Molecular evolution Part I The evolution of macromolecules
![Molecular Evolution AGAMOUS; transcription factor [ Arabidopsis thaliana ] What information can DNA sequences Molecular Evolution AGAMOUS; transcription factor [ Arabidopsis thaliana ] What information can DNA sequences](https://slidetodoc.com/presentation_image/5e085f81e2bc4de2fdaa1d665c9af2a9/image-2.jpg)

- Slides: 56

Molecular evolution Part I: The evolution of macromolecules. Part II: The reconstruction of the evolutionary history of genes and organisms. Biol 336 -12 1
![Molecular Evolution AGAMOUS transcription factor Arabidopsis thaliana What information can DNA sequences Molecular Evolution AGAMOUS; transcription factor [ Arabidopsis thaliana ] What information can DNA sequences](https://slidetodoc.com/presentation_image/5e085f81e2bc4de2fdaa1d665c9af2a9/image-2.jpg)
Molecular Evolution AGAMOUS; transcription factor [ Arabidopsis thaliana ] What information can DNA sequences give us? Evaluating the role of drift/demography vs. selection on trait divergence. Identify function. Looking at genes whose evolutionary history was shared. www. ncbi. nlm. nih. gov Biol 336 -12 2

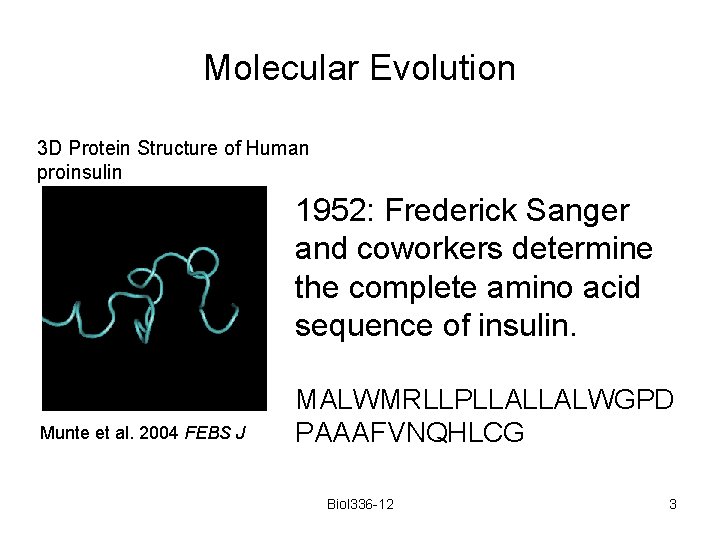
Molecular Evolution 3 D Protein Structure of Human proinsulin 1952: Frederick Sanger and coworkers determine the complete amino acid sequence of insulin. Munte et al. 2004 FEBS J MALWMRLLPLLALLALWGPD PAAAFVNQHLCG Biol 336 -12 3

Molecular Evolution How and why have molecular sequences evolved to be the way they are? Biol 336 -12 4

Molecular Evolution Learning Objectives: 1. 2. 3. 4. Variability within a population Subsitution rates Neutral Theory Detecting selection at the DNA level. Biol 336 -12 5

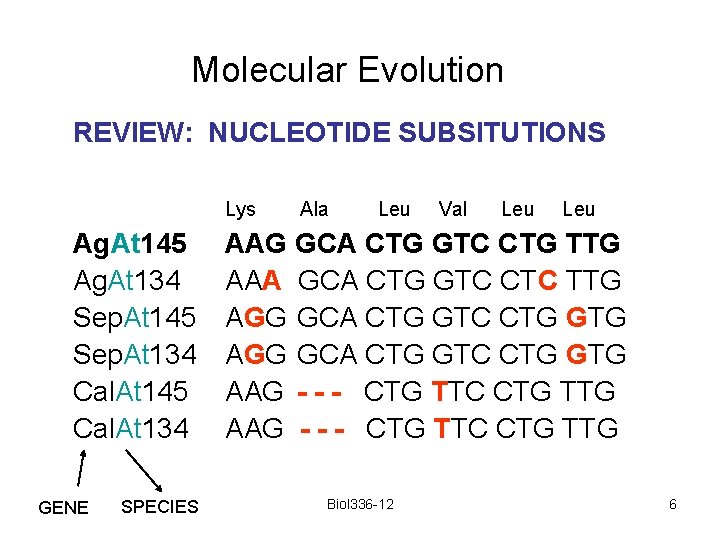
Molecular Evolution REVIEW: NUCLEOTIDE SUBSITUTIONS Lys Ag. At 145 Ag. At 134 Sep. At 145 Sep. At 134 Cal. At 145 Cal. At 134 GENE SPECIES Ala Leu Val Leu AAG GCA CTG GTC CTG TTG AAA GCA CTG GTC CTC TTG AGG GCA CTG GTC CTG GTG AAG - - - CTG TTC CTG TTG Biol 336 -12 6

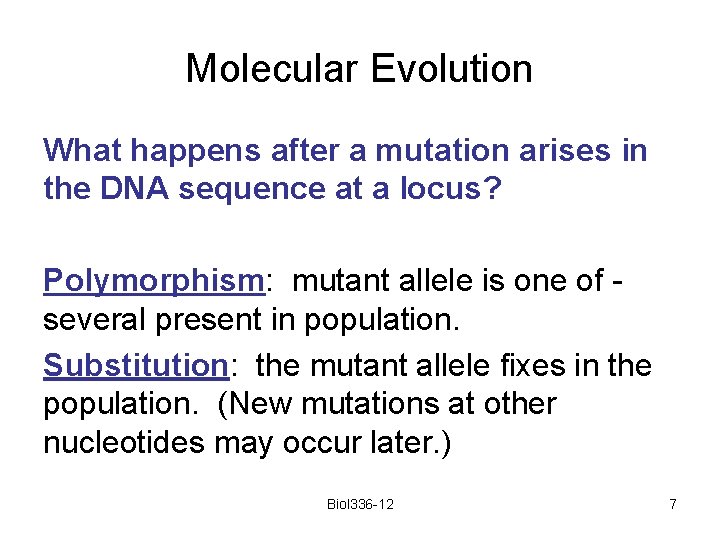
Molecular Evolution What happens after a mutation arises in the DNA sequence at a locus? Polymorphism: mutant allele is one of several present in population. Substitution: the mutant allele fixes in the population. (New mutations at other nucleotides may occur later. ) Biol 336 -12 7

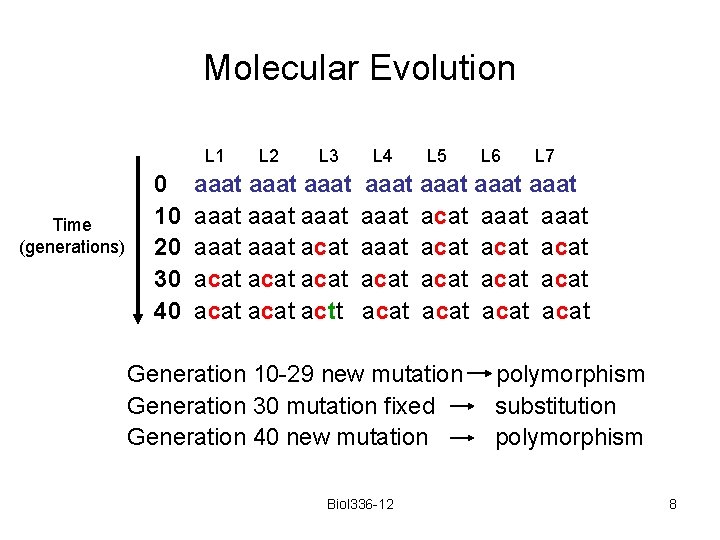
Molecular Evolution L 1 Time (generations) 0 10 20 30 40 L 2 L 3 aaat aaat acat acat actt L 4 L 5 L 6 L 7 aaat aaat acat aaat acat acat acat Generation 10 -29 new mutation Generation 30 mutation fixed Generation 40 new mutation Biol 336 -12 polymorphism substitution polymorphism 8

