NOMENKLATUR DAN KLASIFIKASI VIRUS KLASIFIKASI VIRUS What is














































- Slides: 65

NOMENKLATUR DAN KLASIFIKASI VIRUS

KLASIFIKASI VIRUS

What is the purpose of classification? To make structural arrangement for easy comprehension To communicate taxonomic decisions to the international community of virologist To enable prediction of properties of new viruses Possible evolutionary relationships

Descriptors used in virus Taxonomy Virion properties Morphological properties of virion Size; Shape; presence or absence of envelop or peplomers; capsomeric symmetry & structure Physical properties of the virus Molecular mass Buoyant density Sedimentation coefficient p. H stability, Solvent stability, Radiation stability, detergent stability Cation (Mg 2+, Mn 2+, Ca 2+) stability

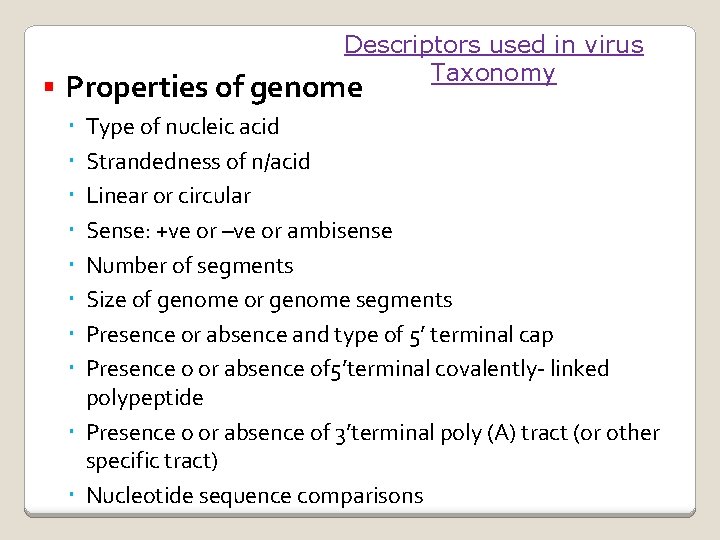
Descriptors used in virus Taxonomy Properties of genome Type of nucleic acid Strandedness of n/acid Linear or circular Sense: +ve or –ve or ambisense Number of segments Size of genome or genome segments Presence or absence and type of 5’ terminal cap Presence o or absence of 5’terminal covalently- linked polypeptide Presence o or absence of 3’terminal poly (A) tract (or other specific tract) Nucleotide sequence comparisons

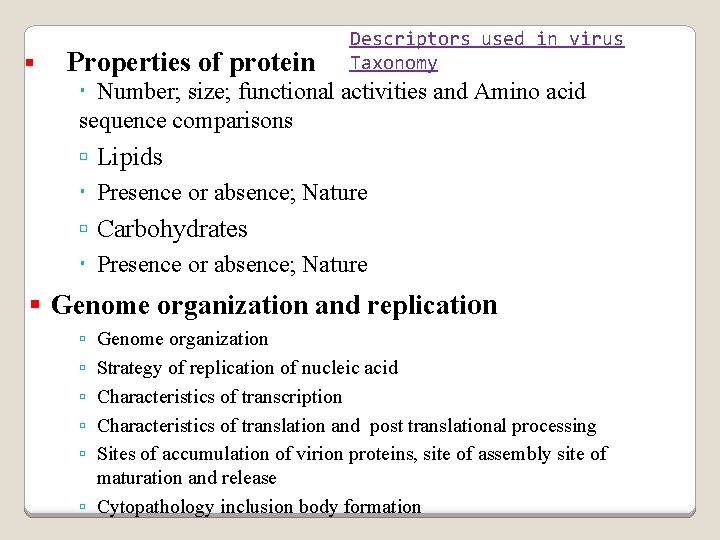
Properties of protein Descriptors used in virus Taxonomy Number; size; functional activities and Amino acid sequence comparisons Lipids Presence or absence; Nature Carbohydrates Presence or absence; Nature Genome organization and replication Genome organization Strategy of replication of nucleic acid Characteristics of transcription Characteristics of translation and post translational processing Sites of accumulation of virion proteins, site of assembly site of maturation and release Cytopathology inclusion body formation

