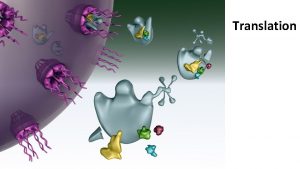
Translation refers to the process of creating proteins





























- Slides: 37

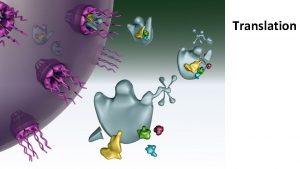
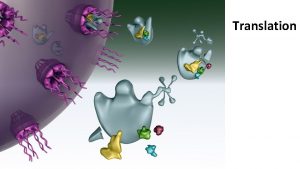
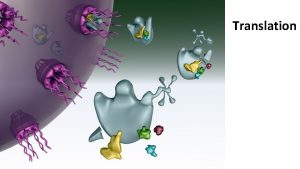
Translation refers to the process of creating proteins from an m. RNA template. The sequence of nucleotides on the RNA is translated into the amino acid sequence of proteins and this reaction is carried out by ribosomes.

Genetic code When nucleic acids were discovered as the primary genetic material, one important question came to the fore. There are only 4 bases in nucleic acids whereas proteins are made of 20 amino acids.

Therefore, it is not possible to have a direct one-to-one correlation between the sequences of nucleotides and amino acids. Even having two nucleotides code for a single amino acid is insufficient.

Therefore, it was suggested that amino acids were coded by stretches of three nucleotides called codons. A series of experiments in the 1960 s confirmed this hypothesis and also showed that these codons do not overlap with each other. In addition, since stretches of 3 nucleotides can give rise to a total of 64 codons, a single amino acid can be coded by multiple codons, a property that is called ‘degeneracy’.

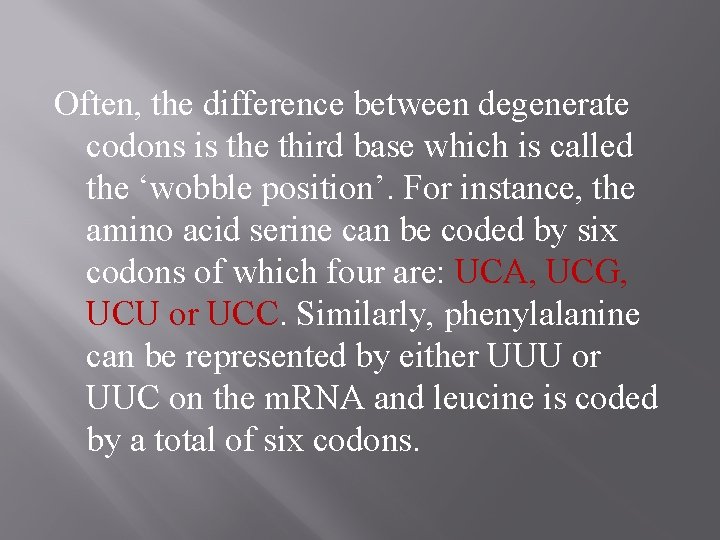
Often, the difference between degenerate codons is the third base which is called the ‘wobble position’. For instance, the amino acid serine can be coded by six codons of which four are: UCA, UCG, UCU or UCC. Similarly, phenylalanine can be represented by either UUU or UUC on the m. RNA and leucine is coded by a total of six codons.

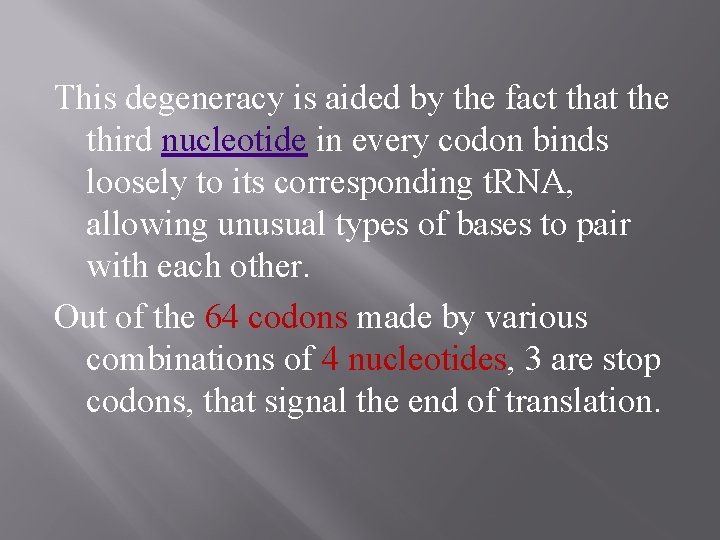
This degeneracy is aided by the fact that the third nucleotide in every codon binds loosely to its corresponding t. RNA, allowing unusual types of bases to pair with each other. Out of the 64 codons made by various combinations of 4 nucleotides, 3 are stop codons, that signal the end of translation.