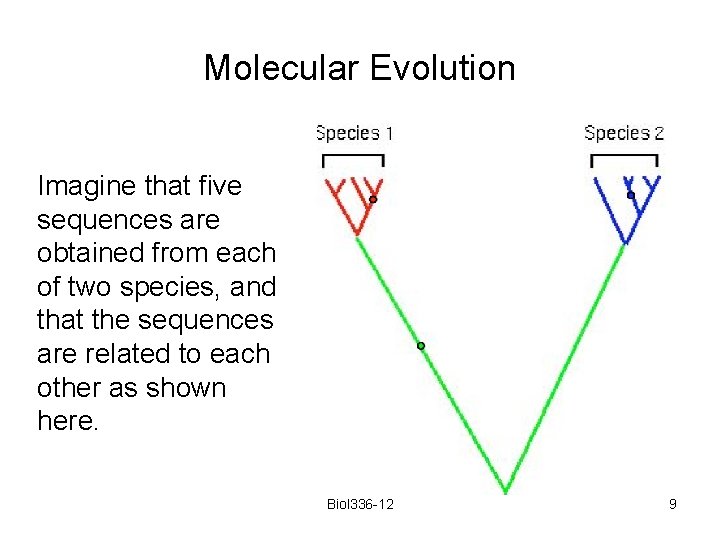
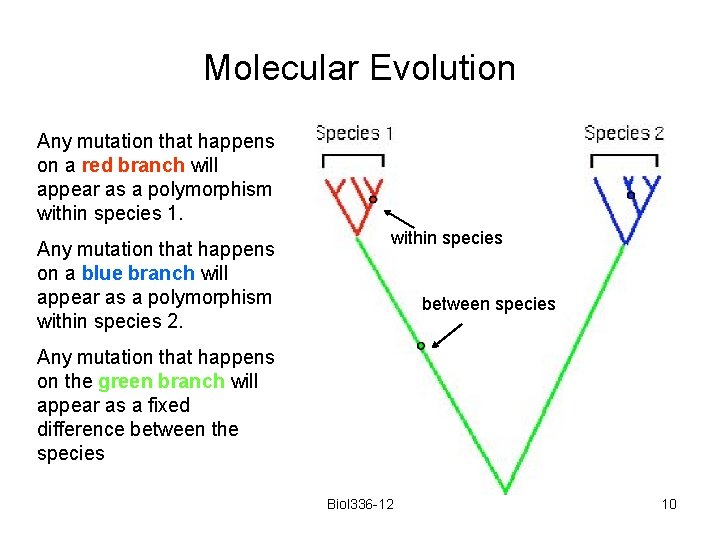
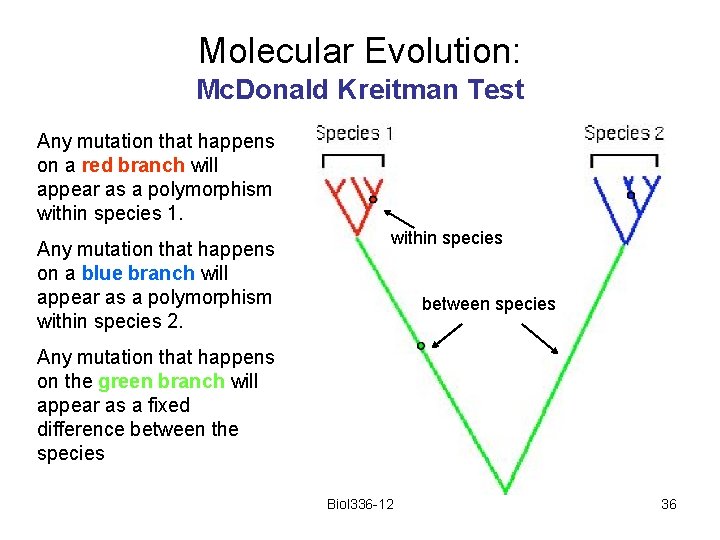
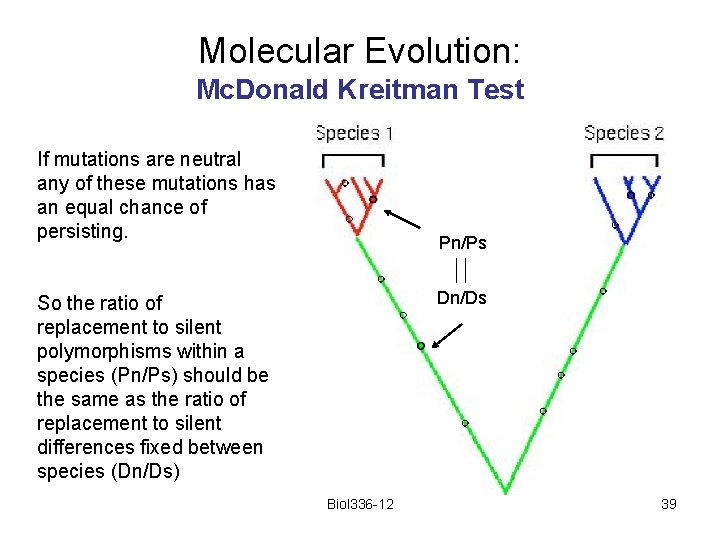
Molecular Evolution Imagine that five sequences are obtained from each of two species, and that the sequences are related to each other as shown here. Biol 336 -12 9

Molecular Evolution Any mutation that happens on a red branch will appear as a polymorphism within species 1. Any mutation that happens on a blue branch will appear as a polymorphism within species 2. within species between species Any mutation that happens on the green branch will appear as a fixed difference between the species Biol 336 -12 10

Molecular Evolution What happens after a mutation arises in the DNA sequence at a locus? Polymorphism: mutant allele is one of several present in population. Substitution: the mutant allele fixes in the population. (New mutations at other nucleotides may occur later. ) Biol 336 -12 11

Molecular Evolution Substitution rate: the rate at which mutant alleles rise to fix within a lineage By comparing DNA sequences from different organisms, we can estimate the rate at which mutations appear and fix, causing basepair substitutions. Biol 336 -12 12

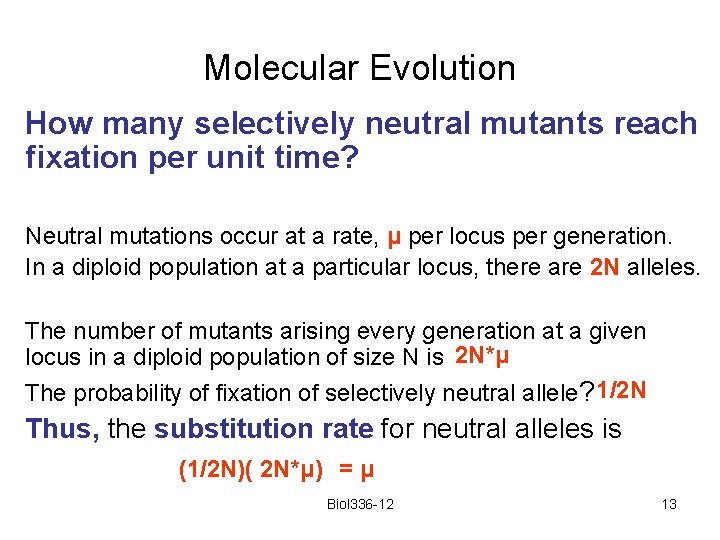
Molecular Evolution How many selectively neutral mutants reach fixation per unit time? Neutral mutations occur at a rate, μ per locus per generation. In a diploid population at a particular locus, there are 2 N alleles. The number of mutants arising every generation at a given locus in a diploid population of size N is 2 N*μ The probability of fixation of selectively neutral allele? 1/2 N Thus, the substitution rate for neutral alleles is (1/2 N)( 2 N*μ) = μ Biol 336 -12 13

