylation B. Histone modification - the histone code 1. Histone acetylation 2. Histone methylation 3. Histone phosphorylation 4. Histone ubiquitilation 5. Different types of histones

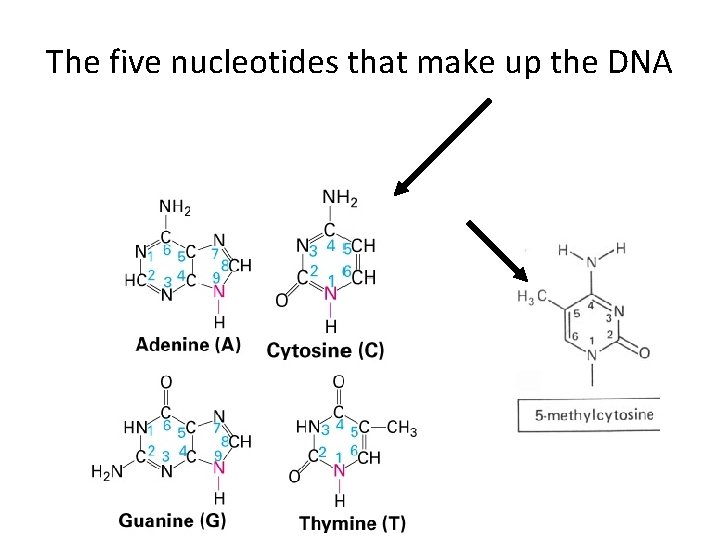
The five nucleotides that make up the DNA

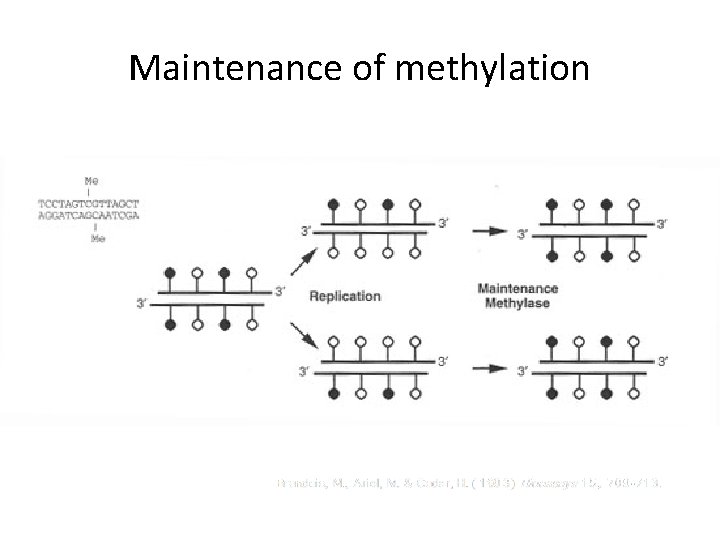
Maintenance of methylation

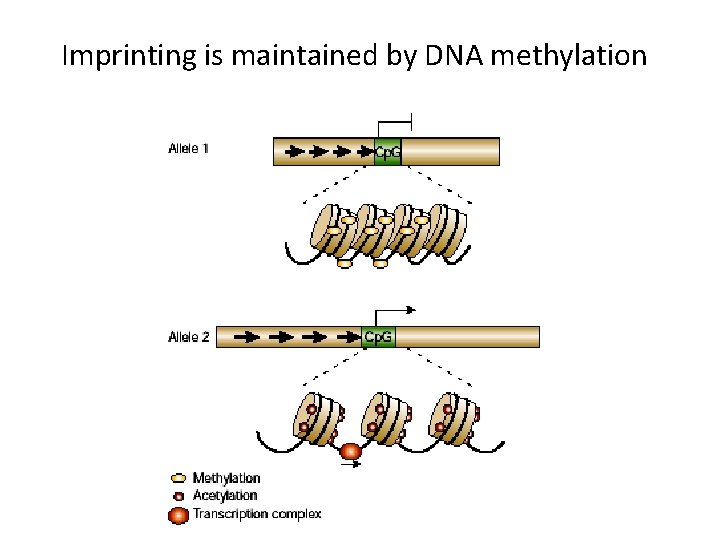
Genomic imprinting Some genes are expressed only from the maternal genome and some only from the paternal genome It is estimated that about 40 genes are imprinted and they can be found on several different chromosomes

Imprinting is maintained by DNA methylation

Why gene imprinting? Parent Offspring Conflict Hypothesis (Haig hypothesis) Conflict between male and female over allocation of maternal resources to offspring Paternally expressed genes would promote growth, maternally expressed genes should slow it down. For example: IGF 2 (growth factor) / IGF 2 receptor (make IGF 2 inactive) IGF 2 gene is paternal or maternal imprinted? How about IFG 2 receptor?

