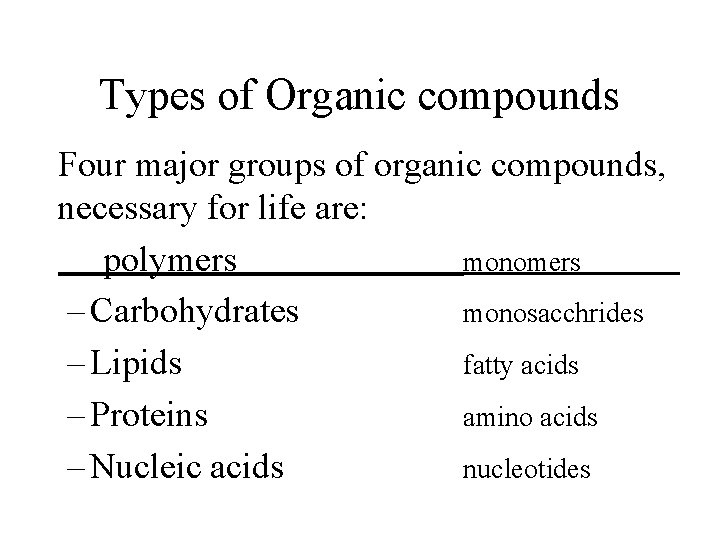
Types of Organic compounds Four major groups of









- Slides: 57

Types of Organic compounds Four major groups of organic compounds, necessary for life are: polymers monomers – Carbohydrates monosacchrides – Lipids fatty acids – Proteins amino acids – Nucleic acids nucleotides

Carbohydrates • Diverse group of substances formed from C, H, and O – ratio of one carbon atom for each water molecule (carbohydrates means “watered carbon”) – glucose is 6 carbon atoms and 6 water molecules (H 20) • Main function is to produce energy

Diversity of Carbohydrates • 3 sizes of carbohydrate molecules – monosaccharides – disaccharides – polysaccharides

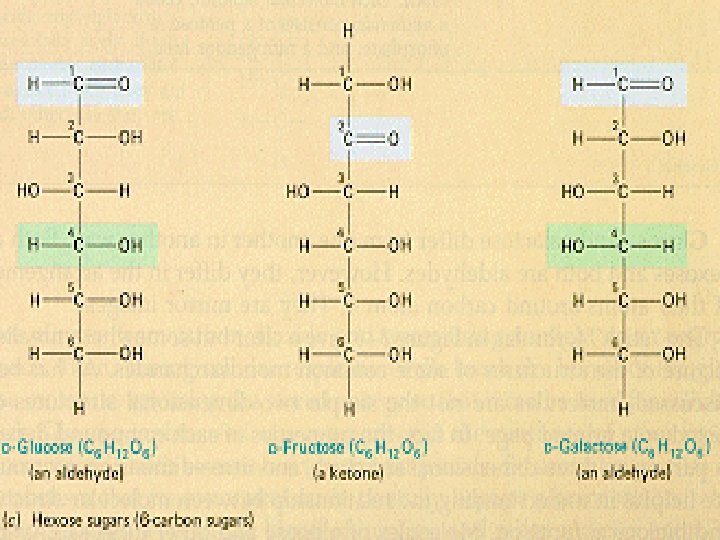
Mono saccharides “one sugar” • • Called simple sugars Contain 3 to 7 carbon atoms (CH 2 O)n We can absorb only 3 simple sugars without further digestion in our small intestine – glucose found in syrup or honey – fructose found in fruit – galactose found in dairy products

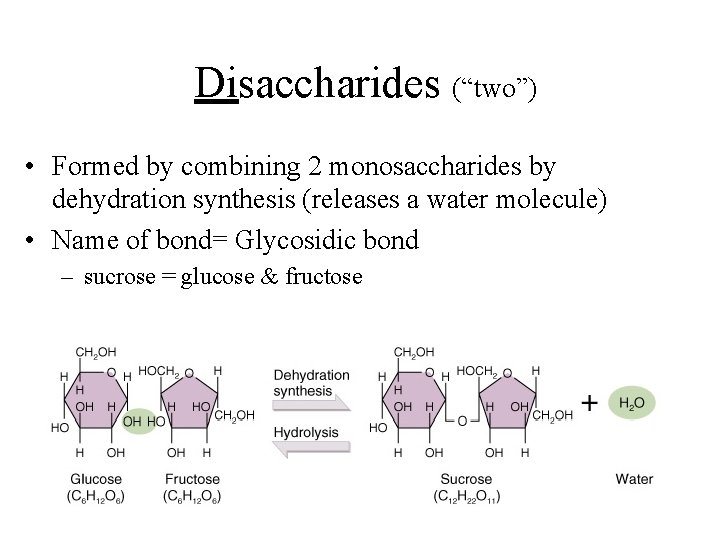
Disaccharides (“two”) • Formed by combining 2 monosaccharides by dehydration synthesis (releases a water molecule) • Name of bond= Glycosidic bond – sucrose = glucose & fructose

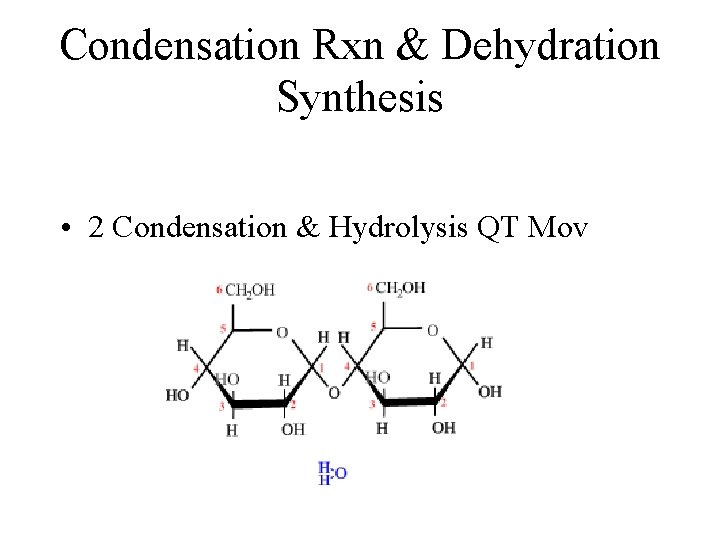
Condensation Rxn & Dehydration Synthesis • 2 Condensation & Hydrolysis QT Mov

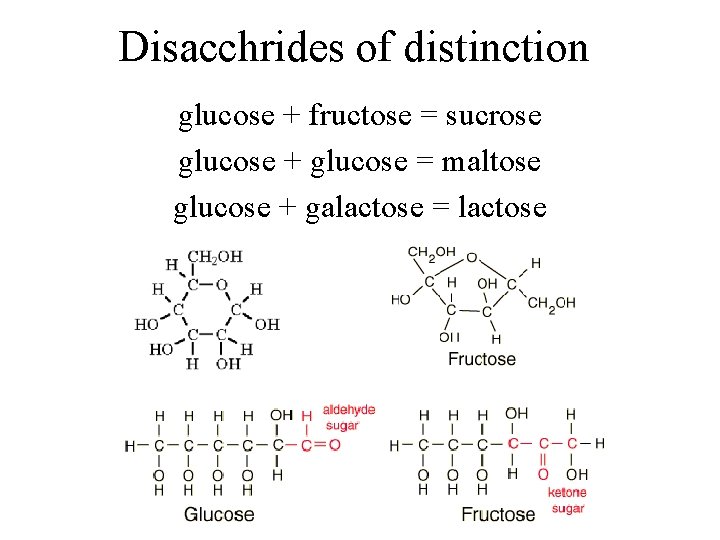
Disacchrides of distinction glucose + fructose = sucrose glucose + glucose = maltose glucose + galactose = lactose