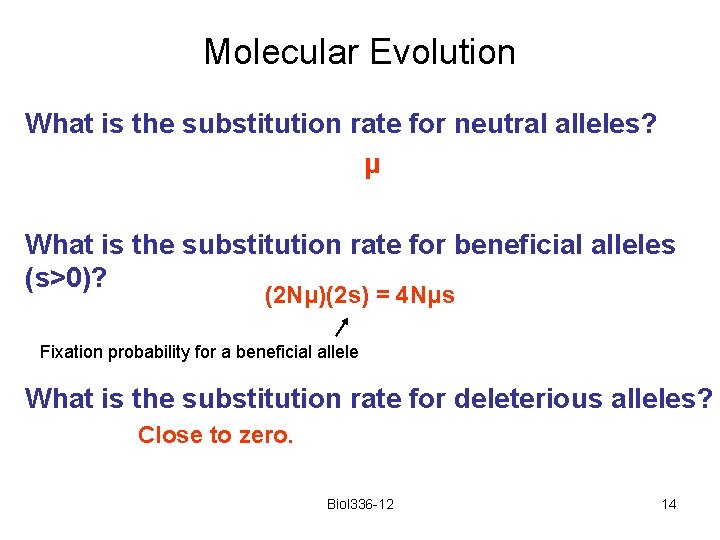
Molecular Evolution What is the substitution rate for neutral alleles? μ What is the substitution rate for beneficial alleles (s>0)? (2 Nμ)(2 s) = 4 Nμs Fixation probability for a beneficial allele What is the substitution rate for deleterious alleles? Close to zero. Biol 336 -12 14

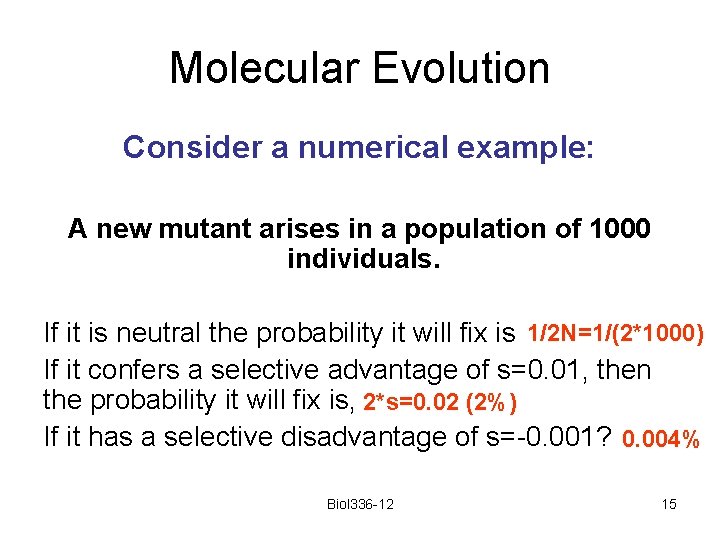
Molecular Evolution Consider a numerical example: A new mutant arises in a population of 1000 individuals. If it is neutral the probability it will fix is 1/2 N=1/(2*1000) If it confers a selective advantage of s=0. 01, then the probability it will fix is, 2*s=0. 02 (2%) If it has a selective disadvantage of s=-0. 001? 0. 004% Biol 336 -12 15

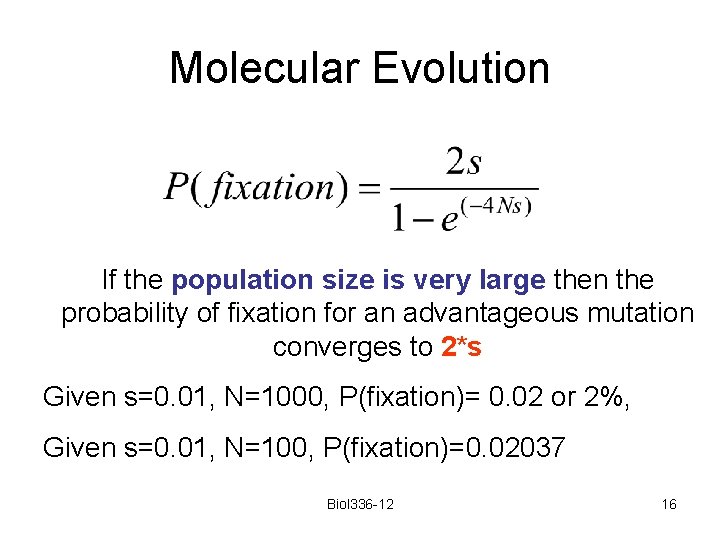
Molecular Evolution If the population size is very large then the probability of fixation for an advantageous mutation converges to 2*s Given s=0. 01, N=1000, P(fixation)= 0. 02 or 2%, Given s=0. 01, N=100, P(fixation)=0. 02037 Biol 336 -12 16

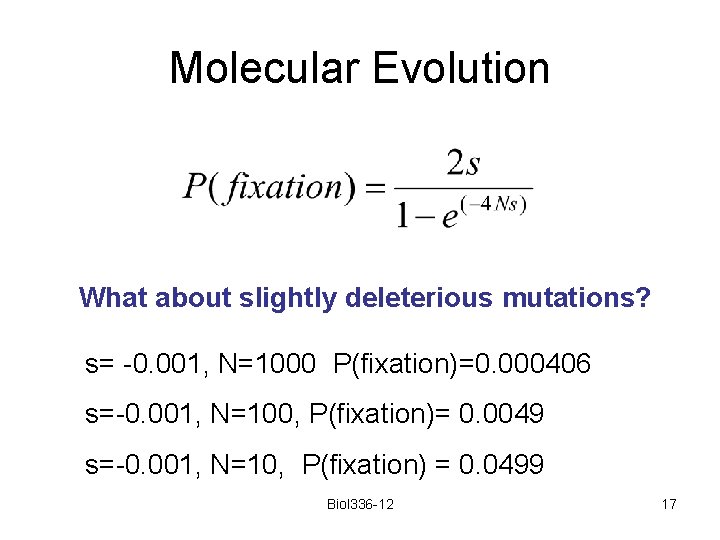
Molecular Evolution What about slightly deleterious mutations? s= -0. 001, N=1000 P(fixation)=0. 000406 s=-0. 001, N=100, P(fixation)= 0. 0049 s=-0. 001, N=10, P(fixation) = 0. 0499 Biol 336 -12 17

Molecular Evolution Are most substitutions (fixed changes) due to drift or natural selection? vs. Agree that: Most mutations are deleterious and are removed. Some mutations are favourable and are fixed. At Dispute: Are most replacement mutations that fix beneficial or neutral? Is observed polymorphism due to selection or drift? Biol 336 -12 18

Molecular Evolution Silent (or synonymous) mutations, where the amino acid remains unchanged, are more likely to be neutral. Replacement (or non-synonymous) mutations causing an amino acid change are more likely to experience selection. – Form and strength depends on gene and its function Biol 336 -12 19