Epigenetic chromatin regulation A. Modification at the DNA level 1. cytosine methylation B. Histone modification - the histone code 1. Histone acetylation 2. Histone methylation 3. Histone phosphorylation 4. Histone ubiquitilation 5. Different types of histones

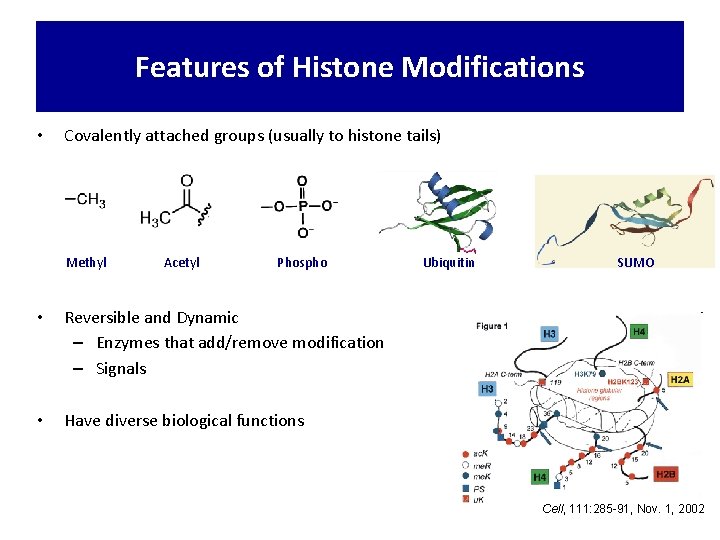
Features of Histone Modifications • Covalently attached groups (usually to histone tails) Methyl Acetyl Phospho • Reversible and Dynamic – Enzymes that add/remove modification – Signals • Have diverse biological functions Ubiquitin SUMO Cell, 111: 285 -91, Nov. 1, 2002

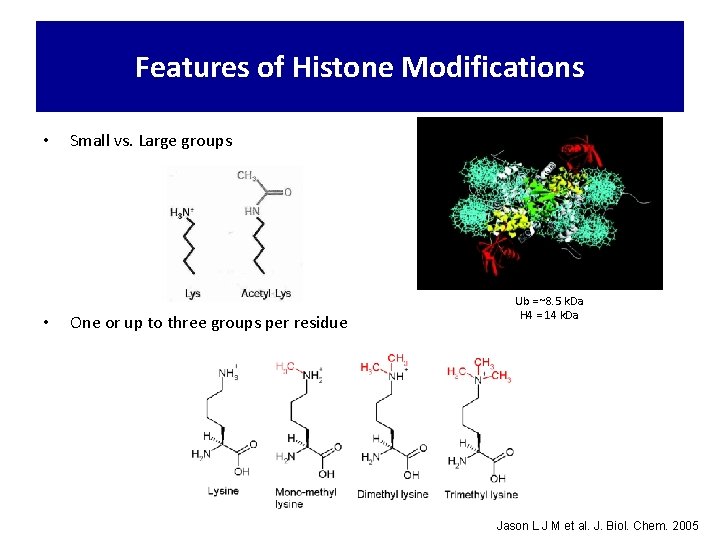
Features of Histone Modifications • • Small vs. Large groups One or up to three groups per residue Ub = ~8. 5 k. Da H 4 = 14 k. Da Jason L J M et al. J. Biol. Chem. 2005


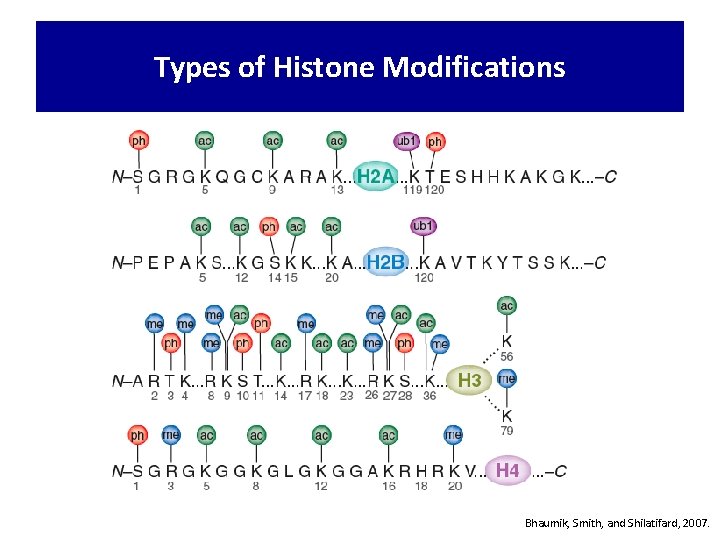
Types of Histone Modifications Bhaumik, Smith, and Shilatifard, 2007.

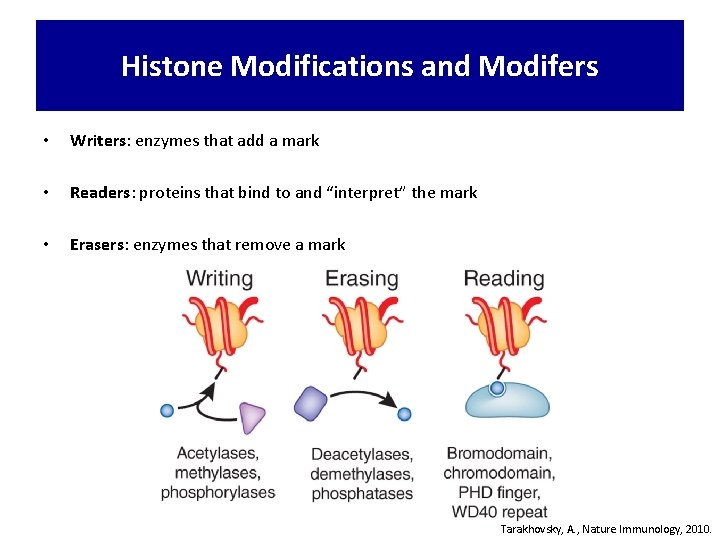
Histone Modifications and Modifers • Writers: enzymes that add a mark • Readers: proteins that bind to and “interpret” the mark • Erasers: enzymes that remove a mark Tarakhovsky, A. , Nature Immunology, 2010.

