Introduction to high throughput sequencing Lecture 1 Introduction

![Genotyping & Consensus Generation haploid strain 1 [A] AACGTTAGCATA strain 2 [C] AACGTTCGCATA strain Genotyping & Consensus Generation haploid strain 1 [A] AACGTTAGCATA strain 2 [C] AACGTTCGCATA strain](https://slidetodoc.com/presentation_image/230aed5150ef5a764db0948beb073a68/image-35.jpg)



- Slides: 75

Introduction to high throughput sequencing

Lecture 1 Introduction to high throughput sequencing Michael Brudno CSC 2431 January 13, 2010 Adapted from presentations by Francis Ouelette, OICR, Michael Stromberg, BC and Asim Siddiqui, ABI

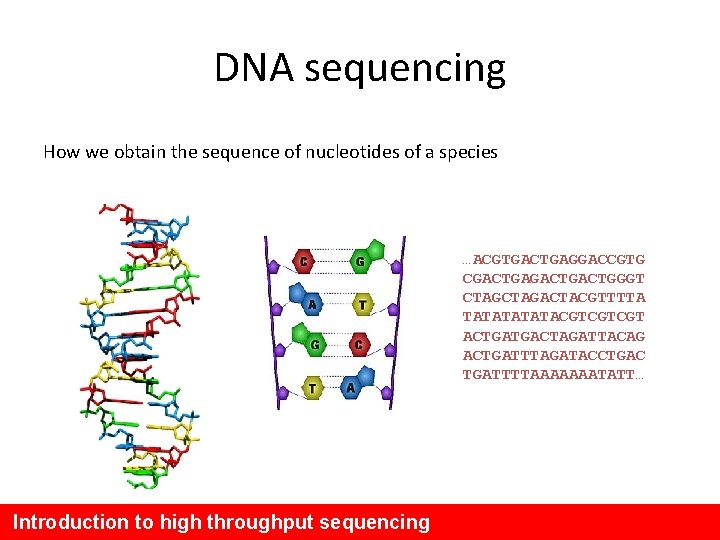
DNA sequencing How we obtain the sequence of nucleotides of a species …ACGTGACTGAGGACCGTG CGACTGACTGGGT CTAGACTACGTTTTA TATATACGTCGTCGT ACTGATGACTAGATTACAG ACTGATTTAGATACCTGAC TGATTTTAAAAAAATATT… Introduction to high throughput sequencing

DNA Sequencing Goal: Find the complete sequence of A, C, G, T’s in DNA Challenge: There is no machine that takes long DNA as an input, and gives the complete sequence as output Can only sequence ~500 letters at a time Introduction to high throughput sequencing

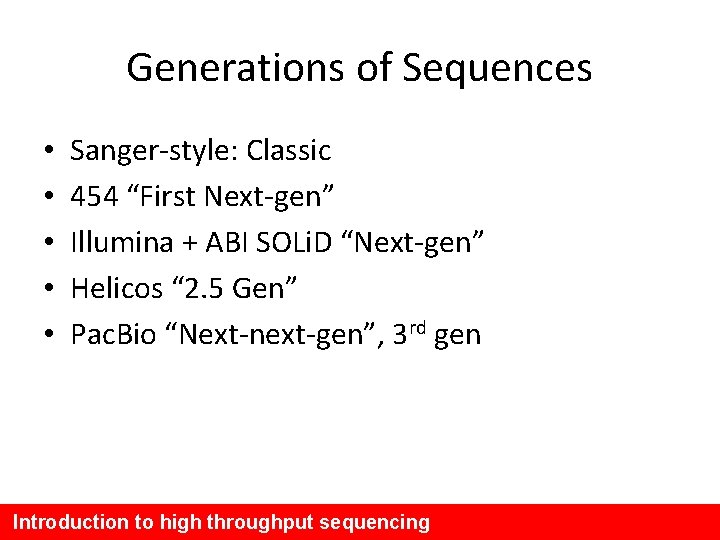
Generations of Sequences • • • Sanger-style: Classic 454 “First Next-gen” Illumina + ABI SOLi. D “Next-gen” Helicos “ 2. 5 Gen” Pac. Bio “Next-next-gen”, 3 rd gen Introduction to high throughput sequencing

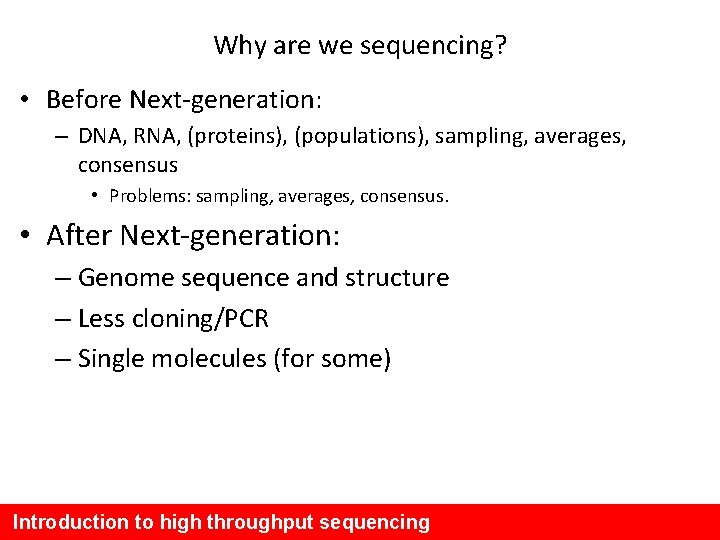
Why are we sequencing? • Before Next-generation: – DNA, RNA, (proteins), (populations), sampling, averages, consensus • Problems: sampling, averages, consensus. • After Next-generation: – Genome sequence and structure – Less cloning/PCR – Single molecules (for some) Introduction to high throughput sequencing

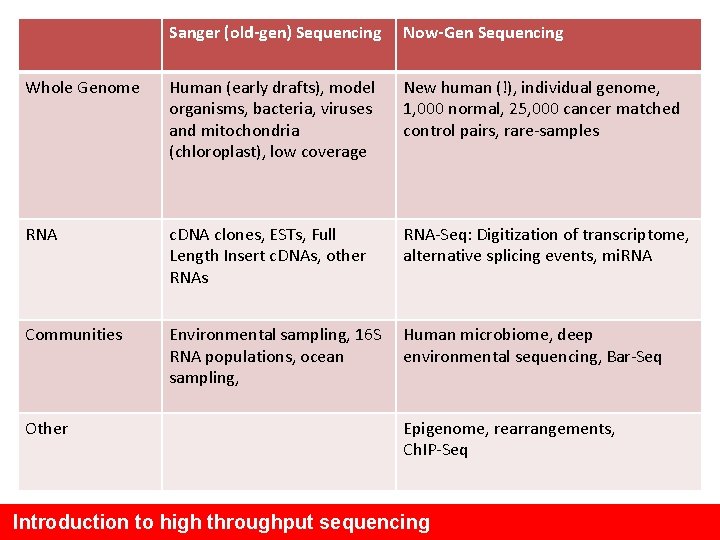
Sanger (old-gen) Sequencing Now-Gen Sequencing Whole Genome Human (early drafts), model organisms, bacteria, viruses and mitochondria (chloroplast), low coverage New human (!), individual genome, 1, 000 normal, 25, 000 cancer matched control pairs, rare-samples RNA c. DNA clones, ESTs, Full Length Insert c. DNAs, other RNAs RNA-Seq: Digitization of transcriptome, alternative splicing events, mi. RNA Communities Environmental sampling, 16 S RNA populations, ocean sampling, Human microbiome, deep environmental sequencing, Bar-Seq Other Epigenome, rearrangements, Ch. IP-Seq Introduction to high throughput sequencing

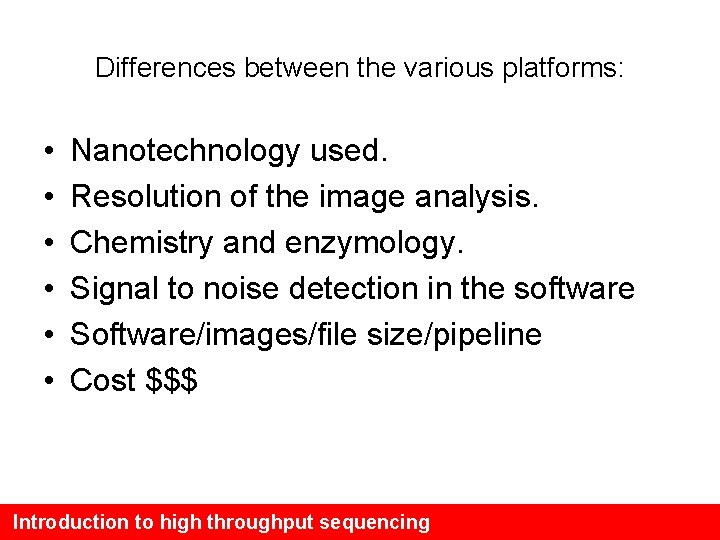
Differences between the various platforms: • • • Nanotechnology used. Resolution of the image analysis. Chemistry and enzymology. Signal to noise detection in the software Software/images/file size/pipeline Cost $$$ Introduction to high throughput sequencing