These are UAA, UAG and UGA and are recognized by proteins called release factors rather than by t. RNAs. When a ribosome encounters a stop codon, it dissociates from the m. RNA through the enzymatic action of release factors.


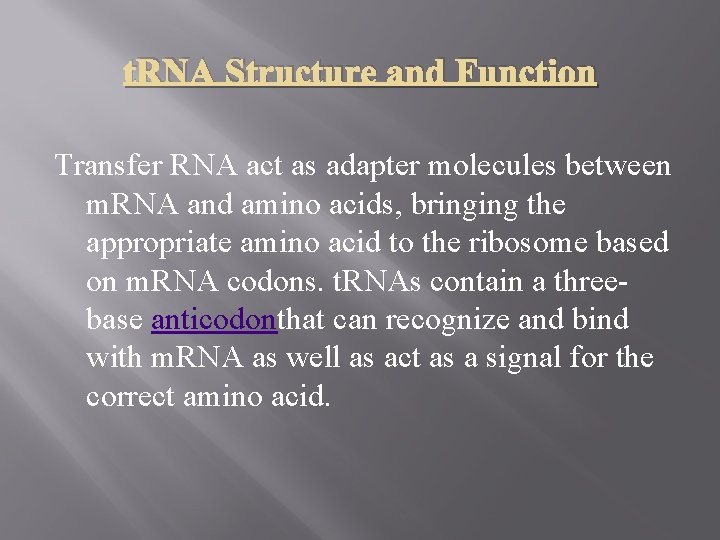
t. RNA Structure and Function Transfer RNA act as adapter molecules between m. RNA and amino acids, bringing the appropriate amino acid to the ribosome based on m. RNA codons. t. RNAs contain a threebase anticodonthat can recognize and bind with m. RNA as well as act as a signal for the correct amino acid.

The anticodon sequence is complementary to the m. RNA codon, and runs in an antiparallel direction, allowing the two molecules to base pair with each other.

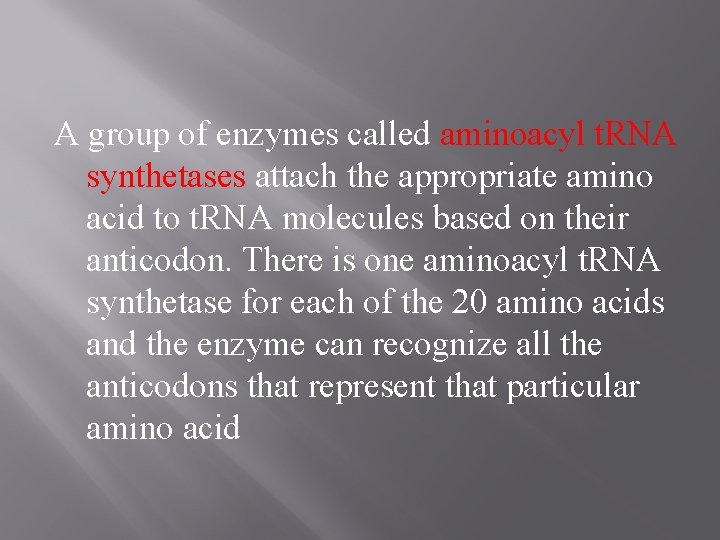
A group of enzymes called aminoacyl t. RNA synthetases attach the appropriate amino acid to t. RNA molecules based on their anticodon. There is one aminoacyl t. RNA synthetase for each of the 20 amino acids and the enzyme can recognize all the anticodons that represent that particular amino acid

These enzymes use the energy from ATP to attach the amino acid to the last nucleotide on the 3′ end of t. RNA. The t. RNA is now considered to be ‘charged’ and can participate in the protein synthesizing reactions on the ribosome.