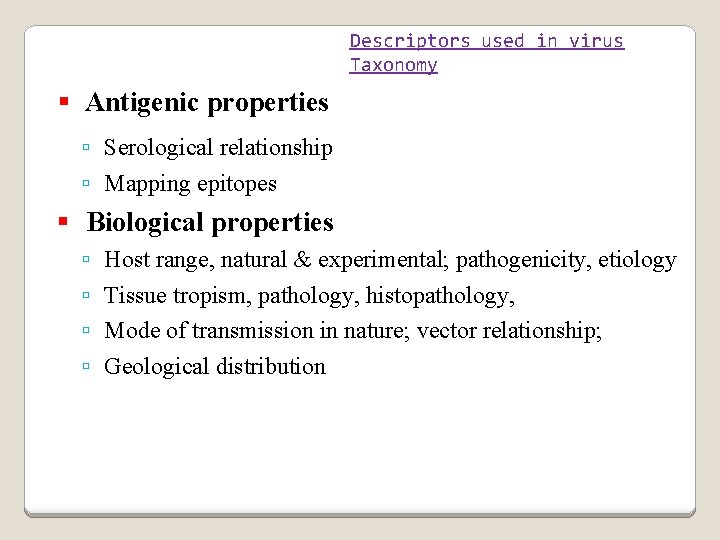
Descriptors used in virus Taxonomy Antigenic properties Serological relationship Mapping epitopes Biological properties Host range, natural & experimental; pathogenicity, etiology Tissue tropism, pathology, histopathology, Mode of transmission in nature; vector relationship; Geological distribution

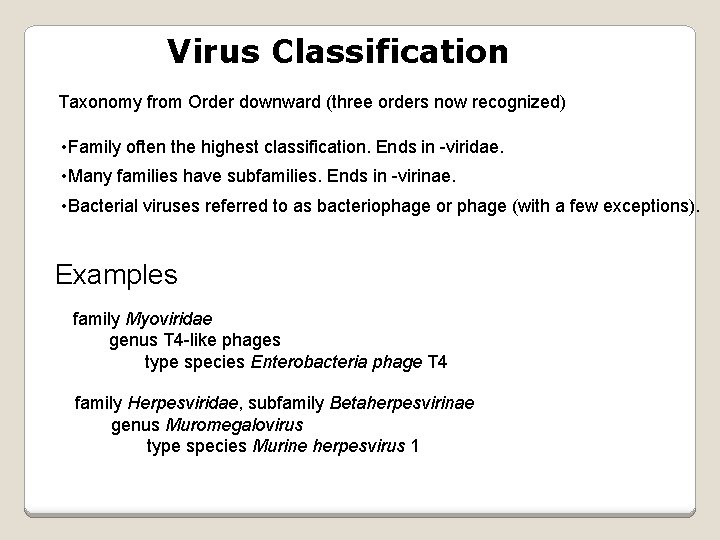
Virus Classification Taxonomy from Order downward (three orders now recognized) • Family often the highest classification. Ends in -viridae. • Many families have subfamilies. Ends in -virinae. • Bacterial viruses referred to as bacteriophage or phage (with a few exceptions). Examples family Myoviridae genus T 4 -like phages type species Enterobacteria phage T 4 family Herpesviridae, subfamily Betaherpesvirinae genus Muromegalovirus type species Murine herpesvirus 1

Virus Classification I - the Baltimore classification • Was crated by American biologist David Baltimore • It is basically based on the method of viral syntesis • It groups virus into families according to their type of genome All viruses must produce m. RNA, or (+) sense RNA A complementary strand of nucleic acid is (–) sense The Baltimore classification has + RNA as its central point Its principles are fundamental to an understanding of virus classification and genome replication, but it is rarely used as a classification system in its own right