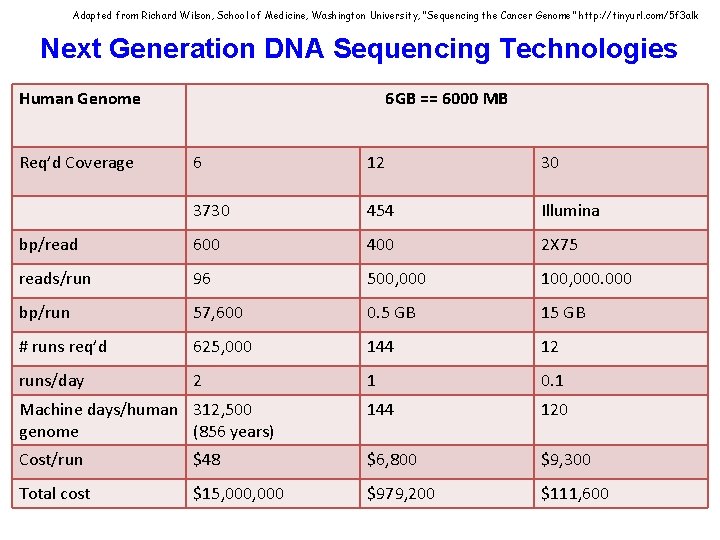
Adapted from Richard Wilson, School of Medicine, Washington University, “Sequencing the Cancer Genome” http: //tinyurl. com/5 f 3 alk Next Generation DNA Sequencing Technologies Human Genome Req’d Coverage 6 GB == 6000 MB 6 12 30 3730 454 Illumina bp/read 600 400 2 X 75 reads/run 96 500, 000 100, 000 bp/run 57, 600 0. 5 GB 15 GB # runs req’d 625, 000 144 12 runs/day 2 1 0. 1 Machine days/human 312, 500 genome (856 years) 144 120 Cost/run $48 $6, 800 $9, 300 Total cost $15, 000 $979, 200 $111, 600

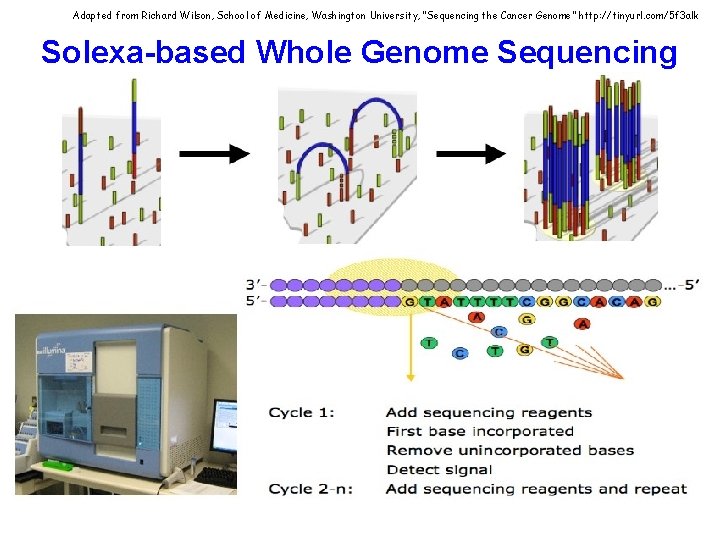
Adapted from Richard Wilson, School of Medicine, Washington University, “Sequencing the Cancer Genome” http: //tinyurl. com/5 f 3 alk Solexa-based Whole Genome Sequencing

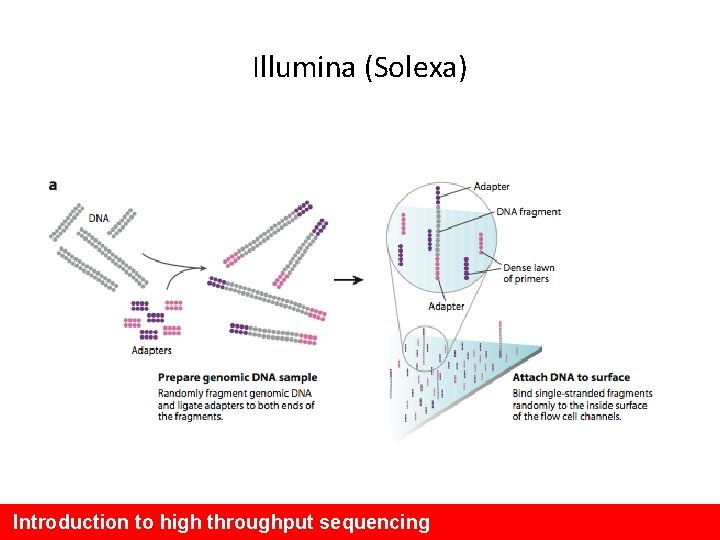
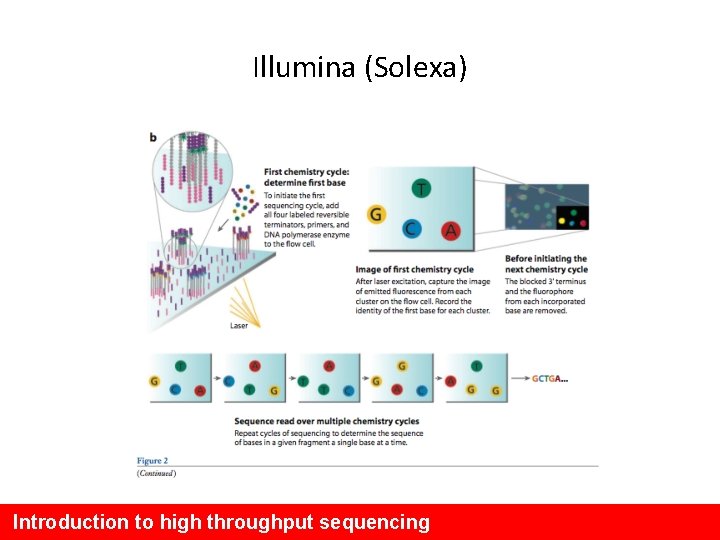
Illumina (Solexa) Introduction to high throughput sequencing

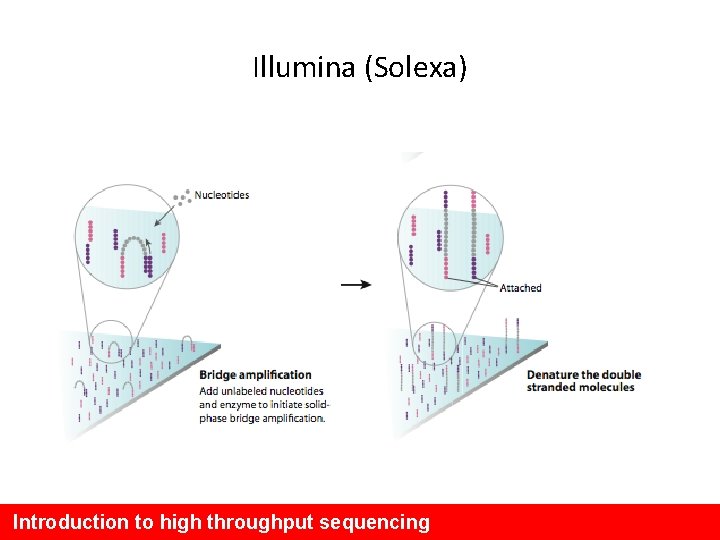
Illumina (Solexa) Introduction to high throughput sequencing

Illumina (Solexa) Introduction to high throughput sequencing

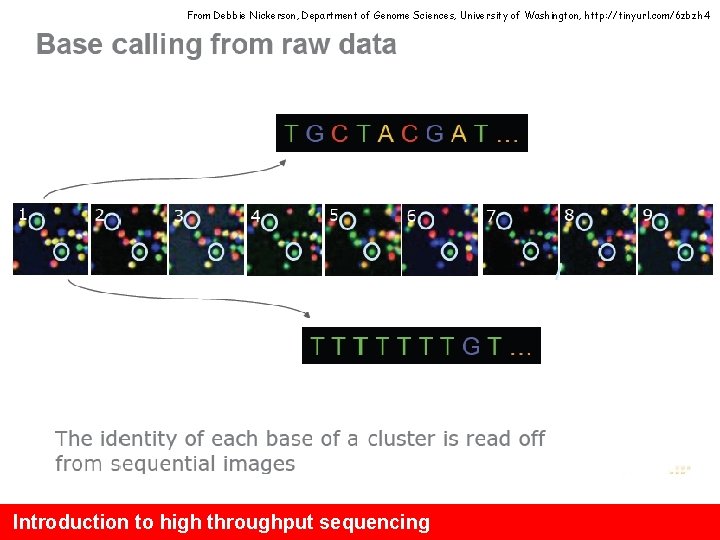
From Debbie Nickerson, Department of Genome Sciences, University of Washington, http: //tinyurl. com/6 zbzh 4 Introduction to high throughput sequencing