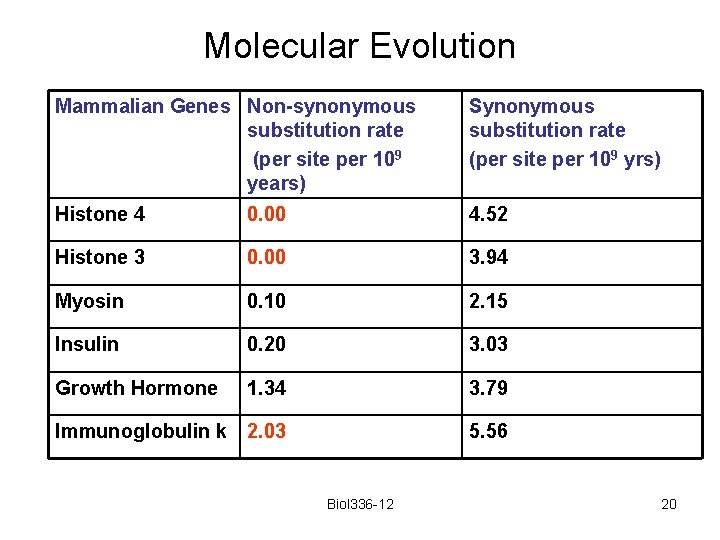
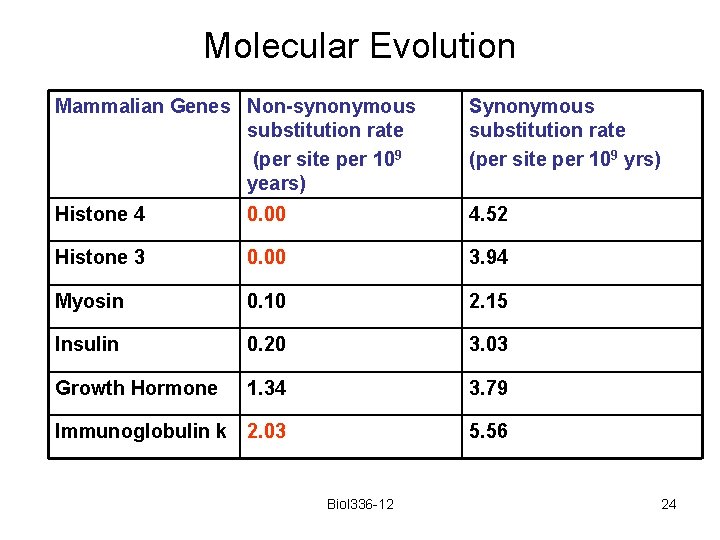
Molecular Evolution Mammalian Genes Non-synonymous substitution rate (per site per 109 years) Synonymous substitution rate (per site per 109 yrs) Histone 4 0. 00 4. 52 Histone 3 0. 00 3. 94 Myosin 0. 10 2. 15 Insulin 0. 20 3. 03 Growth Hormone 1. 34 3. 79 Immunoglobulin k 2. 03 5. 56 Biol 336 -12 20

Molecular Evolution Histones seem to have an unusually low replacement substitution rate. This suggests that mutations causing basepair changes in histones are deleterious WHY? Biol 336 -12 21

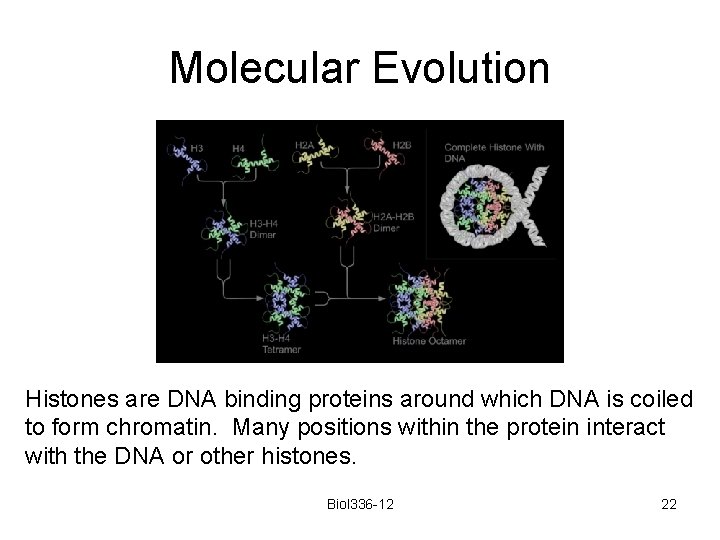
Molecular Evolution Histones are DNA binding proteins around which DNA is coiled to form chromatin. Many positions within the protein interact with the DNA or other histones. Biol 336 -12 22

Molecular Evolution Most amino acid changes in histone proteins may have negative or even lethal consequences. Histone proteins have strong functional constraints. Biol 336 -12 23

Molecular Evolution Mammalian Genes Non-synonymous substitution rate (per site per 109 years) Synonymous substitution rate (per site per 109 yrs) Histone 4 0. 00 4. 52 Histone 3 0. 00 3. 94 Myosin 0. 10 2. 15 Insulin 0. 20 3. 03 Growth Hormone 1. 34 3. 79 Immunoglobulin k 2. 03 5. 56 Biol 336 -12 24

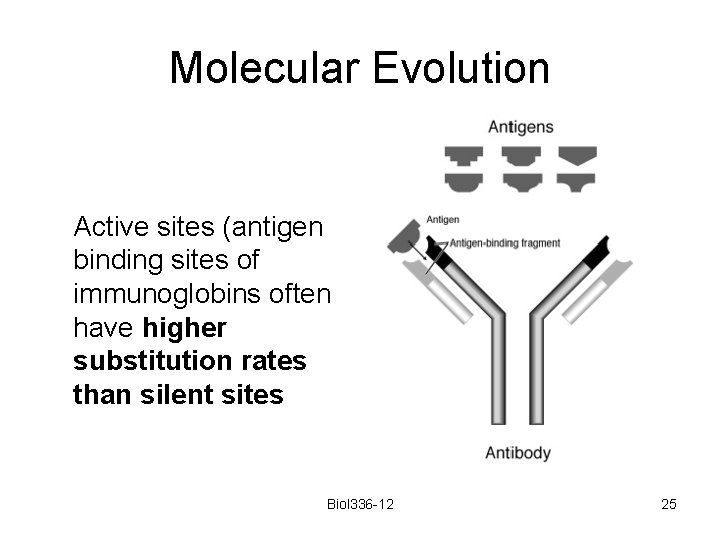
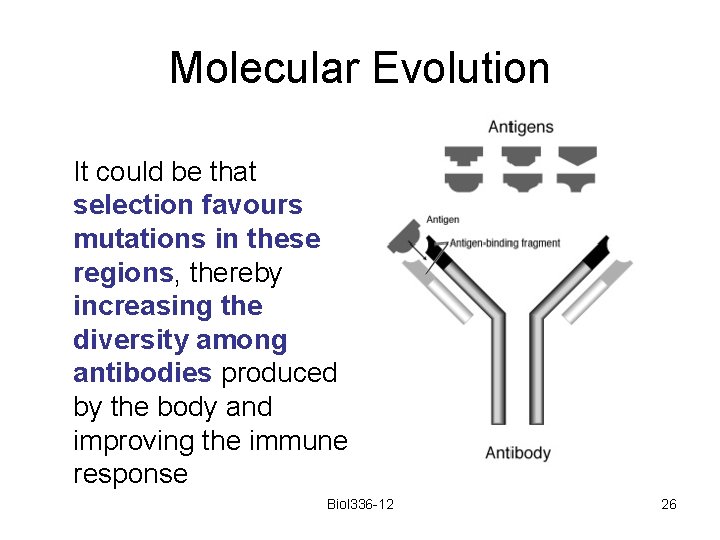
Molecular Evolution Active sites (antigen binding sites of immunoglobins often have higher substitution rates than silent sites Biol 336 -12 25

Molecular Evolution It could be that selection favours mutations in these regions, thereby increasing the diversity among antibodies produced by the body and improving the immune response Biol 336 -12 26