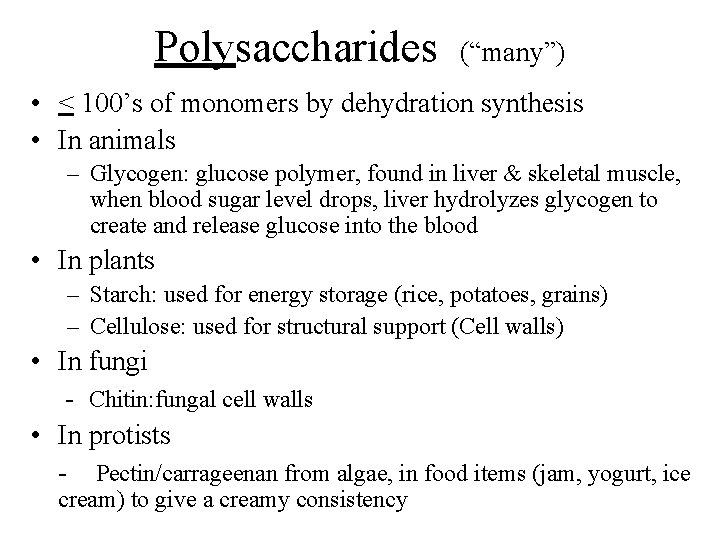
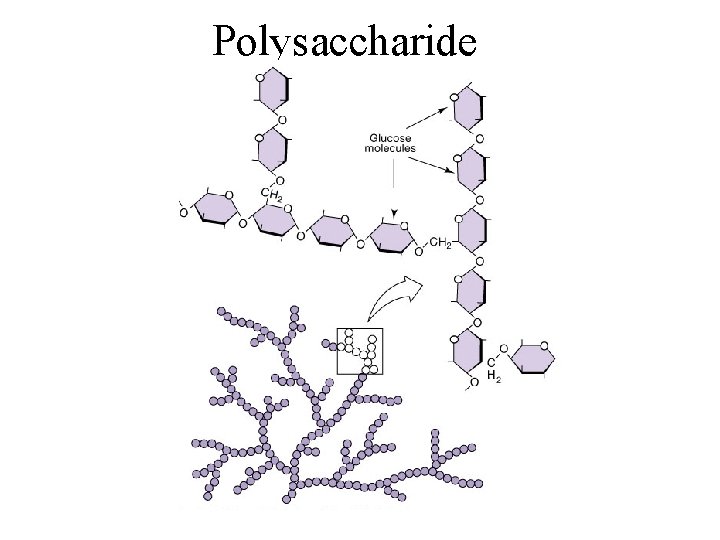
Polysaccharides (“many”) • < 100’s of monomers by dehydration synthesis • In animals – Glycogen: glucose polymer, found in liver & skeletal muscle, when blood sugar level drops, liver hydrolyzes glycogen to create and release glucose into the blood • In plants – Starch: used for energy storage (rice, potatoes, grains) – Cellulose: used for structural support (Cell walls) • In fungi - Chitin: fungal cell walls • In protists - Pectin/carrageenan from algae, in food items (jam, yogurt, ice cream) to give a creamy consistency

Polysaccharide

Lipids = fats, oils, steroids, waxes • Formed from C, H and O • 18 -25% of body weight • Hydrophobic – fewer polar bonds because of fewer oxygen atoms – insoluble in polar solvents like water • Combines with proteins for transport in blood – lipoproteins

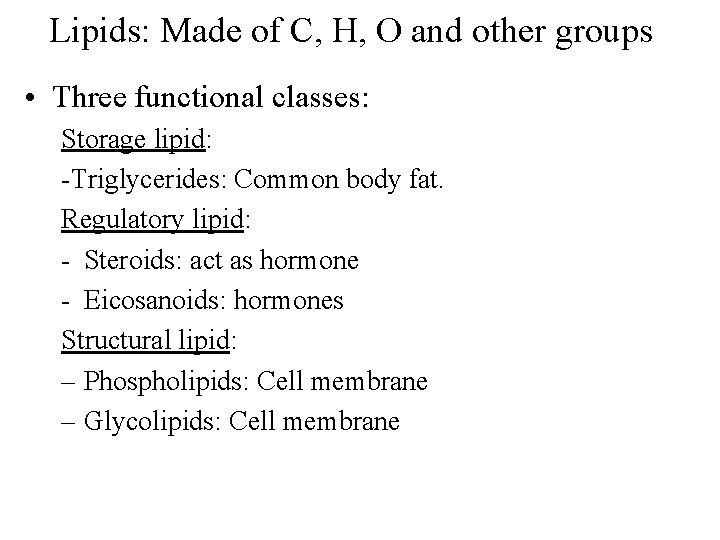
Lipids: Made of C, H, O and other groups • Three functional classes: Storage lipid: -Triglycerides: Common body fat. Regulatory lipid: - Steroids: act as hormone - Eicosanoids: hormones Structural lipid: – Phospholipids: Cell membrane – Glycolipids: Cell membrane

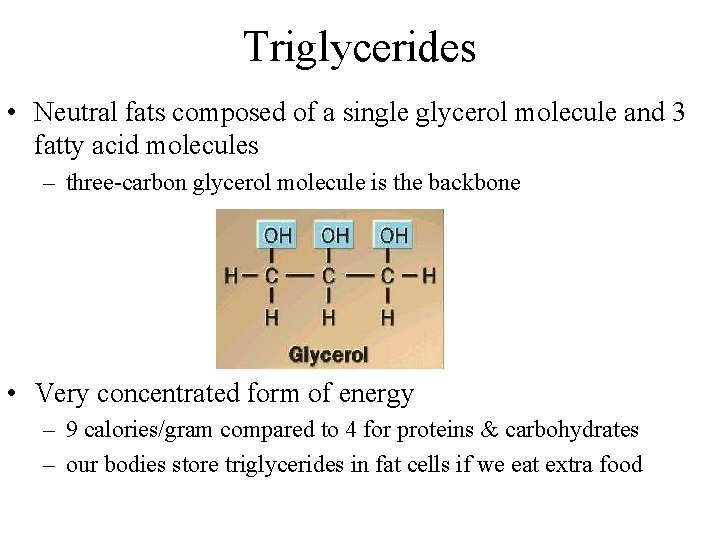
Triglycerides • Neutral fats composed of a single glycerol molecule and 3 fatty acid molecules – three-carbon glycerol molecule is the backbone • Very concentrated form of energy – 9 calories/gram compared to 4 for proteins & carbohydrates – our bodies store triglycerides in fat cells if we eat extra food

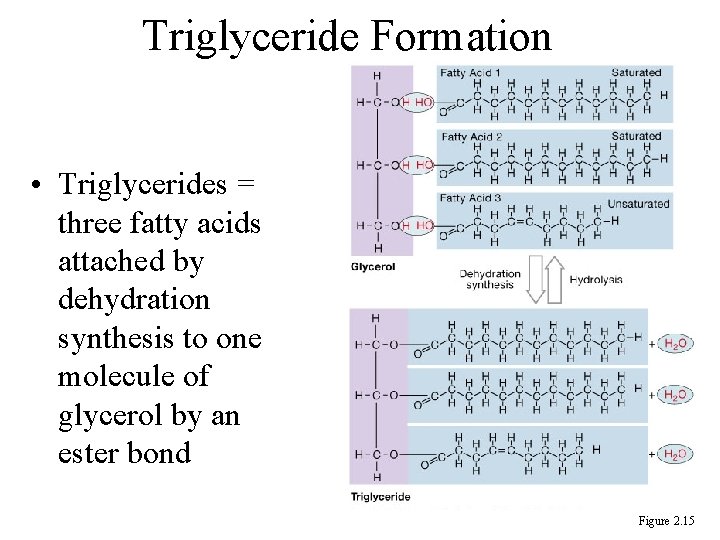
Triglyceride Formation • Triglycerides = three fatty acids attached by dehydration synthesis to one molecule of glycerol by an ester bond Figure 2. 15