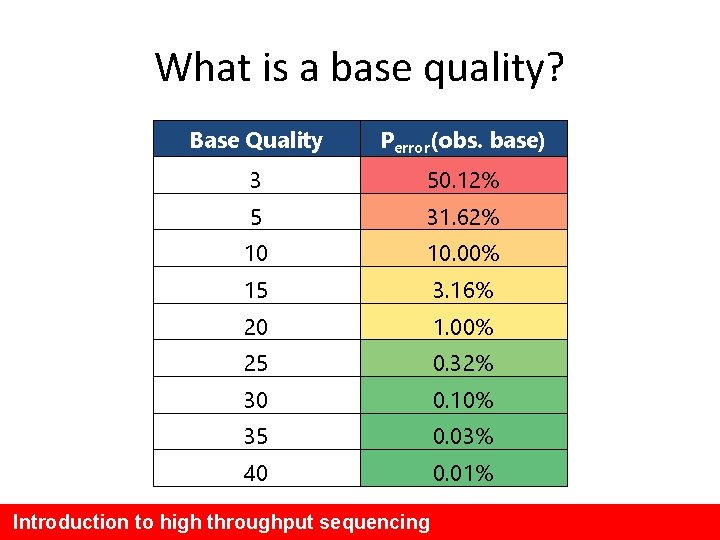
What is a base quality? Base Quality Perror(obs. base) 3 50. 12% 5 31. 62% 10 10. 00% 15 3. 16% 20 1. 00% 25 0. 32% 30 0. 10% 35 0. 03% 40 0. 01% Introduction to high throughput sequencing

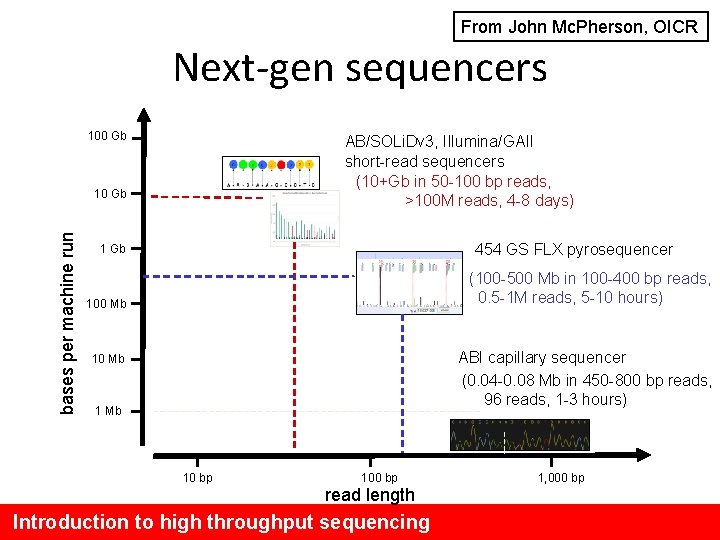
From John Mc. Pherson, OICR Next-gen sequencers 100 Gb AB/SOLi. Dv 3, Illumina/GAII short-read sequencers (10+Gb in 50 -100 bp reads, >100 M reads, 4 -8 days) bases per machine run 10 Gb 454 GS FLX pyrosequencer 1 Gb (100 -500 Mb in 100 -400 bp reads, 0. 5 -1 M reads, 5 -10 hours) 100 Mb ABI capillary sequencer (0. 04 -0. 08 Mb in 450 -800 bp reads, 96 reads, 1 -3 hours) 10 Mb 10 bp 100 bp read length Introduction to high throughput sequencing 1, 000 bp

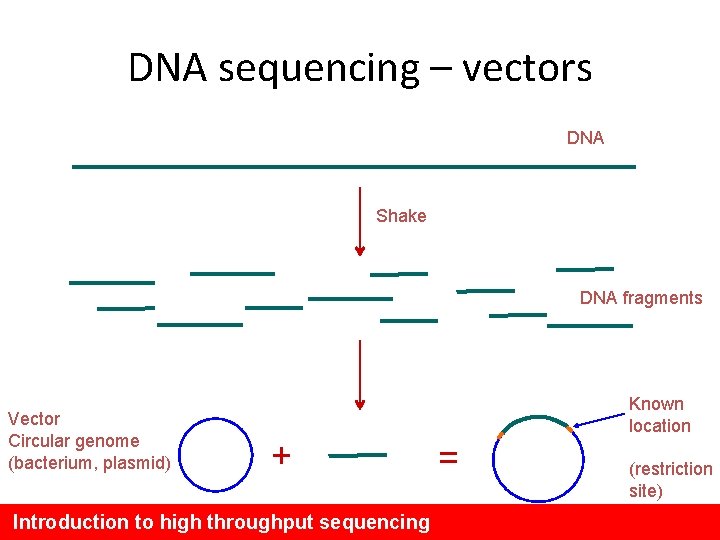
DNA sequencing – vectors DNA Shake DNA fragments Vector Circular genome (bacterium, plasmid) Known location + Introduction to high throughput sequencing = (restriction site)

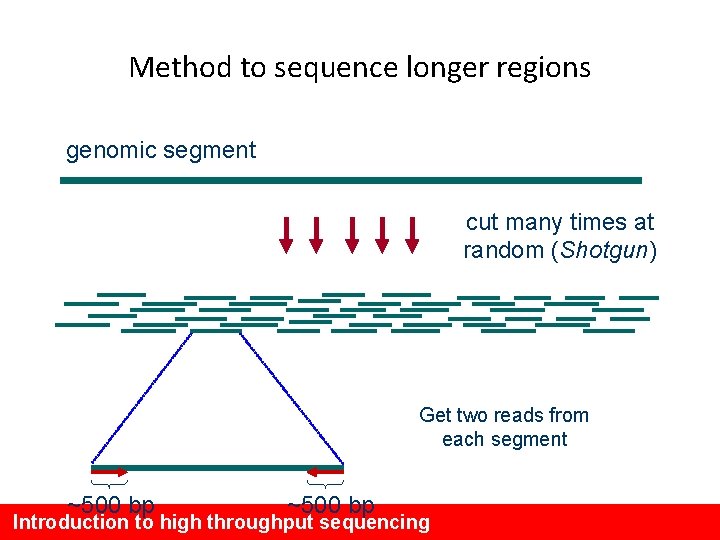
Method to sequence longer regions genomic segment cut many times at random (Shotgun) Get two reads from each segment ~500 bp Introduction to high throughput sequencing

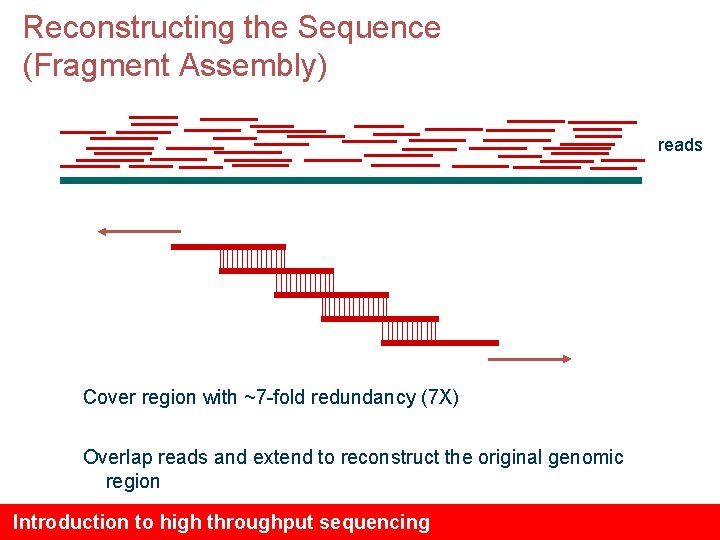
Reconstructing the Sequence (Fragment Assembly) reads Cover region with ~7 -fold redundancy (7 X) Overlap reads and extend to reconstruct the original genomic region Introduction to high throughput sequencing