Molecular Evolution How and why have molecular sequences evolved to be the way they are? Biol 336 -12 27

Molecular Evolution To infer that selection has acted within a genome, one must reject the null hypothesis that no selection has acted. Null hypothesis: describes pattern of sequence evolution under the forces of mutation and drift. Remember from neutral theory: The rate at which one nucleotide is replaced by another nucleotide throughout a population (substitution) equals the rate of mutation (μ) at that site. Biol 336 -12 28

Molecular Evolution How do we detect selection at DNA sequences? Comparing intra-species polymorphism to interspecies differences (Mc. Donald-Kreitman test). Linked/neighbouring neutral markers. Examine genes for Dn/Ds ratios. Biol 336 -12 29

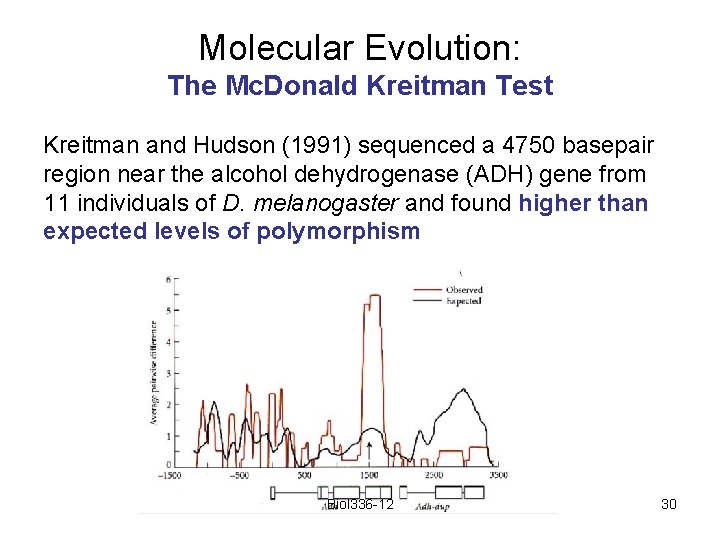
Molecular Evolution: The Mc. Donald Kreitman Test Kreitman and Hudson (1991) sequenced a 4750 basepair region near the alcohol dehydrogenase (ADH) gene from 11 individuals of D. melanogaster and found higher than expected levels of polymorphism Biol 336 -12 30

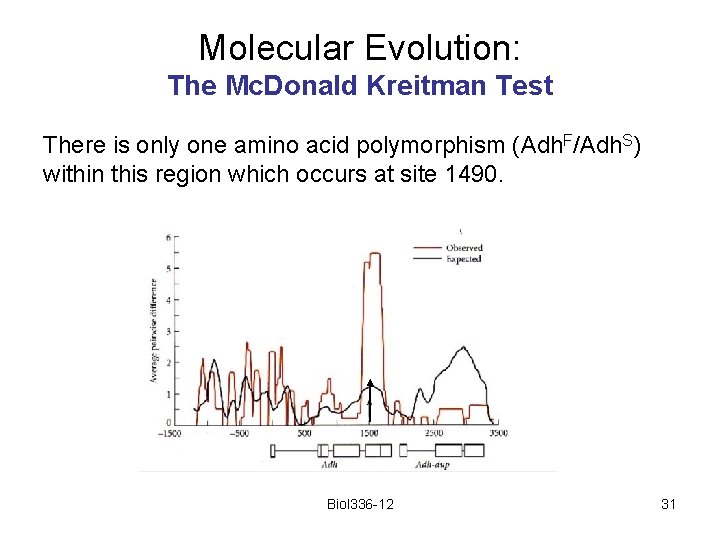
Molecular Evolution: The Mc. Donald Kreitman Test There is only one amino acid polymorphism (Adh. F/Adh. S) within this region which occurs at site 1490. Biol 336 -12 31

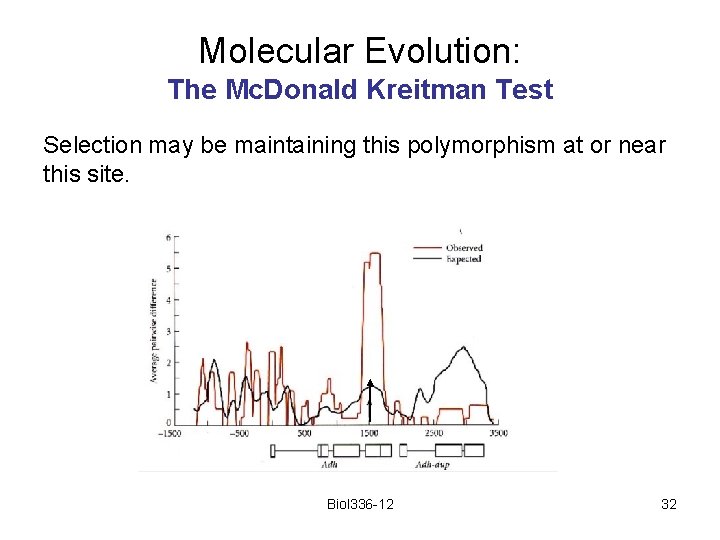
Molecular Evolution: The Mc. Donald Kreitman Test Selection may be maintaining this polymorphism at or near this site. Biol 336 -12 32

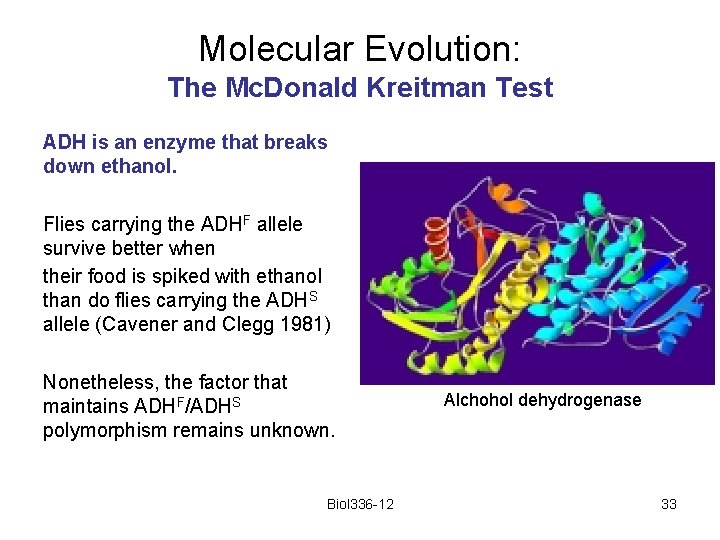
Molecular Evolution: The Mc. Donald Kreitman Test ADH is an enzyme that breaks down ethanol. Flies carrying the ADHF allele survive better when their food is spiked with ethanol than do flies carrying the ADHS allele (Cavener and Clegg 1981) Nonetheless, the factor that maintains ADHF/ADHS polymorphism remains unknown. Biol 336 -12 Alchohol dehydrogenase 33

Molecular Evolution: The Mc. Donald-Kreitman Test How and why have molecular sequences evolved to be the way they are? How do we explain the patterns of variation observed in ADH DNA sequences? Biol 336 -12 34