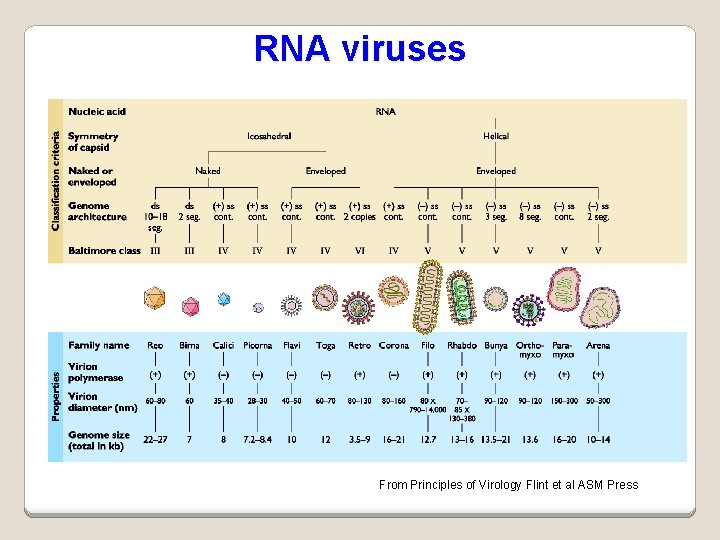
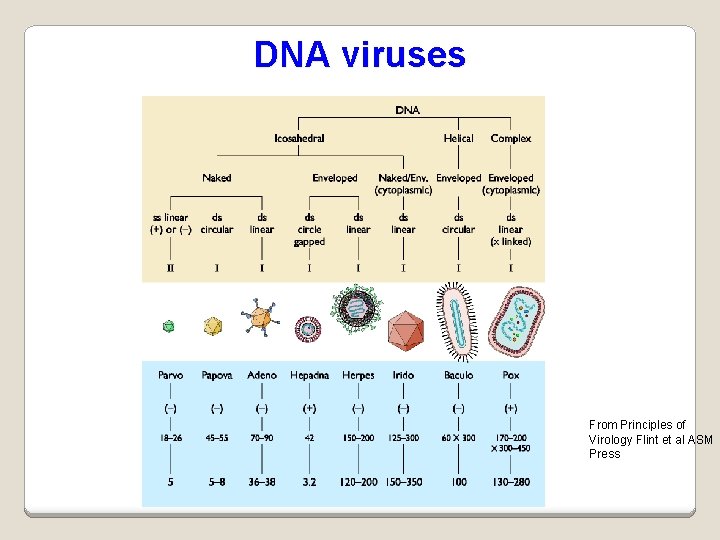
Virus Classification I - the Baltimore classification Based on genetic contents and replication strategies of viruses. According to the Baltimore classification, viruses are divided into the following seven classes: 1. ds. DNA viruses 2. ss. DNA viruses 3. ds. RNA viruses 4. (+) sense ss. RNA viruses (codes directly for protein) 5. (-) sense ss. RNA viruses 6. RNA reverse transcribing viruses 7. DNA reverse transcribing viruses where "ds" represents "double strand" and "ss" denotes "single strand".



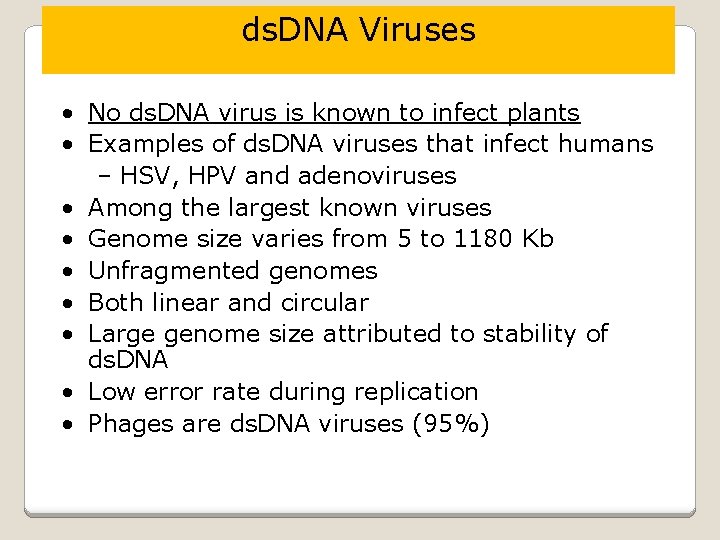
ds. DNA Viruses • No ds. DNA virus is known to infect plants • Examples of ds. DNA viruses that infect humans – HSV, HPV and adenoviruses • Among the largest known viruses • Genome size varies from 5 to 1180 Kb • Unfragmented genomes • Both linear and circular • Large genome size attributed to stability of ds. DNA • Low error rate during replication • Phages are ds. DNA viruses (95%)