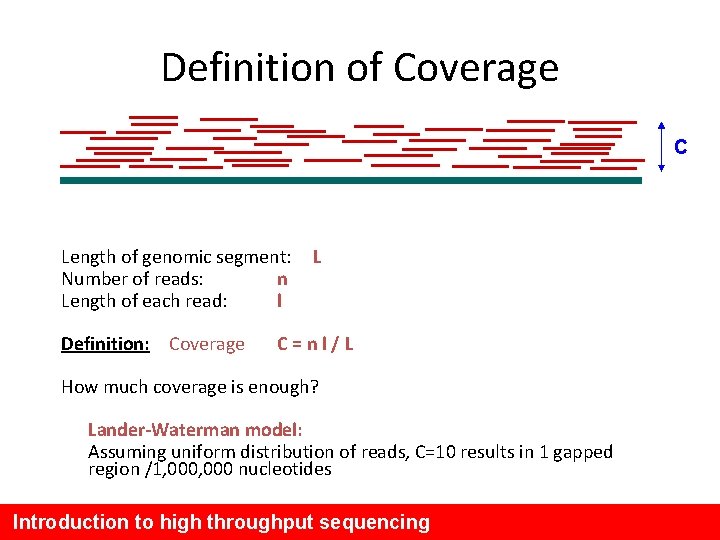
Definition of Coverage C Length of genomic segment: Number of reads: n Length of each read: l Definition: Coverage L C=nl/L How much coverage is enough? Lander-Waterman model: Assuming uniform distribution of reads, C=10 results in 1 gapped region /1, 000 nucleotides Introduction to high throughput sequencing

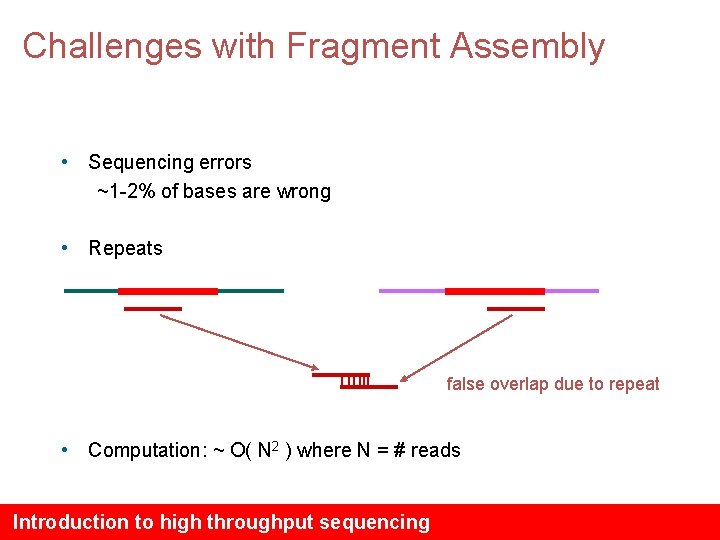
Challenges with Fragment Assembly • Sequencing errors ~1 -2% of bases are wrong • Repeats false overlap due to repeat • Computation: ~ O( N 2 ) where N = # reads Introduction to high throughput sequencing

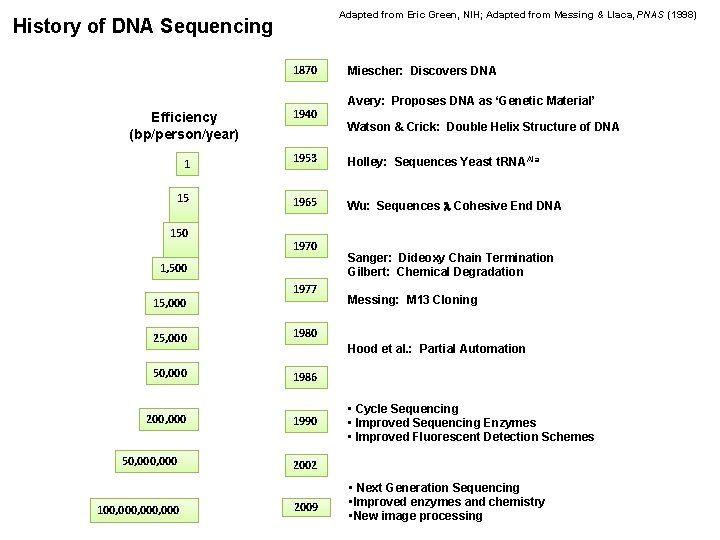
Adapted from Eric Green, NIH; Adapted from Messing & Llaca, PNAS (1998) History of DNA Sequencing 1870 Miescher: Discovers DNA Avery: Proposes DNA as ‘Genetic Material’ Efficiency (bp/person/year) 1940 1 1953 Holley: Sequences Yeast t. RNAAla 15 1965 Wu: Sequences Cohesive End DNA 150 1970 1, 500 15, 000 1977 25, 000 1980 50, 000 1986 200, 000 1990 50, 000 100, 000, 000 Watson & Crick: Double Helix Structure of DNA Sanger: Dideoxy Chain Termination Gilbert: Chemical Degradation Messing: M 13 Cloning Hood et al. : Partial Automation • Cycle Sequencing • Improved Sequencing Enzymes • Improved Fluorescent Detection Schemes 2002 2009 • Next Generation Sequencing • Improved enzymes and chemistry • New image processing

Which representative of the species? Which human? Answer one: Answer two: it doesn’t matter Polymorphism rate: number of letter changes between two different members of a species Humans: ~1/1, 000 – 1/10, 000 Other organisms have much higher polymorphism rates Introduction to high throughput sequencing

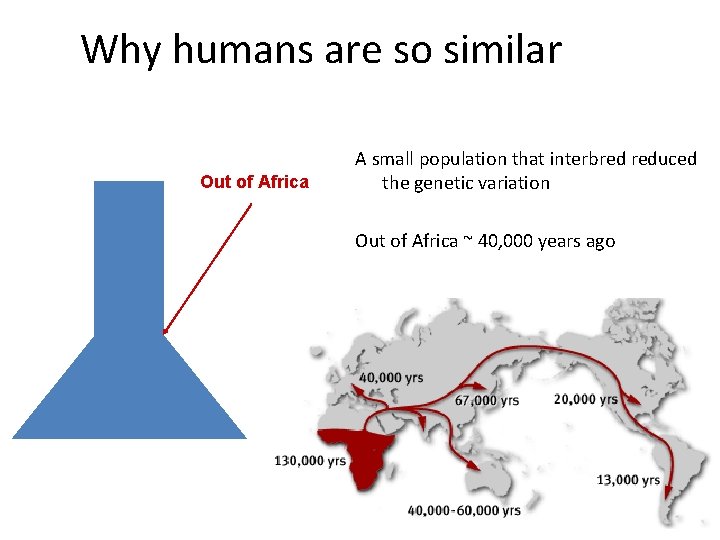
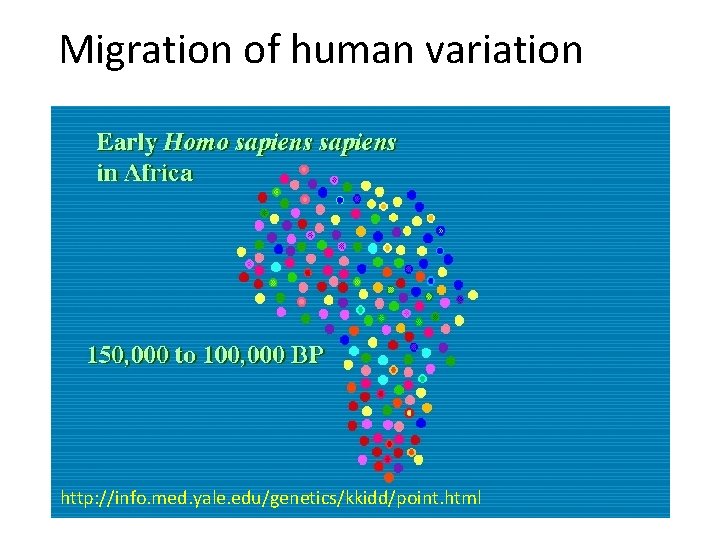
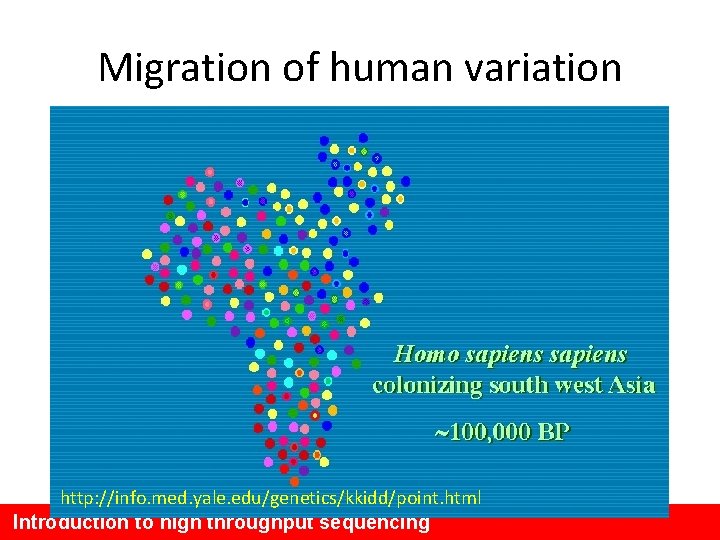
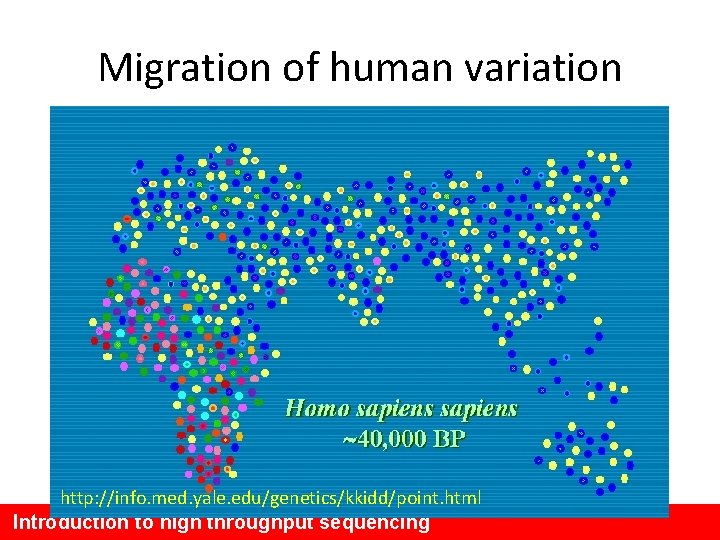
Why humans are so similar Out of Africa A small population that interbred reduced the genetic variation Out of Africa ~ 40, 000 years ago

