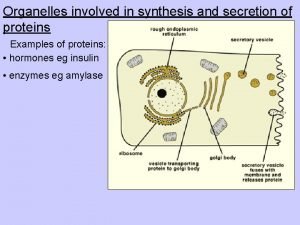
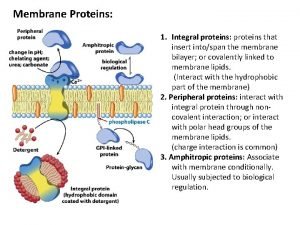
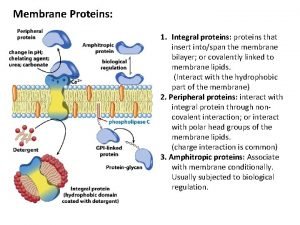
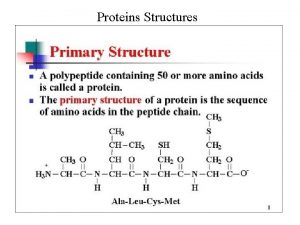
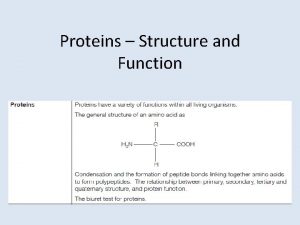
Outline n n The characters of proteins Differences
















- Slides: 67


Outline n n The characters of proteins Differences between protein chemistry & proteomics n Why to study proteome n Proteomics n Introduction to proteomics n Definitions of proteomics n The major techniques in current proteomics n Protein-protein interactions

The characters of proteins


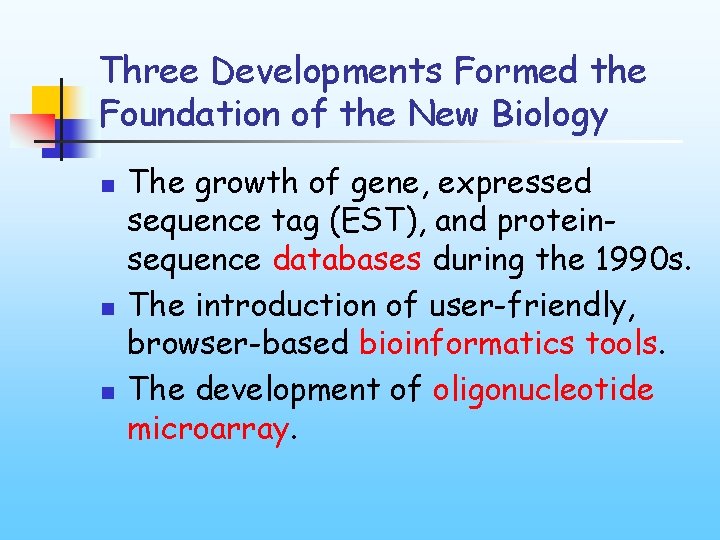
Three Developments Formed the Foundation of the New Biology n n n The growth of gene, expressed sequence tag (EST), and proteinsequence databases during the 1990 s. The introduction of user-friendly, browser-based bioinformatics tools. The development of oligonucleotide microarray.

Why to study proteome ?

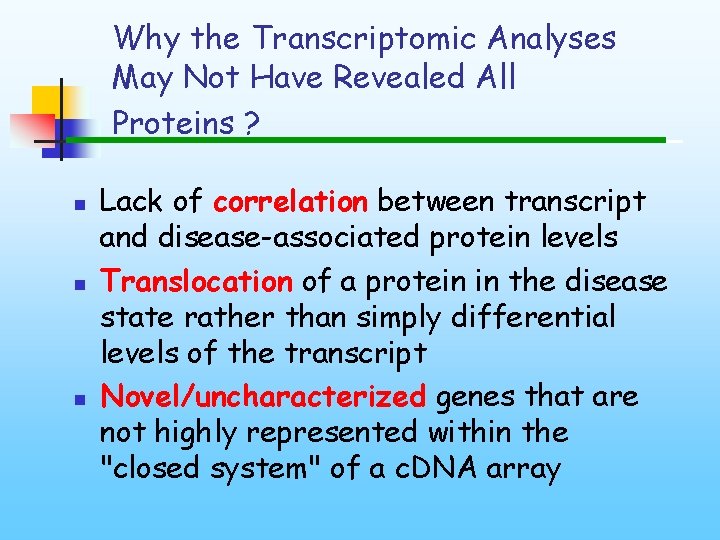
Why the Transcriptomic Analyses May Not Have Revealed All Proteins ? n n n Lack of correlation between transcript and disease-associated protein levels Translocation of a protein in the disease state rather than simply differential levels of the transcript Novel/uncharacterized genes that are not highly represented within the "closed system" of a c. DNA array

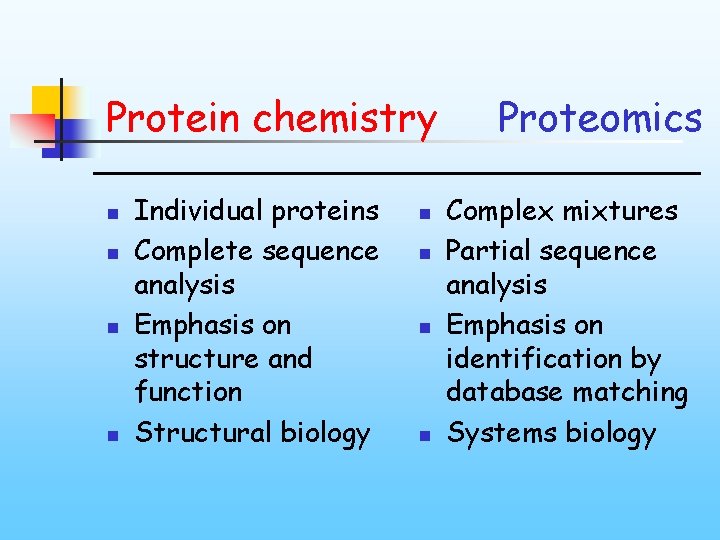
Protein chemistry n n Individual proteins Complete sequence analysis Emphasis on structure and function Structural biology n n Proteomics Complex mixtures Partial sequence analysis Emphasis on identification by database matching Systems biology

Introduction To Proteomics

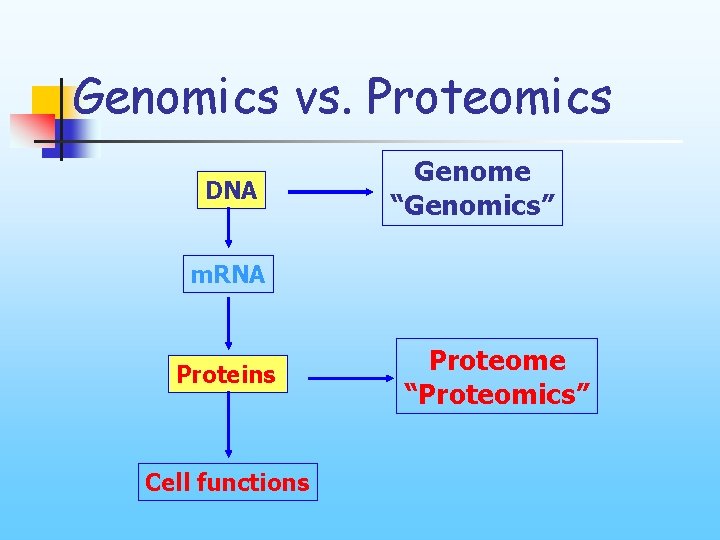
Genomics vs. Proteomics DNA Genome “Genomics” m. RNA Proteins Cell functions Proteome “Proteomics”