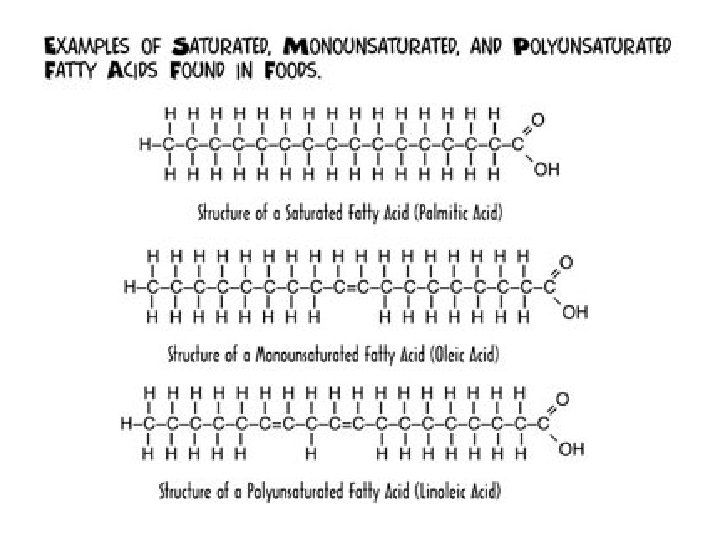
Saturation of Triglycerides • Determined by the number of single or double covalent bonds in fatty acid • Saturated fats contain single covalent bonds and are covered with hydrogen atoms----lard • Unsaturated are not completely covered with hydrogen----safflower oil, corn oil • Polyunsaturated fats contain even less hydrogen atoms----olive and peanut oil


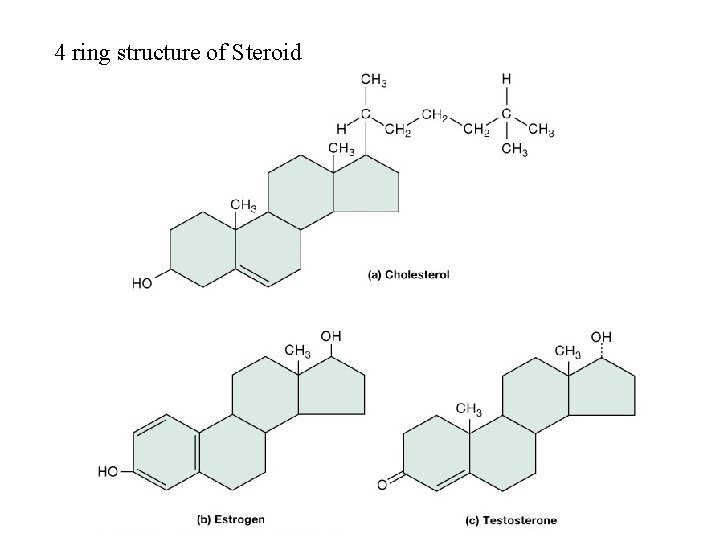
Regulatory lipids: Steroids • Formed from 4 rings of carbon atoms joined together • Common steroids – sex hormones, bile salts, vitamins & cholesterol • Cholesterol found in animal cell membranes – starting material for synthesis of other steroids

4 ring structure of Steroids(Wax)

Eicosanoids • Lipid type derived from a fatty acid called arachidonic acid – prostaglandins = wide variety of functions • • • modify responses to hormones contribute to inflammatory response prevent stomach ulcers dilate airways regulate body temperature influence formation of blood clots – leukotrienes = allergy & inflammatory responses

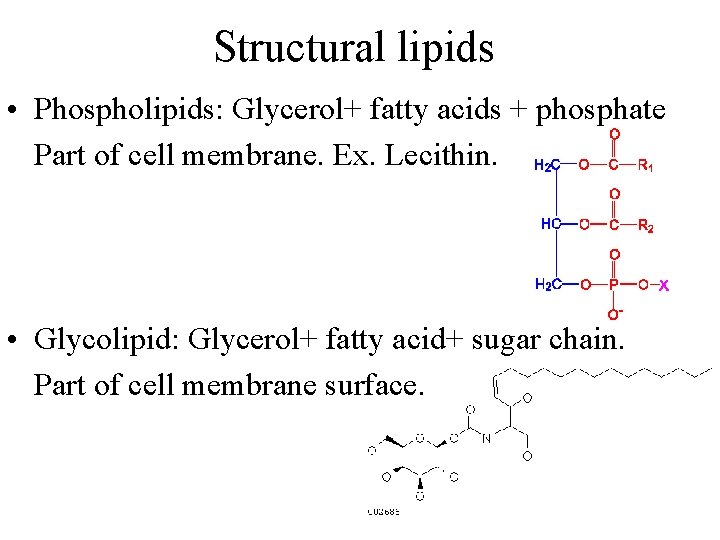
Structural lipids • Phospholipids: Glycerol+ fatty acids + phosphate Part of cell membrane. Ex. Lecithin. • Glycolipid: Glycerol+ fatty acid+ sugar chain. Part of cell membrane surface.

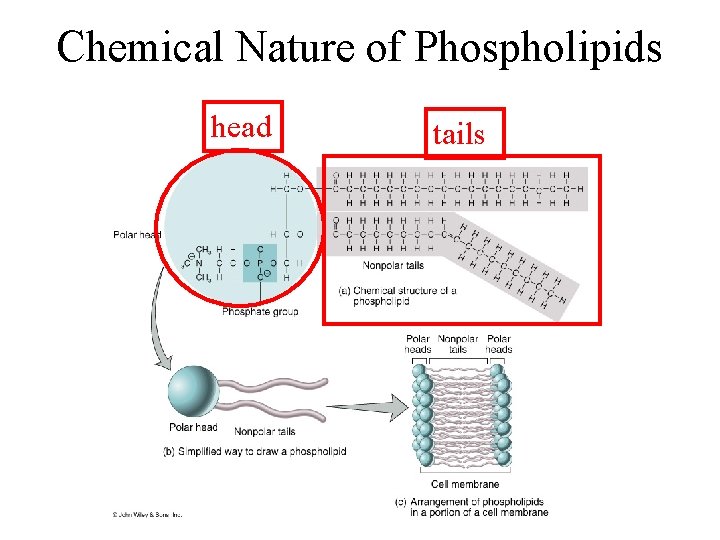
Chemical Nature of Phospholipids head tails

Lipid Behavior in Various Environments

Nucleic acids • Three types: DNA, RNA, ATP • Function: Storage of genetic information and application of genetics (Protein synthesis), and storage of energy

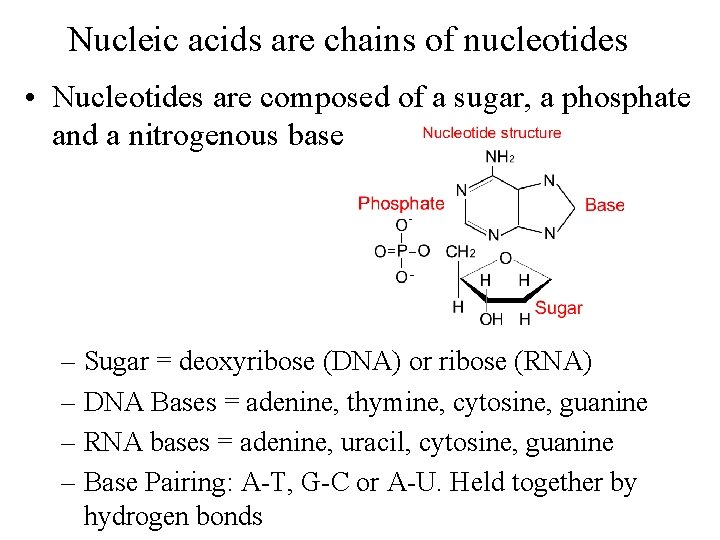
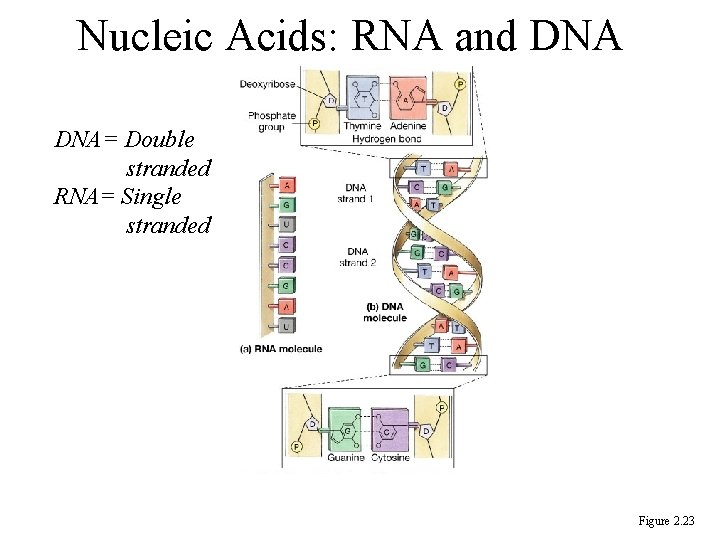
Nucleic acids are chains of nucleotides • Nucleotides are composed of a sugar, a phosphate and a nitrogenous base – Sugar = deoxyribose (DNA) or ribose (RNA) – DNA Bases = adenine, thymine, cytosine, guanine – RNA bases = adenine, uracil, cytosine, guanine – Base Pairing: A-T, G-C or A-U. Held together by hydrogen bonds


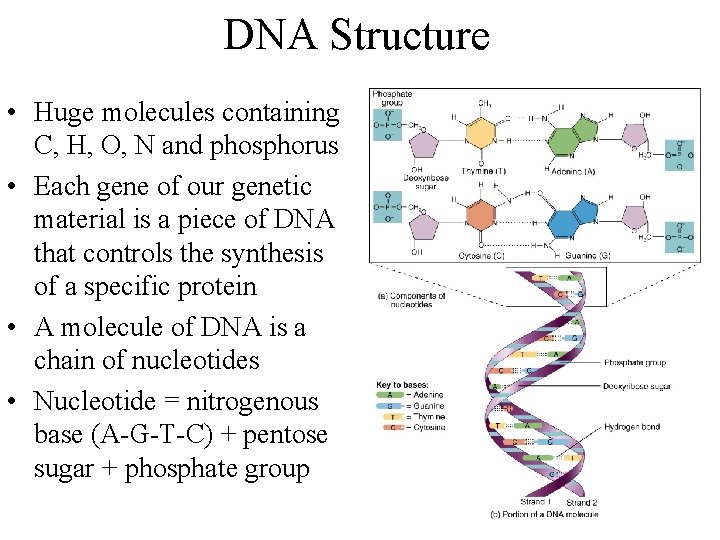
DNA Structure • Huge molecules containing C, H, O, N and phosphorus • Each gene of our genetic material is a piece of DNA that controls the synthesis of a specific protein • A molecule of DNA is a chain of nucleotides • Nucleotide = nitrogenous base (A-G-T-C) + pentose sugar + phosphate group

DNA Fingerprinting • Used to identify criminal, victim or a child’s parents – need only strand of hair, drop of semen or spot of blood • Certain DNA segments are repeated several times – unique from person to person

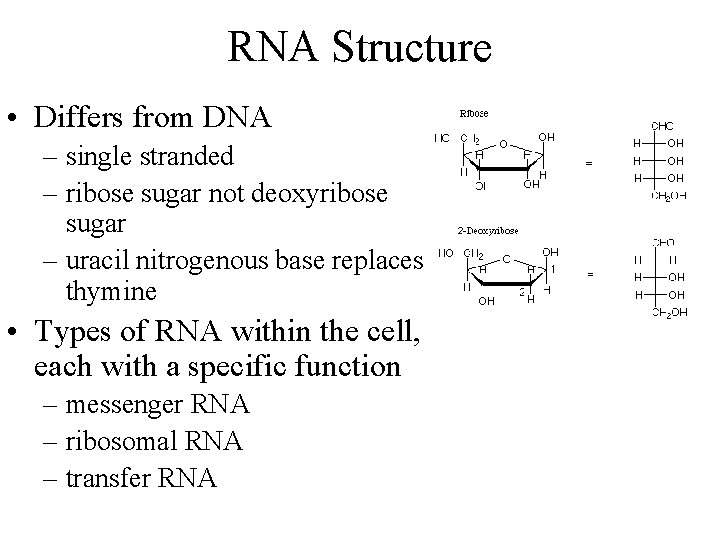
RNA Structure • Differs from DNA – single stranded – ribose sugar not deoxyribose sugar – uracil nitrogenous base replaces thymine • Types of RNA within the cell, each with a specific function – messenger RNA – ribosomal RNA – transfer RNA

Nucleic Acids: RNA and DNA= Double stranded RNA= Single stranded Figure 2. 23

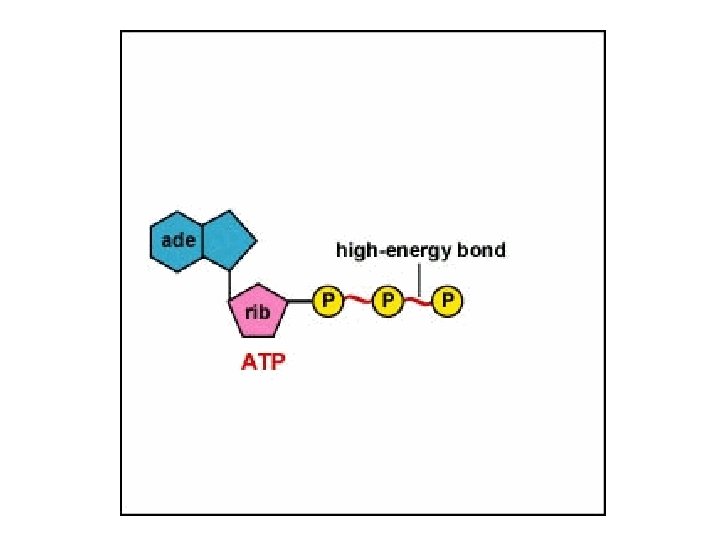
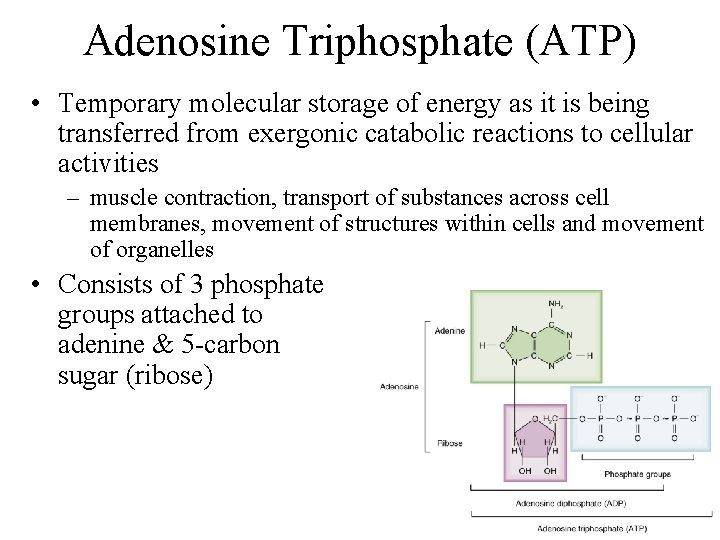
Adenosine Triphosphate (ATP) • Temporary molecular storage of energy as it is being transferred from exergonic catabolic reactions to cellular activities – muscle contraction, transport of substances across cell membranes, movement of structures within cells and movement of organelles • Consists of 3 phosphate groups attached to adenine & 5 -carbon sugar (ribose)

Formation & Usage of ATP • Hydrolysis of ATP (removal of terminal phosphate group by enzyme -- ATPase) – releases energy – leaves ADP (adenosine diphosphate) • Synthesis of ATP – enzyme ATP synthase catalyzes the addition of the terminal phosphate group to ADP – energy from 1 glucose molecule is used during both anaerobic and aerobic respiration to create 36 to 38 molecules of ATP

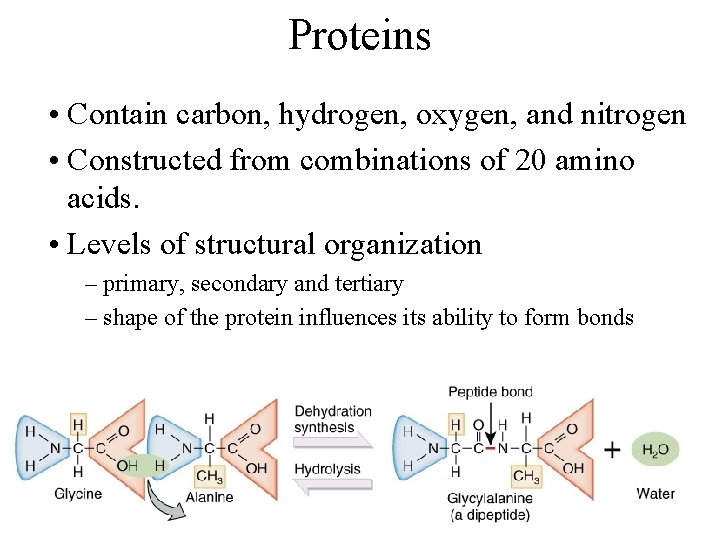
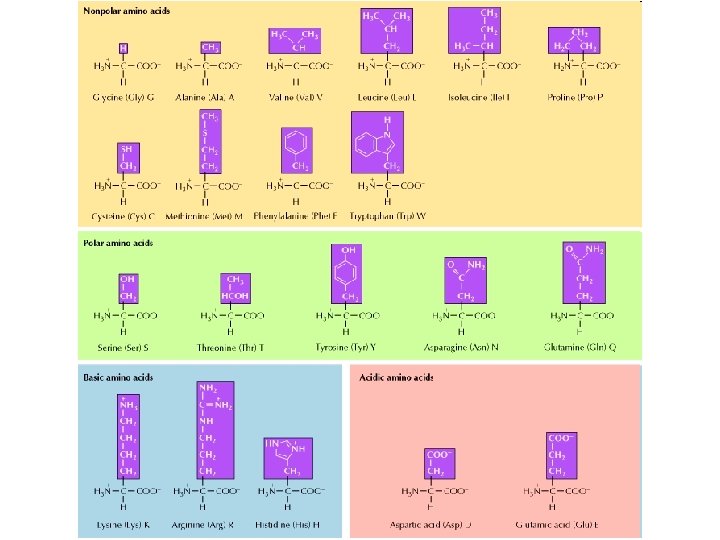
Proteins • Contain carbon, hydrogen, oxygen, and nitrogen • Constructed from combinations of 20 amino acids. • Levels of structural organization – primary, secondary and tertiary – shape of the protein influences its ability to form bonds

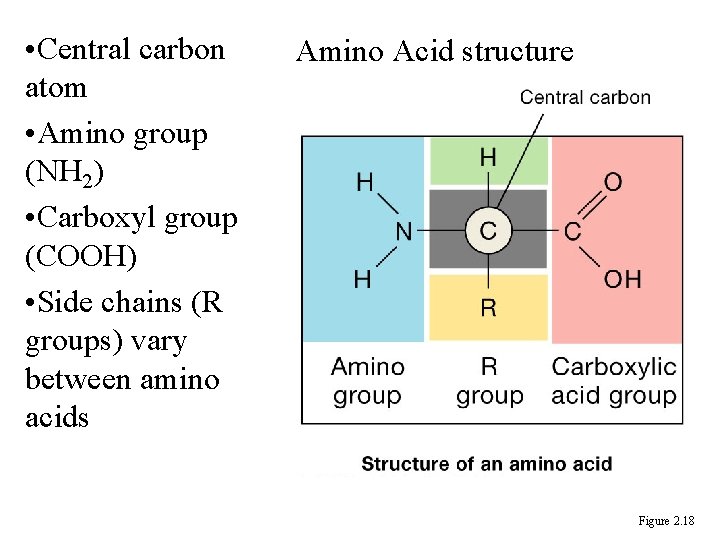
• Central carbon atom • Amino group (NH 2) • Carboxyl group (COOH) • Side chains (R groups) vary between amino acids Amino Acid structure Figure 2. 18


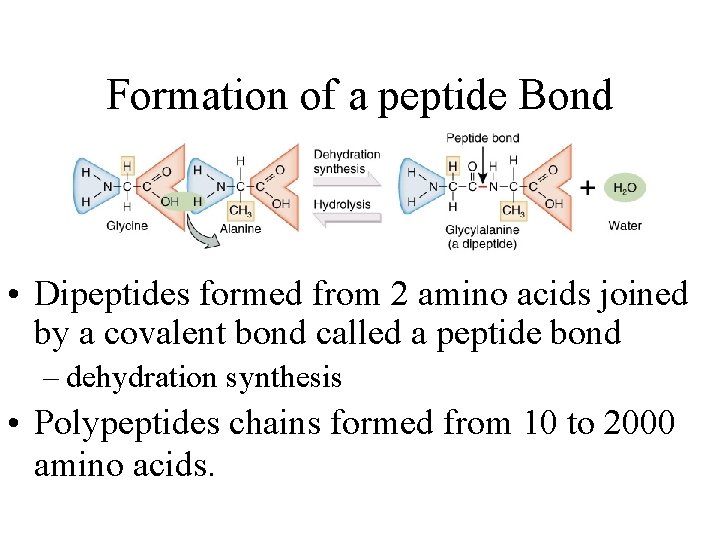
Formation of a peptide Bond • Dipeptides formed from 2 amino acids joined by a covalent bond called a peptide bond – dehydration synthesis • Polypeptides chains formed from 10 to 2000 amino acids.

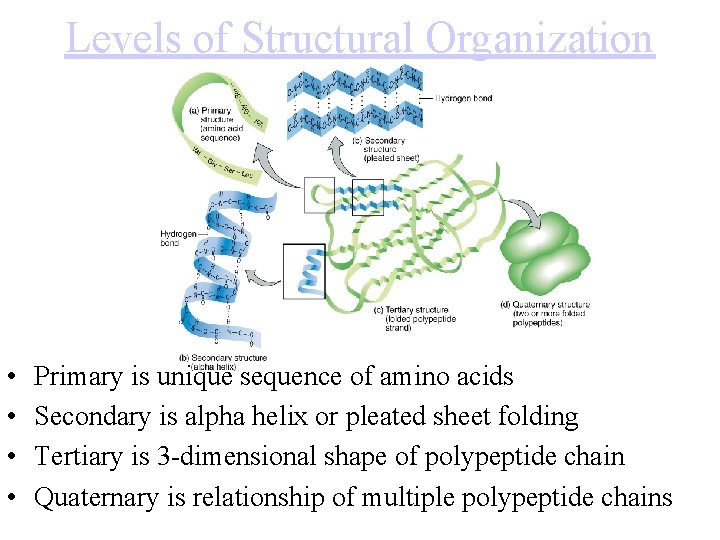
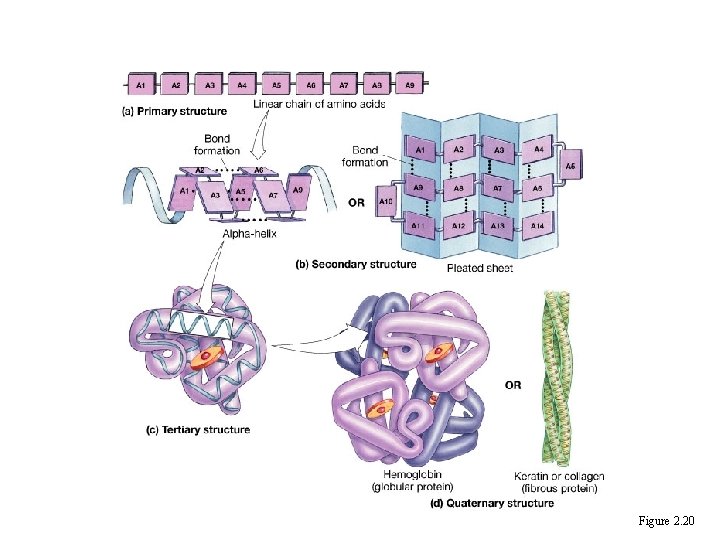
Levels of Structural Organization • • Primary is unique sequence of amino acids Secondary is alpha helix or pleated sheet folding Tertiary is 3 -dimensional shape of polypeptide chain Quaternary is relationship of multiple polypeptide chains

The four levels of protein configuration • • Primary is unique sequence of amino acids Secondary is alpha helix or pleated sheet folding Tertiary is 3 -dimensional shape of polypeptide chain Quaternary is relationship of multiple polypeptide chains • • Protein Folding Tutorial: Protein folding in Water

Protein Structure Figure 2. 20

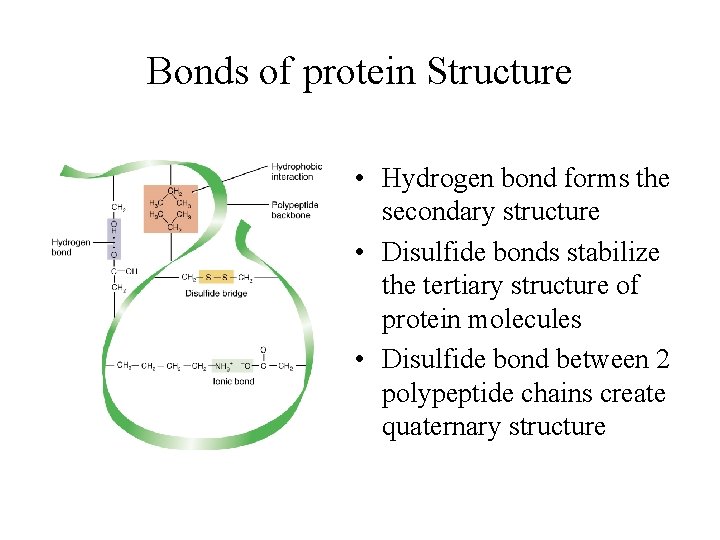
Bonds of protein Structure • Hydrogen bond forms the secondary structure • Disulfide bonds stabilize the tertiary structure of protein molecules • Disulfide bond between 2 polypeptide chains create quaternary structure

Protein Denaturation • Function of a protein depends on its ability to recognize and bind to some other molecule • Hostile environments such as heat, acid or salts will change a proteins 3 -D shape and destroy its ability to function – raw egg white when cooked is vastly different

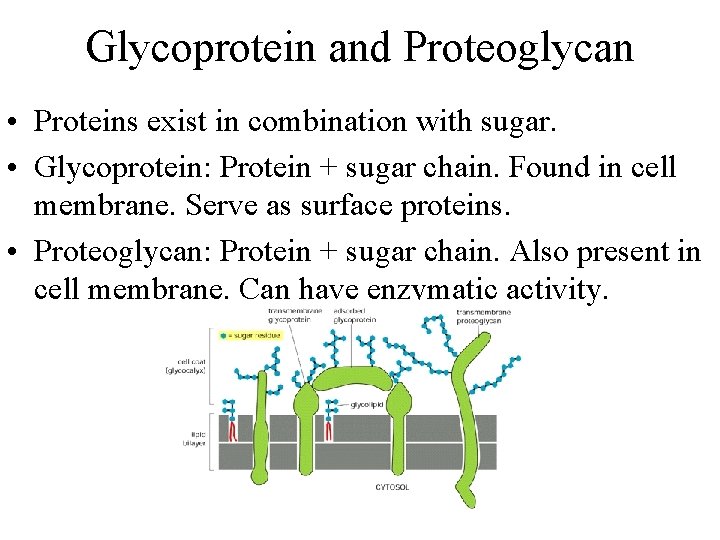
Glycoprotein and Proteoglycan • Proteins exist in combination with sugar. • Glycoprotein: Protein + sugar chain. Found in cell membrane. Serve as surface proteins. • Proteoglycan: Protein + sugar chain. Also present in cell membrane. Can have enzymatic activity.

Enzymes • Enzymes are protein molecules that act as catalysts by lowering Activation Energy • Enzyme = apoenzyme + cofactor – Apoenzymes are the protein portion – Cofactors are nonprotein portion • may be metal ion (iron, zinc, magnesium or calcium) • may be organic molecule derived from a vitamin • Enzymes usually end in suffix -ase and are named for the types of chemical reactions they catalyze • How Enzymes Work • Enzyme Tutorial • Enzyme Catalysis

Enzyme Functions • Bonds made or broken when atoms, ions or molecules collide • Enzymes speed up reactions by properly orienting colliding molecules • 1000 known enzymes speed up metabolic reactions to 10 billion times that in beaker • Composed of protein portion (apoenzyme) & nonprotein portion (cofactor) – cofactors can be metal ions or vitamins

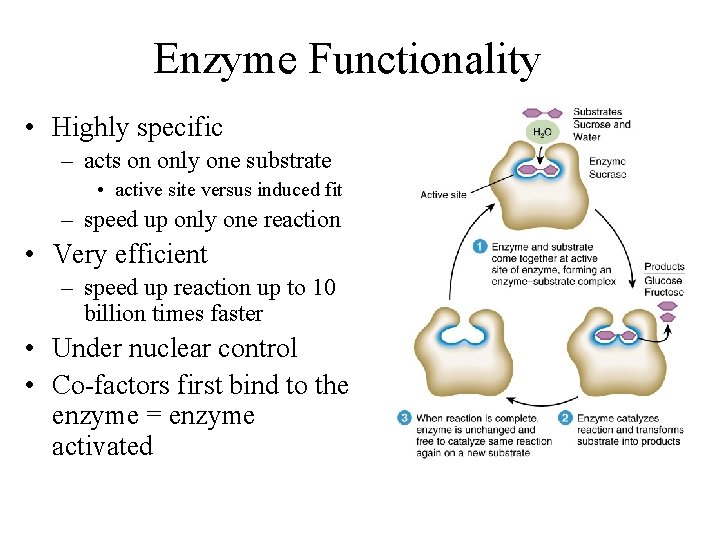
Enzyme Functionality • Highly specific – acts on only one substrate • active site versus induced fit – speed up only one reaction • Very efficient – speed up reaction up to 10 billion times faster • Under nuclear control • Co-factors first bind to the enzyme = enzyme activated

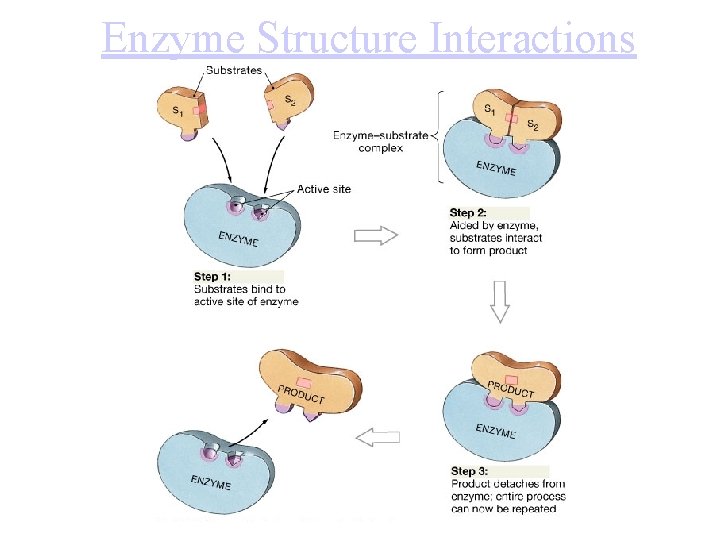
Enzyme Structure Interactions

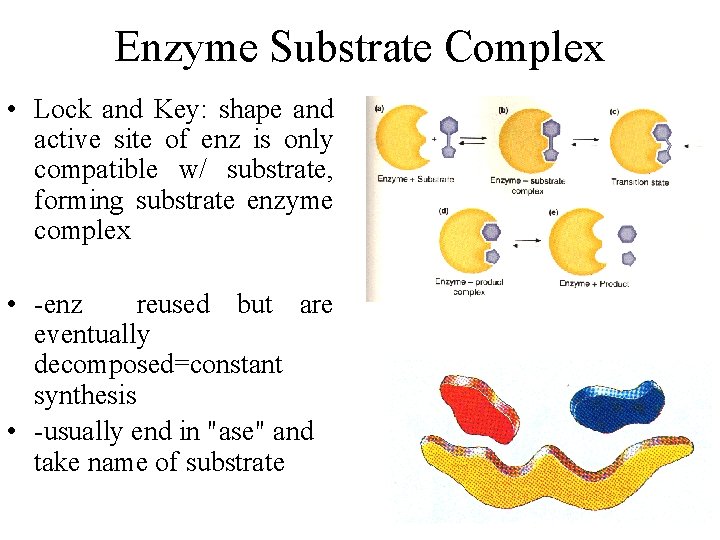
Enzyme Substrate Complex • Lock and Key: shape and active site of enz is only compatible w/ substrate, forming substrate enzyme complex • -enz reused but are eventually decomposed=constant synthesis • -usually end in "ase" and take name of substrate

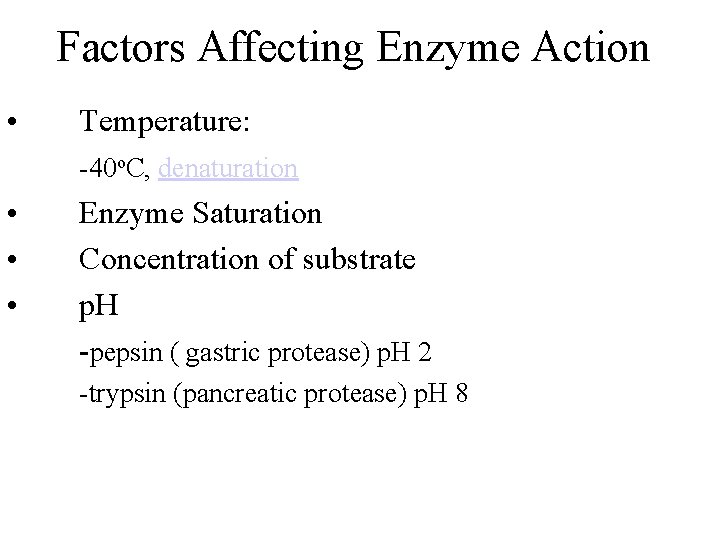
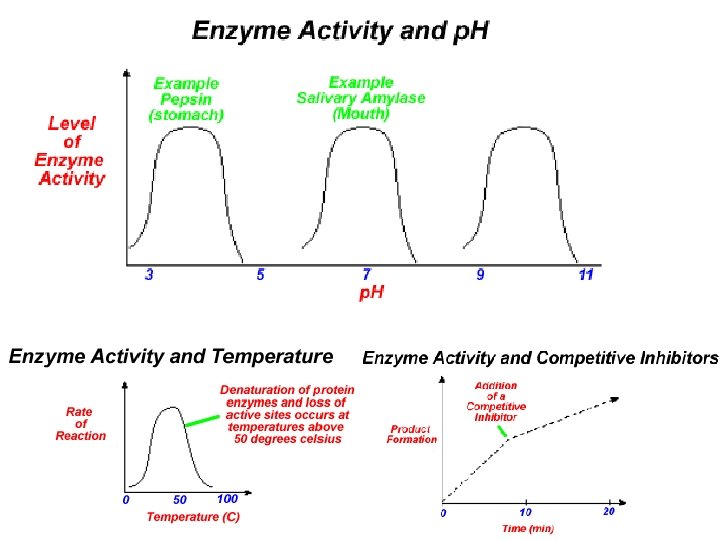
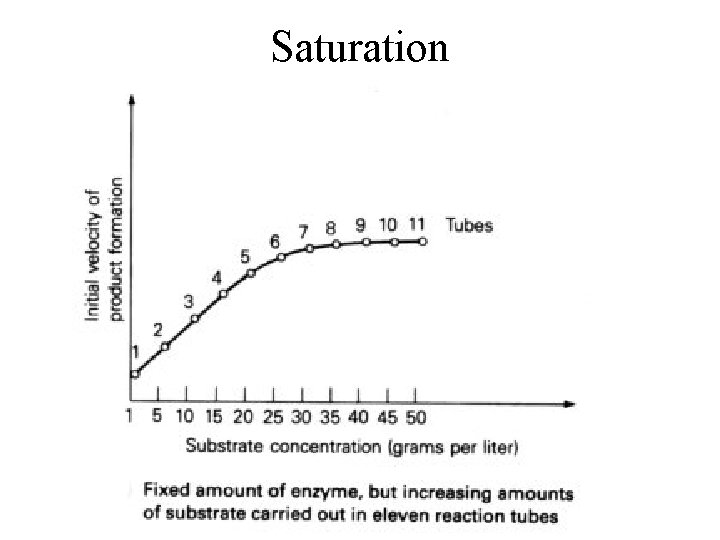
Factors Affecting Enzyme Action • Temperature: -40 o. C, denaturation • • • Enzyme Saturation Concentration of substrate p. H -pepsin ( gastric protease) p. H 2 -trypsin (pancreatic protease) p. H 8


Saturation

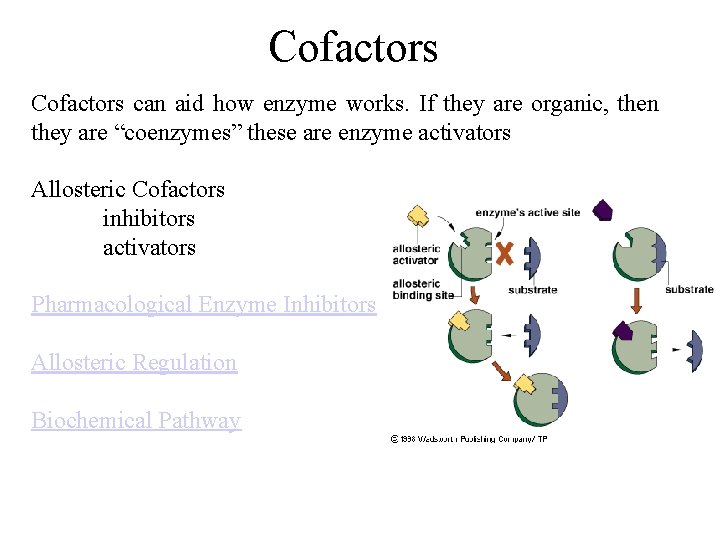
Cofactors can aid how enzyme works. If they are organic, then they are “coenzymes” these are enzyme activators Allosteric Cofactors inhibitors activators Pharmacological Enzyme Inhibitors Allosteric Regulation Biochemical Pathway

• Competitive vs Noncompetitive Inhibitors

In this animation, the enzyme is olive, the substrate is green, the competitive inhibitor is red and the products A & B are yellow and blue. The enzyme has binding site into which either the substrate or the competitive inhibitor may fit. The substrate binds to the enzyme and is then converted to the two products, A & B. The competitive inhibitor binds to active site and prevents the substrate from binding, preventing the enzyme catalyzed reaction from occuring. Sometimes the products of an enzymatic reaction can also be competitive inhibitors of the reaction. Which product, A or B, would most likely be a competitive inhibitor?

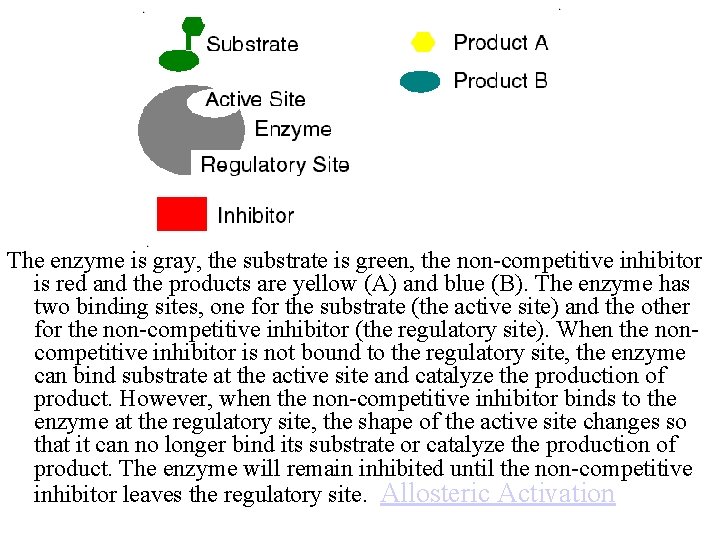
The enzyme is gray, the substrate is green, the non-competitive inhibitor is red and the products are yellow (A) and blue (B). The enzyme has two binding sites, one for the substrate (the active site) and the other for the non-competitive inhibitor (the regulatory site). When the noncompetitive inhibitor is not bound to the regulatory site, the enzyme can bind substrate at the active site and catalyze the production of product. However, when the non-competitive inhibitor binds to the enzyme at the regulatory site, the shape of the active site changes so that it can no longer bind its substrate or catalyze the production of product. The enzyme will remain inhibited until the non-competitive inhibitor leaves the regulatory site. Allosteric Activation

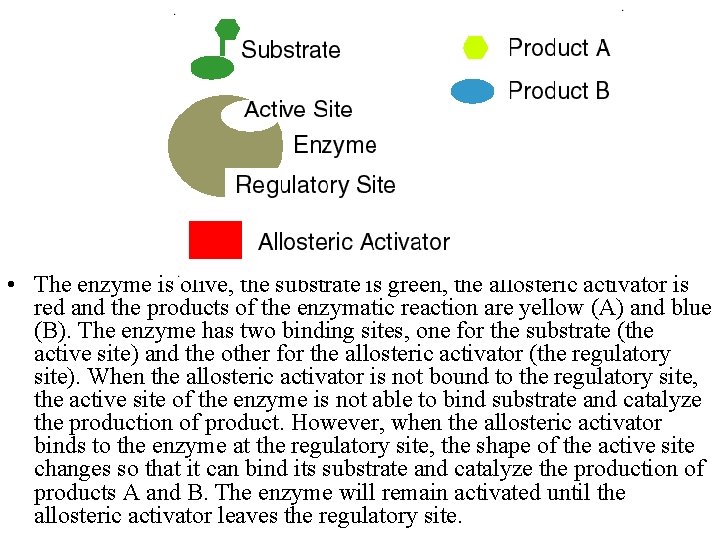
• The enzyme is olive, the substrate is green, the allosteric activator is red and the products of the enzymatic reaction are yellow (A) and blue (B). The enzyme has two binding sites, one for the substrate (the active site) and the other for the allosteric activator (the regulatory site). When the allosteric activator is not bound to the regulatory site, the active site of the enzyme is not able to bind substrate and catalyze the production of product. However, when the allosteric activator binds to the enzyme at the regulatory site, the shape of the active site changes so that it can bind its substrate and catalyze the production of products A and B. The enzyme will remain activated until the allosteric activator leaves the regulatory site.

Negative Feedback/ Feedback Inhibition When the product is in abundance, it binds competitively with its enzyme's active site; as the product is used up, inhibition is reduced and more product can be produced. In this way the concentration of the product is always controlled within a certain range. • Operons in gene regulation • Temperature regulation in animals • Plants response to water limitations

Positive Feedback • Amplification occurs when the stimulus is further activated, which in turn initiates an additional response that produces system change – Lactation in mammals – Onset of labor in childbirth – Ripening of fruit – Blood clotting – Estrogen & Progesterone in Female System

Resources • http: //www. biology. lsu. edu/introbio/tutorial/spotted/Bio. Stu dio/2 ORGANICMOLE/1 Or. Mo. html • Animations: bio. winona. edu/berg/ANIMTNS/cinhban. htm • Microbiology: http: //student. ccbcmd. edu/courses/bio 141/lecguide/unit 6/ • Bio. Topics Contents: http: //www. biotopics. co. uk/conten. html • What is an Enzyme: http: //www. northland. cc. mn. us/biology 1111/animat ions/enzyme. swf
 The four major groups of organic compounds are
The four major groups of organic compounds are Four types of organic molecules
Four types of organic molecules Where are vitamins absorbed
Where are vitamins absorbed Vitamin flowchart
Vitamin flowchart Decomposition of organic matter equation
Decomposition of organic matter equation Organic and inorganic compounds experiment
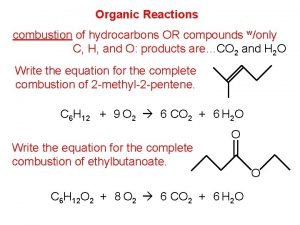
Organic and inorganic compounds experiment Organic compound combustion
Organic compound combustion Organic compound made by living things
Organic compound made by living things All organic compounds must contain the element
All organic compounds must contain the element Organic compounds must contain
Organic compounds must contain Priority order of functional group ncert
Priority order of functional group ncert Organic vs inorganic compounds
Organic vs inorganic compounds What is the difference between organic and inorganic
What is the difference between organic and inorganic Purification and characterization of organic compounds
Purification and characterization of organic compounds Organic compounds such as proteins and starches are too
Organic compounds such as proteins and starches are too Organic vs inorganic compounds
Organic vs inorganic compounds What is the classification of organic compounds
What is the classification of organic compounds Characteristics of organic compounds
Characteristics of organic compounds Principle of distillation
Principle of distillation Reactions of organic compounds
Reactions of organic compounds Organic compounds grade 10 life science
Organic compounds grade 10 life science Organic halogen compounds
Organic halogen compounds Vitamins are organic compounds
Vitamins are organic compounds Metabolic changes of drugs and related organic compounds
Metabolic changes of drugs and related organic compounds All organic compounds contain carbon and ________.
All organic compounds contain carbon and ________. Families of organic compounds
Families of organic compounds Quantitative analysis of organic compounds ppt
Quantitative analysis of organic compounds ppt What is the common name for (ch3)2choch2(ch2)3ch3?
What is the common name for (ch3)2choch2(ch2)3ch3? Dumas method
Dumas method What is cooh in chemistry
What is cooh in chemistry Vitamins are organic compounds that
Vitamins are organic compounds that What is a nucleotide
What is a nucleotide Hydrocarbon prefixes
Hydrocarbon prefixes Site:slidetodoc.com
Site:slidetodoc.com Qualitative analysis of organic functional groups
Qualitative analysis of organic functional groups Functional groups ib chemistry
Functional groups ib chemistry Organic chemistry
Organic chemistry -oate functional group
-oate functional group Soda lime formula
Soda lime formula Venn diagram ionic and covalent bonds
Venn diagram ionic and covalent bonds Body tissues worksheet
Body tissues worksheet What are the four types of author's purpose
What are the four types of author's purpose 4 major tissues
4 major tissues How are ethnic groups and religious groups related
How are ethnic groups and religious groups related Major taxonomic groups
Major taxonomic groups Major groups of microorganisms
Major groups of microorganisms Www youtube com
Www youtube com What are the major ethnic groups in the middle east
What are the major ethnic groups in the middle east Megasporen
Megasporen Four main groups of plants
Four main groups of plants Four main groups of plants
Four main groups of plants Five food groups by icmr
Five food groups by icmr Population distribution
Population distribution Four major population clusters
Four major population clusters Four major spheres of the earth
Four major spheres of the earth Who were the four major allied countries?
Who were the four major allied countries? Buffalo and toronto differ in four major ways
Buffalo and toronto differ in four major ways Four major nursing areas
Four major nursing areas