Migration of human variation http: //info. med. yale. edu/genetics/kkidd/point. html

Migration of human variation http: //info. med. yale. edu/genetics/kkidd/point. html Introduction to high throughput sequencing

Migration of human variation http: //info. med. yale. edu/genetics/kkidd/point. html Introduction to high throughput sequencing

Genetic Variations: Why? Phenotypic differences Inherited diseases Ancestral history Introduction to high throughput sequencing

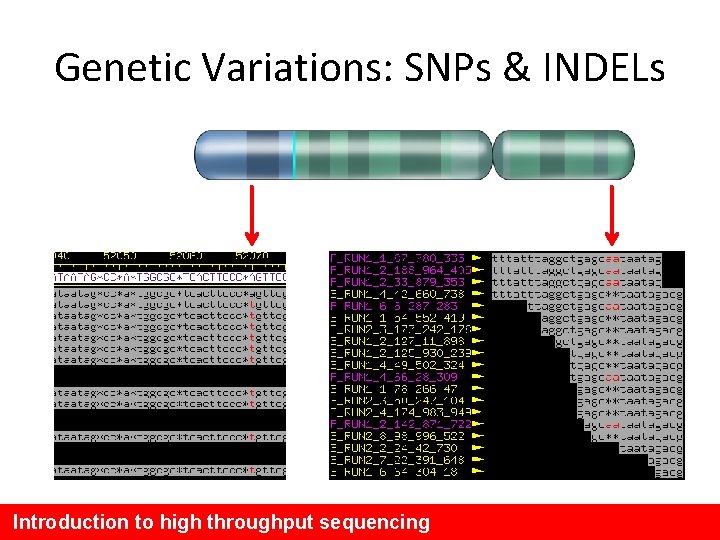
Genetic Variations: SNPs & INDELs Introduction to high throughput sequencing

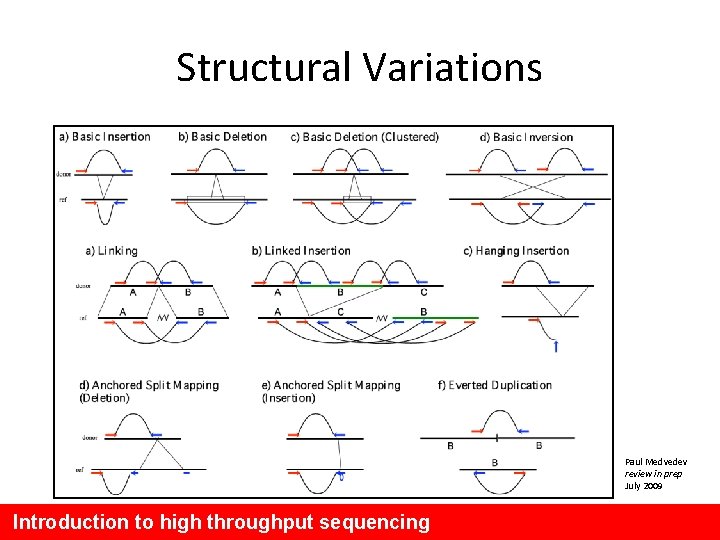
Structural Variations Paul Medvedev review in prep July 2009 Introduction to high throughput sequencing

SNP Discovery: Goal sequencing errors Introduction to high throughput sequencing SNP

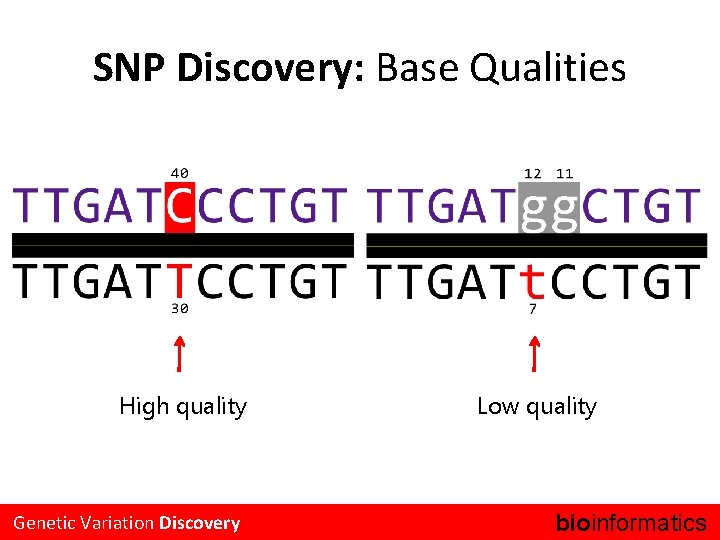
SNP Discovery: Base Qualities High quality Genetic Variation Discovery Low quality bioinformatics.

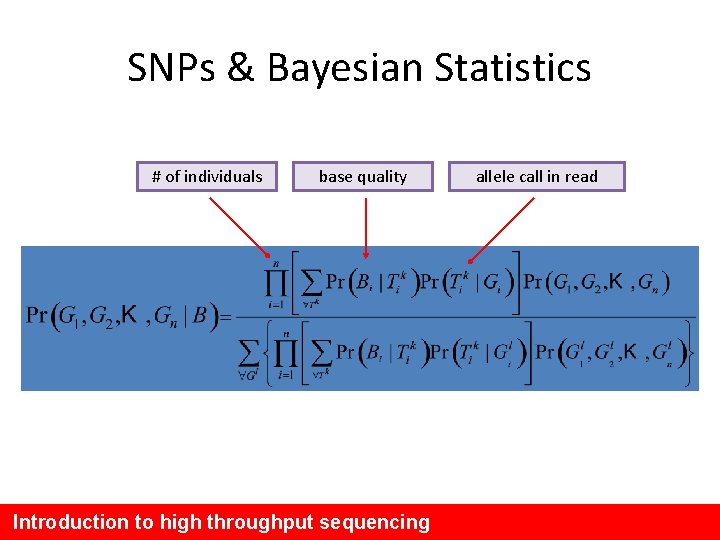
SNPs & Bayesian Statistics # of individuals base quality Introduction to high throughput sequencing allele call in read

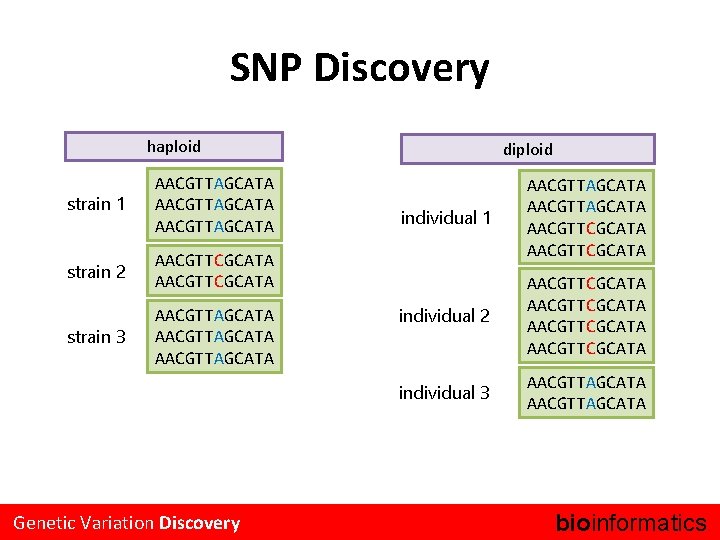
SNP Discovery haploid strain 1 AACGTTAGCATA strain 2 AACGTTCGCATA strain 3 AACGTTAGCATA Genetic Variation Discovery diploid individual 1 AACGTTAGCATA AACGTTCGCATA individual 2 AACGTTCGCATA individual 3 AACGTTAGCATA bioinformatics.
![Genotyping Consensus Generation haploid strain 1 A AACGTTAGCATA strain 2 C AACGTTCGCATA strain Genotyping & Consensus Generation haploid strain 1 [A] AACGTTAGCATA strain 2 [C] AACGTTCGCATA strain](https://slidetodoc.com/presentation_image/230aed5150ef5a764db0948beb073a68/image-35.jpg)
Genotyping & Consensus Generation haploid strain 1 [A] AACGTTAGCATA strain 2 [C] AACGTTCGCATA strain 3 [A] AACGTTAGCATA Genetic Variation Discovery diploid individual 1 [A/C] AACGTTAGCATA AACGTTCGCATA individual 2 [C/C] AACGTTCGCATA individual 3 [A/A] AACGTTAGCATA bioinformatics.