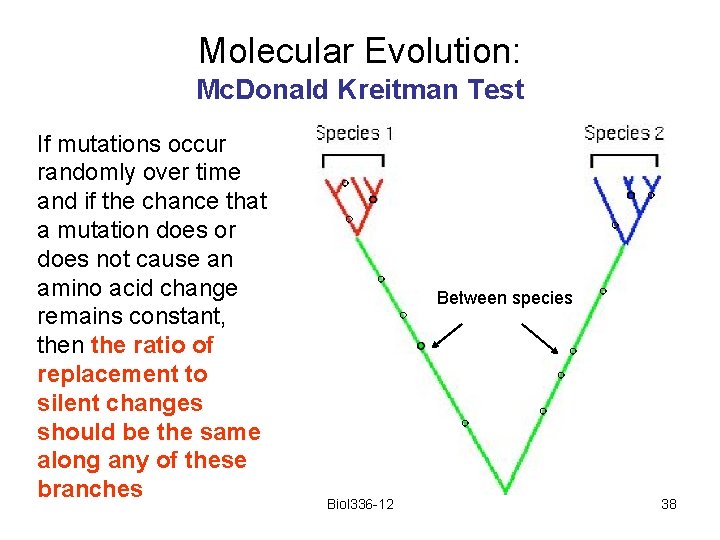
Molecular Evolution: Mc. Donald Kreitman Test Imagine that five sequences are obtained from each of two species, and that the sequences are related to each other as shown here. Biol 336 -12 35

Molecular Evolution: Mc. Donald Kreitman Test Any mutation that happens on a red branch will appear as a polymorphism within species 1. Any mutation that happens on a blue branch will appear as a polymorphism within species 2. within species between species Any mutation that happens on the green branch will appear as a fixed difference between the species Biol 336 -12 36

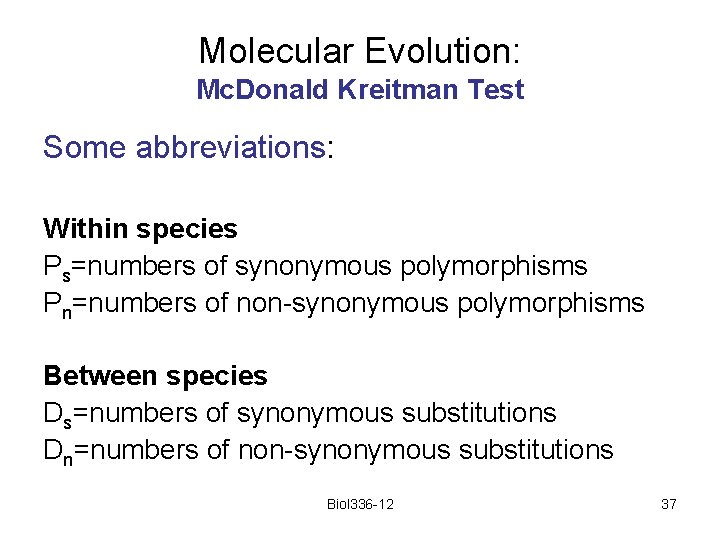
Molecular Evolution: Mc. Donald Kreitman Test Some abbreviations: Within species Ps=numbers of synonymous polymorphisms Pn=numbers of non-synonymous polymorphisms Between species Ds=numbers of synonymous substitutions Dn=numbers of non-synonymous substitutions Biol 336 -12 37

Molecular Evolution: Mc. Donald Kreitman Test If mutations occur randomly over time and if the chance that a mutation does or does not cause an amino acid change remains constant, then the ratio of replacement to silent changes should be the same along any of these branches Between species Biol 336 -12 38

Molecular Evolution: Mc. Donald Kreitman Test If mutations are neutral any of these mutations has an equal chance of persisting. Pn/Ps Dn/Ds So the ratio of replacement to silent polymorphisms within a species (Pn/Ps) should be the same as the ratio of replacement to silent differences fixed between species (Dn/Ds) Biol 336 -12 39

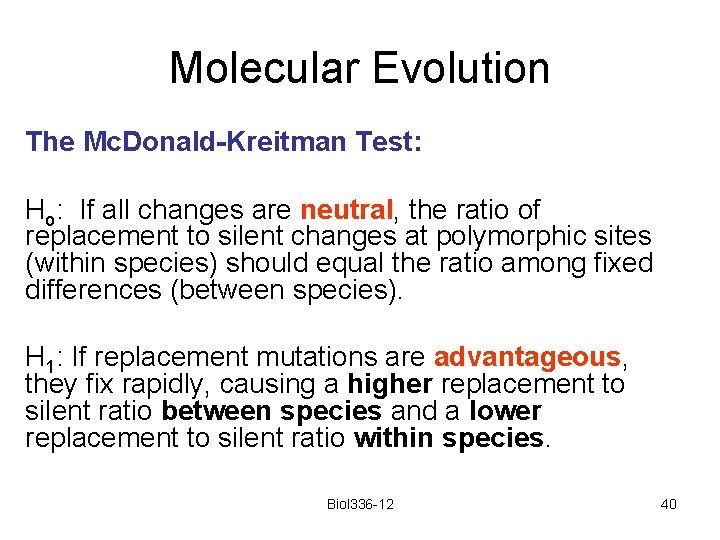
Molecular Evolution The Mc. Donald-Kreitman Test: Ho: If all changes are neutral, the ratio of replacement to silent changes at polymorphic sites (within species) should equal the ratio among fixed differences (between species). H 1: If replacement mutations are advantageous, they fix rapidly, causing a higher replacement to silent ratio between species and a lower replacement to silent ratio within species. Biol 336 -12 40

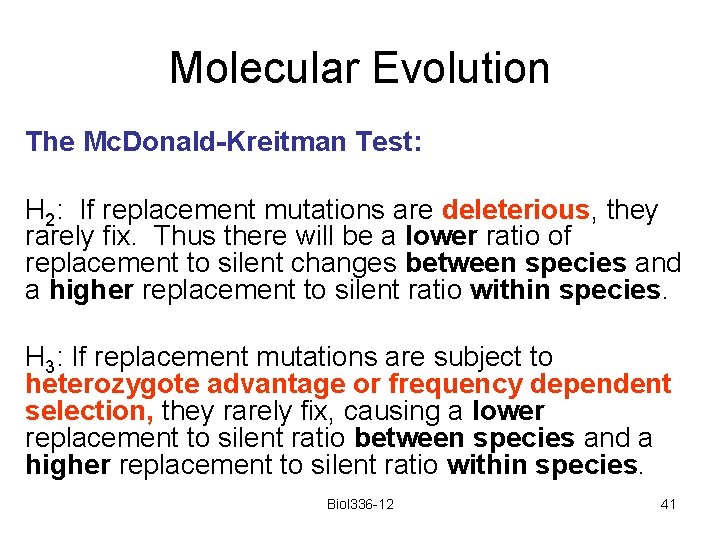
Molecular Evolution The Mc. Donald-Kreitman Test: H 2: If replacement mutations are deleterious, they rarely fix. Thus there will be a lower ratio of replacement to silent changes between species and a higher replacement to silent ratio within species. H 3: If replacement mutations are subject to heterozygote advantage or frequency dependent selection, they rarely fix, causing a lower replacement to silent ratio between species and a higher replacement to silent ratio within species. Biol 336 -12 41

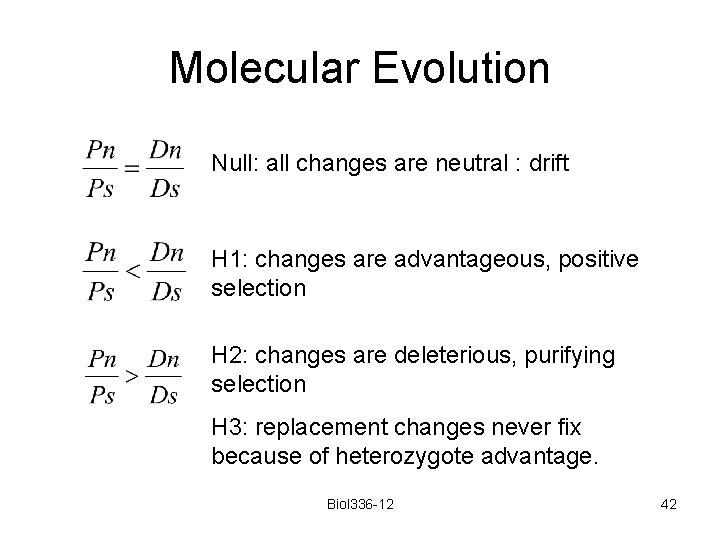
Molecular Evolution Null: all changes are neutral : drift H 1: changes are advantageous, positive selection H 2: changes are deleterious, purifying selection H 3: replacement changes never fix because of heterozygote advantage. Biol 336 -12 42