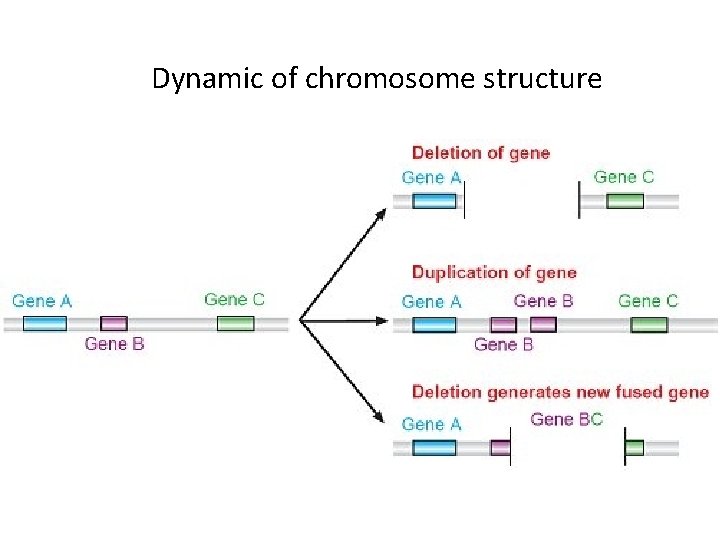
Dynamic of chromosome structure

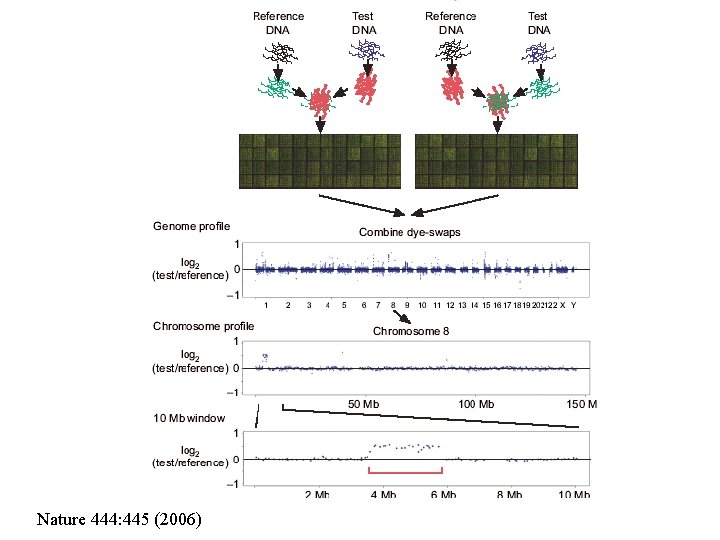
• Copy Number Variation (CNV) in human genome • Total 1, 447 CNV were identified across the 270 samples • Established average length of CNV regions per genome was over 20 million base pairs

Nature 444: 445 (2006)

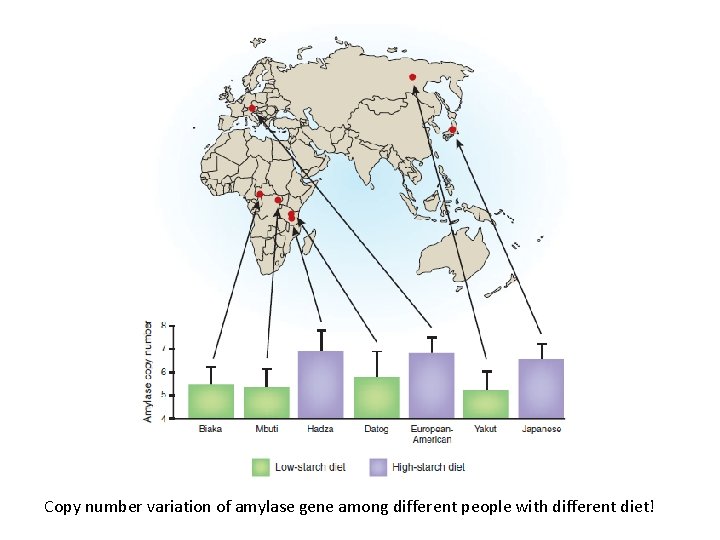
Copy number variation of amylase gene among different people with different diet!