Visualization: Consed Genetic Variation Discovery bioinformatics.

1000 Genomes Project Introduction to high throughput sequencing

1000 G: Goals • Discover genetic variations – 1 % minor allele frequencies across genome – 0. 1 – 0. 5 % MAF across gene regions • Variant alleles – Estimate frequencies – Identify haplotype background – Characterize linkage disequilibrium Introduction to high throughput sequencing

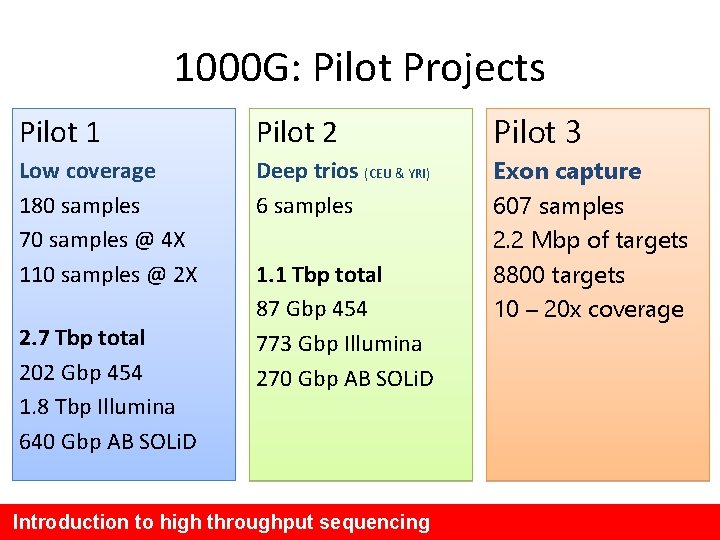
1000 G: Pilot Projects Pilot 1 Pilot 2 Pilot 3 Low coverage 180 samples 70 samples @ 4 X 110 samples @ 2 X Deep trios (CEU & YRI) 6 samples Exon capture 607 samples 2. 2 Mbp of targets 8800 targets 10 – 20 x coverage 2. 7 Tbp total 202 Gbp 454 1. 8 Tbp Illumina 640 Gbp AB SOLi. D 1. 1 Tbp total 87 Gbp 454 773 Gbp Illumina 270 Gbp AB SOLi. D Introduction to high throughput sequencing

Questions about the genome • Obtaining a genome sequence is a one step towards understanding biological processes • Questions that follow from the genome are: – What is transcribed? – Where do proteins bind? – What is methylated? • In other words, how does it work? Introduction to high throughput sequencing

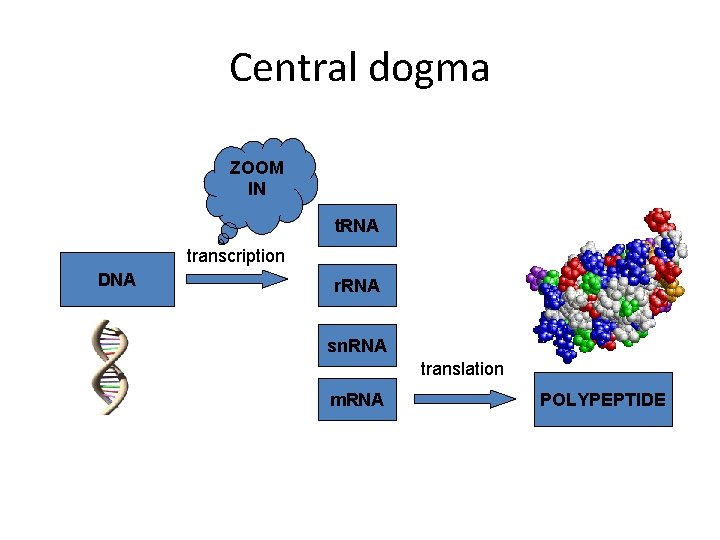
Central dogma ZOOM IN t. RNA transcription DNA r. RNA sn. RNA translation m. RNA POLYPEPTIDE

Transcription • The DNA is contained in the nucleus of the cell. • A stretch of it unwinds there, and its message (or sequence) is copied onto a molecule of m. RNA. • The m. RNA then exits from the cell nucleus. Introduction to high throughput sequencing

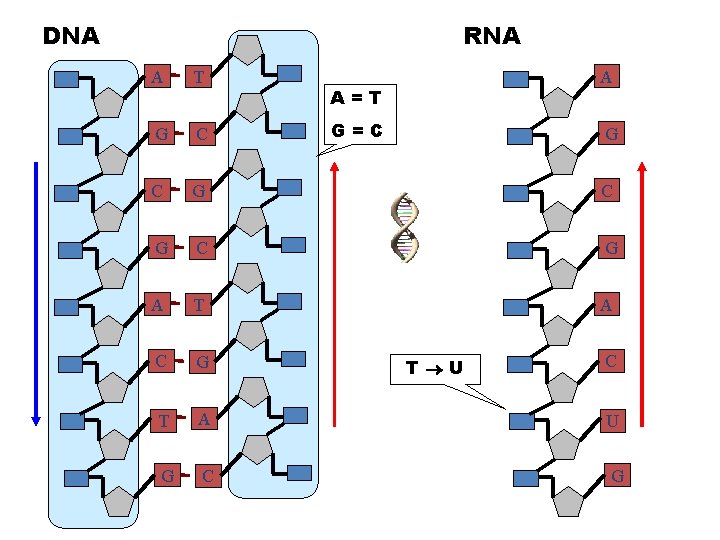
DNA RNA A T A G C C G C G A T A C G T A U G C G A=T G=C G T U C

More complexity • The RNA message is sometimes “edited”. • Exons are nucleotide segments whose codons will be expressed. • Introns are intervening segments (genetic gibberish) that are snipped out. • Exons are spliced together to form m. RNA. Introduction to high throughput sequencing

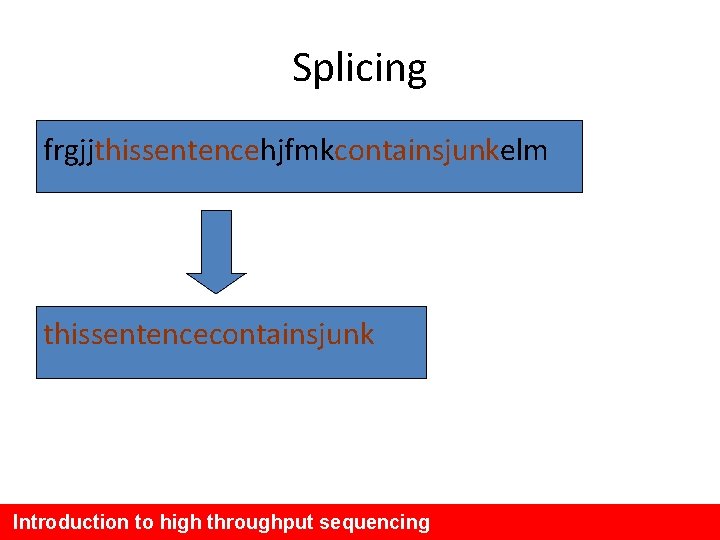
Splicing frgjjthissentencehjfmkcontainsjunkelm thissentencecontainsjunk Introduction to high throughput sequencing

Key player: RNA polymerase • It is the enzyme that brings about transcription by going down the line, pairing m. RNA nucleotides with their DNA counterparts. Introduction to high throughput sequencing