Ribosome Structure and Function Ribosomes are macromolecular, multisubunit structures containing RNA as well as protein and are the primary machines that drive protein synthesis.

The structure of the ribosome derives primarily from it RNA component (ribosomal RNA or r. RNA) and basepairing with m. RNA and t. RNA is crucial to its function.

The ribosome contains two subunits and translation is initiated when the smaller subunit binds to sequences upstream of the coding sequence on the m. RNA.

Prokaryotic translation begins with the r. RNA directly binding to the m. RNA, whereas eukaryotic translation involves other proteins called initiation factors.

The smaller subunit, along with some other proteins recruit the larger subunit of the ribosome, and translation begins.

Primarily, the ribosome contains three important regions – the P site, the A site and the E site – formed by the threedimensional shape of the r. RNA

The P site binds to the growing polypeptide, the A site anchors an incoming charged t. RNA and after peptide bond formation, the t. RNA binds briefly to the E site before leaving the ribosome.


Translation Mechanism Translation proceeds in three stages – initiation, elongation and termination. Each of these is associated with different proteins and at every step, ATP and GTP are used as energy sources.

A single m. RNA can be translated by multiple ribosomes in a process called translatome. These complexes were initially called ergosomes and are now called polysomes or polyribosomes.

STEPS 1. 2. 3. Initiation Elongation Termination

Initiation Translation begins with the mature m. RNA transcript being exported from the nucleus and its 5′ cap being recognized by the smaller subunit of a ribosome. The ribosomal subunit, along with a special t. RNA, scans the m. RNA to find the start site for translation, which is often AUG – the codon for methionine. The sequences around the AUG start codon are also important and can determine how strongly an m. RNA is translated.

Initiation also involves the activity of a number of helper proteins called initiation factors, whose function is to ensure that the various parts of the translation machinery come together in an orderly manner.

The ribosome and initiator t. RNA move slowly along the m. RNA till the start codon is located. The structural features of this t. RNA ensure that it is recognized by initiation factors and discriminated against by translation elongation factors.

The methionine attached to this t. RNA is also adapted to be exclusive for initiation – the amino group is modified to make Nformyl methionine, preventing it from participating in the elongation phase of translation.

Once the larger subunit of the ribosome arrives at the translation start site, and the t. RNA is located at its P site, initiation is said to be complete.


Elongation Binding of the initiator t. RNA to the P site on the ribosome brings it into close contact with the amino or ‘A’ site of the ribosome, where the next codon awaits translation. New t. RNAs carrying amino acids enter the ribosome at the A site.

Through complementary base pairing and the energy from one molecule of GTP, the correct t. RNA binds to the ribosome. Ribosomal RNA then catalyzes the formation of a peptide bond between the first and second amino acids where the first methionine appears to have been ‘transferred’ to the t. RNA on the A site.

Once the peptide bond is formed, the empty t. RNA exits the ribosome and the ribosome itself moves ahead by exactly one codon, so that the t. RNA on the A site moves into the P site. This also exposes the next codon to the A site, ready for the third t. RNA. This process continues till a stop codon is reached.


Termination When a stop codon is present at the A site it is recognized by a set of proteins called release factors. They induce the ribosome to attach a molecule of water to the growing polypeptide chain, rather than another amino acid

This terminates the process of translation, and releases the polypeptide from the ribosome. The two subunits of the ribosome also dissociate from each other, ready for the next cycle of translation.