EXPANDING THE GENE CONCEPT BEYOND THE PROTEIN ENCODING SEQUENCESES of DNA: TRANSCRIPTION OF SOME GENES PRODUCES NONCODING RNAs

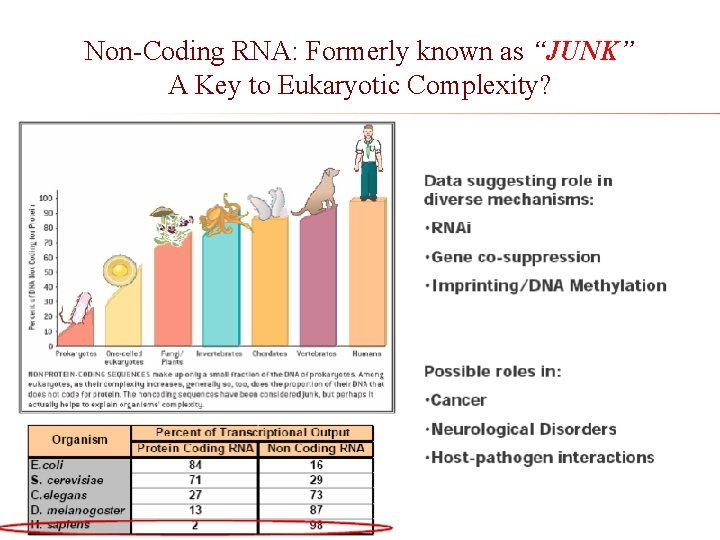
Non-Coding RNA: Formerly known as “JUNK” A Key to Eukaryotic Complexity?

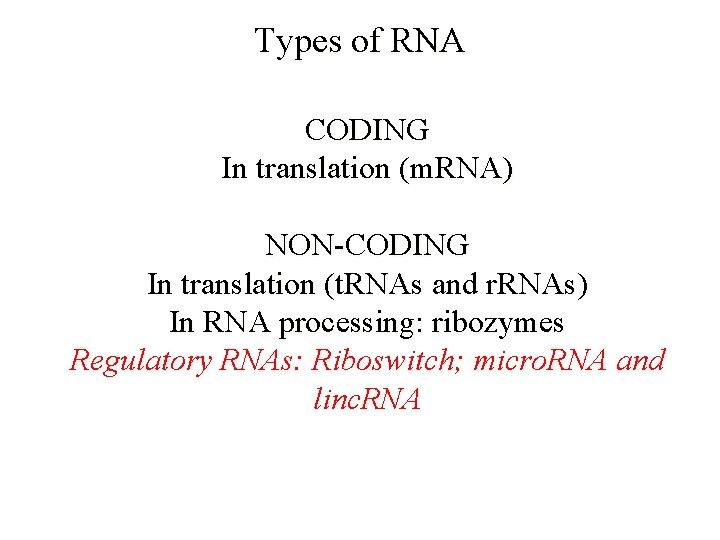
Types of RNA CODING In translation (m. RNA) NON-CODING In translation (t. RNAs and r. RNAs) In RNA processing: ribozymes Regulatory RNAs: Riboswitch; micro. RNA and linc. RNA

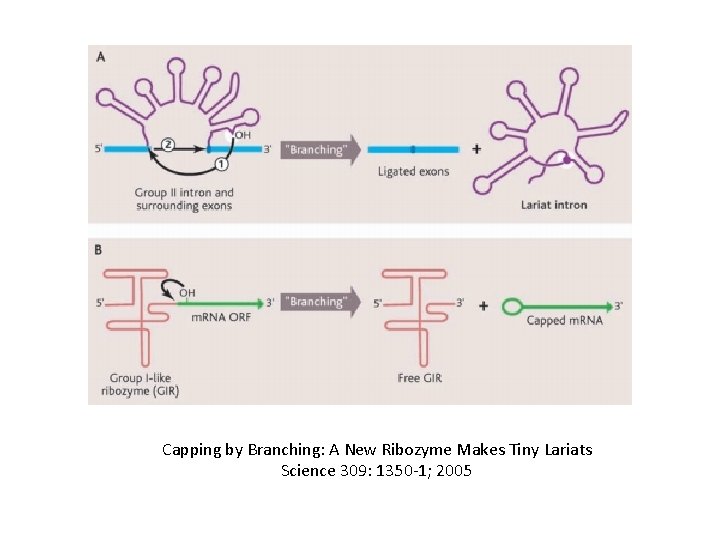
Capping by Branching: A New Ribozyme Makes Tiny Lariats Science 309: 1350 -1; 2005

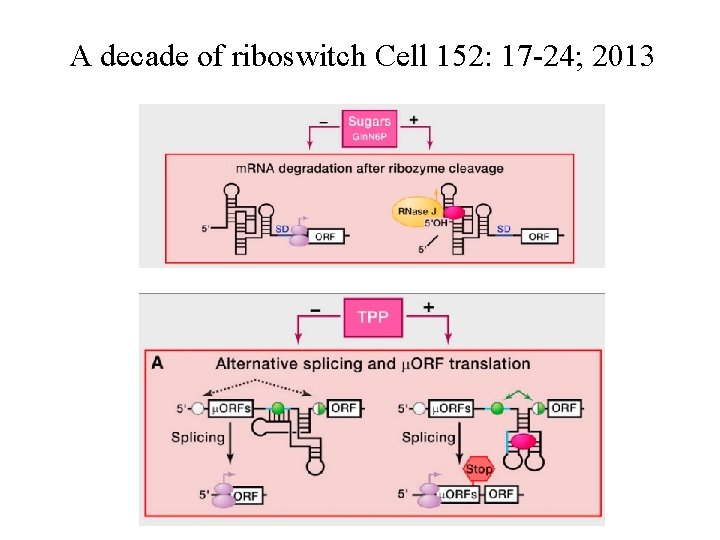
A decade of riboswitch Cell 152: 17 -24; 2013