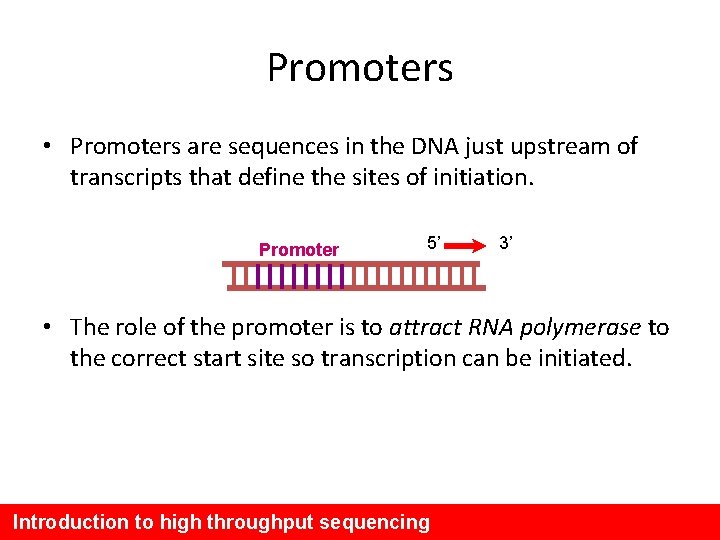
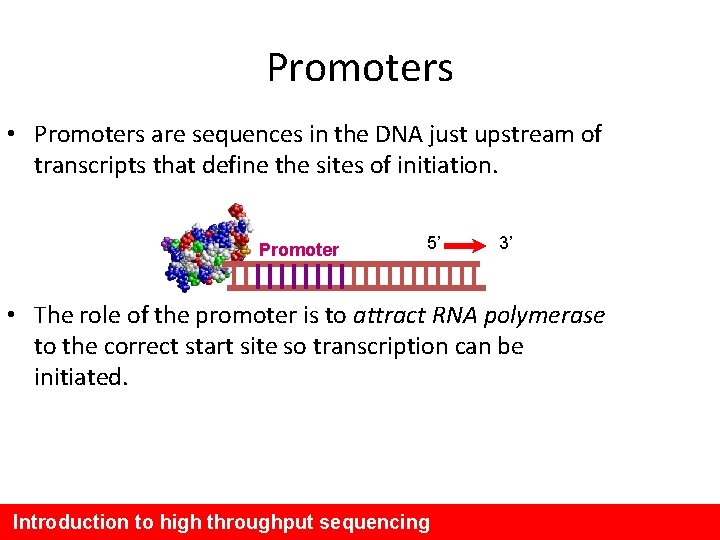
Promoters • Promoters are sequences in the DNA just upstream of transcripts that define the sites of initiation. Promoter 5’ 3’ • The role of the promoter is to attract RNA polymerase to the correct start site so transcription can be initiated. Introduction to high throughput sequencing

Promoters • Promoters are sequences in the DNA just upstream of transcripts that define the sites of initiation. Promoter 5’ 3’ • The role of the promoter is to attract RNA polymerase to the correct start site so transcription can be initiated. Introduction to high throughput sequencing

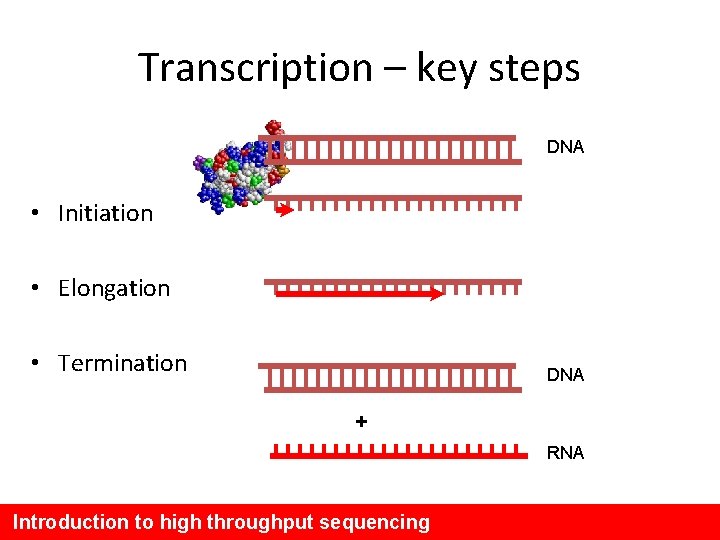
Transcription – key steps DNA • Initiation • Elongation • Termination DNA + RNA Introduction to high throughput sequencing

Genes can be switched on/off • In an adult multicellular organism, there is a wide variety of cell types seen in the adult. eg, muscle, nerve and blood cells. • The different cell types contain the same DNA though. • This differentiation arises because different cell types express different genes. • Promoters are one type of gene regulators Introduction to high throughput sequencing

Transcription (recap) • The DNA is contained in the nucleus of the cell. • A stretch of it unwinds there, and its message (or sequence) is copied onto a molecule of m. RNA. • The m. RNA then exits from the cell nucleus. • Its destination is a molecular workbench in the cytoplasm, a structure called a ribosome. Introduction to high throughput sequencing

The Transcriptome • The transcriptome is the entire set of RNA transcripts in the cell, tissue or organ. • The transcriptome is cell type specific and time dependant i. e. It is a function of cell state • The transcriptome can help us understand how cells differentiate and respond to changes in their environment. Introduction to high throughput sequencing

Transcriptome complexity • Transcripts may be: – Modified – Spliced – Edited – Degraded • Transcriptome is substantially more complex than the genome and is time variant. Introduction to high throughput sequencing

ESTs • ESTs were the first genome wide scan for transcriptional elements • Different library types: – Proportional – Normalized – Subtractive • Can be sequenced from the 5’ or 3’ end Introduction to high throughput sequencing

“Hello Mr Chips” • Microarray chips introduced in 90’s • Parallel way to measure many genes – Probes placed on slides – RNA -> c. DNA, labelled with fluorescent dye and hybridized. – Fluorescence measured • • • Chips have been highly successful Simplified analysis Useful when there is no genome sequence Linear signal across 500 fold variation Standardization has aided use in medical diagnostics – E. g. Mammaprint Introduction to high throughput sequencing

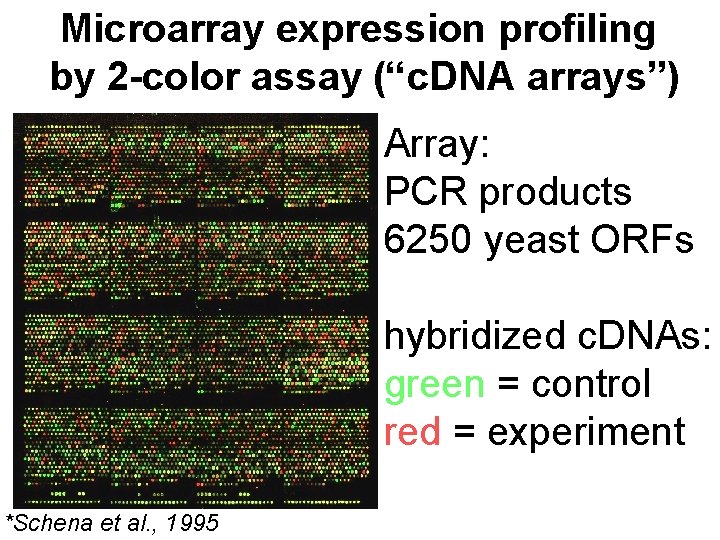
Microarray expression profiling by 2 -color assay (“c. DNA arrays”) Array: PCR products 6250 yeast ORFs hybridized c. DNAs: green = control red = experiment *Schena et al. , 1995

Chips: pros and cons • Advantages – Do not require a genome sequence – Highly characterised, with many s/w packages available – One Affymetrix chip FDA approved • Disadvantages – Measurements limited to what’s on the array – Hard to distinguish isoforms when used for expression – Can’t detect balanced translocations or inversions when used for resequencing Introduction to high throughput sequencing

m. RNA-seq • Basic work flow – Align reads (sometimes to transcriptome first and then the genome) – Tally transcript counts – Align tags to spliced transcripts – Add to transcript counts Introduction to high throughput sequencing

Cloonan et al. 2008 • Used SOLi. D to generate 10 Gb of data from mouse embryonic stem cells and embryonic bodies • Used a library of exon junctions to map across known splice events Introduction to high throughput sequencing

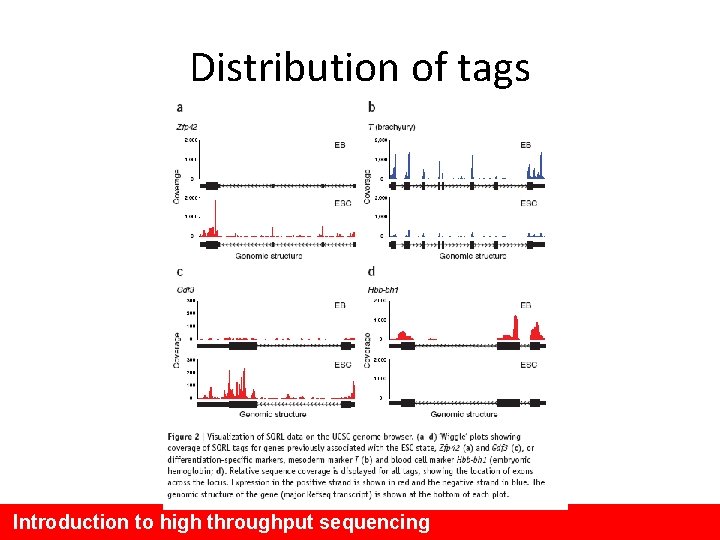
Distribution of tags Introduction to high throughput sequencing