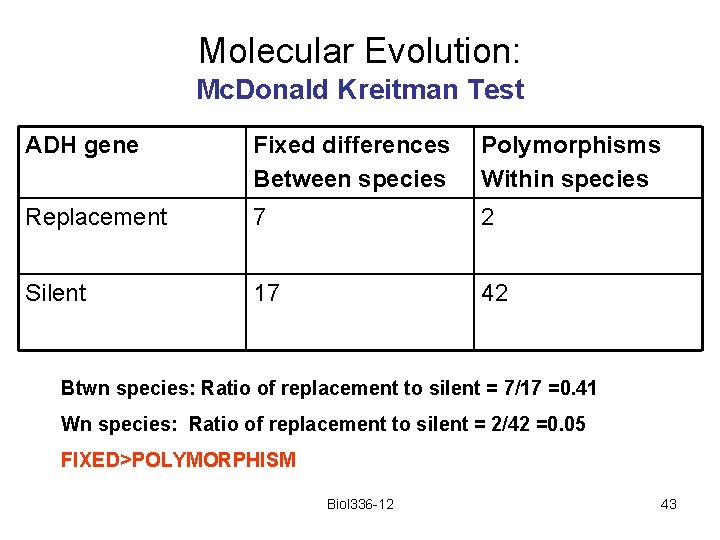
Molecular Evolution: Mc. Donald Kreitman Test ADH gene Fixed differences Between species Polymorphisms Within species Replacement 7 2 Silent 17 42 Btwn species: Ratio of replacement to silent = 7/17 =0. 41 Wn species: Ratio of replacement to silent = 2/42 =0. 05 FIXED>POLYMORPHISM Biol 336 -12 43

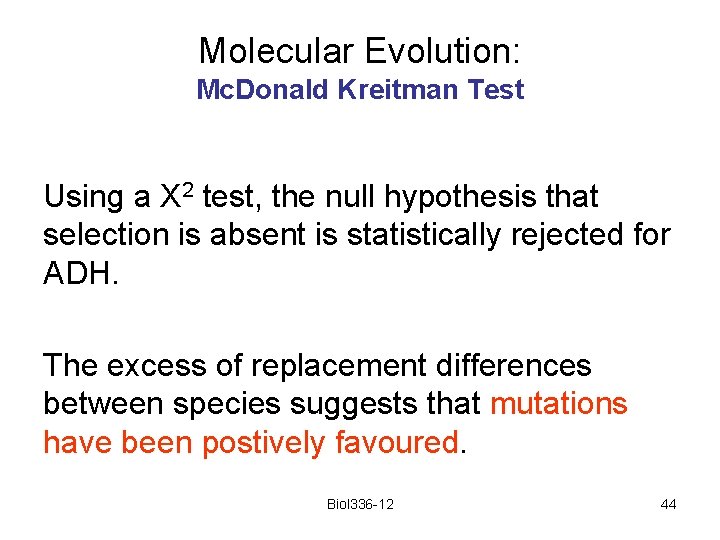
Molecular Evolution: Mc. Donald Kreitman Test Using a X 2 test, the null hypothesis that selection is absent is statistically rejected for ADH. The excess of replacement differences between species suggests that mutations have been postively favoured. Biol 336 -12 44

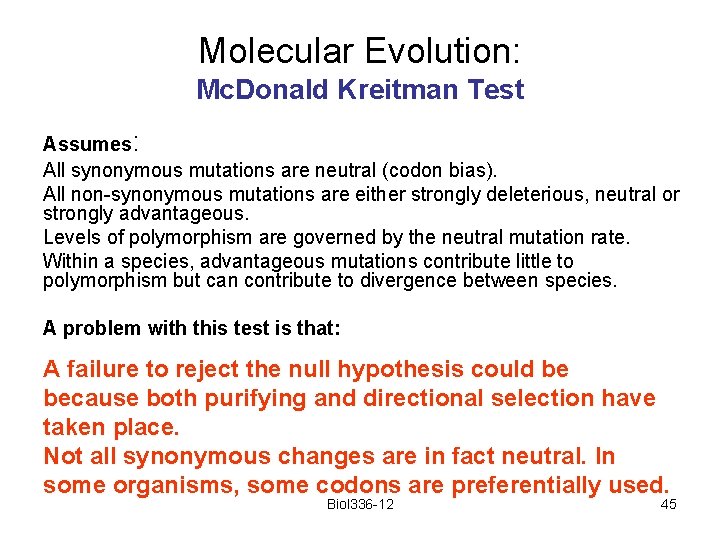
Molecular Evolution: Mc. Donald Kreitman Test Assumes: All synonymous mutations are neutral (codon bias). All non-synonymous mutations are either strongly deleterious, neutral or strongly advantageous. Levels of polymorphism are governed by the neutral mutation rate. Within a species, advantageous mutations contribute little to polymorphism but can contribute to divergence between species. A problem with this test is that: A failure to reject the null hypothesis could be because both purifying and directional selection have taken place. Not all synonymous changes are in fact neutral. In some organisms, some codons are preferentially used. Biol 336 -12 45

Molecular Evolution How else might you detect selection in the genome, in particular the presence of selective sweeps? Biol 336 -12 46

Molecular Evolution: Neighbouring marker sites If a beneficial mutation appears and sweeps through a population, what will happen to the level of polymorphism present at neighbouring DNA sites? Biol 336 -12 47

Molecular Evolution: Neighbouring marker sites If a beneficial mutation appears and sweeps through a population, what will happen to the level of polymorphism present at neighbouring DNA sites? Genetic hitchhiking will decrease variation. Biol 336 -12 48

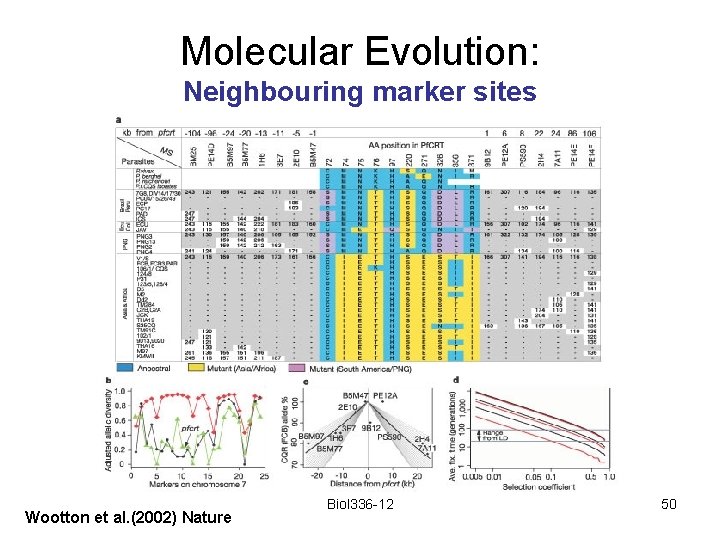
Molecular Evolution: Neighbouring marker sites In the case of Plasmodium falciparum, diversity at neighbouring marker loci decreased. Biol 336 -12 49

Molecular Evolution: Neighbouring marker sites Wootton et al. (2002) Nature Biol 336 -12 50