What is mi. RNA? 51

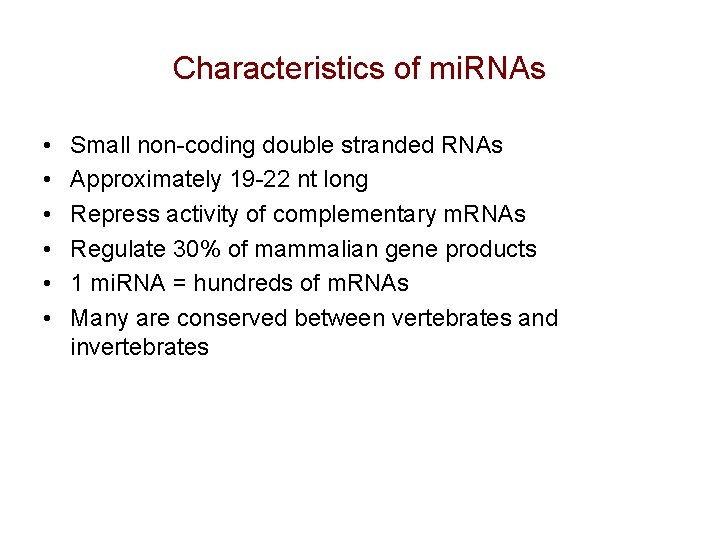
Characteristics of mi. RNAs • • • Small non-coding double stranded RNAs Approximately 19 -22 nt long Repress activity of complementary m. RNAs Regulate 30% of mammalian gene products 1 mi. RNA = hundreds of m. RNAs Many are conserved between vertebrates and invertebrates

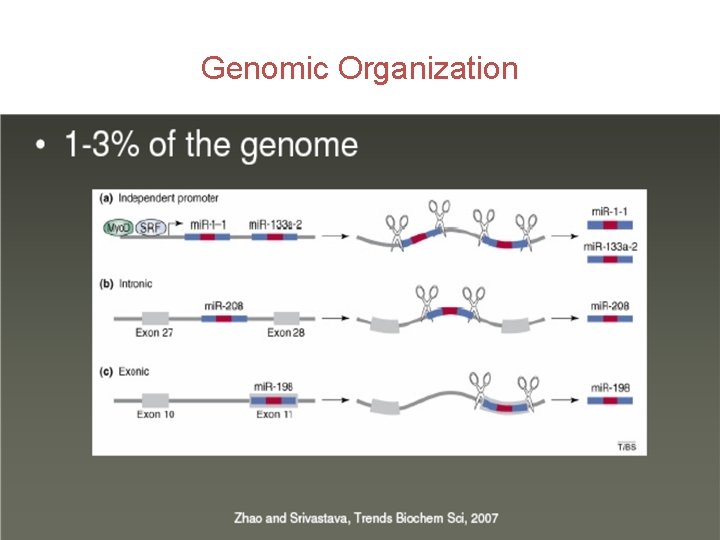
Genomic Organization

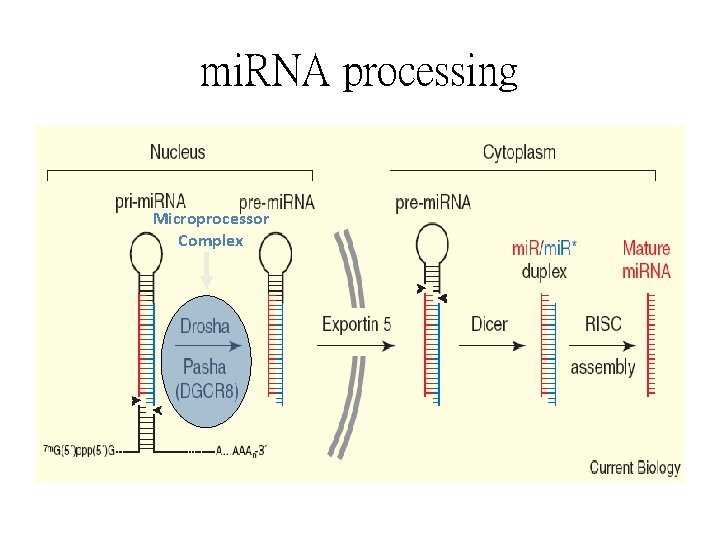
mi. RNA processing Microprocessor Complex