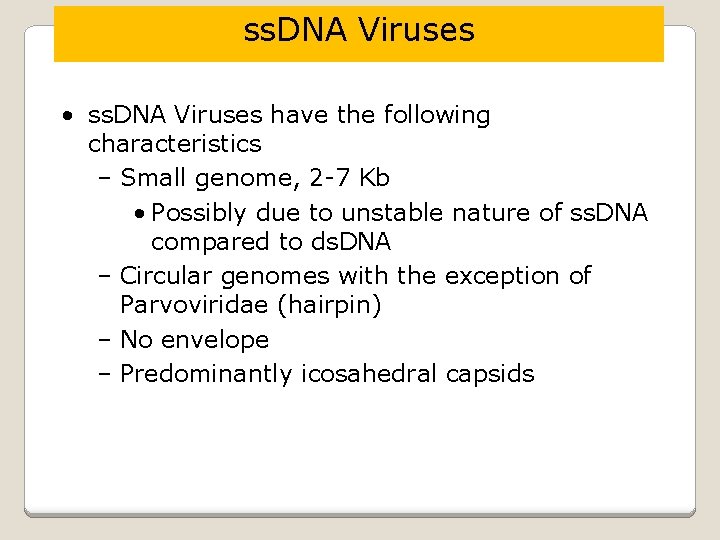
ss. DNA Viruses • ss. DNA Viruses have the following characteristics – Small genome, 2 -7 Kb • Possibly due to unstable nature of ss. DNA compared to ds. DNA – Circular genomes with the exception of Parvoviridae (hairpin) – No envelope – Predominantly icosahedral capsids


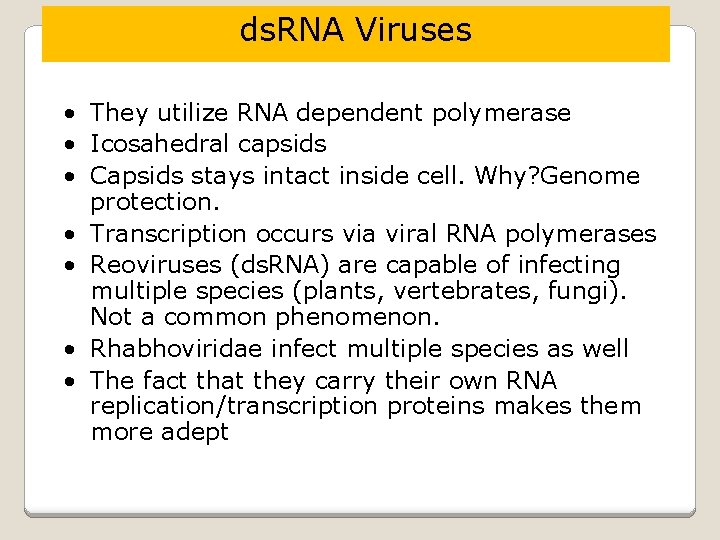
ds. RNA Viruses • They utilize RNA dependent polymerase • Icosahedral capsids • Capsids stays intact inside cell. Why? Genome protection. • Transcription occurs via viral RNA polymerases • Reoviruses (ds. RNA) are capable of infecting multiple species (plants, vertebrates, fungi). Not a common phenomenon. • Rhabhoviridae infect multiple species as well • The fact that they carry their own RNA replication/transcription proteins makes them more adept


Viruses With + strand RNA Genomes • Very common of plant viruses to be + ss. RNA • Only one phage family is + ss. RNA • RNA viruses have linear genomes • Similar to ss. DNA viruses they are susceptible to nucleases and divalant cation degradation • Coronavirus has the largest genome of + ss. RNA virus (16 -30 Kb)