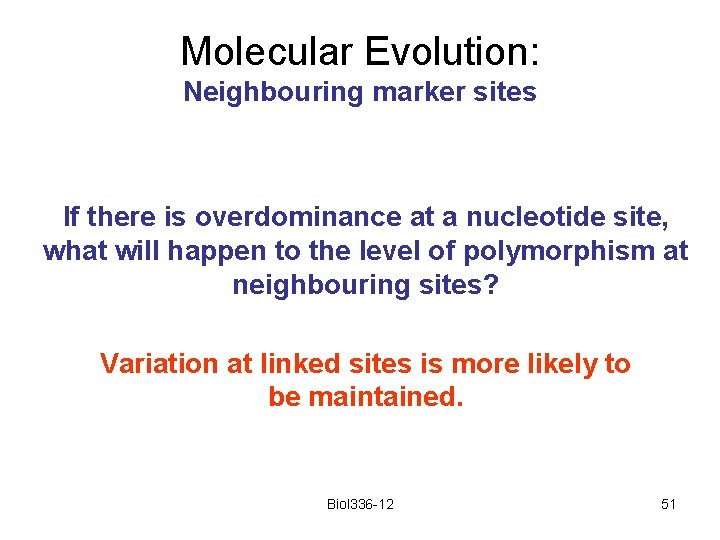
Molecular Evolution: Neighbouring marker sites If there is overdominance at a nucleotide site, what will happen to the level of polymorphism at neighbouring sites? Variation at linked sites is more likely to be maintained. Biol 336 -12 51

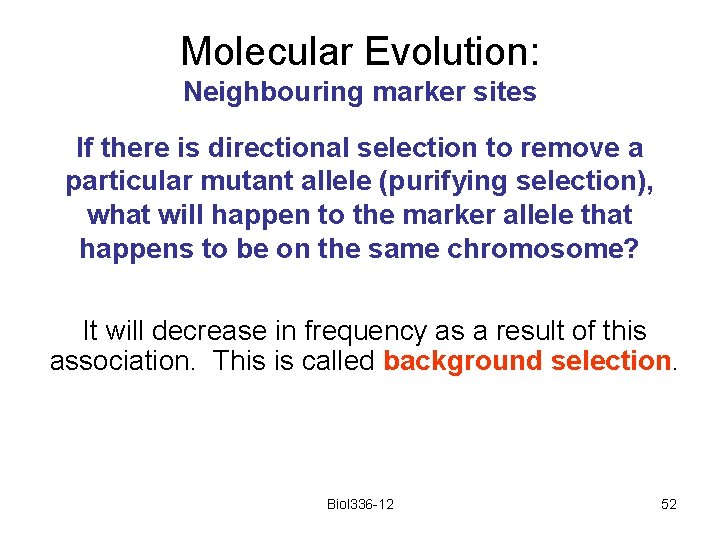
Molecular Evolution: Neighbouring marker sites If there is directional selection to remove a particular mutant allele (purifying selection), what will happen to the marker allele that happens to be on the same chromosome? It will decrease in frequency as a result of this association. This is called background selection. Biol 336 -12 52

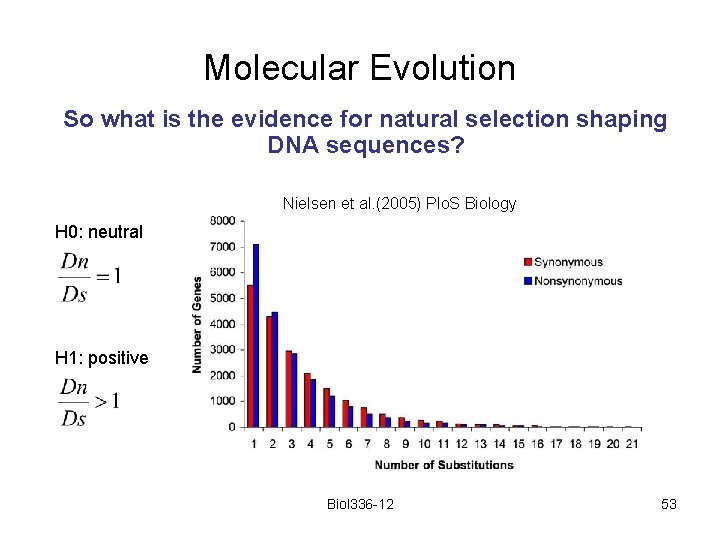
Molecular Evolution So what is the evidence for natural selection shaping DNA sequences? Nielsen et al. (2005) Plo. S Biology H 0: neutral H 1: positive Biol 336 -12 53

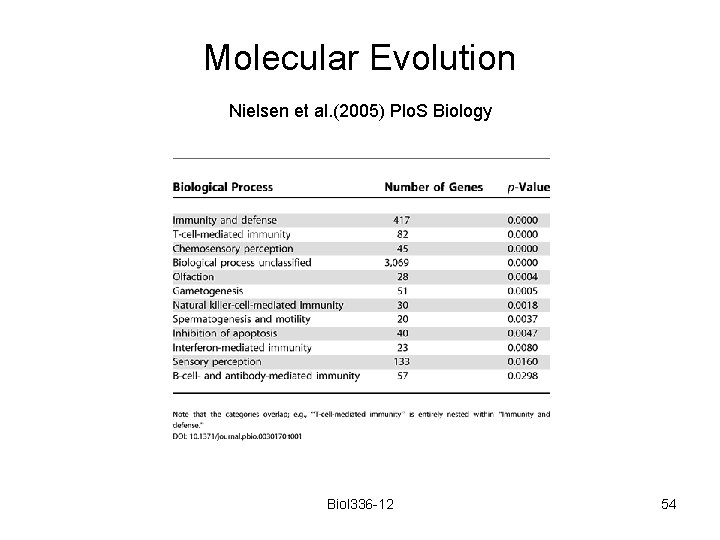
Molecular Evolution Nielsen et al. (2005) Plo. S Biology Biol 336 -12 54

Molecular Evolution How can you detect the signature of selection? Comparing intra-species polymorphism to interspecies differences (Mc. Donald-Kreitman test). Linked/neighbouring neutral markers. Examine genes for Dn/Ds ratios. Biol 336 -12 55

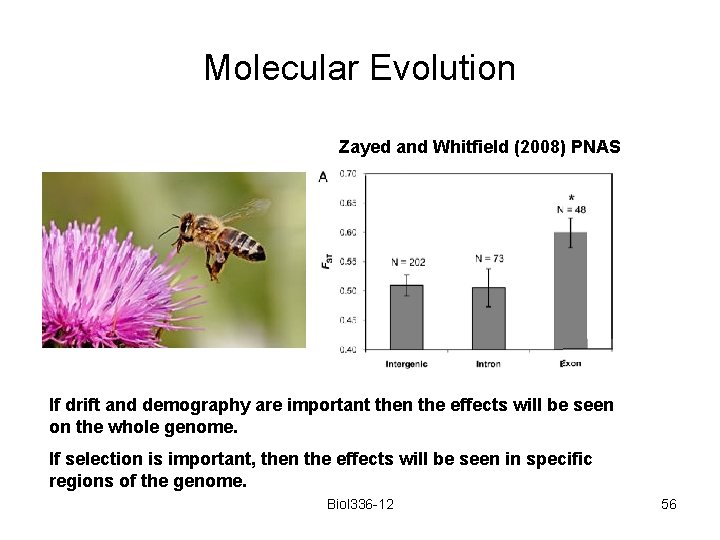
Molecular Evolution Zayed and Whitfield (2008) PNAS If drift and demography are important then the effects will be seen on the whole genome. If selection is important, then the effects will be seen in specific regions of the genome. Biol 336 -12 56