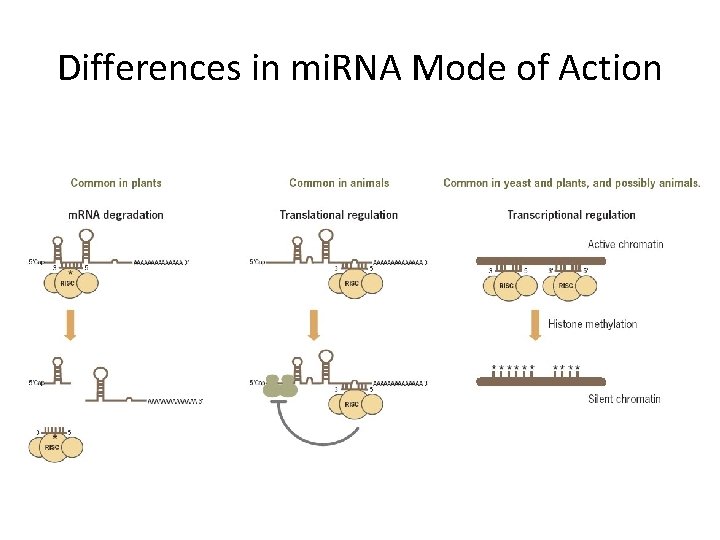
Differences in mi. RNA Mode of Action

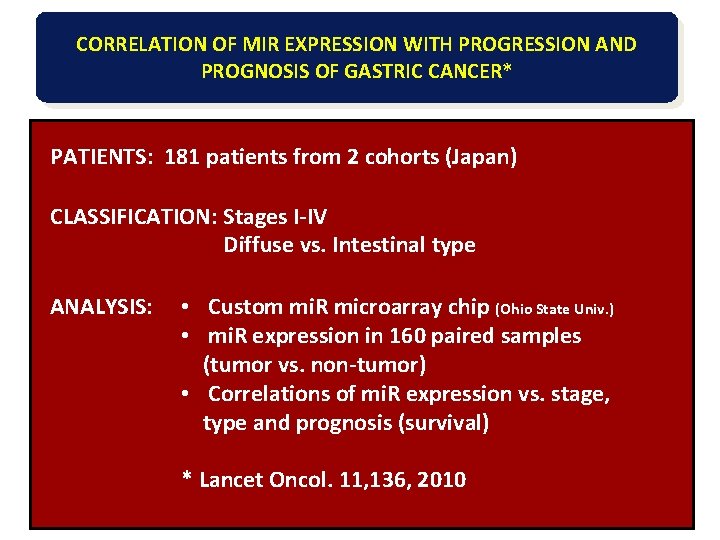
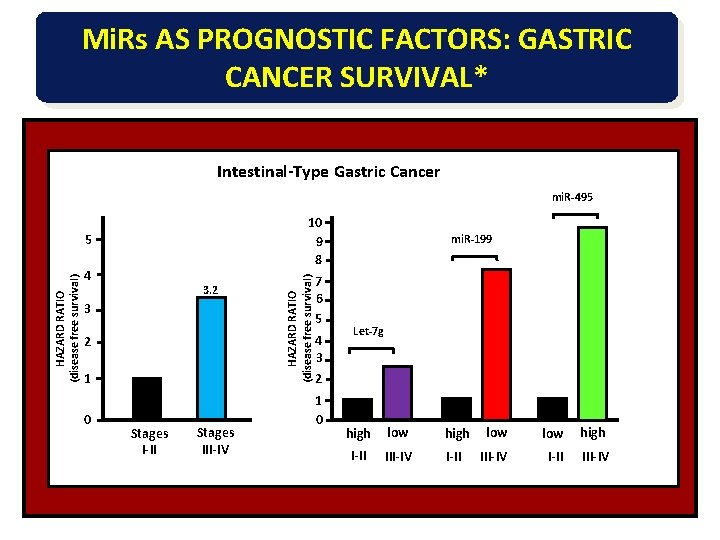
CORRELATION OF MIR EXPRESSION WITH PROGRESSION AND PROGNOSIS OF GASTRIC CANCER* PATIENTS: 181 patients from 2 cohorts (Japan) CLASSIFICATION: Stages I-IV Diffuse vs. Intestinal type ANALYSIS: • Custom mi. R microarray chip (Ohio State Univ. ) • mi. R expression in 160 paired samples (tumor vs. non-tumor) • Correlations of mi. R expression vs. stage, type and prognosis (survival) * Lancet Oncol. 11, 136, 2010

Mi. Rs AS PROGNOSTIC FACTORS: GASTRIC CANCER SURVIVAL* Intestinal-Type Gastric Cancer mi. R-495 HAZARD RATIO (disease free survival) 5 4 3. 2 3 2 1 0 Stages I-II Stages III-IV HAZARD RATIO (disease free survival) 10 9 8 7 6 5 4 3 2 1 0 mi. R-199 Let-7 g high low low high I-II III-IV

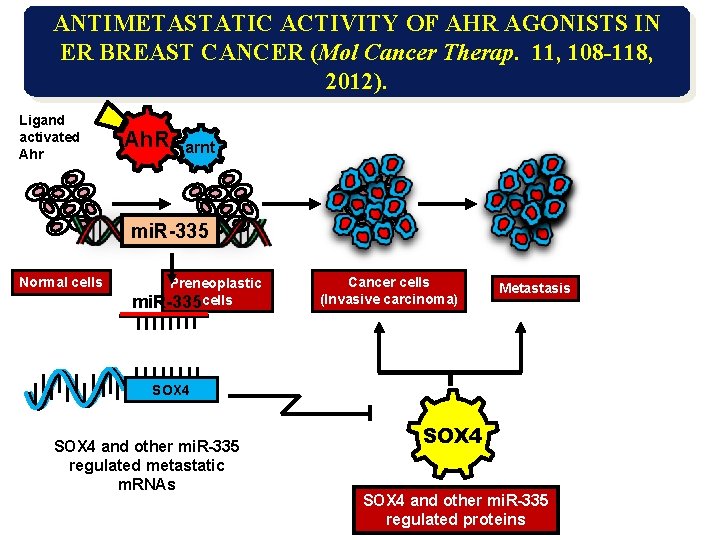
ANTIMETASTATIC ACTIVITY OF AHR AGONISTS IN ER BREAST CANCER (Mol Cancer Therap. 11, 108 -118, 2012). Ligand activated Ahr Ah. R arnt mi. R-335 Normal cells Preneoplastic mi. R-335 cells Cancer cells (Invasive carcinoma) Metastasis SOX 4 and other mi. R-335 regulated metastatic m. RNAs SOX 4 and other mi. R-335 regulated proteins