- ss. RNA Viruses • This group includes some of the deadliest viruses – Ebola, rabies, influenza, measles • Only helical nucleocapsids • Nucleocapsid seems to provide stability for RNA dependent RNA polymerase to generate + ss. RNA • + ss. RNA=m. RNA

Viruses With Reverse Transcription • 3 families belong to this group – Retroviridae, Ex. HIV – Hepadnaviridae, Ex. Hep B – Caulimoviridae, Ex. Cauliflower Mosaic Virus • These families utilize enzyme that uses an RNA template to make DNA template • Reverse transcriptase is packaged in capsid – Similar to + ss. RNA and – ss. RNA that package the RNA dependent polymerase • Retroviruses package 2 copies of their RNA genome in the capsid



Virus classification II the Classical system This is a based on three principles - 1) that we are classifying the virus itself, not the host 2) the nucleic acid genome 3) the shared physical properties of the infectious agent (e. g capsid symmetry, dimensions, lipid envelope)

Virus classification III the genomic system More recently a precise ordering of viruses within and between families is possible based on DNA/RNA sequence By the year 2000 there were over 4000 viruses of plants, animals and bacteria in 71 families, 9 subfamilies and 164 genera

RNA viruses From Principles of Virology Flint et al ASM Press

DNA viruses From Principles of Virology Flint et al ASM Press

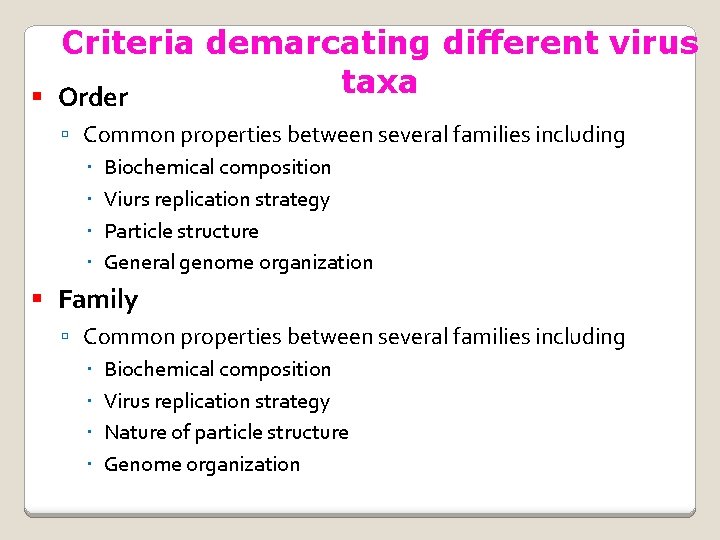
Criteria demarcating different virus taxa Order Common properties between several families including Biochemical composition Viurs replication strategy Particle structure General genome organization Family Common properties between several families including Biochemical composition Virus replication strategy Nature of particle structure Genome organization

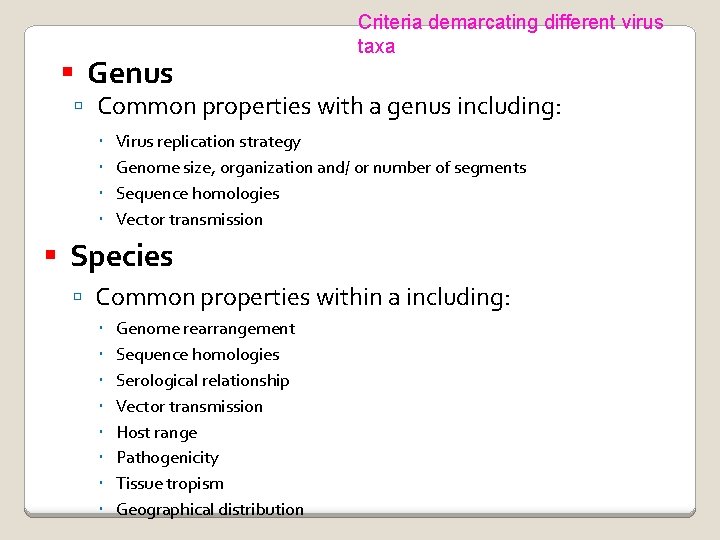
Genus Criteria demarcating different virus taxa Common properties with a genus including: Virus replication strategy Genome size, organization and/ or number of segments Sequence homologies Vector transmission Species Common properties within a including: Genome rearrangement Sequence homologies Serological relationship Vector transmission Host range Pathogenicity Tissue tropism Geographical distribution

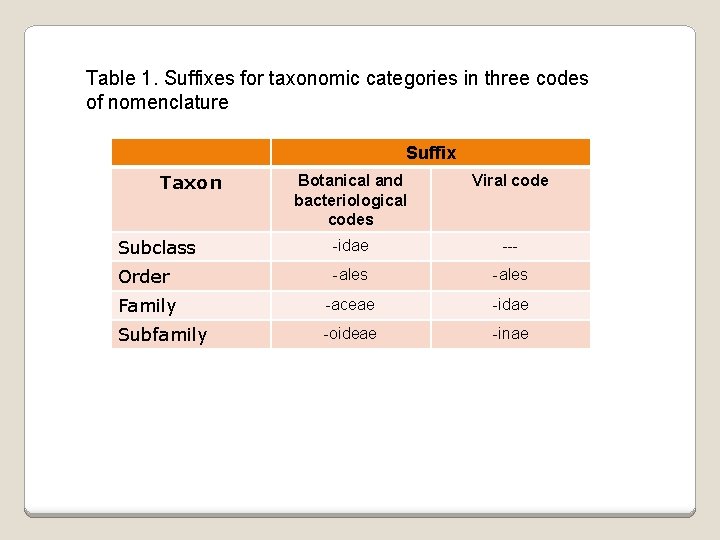
Table 1. Suffixes for taxonomic categories in three codes of nomenclature Suffix Taxon Botanical and bacteriological codes Viral code Subclass -idae --- Order -ales Family -aceae -idae Subfamily -oideae -inae


Criteria for classifying plant virus Structure of the virus particle Physicochemical properties particle Properties of viral nucleic acid Viral proteins 5. Serological relationships 6. Activities in the plant 7. Methods of transmission 1. 2. 3. 4.

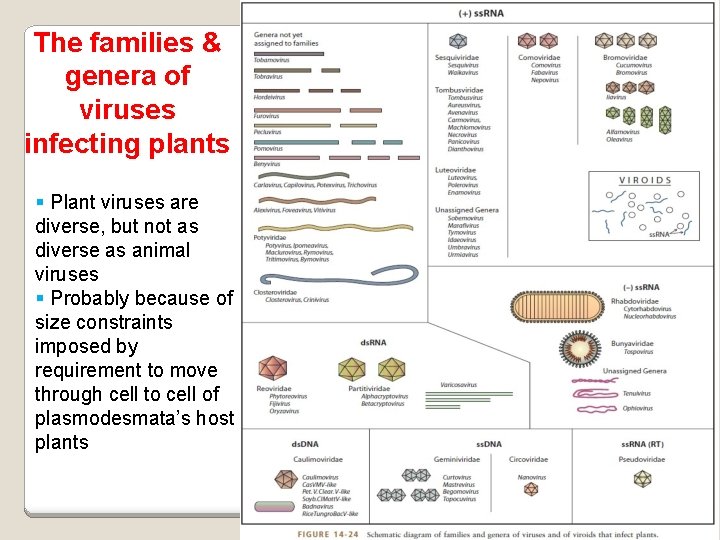
The families & genera of viruses infecting plants Plant viruses are diverse, but not as diverse as animal viruses Probably because of size constraints imposed by requirement to move through cell to cell of plasmodesmata’s host plants








Satellite Viruses that must always be associated with certain typical viruses (helpers) because they depend on the helper for multiplication and plant infections. They often reduce the ability of the helper virus to multiply and cause disease, so the satellite viruses act as parasites. • These viruses require a helper virus • Their genomes encode for capsid proteins • Nucleic acid satellites are either non-coding or encode for noncapsid proteins • Mostly a plant phenomenon • In humans the Hep virus resembles characteristics of satellite virus/viroid


INFECTOUS AGENTS • Virion = a complete virus particle, including envelope (if any) • Viroid = an infectious RNA particle, smaller than a virus, lacking a capsid, that causes various plant diseases • Virusoid (satellite RNA) = same as viroid; small, ss. RNA molecule, usually 500 to 2000 nucleotides in length, lacking a capsid, lack genes required for the replication virusoid require a helper (satellite) virus to replicate, causes various plant diseases.

Example Viroid - avocado sunblotch viroid, peach latent mosaic, potato spindle tuber, coconut cadang-cadang, tomato plant macho viroid, citrus bent leaf viroid, pear blister canker viroid -Virusoid - barley yellow dwarf satellite RNA , tobacco ringspot virus satellite RNA Disease Viroid - cause lethal plant diseases; potato spindle tuber disease, chrysanthemum stunt disease, cucumber pale fruit disease, coconut cadang -cadang disease, chrysanthemum stunt disease, tomato apical stunt disease. Virusoid - cause tobacco necrosis



PRIONS • A unique infectious agent, protein infectious particle • Prion = a small infectious particle consisting of protein and lack nucleic acid. • Prion – features: - Resistant to inactivation by heating to 90 o C, which inactivate virus - The infection is not sensitive to radiation; radiation damages virus genomes - Prions are not destroyed by enzymes that digest nucleic acids - Sensitive to protein denaturing agents; urea, phenol - Prions have direct pairing of amino acids

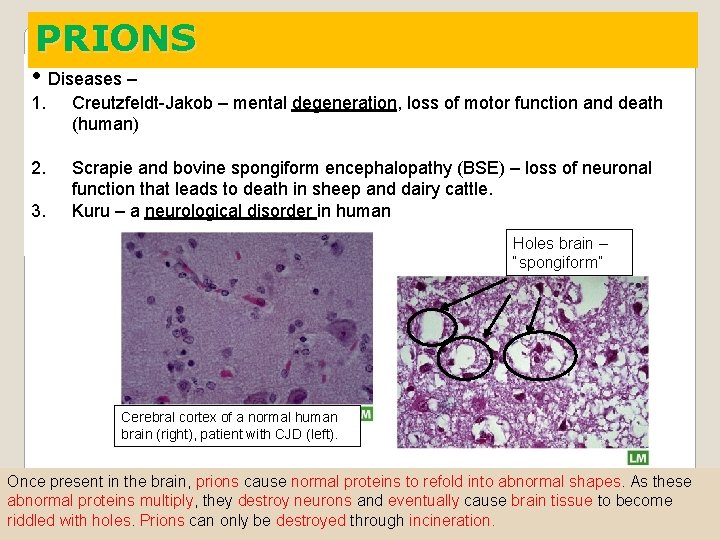
PRIONS • Diseases – 1. Creutzfeldt-Jakob – mental degeneration, loss of motor function and death (human) 2. Scrapie and bovine spongiform encephalopathy (BSE) – loss of neuronal function that leads to death in sheep and dairy cattle. Kuru – a neurological disorder in human 3. Holes brain – “spongiform” Cerebral cortex of a normal human brain (right), patient with CJD (left). Once present in the brain, prions cause normal proteins to refold into abnormal shapes. As these abnormal proteins multiply, they destroy neurons and eventually cause brain tissue to become riddled with holes. Prions can only be destroyed through incineration.

Virus, Viroid and Prion v Characteristic between virus, viroid and prion v. Nucleic acid type, NA strandedness, host range and structural features

NOMENKLATUR VIRUS

How are viruses named? • Based on: - The disease they cause poliovirus, rabies virus - The type of disease murine leukemia virus - Geographic locations Sendai virus, Coxsackie virus - Their discovers Epstein-Barr virus - How they were originally thought to be contracted Tobacco mosaic virus, dengue virus (“evil spirit”), influenza virus (the “influence” of bad air) - Combinations of the above Rous Sarcoma virus

Latin binomials were proposed first by Holmes in 1939 Various other schemes proposed between 1940 and 1966 International Committee on the Nomenclature of Viruses (ICNV) (1966) Naming and cataloguing of viruses. Renamed: International Committee Taxonomy of Viruses (ICTV) in 1973.

The International Committee on Taxonomy of Viruses (ICTV) has framed 20 rules for virus nomenclature. Among others it recognizes common names, their international meaning and use of existing names. ICTV database (ICTVdb)- A centralized repository of virus information Collect information from databases around the world. Facilitated the job of accurate identification and diagnosis of new and important virus diseases.











Terima Kasih
 Permendagri tentang klasifikasi kodefikasi dan nomenklatur
Permendagri tentang klasifikasi kodefikasi dan nomenklatur Esterbindung
Esterbindung Aldehyde nomenklatur
Aldehyde nomenklatur Nomenklatur ester
Nomenklatur ester Nomenklatur roda gigi
Nomenklatur roda gigi Nomenklatur übertragungsglieder
Nomenklatur übertragungsglieder Benennung organischer verbindungen übungen
Benennung organischer verbindungen übungen E z nomenklatur
E z nomenklatur Tata nama senyawa kimia
Tata nama senyawa kimia Klasifikasi virus
Klasifikasi virus Klasifikasi virus
Klasifikasi virus Virusmax
Virusmax Klasifikasi produk
Klasifikasi produk Klasifikasi sistem informasi manajemen
Klasifikasi sistem informasi manajemen Taksonomi bakteri
Taksonomi bakteri Diffrential cost
Diffrential cost Klasifikasi bimbingan konseling
Klasifikasi bimbingan konseling Perkembangan dan klasifikasi akuntansi internasional
Perkembangan dan klasifikasi akuntansi internasional Perkembangan dan klasifikasi komputer
Perkembangan dan klasifikasi komputer Perbedaan estimasi dan klastering
Perbedaan estimasi dan klastering Pencernaan lemak
Pencernaan lemak Klasifikasi dan karakteristik anggaran negara
Klasifikasi dan karakteristik anggaran negara Kriteria usaha mikro
Kriteria usaha mikro Perbedaan definisi dan klasifikasi
Perbedaan definisi dan klasifikasi Trojanski konj virus
Trojanski konj virus Dermotropos
Dermotropos Virus dermatrópicos ejemplos
Virus dermatrópicos ejemplos Virus informatic definitie
Virus informatic definitie Veoh virus
Veoh virus Blended threat virus
Blended threat virus Rabies virus incubation period
Rabies virus incubation period Lytta virus
Lytta virus Poxviridae ailesi
Poxviridae ailesi Mumps virus
Mumps virus Mumps medicine
Mumps medicine Adware virus informatico
Adware virus informatico Virus rna jenis picornaviridae
Virus rna jenis picornaviridae Como se clasifica los virus
Como se clasifica los virus Estructuras reproductivas de los hongos
Estructuras reproductivas de los hongos Pepino mozaïek virus
Pepino mozaïek virus Virions
Virions Viral inoculation in embryonated egg
Viral inoculation in embryonated egg Mcafee anti-virus
Mcafee anti-virus Virus polimorfos o mutantes
Virus polimorfos o mutantes Jason
Jason Contoh peta konsep virus
Contoh peta konsep virus Stomach virus
Stomach virus Sofiacruzv
Sofiacruzv Polihidris
Polihidris Rage
Rage Growth curve of virus
Growth curve of virus Sobia canyon pricing
Sobia canyon pricing Parantral
Parantral Merilyn merisalu
Merilyn merisalu Trap door virus
Trap door virus Virus trojanski konj
Virus trojanski konj Que son los virus
Que son los virus Virus multiple sclerosis
Virus multiple sclerosis Coder un virus
Coder un virus Virus
Virus Japanese encephalitis virus
Japanese encephalitis virus Sonia disowns rahul
Sonia disowns rahul Virus mnemonic
Virus mnemonic Chemical nature of virus
Chemical nature of virus Tuberculose é um virus
Tuberculose é um virus Concatenamero significato
Concatenamero significato