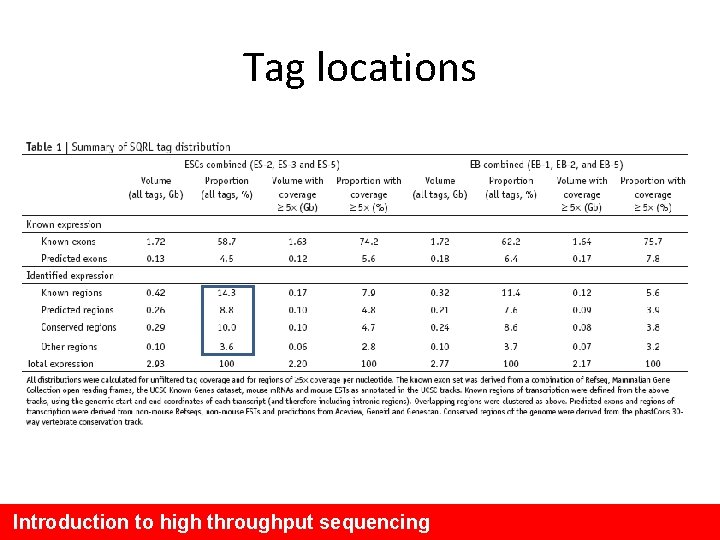
Tag locations Introduction to high throughput sequencing

General issues • Coverage across the transcript may not be random • Some reads map to multiple locations • Some reads don’t map at all • Reads mapping outside of known exons may represent – New gene models – New genes Introduction to high throughput sequencing

Size of the transcriptome • Carter et al (2005) – Using arrays estimated 520, 000 to 850, 000 transcripts per cell. – Use upper limit and estimate average transcript size of 2 kb – Transcriptome ~2 GB • Transcriptome cost ~ genome cost Introduction to high throughput sequencing

The Boundome • DNA binding proteins control genome function • Histones impact chromatin structure • Activators and repressors impact gene expression • The location of these proteins helps us understand how the genome works Introduction to high throughput sequencing

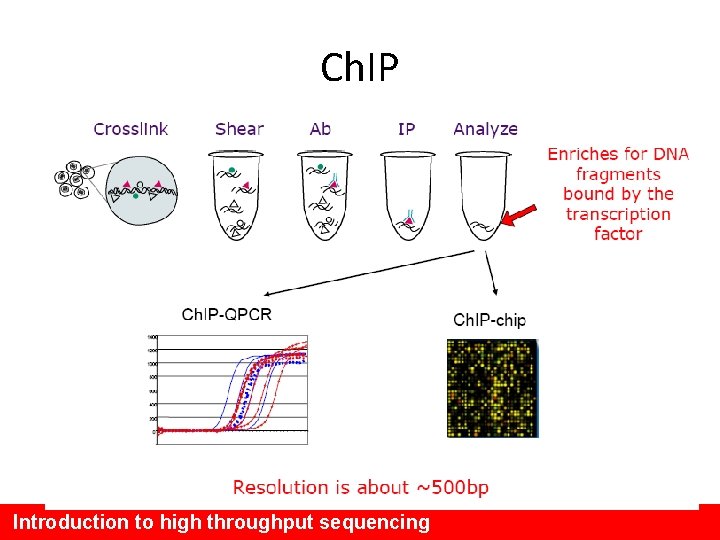
Ch. IP Introduction to high throughput sequencing

Chip-Seq • Instead of probing against a chip, measure directly • Basic work flow – Align reads to the genome – Identify clusters and peaks – Determine bound sites Introduction to high throughput sequencing

Robertson et al. 2007 • Used Illumina technology to find STAT 1 binding sites • Comparisons with two Ch. IP-PCR data sets suggested that Ch. IP-seq sensitivity was between 70% and 92% and specificity was at least 95%. Introduction to high throughput sequencing

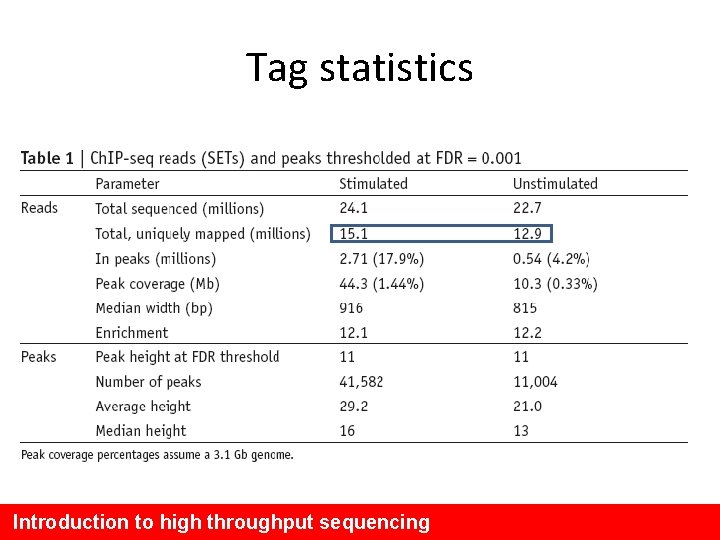
Tag statistics Introduction to high throughput sequencing

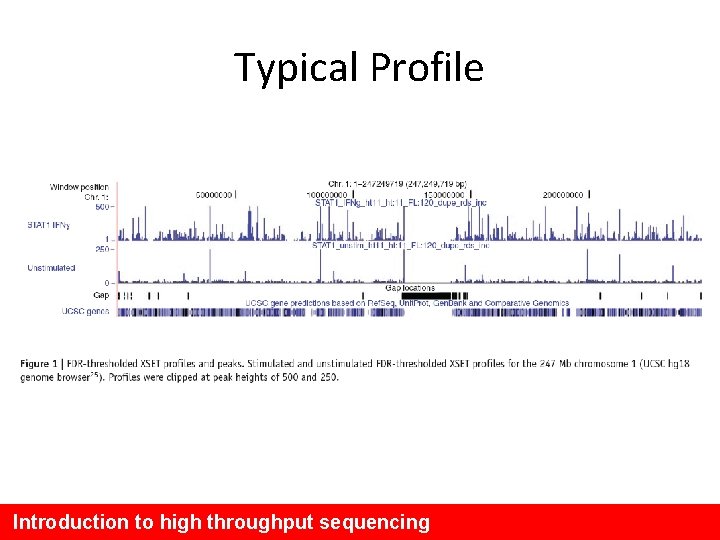
Typical Profile Introduction to high throughput sequencing

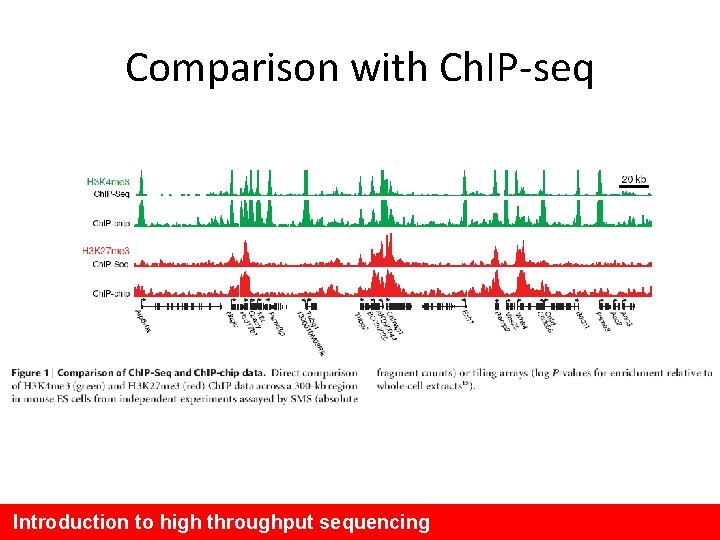
Mikkelsen et al. , 2007 • Performed a comparison with Ch. IP-chip methods ~98% concordance Introduction to high throughput sequencing

Comparison with Ch. IP-seq Introduction to high throughput sequencing

The Methylome • In methylated DNA, cytosines are methylated. • This leads to silencing of genes in the region e. g. X inactivation • It is yet another form of transcriptional control and together with histone modifications a key component of epigenetics Introduction to high throughput sequencing

Bi-sulphite sequencing • Converts un-methylated cytosines to uracil (which becomes thymine when converted to c. DNA) • Experimental procedure is difficult • Sequence alignment is tricky, but the basic concepts hold Introduction to high throughput sequencing

Taylor et al, 2007 • Targeted sequencing reduced alignment difficulties • Used dynamic programming to identify alignments of sequences against an in silico bisulphate converted sequence of the target amplicon regions Introduction to high throughput sequencing

Metagenomics • Craig Venter’s sequencing of the sea one of the earliest and most well known examples – Used Sanger sequencing • Many recent studies including – Angly et al – studied ocean virome – Cox-Foster et al – studied colony collapse disorder • All use 454 for its longer read length and target amplification of 16 S or 18 S ribsomal subunits Introduction to high throughput sequencing