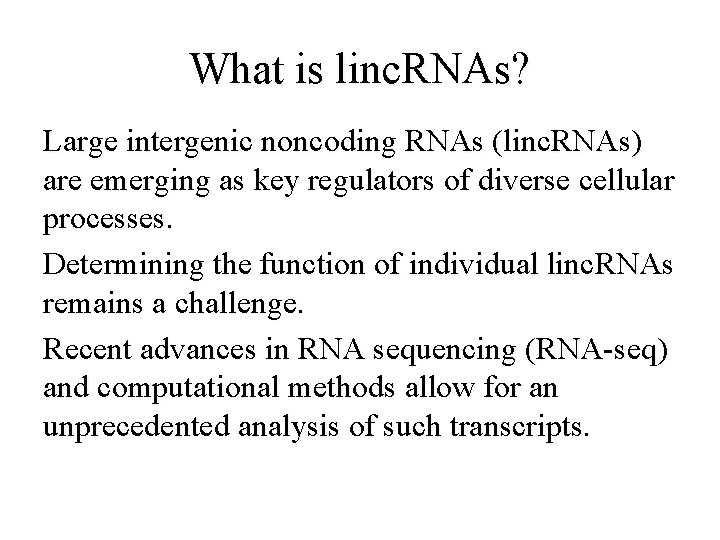
What is linc. RNAs? Large intergenic noncoding RNAs (linc. RNAs) are emerging as key regulators of diverse cellular processes. Determining the function of individual linc. RNAs remains a challenge. Recent advances in RNA sequencing (RNA-seq) and computational methods allow for an unprecedented analysis of such transcripts.


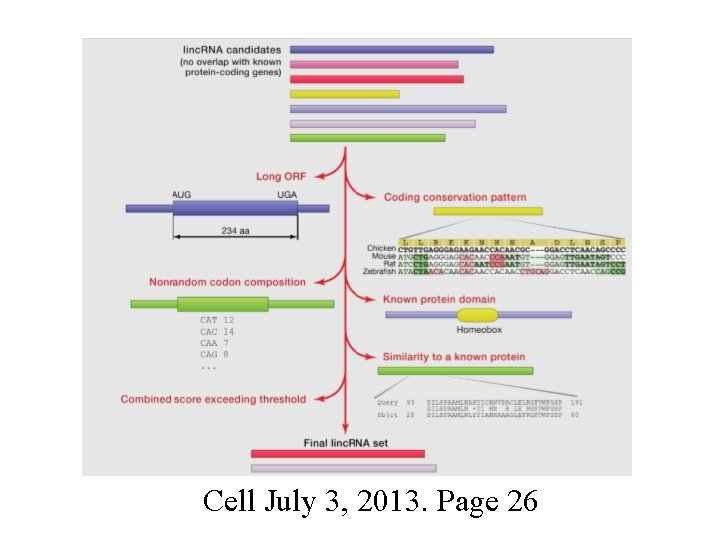
Cell July 3, 2013. Page 26


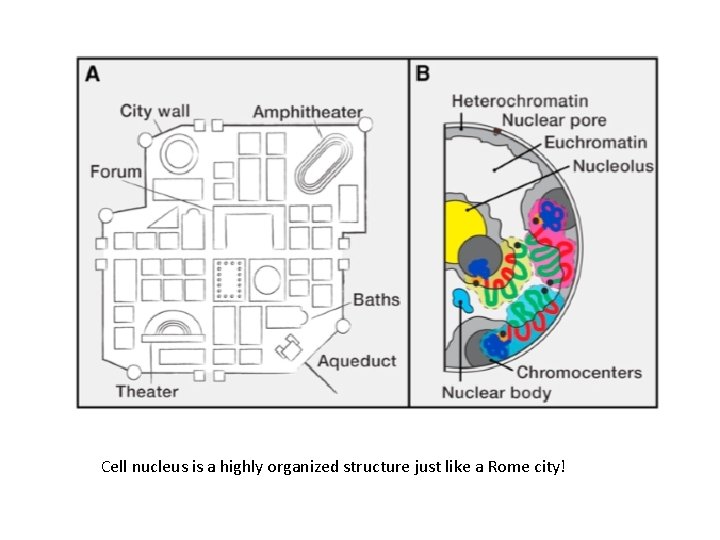
Cell nucleus is a highly organized structure just like a Rome city!