Thank you
 Translation refers to the
Translation refers to the Semantic translation method
Semantic translation method Cisco voice translation-rule
Cisco voice translation-rule Parent function of linear
Parent function of linear Noun phrase example
Noun phrase example Carrier vs channel proteins
Carrier vs channel proteins Globular vs fibrous proteins
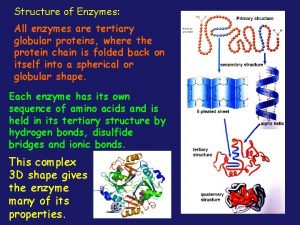
Globular vs fibrous proteins All enzymes are globular proteins
All enzymes are globular proteins Section 8-1 carbohydrates fats and proteins answer key
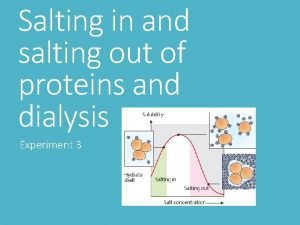
Section 8-1 carbohydrates fats and proteins answer key Salting in
Salting in Complementary proteins
Complementary proteins Qualitative test for protein
Qualitative test for protein What are derived proteins
What are derived proteins Monomers of proteins
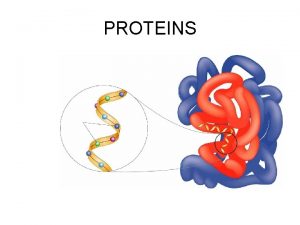
Monomers of proteins Protein definition
Protein definition Protein metabolism
Protein metabolism How do proteins fold
How do proteins fold Protein databases
Protein databases Bradford method
Bradford method Dr nutrition
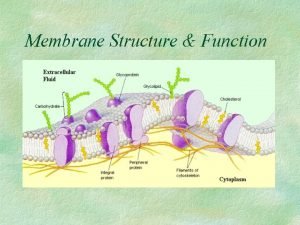
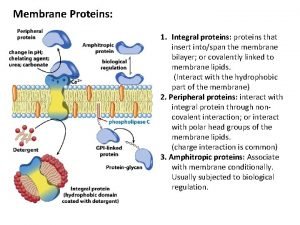
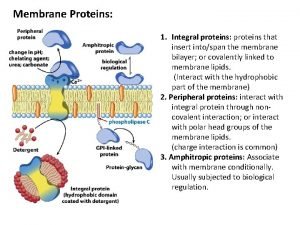
Dr nutrition Functions of membrane proteins
Functions of membrane proteins Motifs and domains of proteins
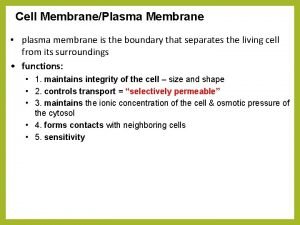
Motifs and domains of proteins Cell membrane function
Cell membrane function Peripheral vs integral proteins
Peripheral vs integral proteins Functions of membrane proteins
Functions of membrane proteins Precipitation of proteins by strong mineral acids
Precipitation of proteins by strong mineral acids Purification yield calculation
Purification yield calculation Fibrous protein collagen
Fibrous protein collagen Not all enzymes are proteins
Not all enzymes are proteins Salting in salting out
Salting in salting out Salt bridge amino acids
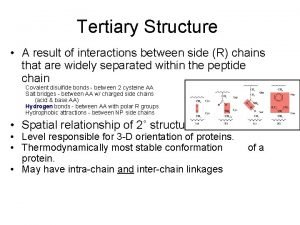
Salt bridge amino acids Higher order structure of proteins
Higher order structure of proteins Difference between glycoprotein and glycolipid
Difference between glycoprotein and glycolipid Membrane proteins functions
Membrane proteins functions Salting in salting out
Salting in salting out Biomedical importance of protein
Biomedical importance of protein Organic compounds such as proteins and starches are too
Organic compounds such as proteins and starches are too Primary vs secondary vs tertiary vs quaternary structures
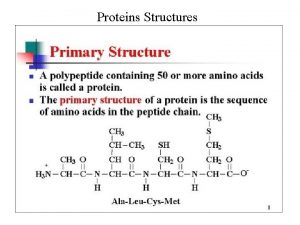
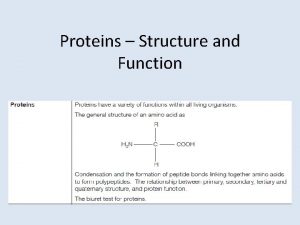
Primary vs secondary vs tertiary vs quaternary structures