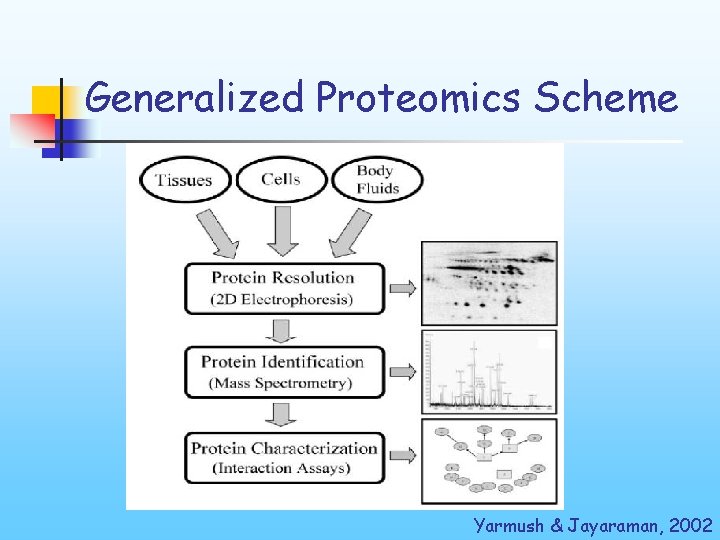
Generalized Proteomics Scheme Yarmush & Jayaraman, 2002

Definitions of Proteomics

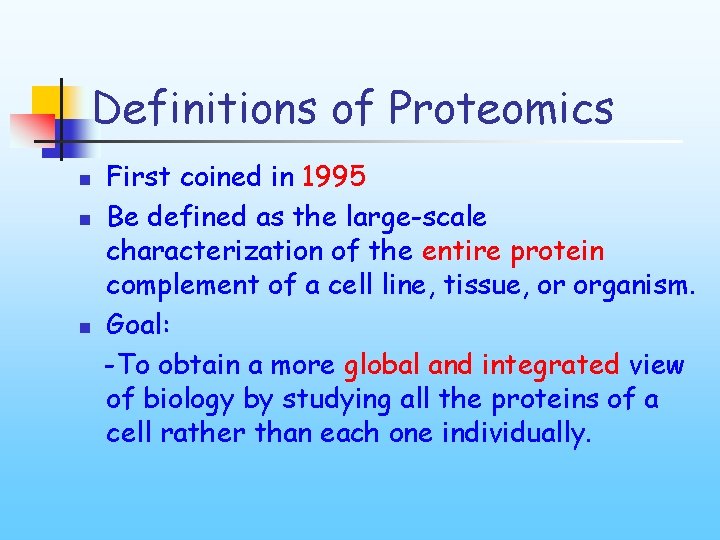
Definitions of Proteomics n n n First coined in 1995 Be defined as the large-scale characterization of the entire protein complement of a cell line, tissue, or organism. Goal: -To obtain a more global and integrated view of biology by studying all the proteins of a cell rather than each one individually.

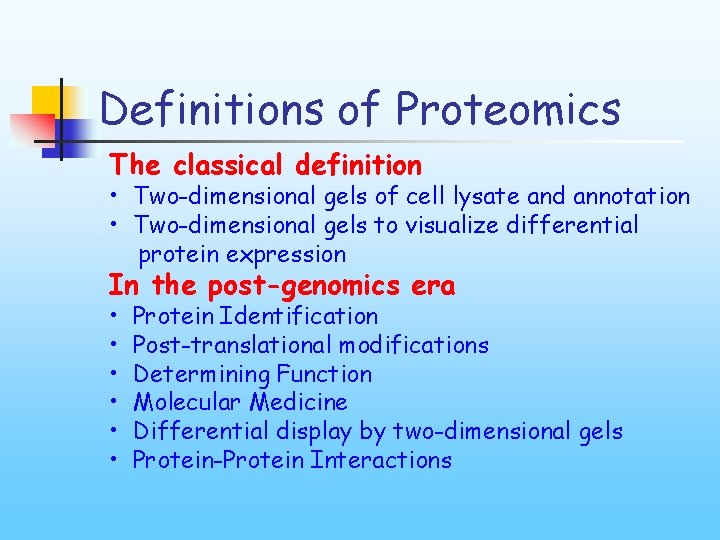
Definitions of Proteomics The classical definition • Two-dimensional gels of cell lysate and annotation • Two-dimensional gels to visualize differential protein expression In the post-genomics era • • • Protein Identification Post-translational modifications Determining Function Molecular Medicine Differential display by two-dimensional gels Protein-Protein Interactions

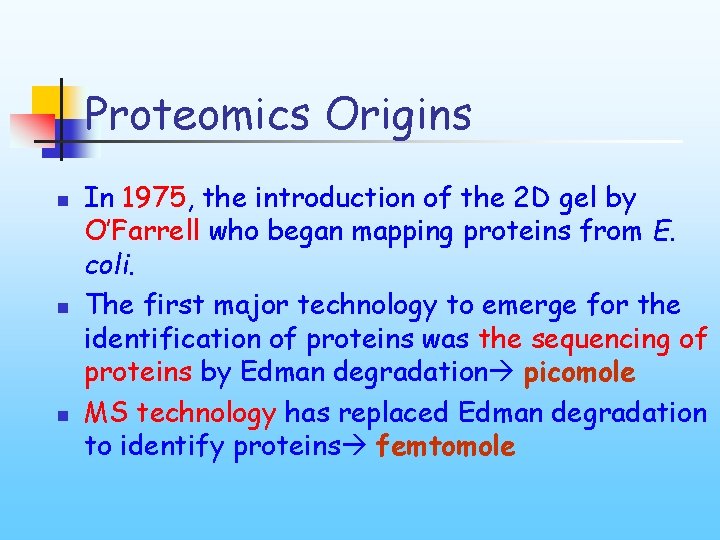
Proteomics Origins n n n In 1975, the introduction of the 2 D gel by O’Farrell who began mapping proteins from E. coli. The first major technology to emerge for the identification of proteins was the sequencing of proteins by Edman degradation picomole MS technology has replaced Edman degradation to identify proteins femtomole

How Proteomics Can Help Drug Development http: //www. sciam. com. tw/readshow. asp? FDoc. No=63&CL=18

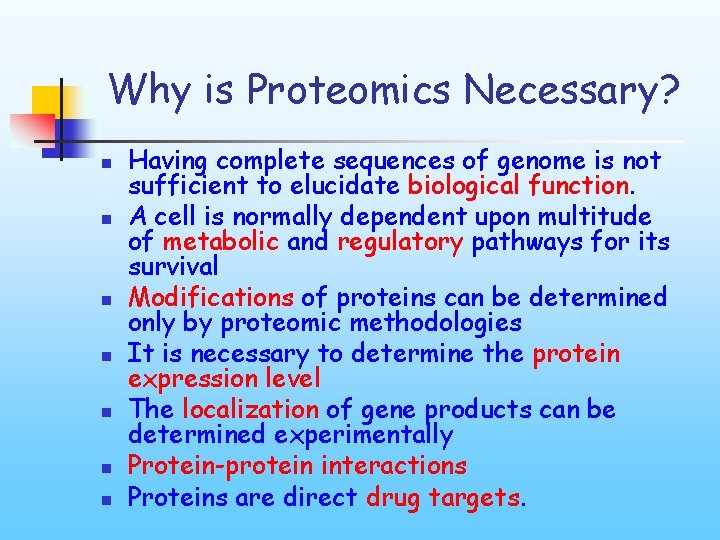
Why is Proteomics Necessary? n n n n Having complete sequences of genome is not sufficient to elucidate biological function. A cell is normally dependent upon multitude of metabolic and regulatory pathways for its survival Modifications of proteins can be determined only by proteomic methodologies It is necessary to determine the protein expression level The localization of gene products can be determined experimentally Protein-protein interactions Proteins are direct drug targets.


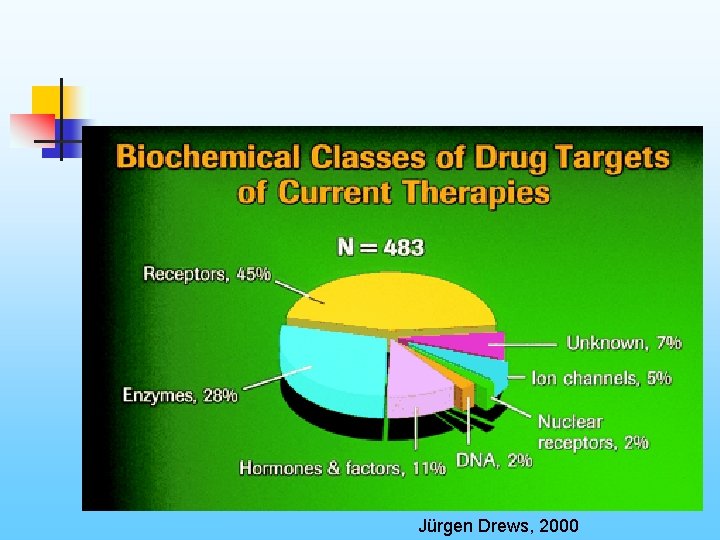
Jürgen Drews, 2000

Amgen(Applied Molecular Genetics) 成立日期: 1980 年 4 月8 日 CEO:Kevin W. Sharer 員 人數: 6342 市場總值: 698. 4 億美元 產品項目:重組蛋白藥物 EPOGENR (Epoetin alfa) NEUPOGENR (Filgrastim) INFERGENR (Interferon alfacon-1) 資料來源:彭博資訊社、Zacks. com,6/14/2001


The Major Techniques in Current Proteomics

The Major Techniques in Current Proteomics n Two-dimensional electrophoresis n n n Mass Spectrometry n n n IEF strip separation SDS-PAGE gel separation Protein sequencing Peptide mapping Others n n n ICAT Yeast two hybrid assay Protein chips

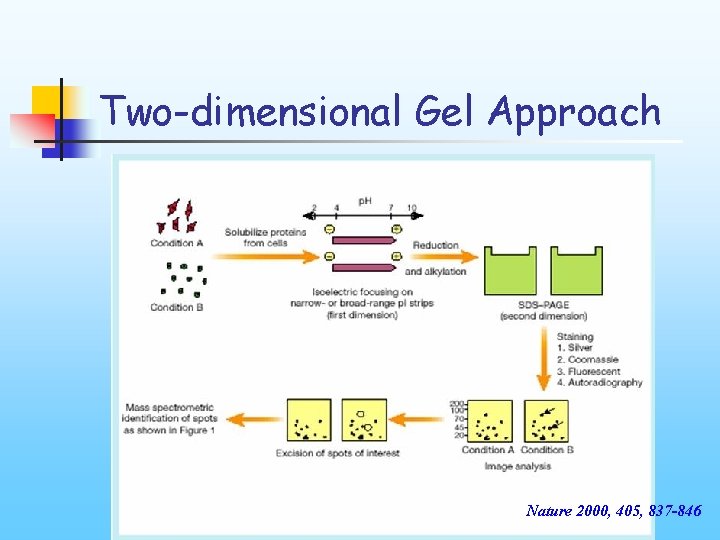
Two-dimensional Gel Approach Nature 2000, 405, 837 -846

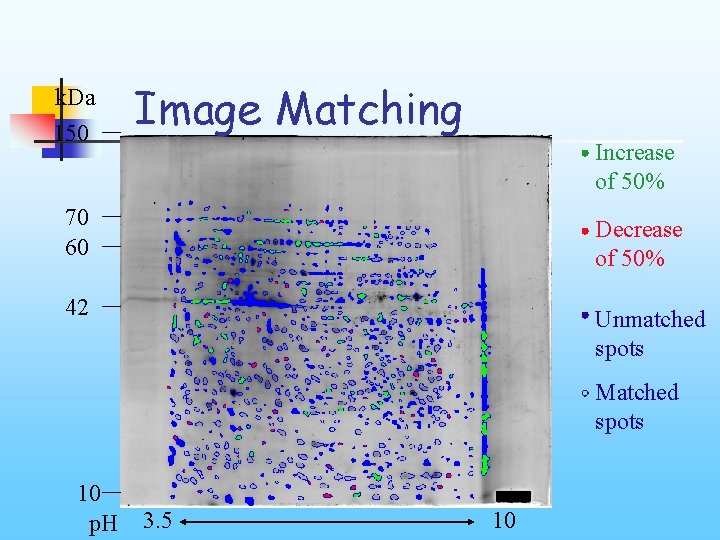
k. Da 150 Image Matching Increase of 50% 70 60 Decrease of 50% 42 Unmatched spots Matched spots 10 p. H 3. 5 10

www. expasy. ch/ch 2 d


http: //www. expasy. ch/melanie/


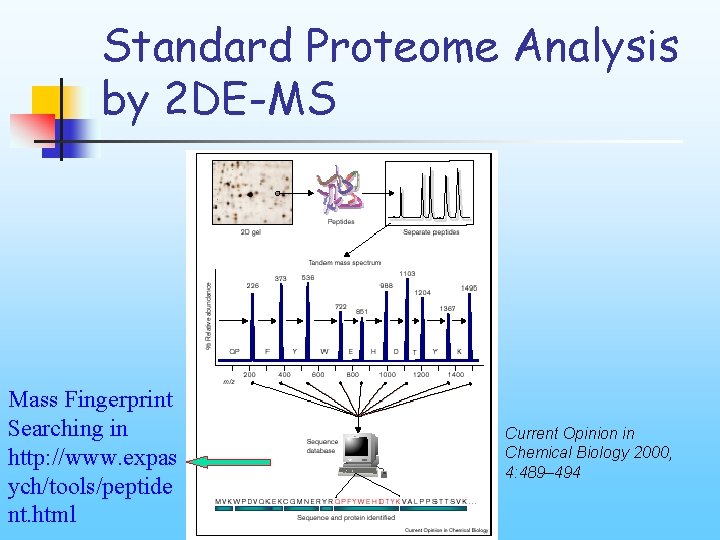
Standard Proteome Analysis by 2 DE-MS Mass Fingerprint Searching in http: //www. expas ych/tools/peptide nt. html Current Opinion in Chemical Biology 2000, 4: 489– 494

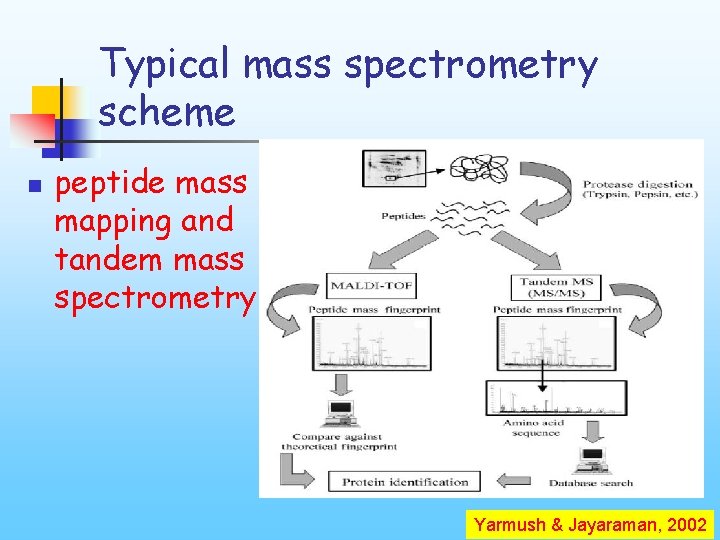
Typical mass spectrometry scheme n peptide mass mapping and tandem mass spectrometry Yarmush & Jayaraman, 2002

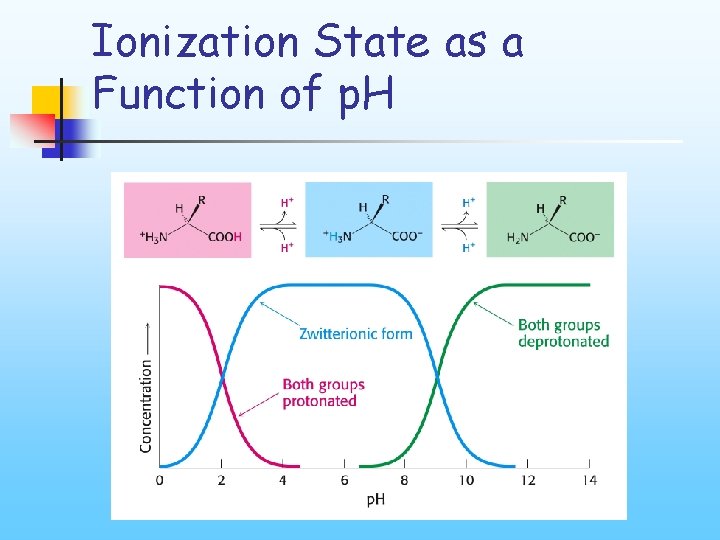
Ionization State as a Function of p. H

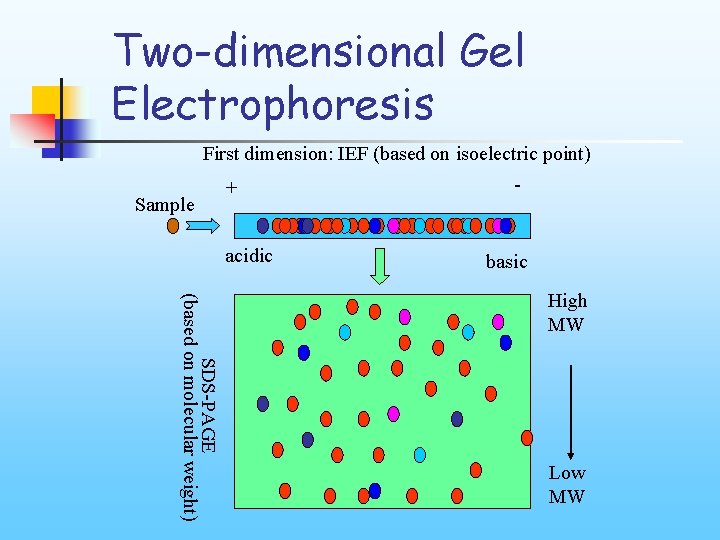
Two-dimensional Gel Electrophoresis First dimension: IEF (based on isoelectric point) Sample + acidic - basic SDS-PAGE (based on molecular weight) High MW Low MW

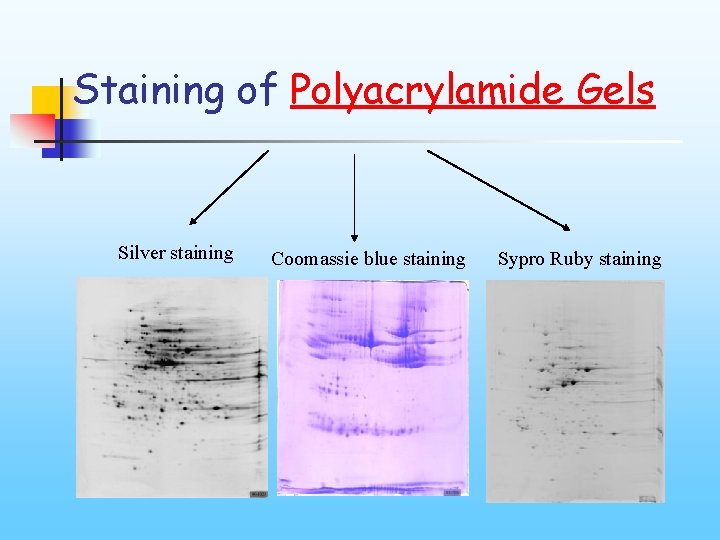
Staining of Polyacrylamide Gels Silver staining Coomassie blue staining Sypro Ruby staining

Image Analysis

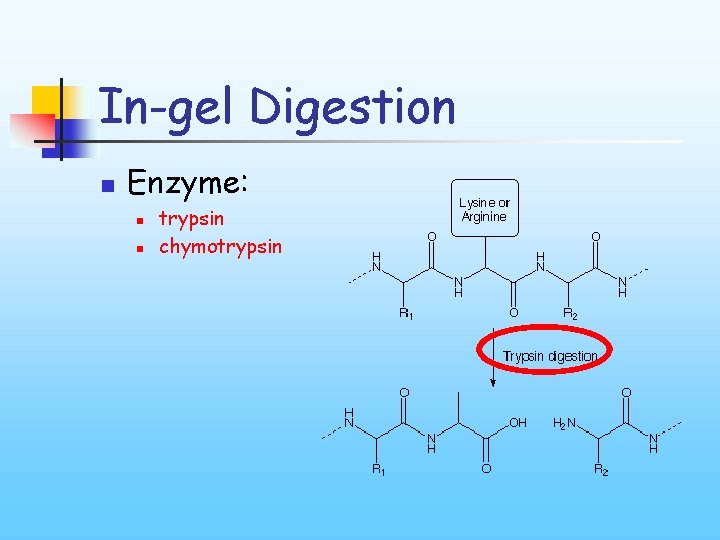
In-gel Digestion n Enzyme: n n trypsin chymotrypsin

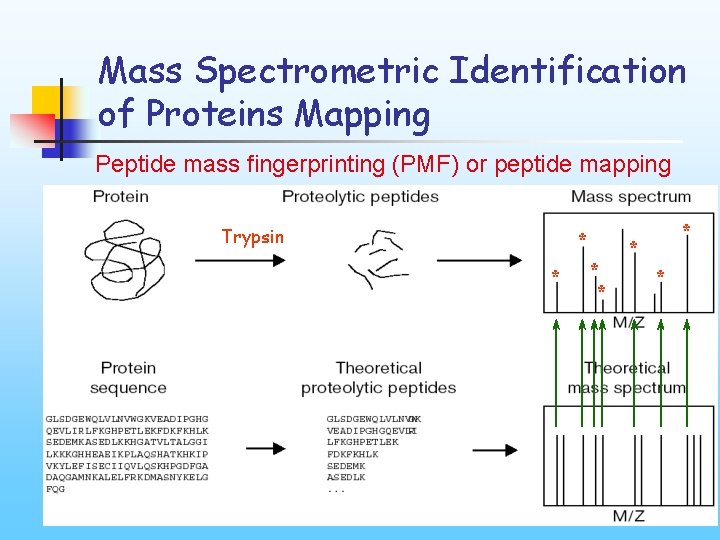
Mass Spectrometric Identification of Proteins Mapping Peptide mass fingerprinting (PMF) or peptide mapping Trypsin * * * *

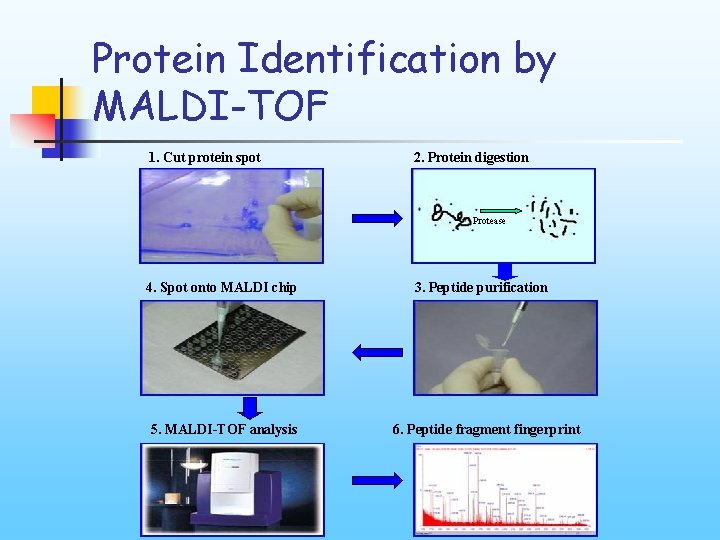
Protein Identification by MALDI-TOF 1. Cut protein spot 2. Protein digestion Protease 4. Spot onto MALDI chip 5. MALDI-TOF analysis 3. Peptide purification 6. Peptide fragment fingerprint

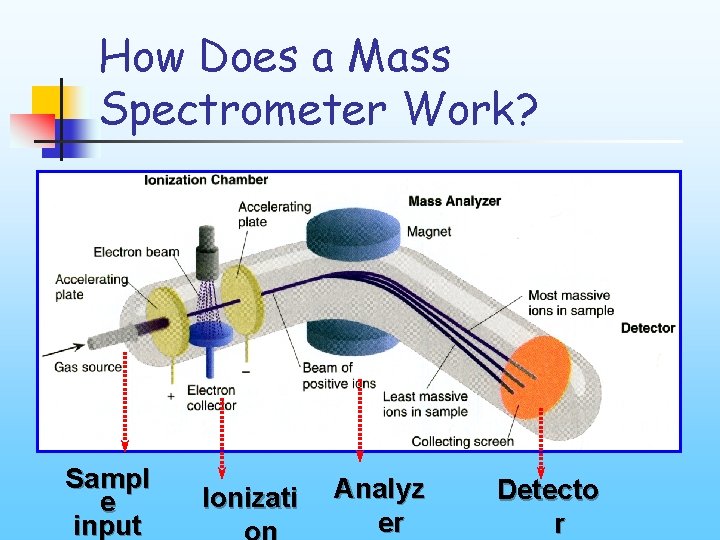
How Does a Mass Spectrometer Work? Sampl e input Ionizati Analyz er Detecto r

How Does a Mass Spectrometer Work? • Sample Input: Gas Chromatography (GC), Liquid Chromatography (LC), Capillary Electrophoresis (CE), Solid crystal etc. • Ionization: Electrospray, Matrix-assisted Laser Desorption/Ionization (MALDI) etc • Analysis: quadrupole, time of flight(TOF), ion trap etc.

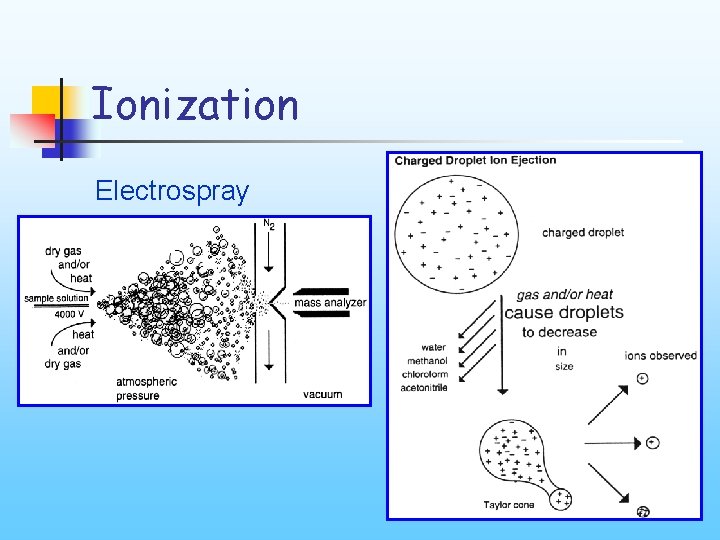
Ionization Electrospray

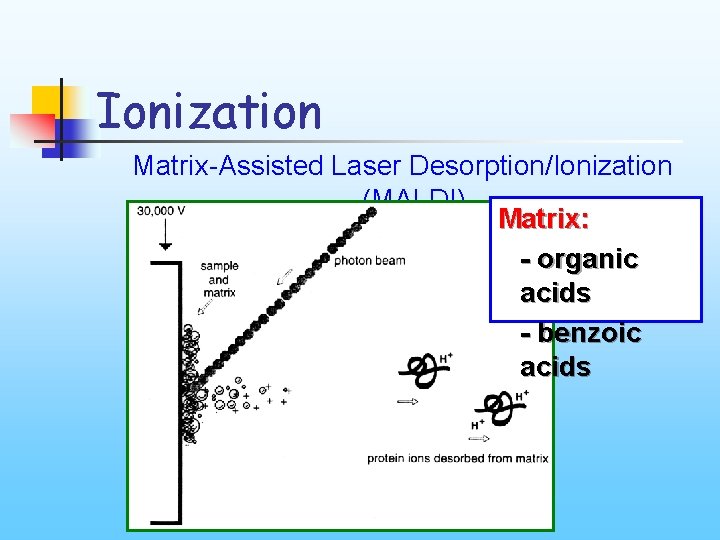
Ionization Matrix-Assisted Laser Desorption/Ionization (MALDI) Matrix: - organic acids - benzoic acids

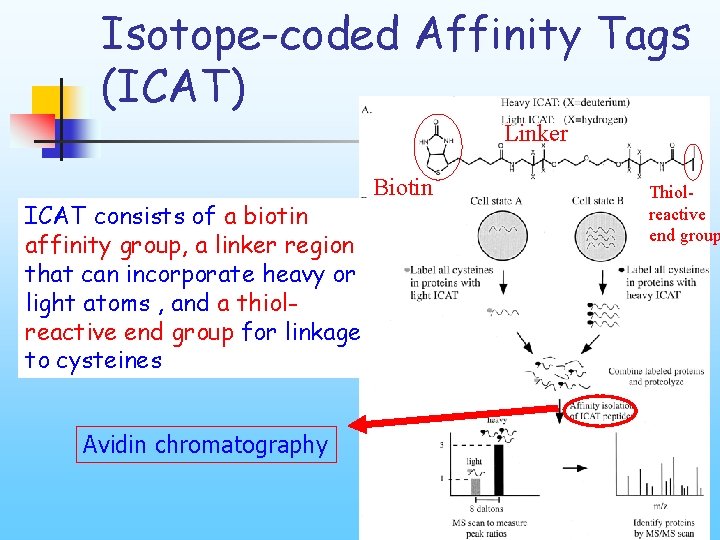
Isotope-coded Affinity Tags (ICAT) Linker ICAT consists of a biotin affinity group, a linker region that can incorporate heavy or light atoms , and a thiolreactive end group for linkage to cysteines Avidin chromatography Biotin Thiolreactive end group

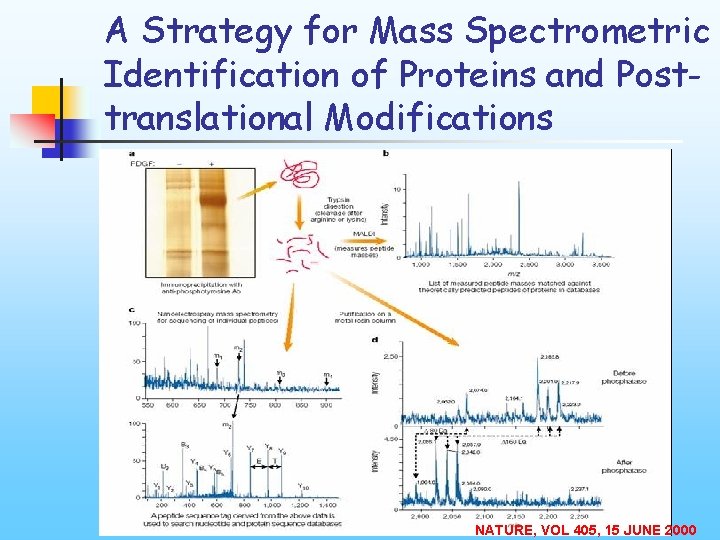
A Strategy for Mass Spectrometric Identification of Proteins and Posttranslational Modifications NATURE, VOL 405, 15 JUNE 2000

Proteome chip ‘proteome chip’ composed of 6, 566 protein samples representing 5, 800 unique proteins, which are spotted in duplicate on a single nickelcoated glass microscope slide 39. The immobilized GST fusion proteins were detected using a labeled antibody against GST. (Mac. Beath G. Nat Genet 2002 Dec; 32 Suppl 2: 526 -32 )

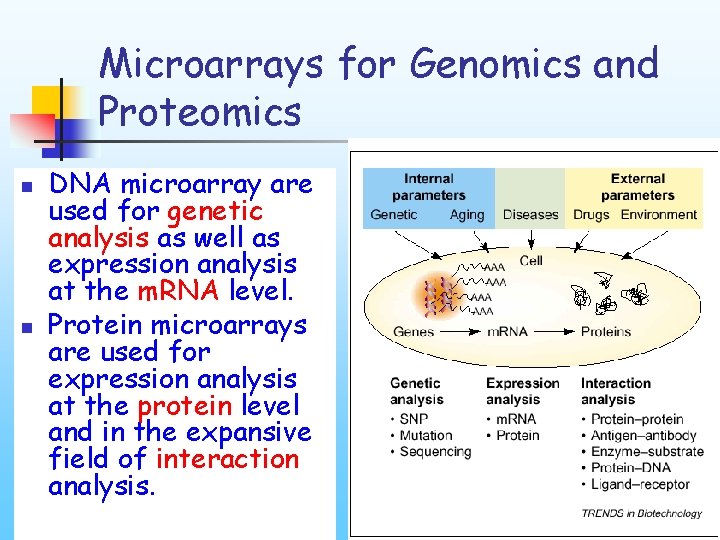
Microarrays for Genomics and Proteomics n n DNA microarray are used for genetic analysis as well as expression analysis at the m. RNA level. Protein microarrays are used for expression analysis at the protein level and in the expansive field of interaction analysis.

Protein Microarrays In Medical Research n n n Accelerate immune diagnostics. The reduction of sample volume----the analysis of multiple tumor markers from a minimun amount of biopsy material. New possibilities for patient monitoring during disease treatment and therapy will be develpoed based on this emerging technology.

Clinical and Biomedical Applications of Proteomics n n An approach complementary to genomics is required in clinical situations to better understand epigenetic regulation and get closer to a "holisitic" medical approach. The potential clinical applications of 2 -D PAGE, especially to the analysis of body fluids and tissue biopsies. n n n Identifying the origin of body fluid samples or the origin of a tissue biopsy. Analyzing protein phenotypes and protein posttranslational modifications in fluid, cells, or tissues. Examining the clonality of immunoglobulins and detecting clones which are not seen with conventional techniques. Monitoring disease processes and protein expression. Discovering new disease markers and/or patterns in body fluids, cells, or tissues.

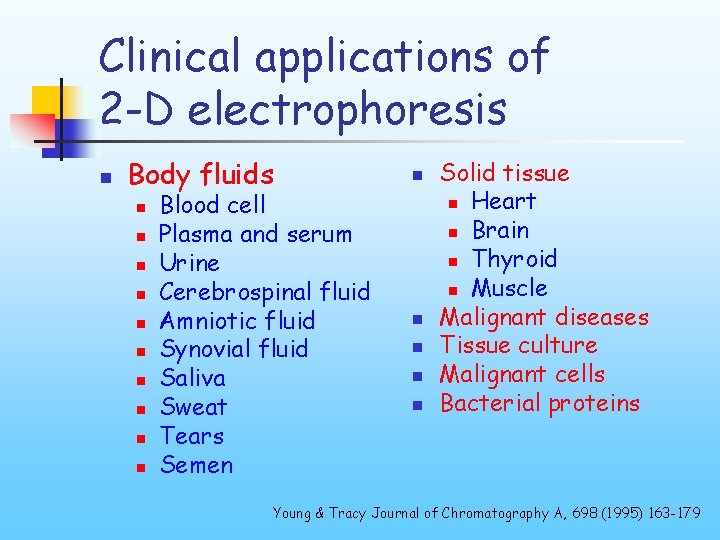
Clinical applications of 2 -D electrophoresis n Body fluids n n n n n Blood cell Plasma and serum Urine Cerebrospinal fluid Amniotic fluid Synovial fluid Saliva Sweat Tears Semen n n Solid tissue n Heart n Brain n Thyroid n Muscle Malignant diseases Tissue culture Malignant cells Bacterial proteins Young & Tracy Journal of Chromatography A, 698 (1995) 163 -179

Protein-protein Interactions

Protein-protein Interactions n n n Introduction Mass Spectrometry Yeast Two-hybrid Assay

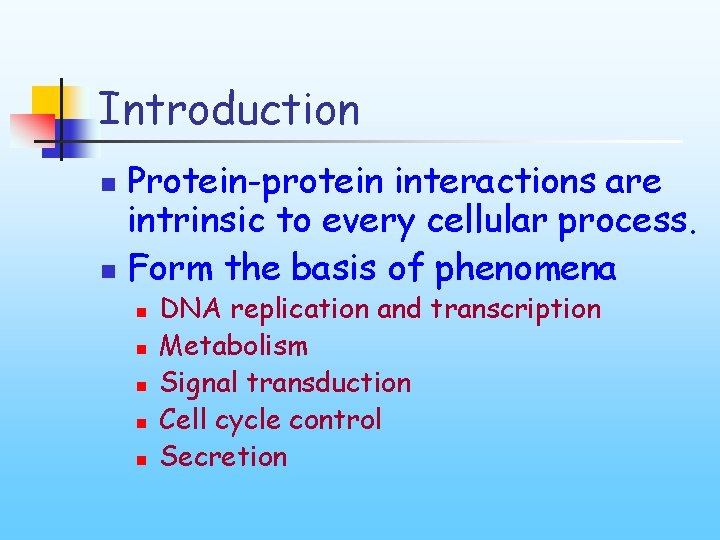
Introduction Protein-protein interactions are intrinsic to every cellular process. n Form the basis of phenomena n n n DNA replication and transcription Metabolism Signal transduction Cell cycle control Secretion

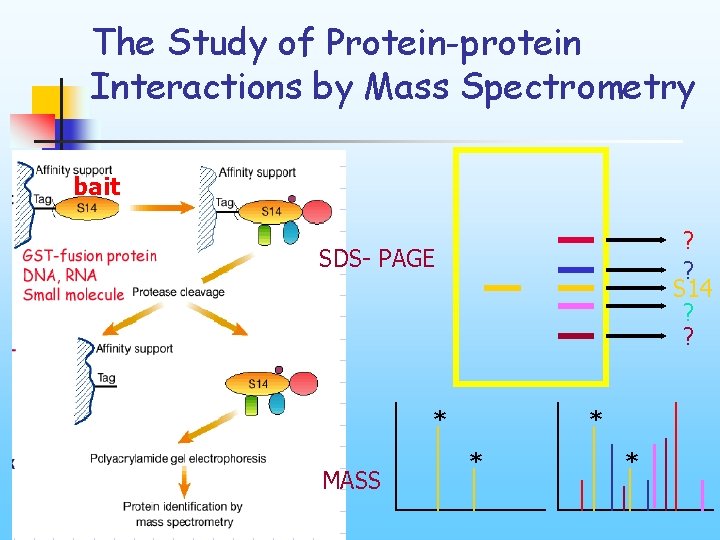
The Study of Protein-protein Interactions by Mass Spectrometry bait ? ? S 14 ? ? SDS- PAGE * MASS * * *

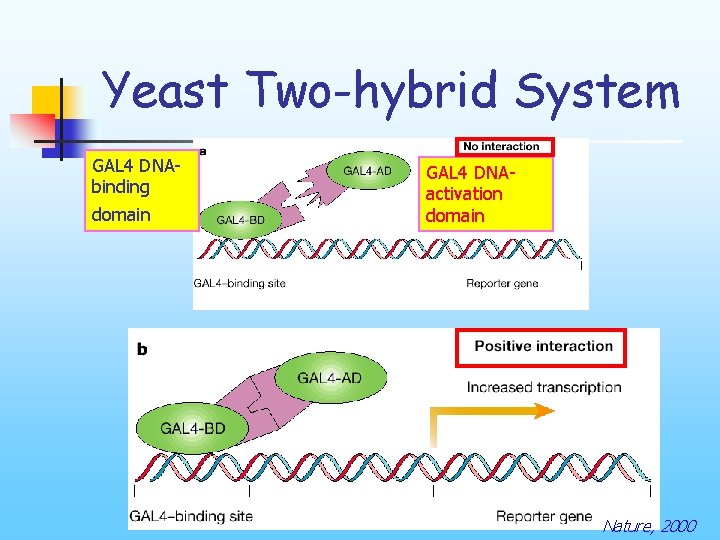
Yeast Two-hybrid System n n Useful in the study of various interactions The technology was originally developed during the late 1980's in the laboratory Dr. Stanley Fields (see Fields and Song, 1989, Nature).

Yeast Two-hybrid System GAL 4 DNAbinding domain GAL 4 DNAactivation domain Nature, 2000

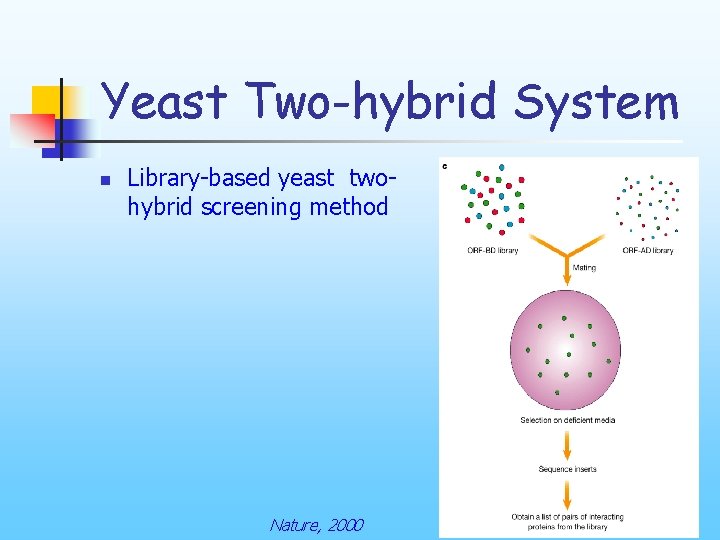
Yeast Two-hybrid System n Library-based yeast twohybrid screening method Nature, 2000

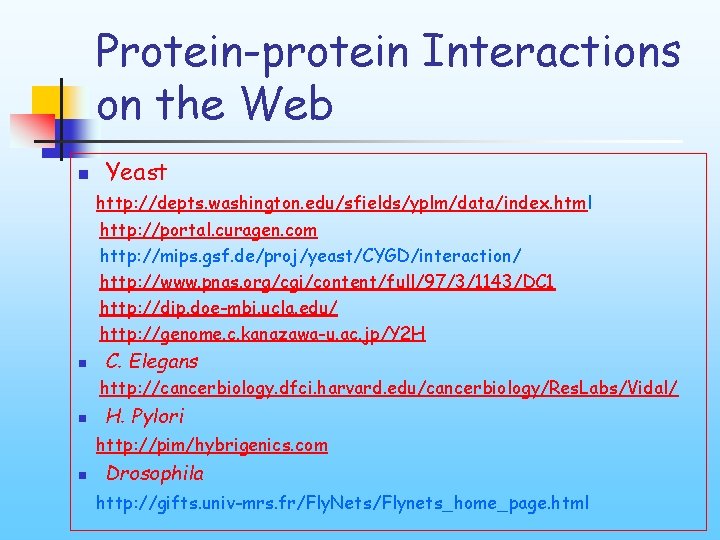
Protein-protein Interactions on the Web n Yeast http: //depts. washington. edu/sfields/yplm/data/index. html http: //portal. curagen. com http: //mips. gsf. de/proj/yeast/CYGD/interaction/ http: //www. pnas. org/cgi/content/full/97/3/1143/DC 1 http: //dip. doe-mbi. ucla. edu/ http: //genome. c. kanazawa-u. ac. jp/Y 2 H n C. Elegans http: //cancerbiology. dfci. harvard. edu/cancerbiology/Res. Labs/Vidal/ n H. Pylori http: //pim/hybrigenics. com n Drosophila http: //gifts. univ-mrs. fr/Fly. Nets/Flynets_home_page. html

Pathway Software BIOCARTA http: //biocarta. com/ Browse all pathway

Pathway Software BIOCARTA

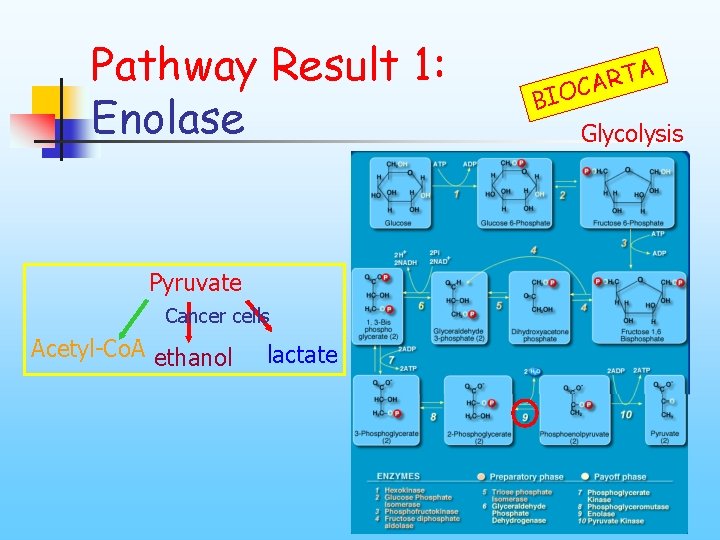
Pathway Result 1: Enolase Pyruvate Cancer cells Acetyl-Co. A ethanol lactate A T R CA BIO Glycolysis

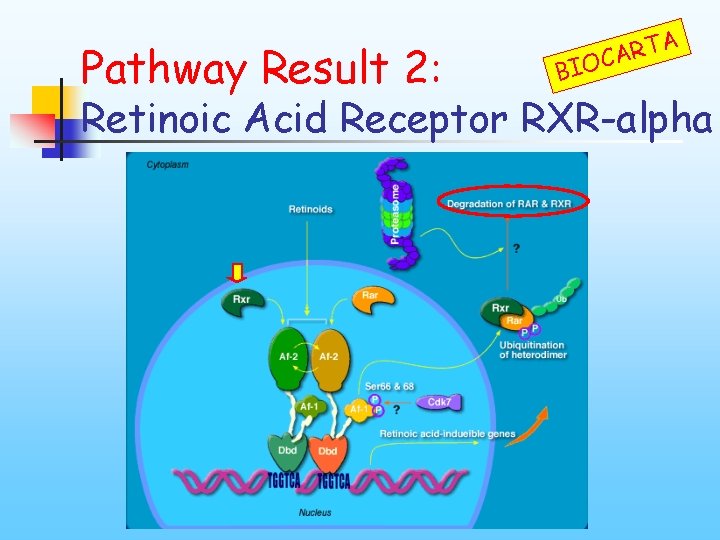
Pathway Result 2: BIO A T R CA Retinoic Acid Receptor RXR-alpha

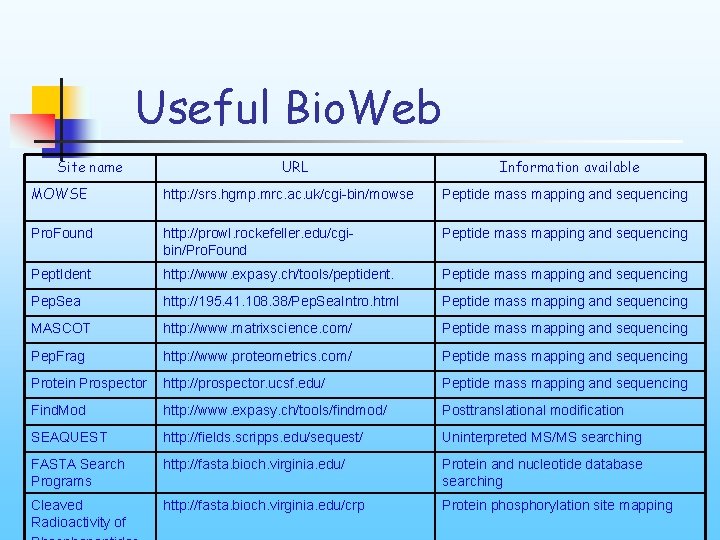
Useful Bio. Web Site name URL Information available MOWSE http: //srs. hgmp. mrc. ac. uk/cgi-bin/mowse Peptide mass mapping and sequencing Pro. Found http: //prowl. rockefeller. edu/cgibin/Pro. Found Peptide mass mapping and sequencing Pept. Ident http: //www. expasy. ch/tools/peptident. Peptide mass mapping and sequencing Pep. Sea http: //195. 41. 108. 38/Pep. Sea. Intro. html Peptide mass mapping and sequencing MASCOT http: //www. matrixscience. com/ Peptide mass mapping and sequencing Pep. Frag http: //www. proteometrics. com/ Peptide mass mapping and sequencing Protein Prospector http: //prospector. ucsf. edu/ Peptide mass mapping and sequencing Find. Mod http: //www. expasy. ch/tools/findmod/ Posttranslational modification SEAQUEST http: //fields. scripps. edu/sequest/ Uninterpreted MS/MS searching FASTA Search Programs http: //fasta. bioch. virginia. edu/ Protein and nucleotide database searching Cleaved Radioactivity of http: //fasta. bioch. virginia. edu/crp Protein phosphorylation site mapping

Major Directions in Coming Proteomics n n Chemical proteomics (screens for activity and binding) Structural proteomics (target validation and development) Interaction proteomics (identification of new protein targets) Bioinformatics (annotation of the proteome)

Major Directions in Coming Proteomics n n Protein structure prediction and modeling Assignment of protein structure to genomes Classifications of protein structures Drug discovery and development

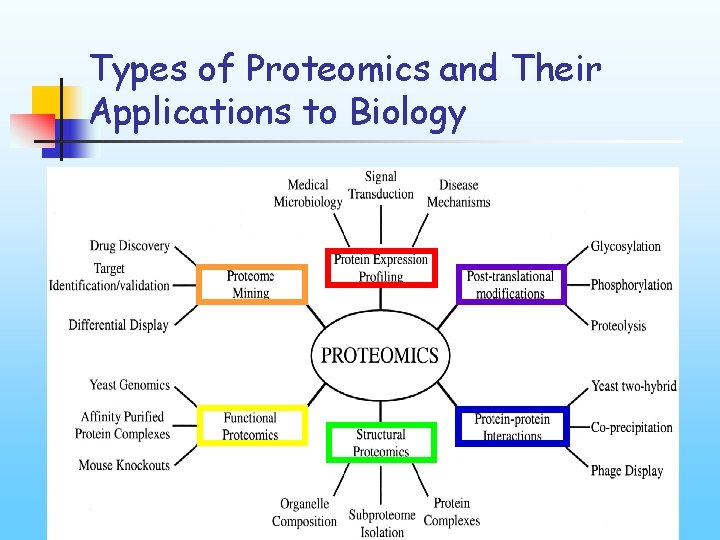
Types of Proteomics and Their Applications to Biology

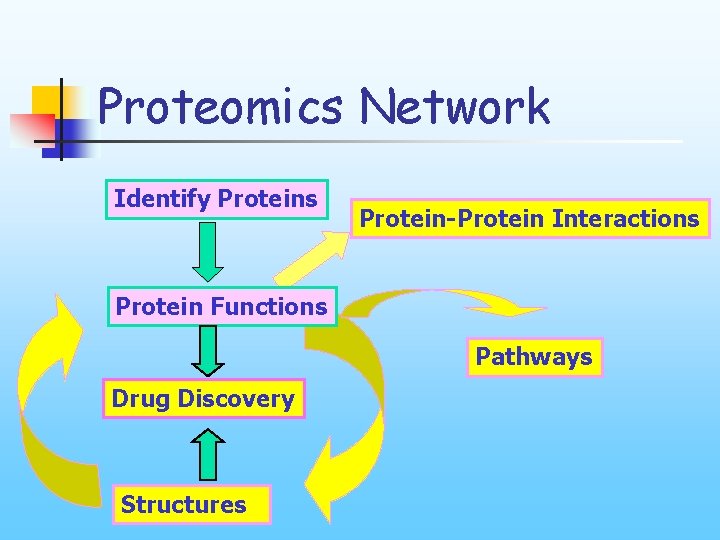
Proteomics Network Identify Proteins Protein-Protein Interactions Protein Functions Pathways Drug Discovery Structures

Thank You!
 Quotation sandwich
Quotation sandwich Flat characters in hamlet
Flat characters in hamlet Protein metabolism notes
Protein metabolism notes Three peptide chains woven like a rope
Three peptide chains woven like a rope Transmembrane
Transmembrane What is protein
What is protein Section 8-1 carbohydrates fats and proteins answer key
Section 8-1 carbohydrates fats and proteins answer key Ribosome factory part or worker
Ribosome factory part or worker Fibrous function
Fibrous function Qualitative test for proteins
Qualitative test for proteins What role does the ribosome play in assembling proteins?
What role does the ribosome play in assembling proteins? Glycoprotein vs glycolipid
Glycoprotein vs glycolipid Amphoteric proteins
Amphoteric proteins Food sources of nucleic acids
Food sources of nucleic acids Where are proteins found
Where are proteins found Section 8-1 carbohydrates fats and proteins answer key
Section 8-1 carbohydrates fats and proteins answer key E.r function
E.r function Higher order structure of proteins
Higher order structure of proteins What are derived proteins
What are derived proteins Salting out proteins
Salting out proteins Functions of muscle tissue
Functions of muscle tissue Exocytosis definition biology
Exocytosis definition biology Alkaloidal reagents denature proteins
Alkaloidal reagents denature proteins Protein pump vs protein channel
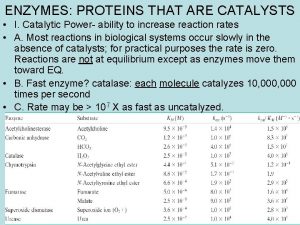
Protein pump vs protein channel Proteins with catalytic power are called
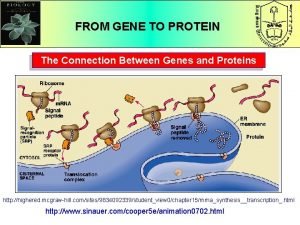
Proteins with catalytic power are called What is the connection between genes and proteins
What is the connection between genes and proteins Functions of plasma proteins
Functions of plasma proteins Hierarchy of protein structure
Hierarchy of protein structure Translation refers to the
Translation refers to the What is protien
What is protien How do proteins fold
How do proteins fold Hbv and lbv
Hbv and lbv Peripheral proteins
Peripheral proteins Not all enzymes are proteins
Not all enzymes are proteins Salting in
Salting in Does exocytosis require energy
Does exocytosis require energy The smallest living unit within the human body is
The smallest living unit within the human body is Protein domains and motifs
Protein domains and motifs Proteins are divided into two groups
Proteins are divided into two groups Describe a picture
Describe a picture Amphoteric proteins
Amphoteric proteins Ribosomal proteins
Ribosomal proteins Alhusseini
Alhusseini Protein elements
Protein elements Structural role of proteins
Structural role of proteins Cell membrane function
Cell membrane function Monomers protein
Monomers protein Biomedical importance of protein
Biomedical importance of protein Carbohydrates in milk
Carbohydrates in milk Precipitation of proteins by strong mineral acids
Precipitation of proteins by strong mineral acids Globular vs fibrous proteins
Globular vs fibrous proteins Has passageways that carry proteins
Has passageways that carry proteins Metodo di lowry
Metodo di lowry Structural proteins function
Structural proteins function Diabetic diet chart
Diabetic diet chart Protein polymer example
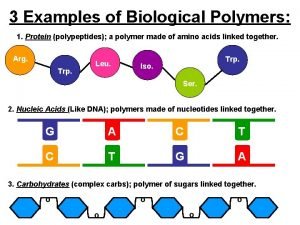
Protein polymer example Protein hydrolysis
Protein hydrolysis Biological significance of protein
Biological significance of protein Protein databas
Protein databas Three main types of rna
Three main types of rna Euchromatin
Euchromatin Salting out proteins
Salting out proteins Building blocks of protein
Building blocks of protein Non collagenous proteins of periodontium
Non collagenous proteins of periodontium Ups trucks means which organelle?
Ups trucks means which organelle? Cummings
Cummings Facts about proteins biology
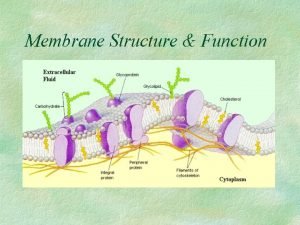
Facts about proteins biology Membrane synthesis
Membrane synthesis