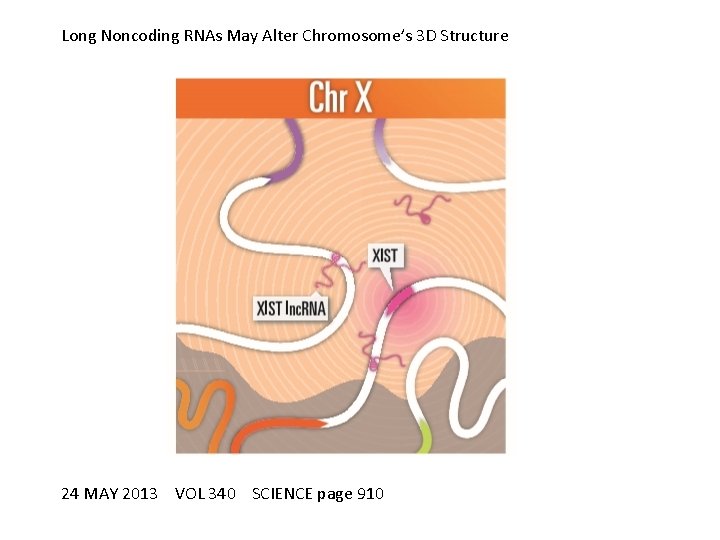
Long Noncoding RNAs May Alter Chromosome’s 3 D Structure 24 MAY 2013 VOL 340 SCIENCE page 910

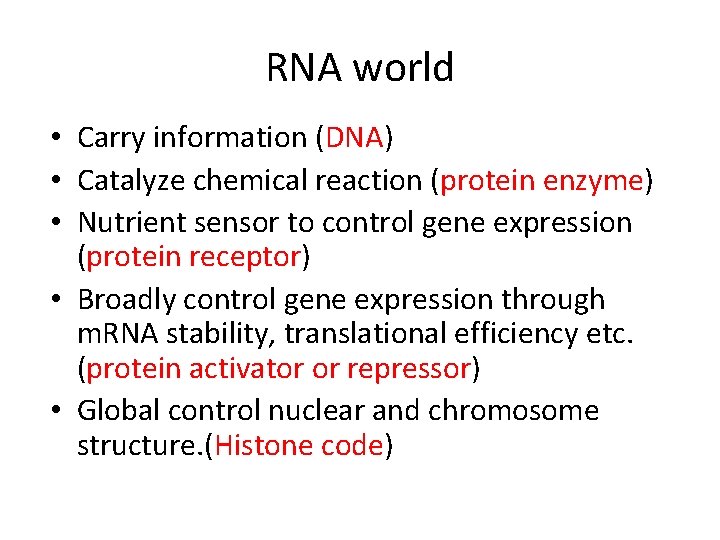
RNA world • Carry information (DNA) • Catalyze chemical reaction (protein enzyme) • Nutrient sensor to control gene expression (protein receptor) • Broadly control gene expression through m. RNA stability, translational efficiency etc. (protein activator or repressor) • Global control nuclear and chromosome structure. (Histone code)
 Cdc whole school whole community whole child
Cdc whole school whole community whole child Genome-to-genome distance calculator
Genome-to-genome distance calculator How to read chromosome
How to read chromosome Perpartes
Perpartes Shotgun sequencing
Shotgun sequencing Shotgun sequencing
Shotgun sequencing Whole part whole practice
Whole part whole practice Genome assembly
Genome assembly Difference between autosomes and sex chromosome
Difference between autosomes and sex chromosome Chromosome vs dna vs gene
Chromosome vs dna vs gene Chromosomes and their functions
Chromosomes and their functions Chromosome in dna
Chromosome in dna Chromosome sets (=n) in mitosis and meiosis
Chromosome sets (=n) in mitosis and meiosis Anaphase meaning
Anaphase meaning Non homologous chromosomes
Non homologous chromosomes Difference between meiosis 1 and meiosis 2
Difference between meiosis 1 and meiosis 2 What is genome
What is genome Plant genome research program
Plant genome research program Human genome consists of
Human genome consists of Stanford
Stanford Human genome size
Human genome size Mash bioinformatics
Mash bioinformatics Human genome size
Human genome size Bacterial artificial chromosome slideshare
Bacterial artificial chromosome slideshare Vntrs vs strs
Vntrs vs strs Repeated sequences
Repeated sequences Dna
Dna Human genome project source code
Human genome project source code Sickle cell karyotype
Sickle cell karyotype Patric bioinformatics
Patric bioinformatics National human genome research institute
National human genome research institute Genome modification ustaz auni
Genome modification ustaz auni National human genome research institute
National human genome research institute Sequencing human genome
Sequencing human genome Genome klick
Genome klick History of sequencing
History of sequencing Chapter 15 the human genome answer key
Chapter 15 the human genome answer key Chapter 14 the human genome
Chapter 14 the human genome Human genome project
Human genome project Codom
Codom National human genome research institute
National human genome research institute Ucsc genome browser tutorial
Ucsc genome browser tutorial Genome
Genome Genome identification
Genome identification Genome sequencing
Genome sequencing Savant genome browser
Savant genome browser Yale university poster
Yale university poster Ap biol
Ap biol Alternate splicing
Alternate splicing Gene annotation
Gene annotation Genome.gov
Genome.gov Genome research limited
Genome research limited Innovation genome project
Innovation genome project Genome mapping
Genome mapping Igv genome browser
Igv genome browser Img genome
Img genome Artemis genome
Artemis genome Genome project
Genome project Maker genome annotation
Maker genome annotation Section 14-3 human molecular genetics answer key
Section 14-3 human molecular genetics answer key Genome definition
Genome definition Who discovered the structure of dna
Who discovered the structure of dna Tomato genome browser
Tomato genome browser Genome adalah
Genome adalah Integrative genomics viewer
Integrative genomics viewer Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome